Abstract
Advanced research requires intensive interaction among a multitude of actors, often possessing different expertise and usually working at a distance from each other. The field of collaborative research aims to establish suitable models and technologies to properly support these interactions. In this article, we first present the reasons for an interest of Bioinformatics in this context by also suggesting some research domains that could benefit from collaborative research. We then review the principles and some of the most relevant applications of social networking, with a special attention to networks supporting scientific collaboration, by also highlighting some critical issues, such as identification of users and standardization of formats. We then introduce some systems for collaborative document creation, including wiki systems and tools for ontology development, and review some of the most interesting biological wikis. We also review the principles of Collaborative Development Environments for software and show some examples in Bioinformatics. Finally, we present the principles and some examples of Learning Management Systems. In conclusion, we try to devise some of the goals to be achieved in the short term for the exploitation of these technologies.
Keywords: social networks, open source, collaborative research, collaborative development, collaborative learning
INTRODUCTION
A short historical introduction
Telecommunication networks are meant to enable data exchange and collaboration among people. At the dawn of the Internet, network tools and applications varied widely and did not interoperate. Tools available at that time were merely classified as either network information retrieval (NIR) or computer-mediated communication (CMC) tools. While the former mainly served to distribute documents and to allow free access to electronic archives, the latter were meant to allow network users to communicate with each other, thereby constituting the first true chance to collaborate through networks.
CMC tools were initially asynchronous and based on electronic mail and newsgroups. E-mail systems soon generated mailing lists, while newsgroups spawned electronic fora. Synchronous communication was introduced with the advent of chat services and instant messaging; an offshoot of these tools was the multimedia teleconferencing systems that are currently in use. Virtual reality was first introduced with multi-user domain (MUD), and especially by MUD object-oriented (MOO) systems. These in turn generated mainstream virtual reality environments, such as the second life system.
Life sciences researchers originally profited above all from CMC tools. The Bionet newsgroups hierarchy remains one of the most famous and useful CMC systems supporting life sciences research. Many mailing lists born in that context are still in use.
The development of open source software greatly enhanced the possibility to effectively and efficiently exchange knowledge, practices, skills and, of course, software source. Websites dedicated to communities of scientists have been launched, and these often create the grounds for real collaborative research and development.
Bioinformatics in this context
Bioinformatics is an established, highly interdisciplinary, field that aims to analyze biological data through the use of methods and technologies from mathematics, statistics, computer sciences, physics and, of course, biology and medicine.
Bioinformatics deals with heterogeneous data, ranging from structured and unstructured text, natural and synthetic images, diagrams and schema, and including data such as raw sequences, annotated genomes, protein structures, expression profiles, deep-sequencing data, networks and pathways, ontology relation diagrams, and so on. Moreover, the amount of available information is growing exponentially, together with the means to store and analyse it. Data are available online from different repositories with heterogeneous formats, and algorithms to analyse them are rarely able to inter-communicate and inter-operate.
Extracting knowledge from biological data has become a very complex task. In addition, expertise and skills are now increasingly more specialized and widely distributed: indeed, very few groups possess by themselves all the knowledge and skills needed to solve emerging problems. Groups naturally tend to collaborate in order to tackle unsolved issues and/or to gain insight into not yet understood biological mechanisms.
There is no shortage of life science projects that could exploit and benefit from collaboration among scientists: prediction and analysis of interaction networks (which involve various elements, like DNA, RNA, proteins and other molecules), design and discovery of microRNAs to alter protein function or gene expression and development of ontologies for coding and annotating biological data and knowledge, to name just a few.
Moreover, each of the above problems requires, in addition to computational (in silico) analysis, experimental (in vivo) biological analysis. The need to induce close interaction between in silico and in vivo researchers from different groups has recently prompted the development of new methods and tools (mostly domain independent) for bioinformatics collaboration [1,2].
What follows is a review of some of the technologies, tools and applications available for collaborative work, and a discussion of the prospects for their use to support bioinformatics.
TECHNOLOGIES AND APPLICATIONS FOR COLLABORATIVE RESEARCH AND DEVELOPMENT
The most recent network tools for collaborative research and development are impressive. Not only are researchers now closely and continuously in touch via email and instant messaging, but they can also jointly develop software, discuss publication contents, compare development strategies, write documents and build databases and knowledge bases.
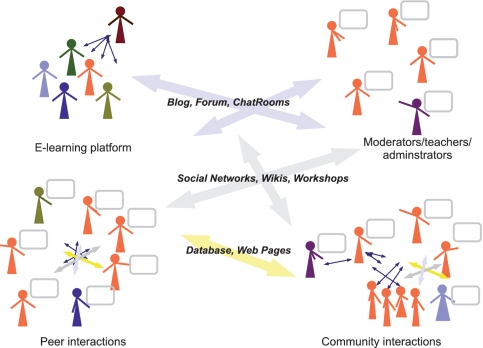
Figure 1 depicts some of the possible interactions among researchers. Collaboration allows sharing information or objects that may be stored in web pages or databases. It may be established between two researchers (peer-to-peer interactions) or among groups (many-to-many interactions), in which case it may be implemented by using collaborative systems. Communications and collaborations may be carried out through such technologies such as instant messaging, chat, blogs, forums, social networking and so on.
Figure 1:
Graphical representation of some of the possible interactions among researchers that may leverage on ICT technologies.
The direct applications in support of life sciences research are discussed below.
Social networking
Collaborative web sites were the first basic tool for cooperative development. Since they were meant to allow researchers to implement their systems in a shared place, collaboration features were limited. Bioinformatics.org (http://www.bioinformatics.org/) and the Open Bioinformatics Foundation (O|B|F, http://www.open-bio.org/wiki/Main_Page), home of bio* projects (BioPerl, BioJava, Biopython, BioRuby, and more), were two of the most interesting and stimulating examples of this kind.
People who have common interests and/or needs tend to form communities in order to communicate and to share knowledge. Social networks, also known as online communities, are now very popular and widely accessible. Based on the so-called Web 2.0 philosophy, which predicates a direct and close interaction between the user and the network service, users may interact and collaborate with each other as content creators, instead of viewing content that was created for them.
Interaction mainly entails authoring, i.e. the ability to add both original content and comments, and tagging the possibility to assign short textual tags to content to facilitate searching without the need for predefined categories. The collection of tags is referred to as a ‘folksonomy’ (i.e. folk taxonomy). A user may access a social network by creating a personal profile (an online identity), in which he/she provides private details, uploads objects (files) and posts opinions to be shared. Sharing may be public or restricted to a sub-network of users belonging to the same community.
Well-known examples of social networks are LinkedIn (http://linkedin.com), mainly a professional, business-related network, and Facebook (http://facebook.com) and Orkut (http://orkut.com), which are designed to connect friends and family, users with mutual interests (e.g. fans of sports teams or followers of a social campaign), and business owners with possible clients. Researchers, too, willing to compare or discuss theories, experiments or results, have become avid users. Other social networks, such as Flickr (http://flickr.com), dedicated to photography, YouTube (http://youtube.com) to videos, and MySpace (http://myspace.com) to music, do not require the creation of profiles, and content is shared with whomever accesses it.
Social networks and scientific collaboration
Many social networks have been deployed in the field of scientific collaboration. These are often devoted to sharing, commenting and tagging scientific publications. This is the case for BiomedExperts.com from Elsevier (http://www.biomedexperts.com/), which points out co-authorships of articles and allows graphical navigation inside collaborative networks, SlideShare (http://www.slideshare.net/), dedicated to sharing presentation slides, and CiteULike (http://www.citeulike.org/), Connotea (http://www.connotea.org/) and Mendeley (http://www.mendeley.com/).
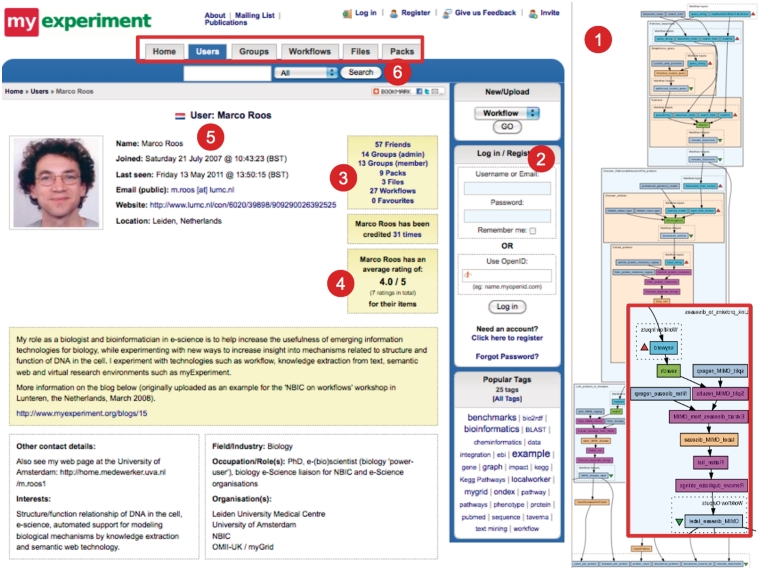
myExperiment (http://myexperiment.org) [3] is a social network for sharing and retrieving automated scientific workflows. To gain new knowledge bioinformatics research often requires applying analysis processes that are composed of many interrelated steps. The automation of such a process constitutes a workflow. Researchers may also reuse parts of workflows, and new workflows can be built on top of existing ones. Figure 2 shows the interface of myExperiment. myExperiment is based on a community of registered users. Participants may use, modify and re-upload any existing workflow. They can then create or join groups, while the system keeps track of friends/colleagues and workflows. A user can also add personal and working information. Users may recommend the professional ‘credibility’ of any participant, which is then reported to the community. Workflows are protected by copyright, so that rights of users who contributed to their release are guaranteed.
Figure 2:
The myExperiment interface. myExperiment allows to up- and download, analyse and run workflows. The pictured workflow (1) looks for diseases relevant to a query string. It finds documents related to the words in the query string, proteins from the abstract of the retrieved papers, filter false positive by requiring that they have a valid UniProt ID. Finally, it links proteins to diseases contained in the OMIM database (highlighted in the red box). A user must register (2) and he can then create or join some groups (3). The system keeps trace of his friends and workflows (3) and personal information (5). Other users can recommend his work ‘credibility’ (4). A web navigator can search for workflows, users and groups by inserting key words (6).
Other examples make use of social tagging. Annotea (http://annotea.org/) is a knowledge base that allows the sharing of web-based metadata. Annotations may include comments, notes or remarks that can be associated with a web page or to a part of it. Once a user retrieves the document, the attached annotations are also loaded and the user obtains the opinion of peers about it. These knowledge bases may also be used to automatically tag sentences [4] (http://tagme.di.unipi.it/).
Critical issues concerning social networks
Despite their popularity, social networks are still beset with several critical issues. Beyond the possible uncontrolled spread of incorrect information and the impossibility to check the credibility of information and to guarantee safe communications, it is noteworthy that networks are not inter-connected. More precisely, a user needs to identify himself in each network in which he participates, and communities may rarely merge [5]. Moreover, people do not have any control on their own personal data (e.g. images that other users publish online depicting them) [6].
A possible step forward to a better identification of users is OpenID, an open, decentralized authentication standard that allows users to log on to different services with the same digital identity. These services, however, must allow and implement the OpenID standard. myOpenID (https://www.myopenid.com/) is the first and largest independent OpenID provider.
Therefore, from the current centralized view of the web, that is seen as a set of isolated communities with some common members, researchers are migrating to decentralized web models [7], where users may select a trusted server as a repository for his/her data, where his/her own main ID is established, and grant access to these data to selected networks only. Such models [8,9] make use of tools allowing the standardization of formats, such as RDF, and ontologies for web content and users, such as FOAF (friend-of-a-friend) [10] and SIOC (semantically interlinked online communities, http://sioc-project.org/).
Documentation development tools
Google docs (http://docs.google.com/) and Windows Live Office (http://login.live.com/) are two of the best-known tools enabling Internet users to share and collectively edit documents. They facilitate the online creation, storage and sharing of text documents, spreadsheets, presentations and images. In addition, numerous users may simultaneously edit documents. Windows Live Office is built on top of SkyDrive, a password-protected file storage and sharing system: users are authenticated by Windows Live ID. A tight integration with the MS Office software suite is available, so that files may easily be downloaded, edited and re-uploaded.
Wiki systems have recently emerged as a network tool able to stimulate users to collaboratively contribute to the building of a common knowledge base. Well-known examples are proof of this concrete opportunity, first and foremost of which is the Wikipedia system (http://www.wikipedia.org/). The variety of advantages that wiki systems offer for the management of biological data and information have become evident. Some of the specific aims of wikis for biology (biological wikis) include collaborative efforts for the development and sharing of knowledge, and the creation and annotation of database contents.
The collaborative development and sharing of documentation and knowledge allows communities to promote, exploit, discuss and reach consensus on procedures, experiences and other varied information. Indeed, valuable expertise on and interests in special topics are usually distributed and are rarely concentrated in a unique site or research group.
The collaborative annotation of biological databases is increasingly under consideration because extended and accurate curation of an ever-increasing volume of data is both expensive and time consuming. Such distributed networks can help enhance and extend database curation beyond what it is usually possible because of limited numbers of dedicated staff. It allows users to contribute their expertise and observations independently of the database's centralized organization. Although the contents of the database are collaboratively annotated, the underlying database is left unchanged.
However, before these innovations may actually be implemented, some issues need to be addressed. The authoritativeness of contributions is essential and their quality must be assured. The open edition model of many wiki systems, e.g. Wikipedia, does not appear to be completely adequate, and some forms of user identification, as well as peer-evaluation of contributions, must be defined. Also, special features are needed in order to accommodate for the specific nature of the information in question, since textual information constitutes only a small part of biological data and many other heterogeneous data types, such as images, plots and diagrams, must be taken into account and properly managed.
Biological wikis
Some wiki systems devoted to biological research have already been developed, many of which were presented at the NETTAB/BBCC 2011 workshop on ‘Biological Wikis’ [11]. Here, we introduce some biological wikis that try to respond with above issues.
Gene Wiki [12,13] (http://en.wikipedia.org/wiki/Gene_Wiki and http://en.wikipedia.org/wiki/Portal:Gene_Wiki) is a specialized section of Wikipedia aimed at re-organizing, extending and completing its articles related to human genes. Wikipedia is indeed very popular and its articles often appear among first Google search results. The goal of Gene Wiki is to provide qualified information to a wide audience by making available high-quality articles for every notable human gene via one of the most widely used information systems. In 2008, Gene Wiki already counted more than 10 000 pages that were built starting from existing protein databases and improved through the contribution of an increasingly large user base. According to calculations by the maintainers of Gene Wiki, about the 86% of all its articles appear in the first page of the related Google search by gene symbol.
In order to verify this statement, we randomly selected a set of 9968 gene symbols from the HUGO Gene Nomenclature Committee (HGNC) database and searched all these terms with Google. As a result, we got 3709 links to the main Wikipedia site (http://en.wikipedia.org/) in the first page, i.e. about the 37% of searches returned a link to Wikipedia. By taking into account that about one-third of human genes are currently represented in Gene Wiki, this test tends to confirm the above statement. A similar test was carried out with the Bing search engine. In this case, we searched 11 494 symbols that returned 4247 hits to Wikipedia, with the same percentage as Google. We also had a closer look at results of those genes that are listed in the Gene Wiki site as the biggest by size of the description or by recent growth (Table 1).
Table 1:
Results of on-line searches of gene symbols referring to ‘Top Gene Wiki articles’, as shown in the Gene Wiki portal page (http://en.wikipedia.org/wiki/Portal:Gene_Wiki) by using Google and Bing
| Gene Symbol | Rank (size) | Rank (growth) | Bing | |
|---|---|---|---|---|
| RELN | 1 | 6 | 4 | 2 |
| HSPG2 | 2 | 1 | 2 | 1 |
| BIRC5 | 3 | – | 2 | 2 |
| SULF1 | 4 | 2 | 2 | 2 |
| INS | 5 | – | 3a | >50 |
| SFRP1 | 6 | – | 2 | 1 |
| HTR2A | 7 | 7 | 2 | 1 |
| CST3 | 8 | – | 2 | 7a |
| H19 | 9 | – | 1 | 28 |
| GCK | 10 | – | 5 | 32a |
| KCNA3 | – | 3 | 2 | 1 |
| ADORA2A | – | 4 | 2 | 4 |
| HTR1A | – | 5 | 2 | 1 |
| KITLG | – | 8 | 2 | 2 |
| TYK2 | – | 9 | 1 | 1 |
| MAOA | – | 10 | 3 | >50 |
When searching with Google, a link to the related Gene Wiki article was found in the first page for all 16 gene symbols. A similar result was achieved by using Bing, although in this case links to Gene Wiki did not appear in the first result page for four symbols. aLink to a disambiguation page of Wikipedia.
Wikipedia is implemented using MediaWiki (http://www.mediawiki.org/), a wiki development tool that has the great advantage of being based on a modular structure, with a simple extension mechanism that allows implementing new features. Semantic MediaWiki (http://semantic-mediawiki.org/wiki/Semantic_MediaWiki) is an extension that allows storing and querying wiki pages, and it is especially useful for biological wikis linking to biological databases.
WikiGenes [14] (http://www.wikigenes.org/) is a wiki system whose main goal is to encourage the collaborative creation of scientific papers by taking into account all contributions, even minor ones. In each article, every text is associated with its author. Moreover, a page is defined for each author where his/her publications, expertise, and contributions to WikiGenes are listed. Other researchers may then evaluate authors as in peer-review systems and scores may be associated with contributions. The result of this approach is that users may examine each single contribution, verify who provided which contents and assess their accuracy and viability. WikiGenes also includes a feature that allows authors to add annotations and links to external systems, such as PubChem, NCBI Gene, Uniprot and Pubmed.
WikiPathways [15] (http://www.wikipathways.org/) is a wiki system aimed at complementing some existing databases of metabolic pathways (KEGG, Reactome, Pathway Commons). A large community of researchers, not restricted to the most expert in the field, may comment, annotate and suggest changes, without directly affecting the databases. Administrators may take advantage of these annotations and possibly correct and/or update their databases. Within WikiPathways, each pathway is represented in a distinct page, where its diagram, overall description, components and history of changes are included. A graphical editor allows making some changes to the diagram. Pathways may be searched by names of components and by free text descriptions and annotations. Browsing by species and by ontology terms is also allowed. Pathways may be downloaded in various standard formats.
WikiProteins [16] (http://www.wikiprofessional.org/) is based on the ‘Concept Web’ idea. Millions of biomedical ‘concepts’ are currently available and distributed in databases, reference thesauri and ontologies. Many of these concepts were extracted from UMLS, UniProtKB, IntAct and Gene Ontology, and stored, together with their inter-relations, using an original technology based on basic knowledge units, so-called knowlets that specify a pair of concepts and their relation, which is also annotated by its evidence category. The ‘concept space’ is then populated by all knowlets and can be displayed using proper filters based on concepts or evidence categories. The concept space can also be converted to RDF and consequently searched by using SPARQL query language.
For each concept, WikiProteins presents one page. All information connected to the concept is automatically included by extracting it from the concept space. All other concepts present in the page are highlighted and may be used as a link to the related WikiProteins page, thus allowing end users to navigate the wiki (and the concept space). Registered users may update WikiProteins pages. These changes, however, are not automatically converted into the concept space: they are examined and assessed by the administrator of the system and may be incorporated into the concept space only at a later stage.
Collaborative ontology development
In the development of biological ontologies, collaborative editing is crucial. Ontologies are defined as ‘formal, explicit specifications of shared conceptualizations’ [17]. They are often the result of an effort that is carried out by a community of experts. For this, it is important that they access a common editing tool. Collaborative development has been featured by various ontology editors. Noy et al. [18] conducted a study to compare features and tools for collaborative knowledge construction.
Protégé (http://protege.stanford.edu/) is an ontology editing and knowledge acquisition tool under development at Stanford University [19] with an active, international user community, adopted by many projects (a list is available at http://protege.cim3.net/cgi-bin/wiki.pl?ProjectsThatUseProtege). Collaborative Protégé [20] is an extension that supports collaborative ontology editing as well as annotation of ontology components and changes. Its main features are the ability to create notes and attach them to different components (classes, properties and instances) and to track changes, so that the history of changes may be managed. Notes may be classified according to a classification including, e.g. advice, comment, example, explanation and question. Collaborative Protégé also includes features for communicating, discussing and voting among participants. WebProtégé [21] is a web-client for Collaborative Protégé that allows collaborative ontology development in a web environment.
Software development tools
Software development relies heavily on collaboration. Software engineers within and outside project teams (co-located or remotely located) need to properly interact and coordinate their work in the production of complex systems. Establishing a suitable collaborative infrastructure that allows the maintenance of a shared understanding of artefacts, modules and activities is a difficult task [22–24]. Several factors, such as the structure of the team and the application domain, must be taken into account. Furthermore, developer teams usually have their favourite collections of legacy tools, which are commonly determined by a historical usage.
Principles behind collaborative development environments
In literature, some frameworks, which allow categorizing tools with respect to their application area, functionalities and approaches to collaboration are described [22–27].
In Ref. [24], a categorization of tools based on implementation effort, defined as the time spent by the user to setup the tool, is introduced. Authors introduce a pyramid framework, which recognizes five levels of coordination support and three critical crosscutting tools categories (artefacts management, task management and communication). Tools that are located higher in the pyramid layer provide more sophisticated automated support, thereby reducing the user effort required in collaborating.
In Ref. [27], the authors provide a taxonomy of current collaboration tools [Table 2, adaptation from (27)]. These are categorized in a practical manner as version control systems that allow users to share artefacts, web accessible trackers able to manage issues such as tickets or bugs, remote building tools, modellers allowing the creation of formal artefacts including UML, knowledge centres that permit users to share knowledge through the web, and communication tools which support remote interactions.
Table 2:
A taxonomy of collaboration tools and a list of some representative systems with web site addresses [adapted from Ref. (27)]
| Category | Goal | System | Website |
|---|---|---|---|
| Version control systems | Allowing to share artefacts | CVS | http://savannah.nongnu.org/projects/cvs |
| Subversion | http://subversion.apache.org/ | ||
| Git | http://git-scm.com/ | ||
| Bazar | http://bazaar.canonical.com/ | ||
| Darcs | http://darcs.net/ | ||
| Mercurial | http://mercurial.selenic.com/ | ||
| Web accessible trackers | Managing issues such as tickets or bugs | Jira | http://www.atlassian.com/ |
| Mantis | http://www.mantisbt.org/ | ||
| Bugzilla | http://www.bugzilla.org/ | ||
| Remote building tools | Supporting application deployment | Maven | http://maven.apache.org/ |
| Ant | http://ant.apache.org/ | ||
| CruiseControl | http://cruisecontrol.sourceforge.net/ | ||
| Premake | http://industriousone.com/premake | ||
| Modelers | Allowing model-based collaborations to create | Visible Analyst | http://www.visible.com |
| formal artefacts | Collaborative UML | http://sourceforge.net/projects/rtumldesigner/ | |
| Knowledge centers | Sharing knowledge through the web | KnowledgeTree | http://www.knowledgetree.com/ |
| Communication tools | Managing remote interactions | eConference | http://code.google.com/p/econference4/ |
| Google Wave | http://wave.google.com/ |
Those categories are then plugged into the more general Collaborative Development Environment (CDE) that yields a workspace composed of a set of standardized tools suitable for global software development teams. A comparison of open source hosting facilities conceived as CDEs can be found in Wikipedia (http://en.wikipedia.org/wiki/Comparison_of_open_source_software_hosting_facilities).
Following the definition of awareness given by Dourish and Bellotti [28] (‘an understanding of the activities of others, which provides a context for your own activities’), Omoronyia et al. [29] identified five types of high-level awareness that are suitable to model collaborative software development tools. ‘Workspace or activity awareness’ allows defining a model to track interactions in the shared workspace. ‘Informal awareness’, which is commonly employed by instant messaging systems, provides the knowledge about who is around and who could be available for a task. ‘Group-structural awareness’ establishes roles, responsibilities and positions. ‘Social awareness’ measures the user-interest in the collaborative tasks. Finally, ‘context awareness’ is a cross-section of all the other categories of awareness, including issues such as the workspace context of tasks and artefacts, their changing states over time, and collaborators. Improvements of awareness in distributed software, mainly based on Web 2.0 applications, can be found within Integrated Development Environments (IDE) and related tools [27].
Jazz (http://www.jazz.net/), a real-time team collaboration platform built on top of the Eclipse IDE, allows integrating work spread across distributed development sites. Jazz supports the tagging of development tasks by user-defined keywords. TagSEA (Tags for Software Engineering Activities in Eclipse, http://tagsea.sourceforge.net/), which is based on the concept of Waypoints (locations of interest) and social tagging (social bookmarking), facilitates the collaborative annotation during software development. CASSIUS [30], a notification server, allows users to model software hierarchies so that an end user can subscribe and browse through those hierarchies he/she is interested in.
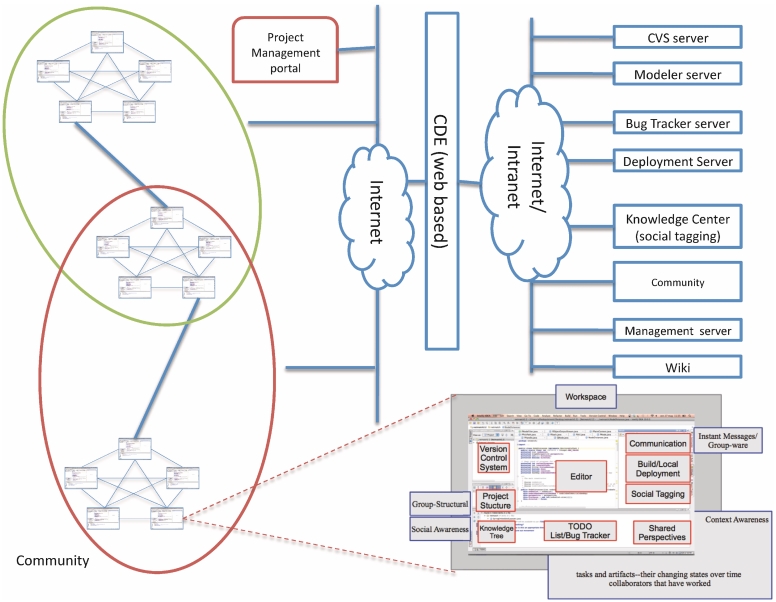
In Refs [31,32], mining algorithms, such as the HITS algorithm [33] for recommendation, are applied among software project entities. Rational Team Concert (http://jazz.net/projects/rational-team-concert/), implemented on top of the Jazz Framework, allows mining relations of awareness keys within shared software projects. Ariadne [34] (http://awareness.ics.uci.edu/~ariadne/), a plug-in for Eclipse, analyses dependences in software projects by collecting authorship information. The tool translates technical dependences among components into social dependences among developers and graphically describes the dependence information (the general architecture of a CDE Figure 3).
Figure 3:
The general architecture of a Collaborative Development Environment (CDE). Integrated Development Environments (IDEs) are equipped with a set of integrated tools allowing awareness and interaction among users communities.
Collaborative development environments in bioinformatics
Many CDEs are used to build bioinformatics software. Freshmeat (http://freshmeat.net/), OpenSymphony (http://www.opensymphony.com/), GitHub (http://github.com/), CodePlex (http://www.codeplex.com/) and launchpad (https://launchpad.net/) host several projects for the analysis of biological data. Although we are only at the beginning of such development software in the field of bioinformatics, several successful initiatives are already present.
Bioconductor [35] implements many tools for the analysis of high-throughput genomic data on top of R programming language. It is open source and open development. It has two releases per year, more than 460 packages and an active user community. Cytoscape [36] is a bioinformatics tool for the visualization and analysis of biological networks. A ‘Core’ tool provides basic functionality for network layout and query and for visually integrating the network with data. The Core is extensible through a plug-in architecture, allowing rapid development of additional computational analyses and features.
In Ref. [37], the authors propose a model-driven approach to the collaborative design of distributed web services based on jABC (http://www.jabc.de/), a framework for service development based on lightweight process coordination. Extensions can be found in Refs [38,39].Confucius [40], previously named Co-Taverna [41], allows the collaborative composition of scientific workflows. It is based on an ontology of scientific collaboration based on a set of primitives and patterns. Collaboration protocols are then applied to support effective concurrency control in the process of collaborative workflow composition. Biocep-R [42] is an open source for the virtualization of scientific computing environments (SCEs) such as R and Scilab. It allows the collaborative analysis of computation tools running on the Cloud.
Education and training tools
In the connected era, human knowledge is growing exponentially. This results in the paradox that the more we have to learn, the less time we have to learn it. We are thus faced with the challenge keep pace with everything we must know, when we must know it [43]. One strategy relies on capturing knowledge so that it can be instantaneously accessed and shared.
The technological revolution underpinned by a strong pedagogical theory, based on constructivism, connection and separations concepts, allows us to reach such a target.
Pedagogical principles
According to the theory of constructivism [44], interaction of human experiences and ideas generates knowledge: we learn from the environment and from each other. The implications in e-learning are remarkable. Commonly, groups rank what is knowledge and at the same time determine what is not considered knowledge at all.
Constructivism derives from a more general concept called social constructionism [45], which is based on the idea that the best way for people to learn is being involved in a social process of constructing knowledge for others. The process of negotiating semantics and utilizing shared artefacts is a process of constructing knowledge too. This results in the fact that learning is something we do mainly in groups. Thus, learning can be viewed as a process of negotiating meaning in a culture of shared artefacts and symbols [45,46].
Moreover, concepts such as connections and separations reveal that the sharing of information among communities stimulates the behaviour of a single user. However, the single user should carefully retain his individualism and his own ideas.
In the field of bioinformatics, preliminary studies in small communities have shown the effectiveness of such an approach, compared to traditional methods, in the cooperative learning of students of biochemistry classes [47]. Those outcomes were subsequently confirmed by a combination of a standard bioinformatics course with a web-based virtual laboratory aimed at stimulating collaboration and peer support on technical questions [48].
Collaboration may be across classrooms, communities and countries and may make use of tools such as blogs, sharing of videos and so on. These also guarantee peer-to-peer communication, which is at the heart of a collaborative learning process (Figure 1). However, important to the success of collaborations, in terms of quality and duration over time, is the environment, which needs to be flexible, easy to use and adaptable to suit the needs of members.
Learning management systems
Learning Management Systems (LMSs) are software that automates the administration of training events. The LMS approach, which is increasingly used for university courses, particularly for small groups [47], is able to assist students by guaranteeing a variety of learning outcomes, including working collaboratively with others, taking responsibility for their own learning and deepening their understanding of course contents. Moodle and Drupal [49–51] are two successful examples of LMSs (other more general purpose software packages are available at wordpress.com, dotnetnuke.com, educommons.com, atutor.ca).
Moodle stands for modular object-oriented dynamic learning environment, but used as a verb it denotes a process of enjoyable tinkering that often leads to increased knowledge, insight and creativity. This fits both the philosophy underpinning Moodle's development and the way it is used to teach and learn. Its main goal is to create rich interactions between teachers and learners. Its main features are: store, communicate, evaluate and collaborate. Users can
store files, web pages, folders, links and digital documents;
communicate through fora, messaging and chat rooms, thereby allowing class discussions and debates, instant feedback to solve problems, private conversations and subscription to blogs, fora and Wikis;
collaborate through blogs, Wikis, glossaries, social networks, fora, workshops, databases and lessons;
correct quizzes and grade assignments.
Users may act as administrators, teachers, students, parents and guests. Students may share notes, see and debate on line the correction and grading of their homework and watch lessons. Teachers may collect all their lessons, grades and corrected assignments in one place, cumulate scores, disciplinary actions and notes, and learn from the feedback and interactions with and among their students.
Drupal is not a traditional LMS, but contains viable modules that can manage the learning process [52]. It is modular, in that its basic features are included in the ‘core’ package, while thousands of community developed modules make it possible to construct a dynamic web site for any application. Everything a user creates in Drupal is a node, which is a piece of content of the web site. Drupal is also flexible: when creating a web site, one can choose from among several different content structures. One of the many uses of Drupal is the creation of a collaborative book in which chapters, sections and subsections may be managed as pages. A group of users may work together in writing, modifying and organizing pages. Examples of Drupal's use come from Economist.com, the weekly magazine focusing on international politics and business news, HowToDoThings.com, which aims at solving everyday problems, and the World Wild Fund for Nature (panda.org), the leading international organization dedicated to conservation and protection of the environment.
Due to the boom of heterogeneous e-learning systems, rules to ensure compatibility (standardization) are needed. One of the first efforts in this direction is SCORM (Shareable Content Object Reference Model, http://scorm.com), which provides standard objects to be shared among LMSs. Projects such as DotNetScorm (http://dotnetscorm.codeplex.com) are aimed at creating SCORM standards.
DISCUSSION AND CONCLUSION
Technologies and applications for collaborative research and development, including those supporting document creation, software development and education and training, are evolving intensively. These new tools are often based on the principles of social networks and thus introduce into a researcher's daily activities continuous interaction with peers through large communities of users.
Although the fall-out of these collaborative environments in bioinformatics research is still limited to a few, but enlightening, cases, there are clear prospects for their utilization in the short- to mid-term. These include the creation of coherent and comprehensive knowledge bases supported by highly qualified experts, the development of modular and interoperable software based on common data models and structures, the carrying out of standardized, public, comprehensive online courses aimed at shared education and training in bioinformatics given by the most distinguished scientists and professors. Before these goals may be reached, however, a number of issues must be faced and solved.
Assessing and ensuring a digital identity is still difficult, if not impossible. Instead, it should be granted in order to guarantee privacy and to prevent impostors. User names and passwords alone cannot authenticate the identity of researchers, who should be urged to adopt unique open identities for their participation in collaborative activities. Authentication of researchers is indeed essential: knowing who is who prevents fraud, assigns rights on functions, actions and documents, and attributes the origin of annotations, comments and information. Also, knowing who actually did what, that is disambiguating authorship, is needed in order to assign credits to users for their contributions. This can be extremely relevant to stimulate the broadest and most qualified participation in collaborative efforts.
Development of modular open source tools is still far from being satisfactory. Additional common data models and structures are needed so that software tools may be developed and updated faster and easily reused.
Semantic Wiki systems could provide the grounds for the construction of a shared knowledge base. A survey of existing systems, and of current developments, would be useful in order to identify possible synergies and acknowledge the best efforts achieved by relevant communities, as well as to ensure a coherent set of interoperable biological wikis and to support the majority of biological databases.
Solving these problems and developing more advanced tools for collaborative research would no doubt bring about a change in scientists’ attitude and outlook, leading towards what we could call Science 2.0: a new paradigm of research based on the free and widespread availability of data, the sharing and reuse of methods and tools and the collaborative pursuit of common goals and objectives.
For this to happen, a major effort is needed. Interested communities should meet and discuss possible collaborations, interactions and convergence on common technologies and tools. Public courses on tools and technologies for collaborative work in support of bioinformatics should be designed, implemented and promoted.
Key points.
At present, biological research projects may greatly benefit from a broad collaboration of scientists, from different domains and with different expertise and skills.
Researchers are now closely connected through networks in which they can develop software, discuss publication content, compare research strategies, write documents and collectively build data and knowledge bases.
The adoption of Web 2.0 approaches, which implies a close interaction between users and network services and enables researchers to interact and collaborate with each other as content creators, may be the basis for a new generation of collaborative tools for research.
FUNDING
This work was partially funded by the Italian Ministry of Education, University and Scientific and Technology Research (MIUR), project Laboratory for Interdisciplinary Technologies in Bioinformatics (LITBIO), and by the Italian Ministry of Health, project National Network for Oncology Bioinformatics (Rete Nazionale di Bioinformatica Oncologica – RNBIO).
Acknowledgements
Authors wish to thank Tom Wiley for his precious support in the preparation of the final version of the article.
Biographies
Paolo Romano obtained his PhD in bioengineering degree from the Polytechnic of Milan. Since 1993 he has been a researcher at the National Cancer Research Institute of Genoa. His interests include biological databases, data modelling and integration, automation of retrieval and analysis processes through semantic tools and programming interfaces.
Rosalba Giugno is Assistant Professor in Computer Science at the University of Catania. She has been a visiting researcher at Cornell University, the University of Maryland and New York University. Her research interests include data mining and algorithms for bioinformatics.
Alfredo Pulvirenti is an Assistant Professor of Computer Science at the University of Catania. He has been a visiting researcher at New York University. His research interests include data mining and machine learning, and algorithms for bioinformatics.
References
- 1.Marcus FB. Bioinformatics and Systems Biology. Collaborative Research and Resources. Berlin Heidelberg: Springer; 2008. [Google Scholar]
- 2.Parslow GR. Multimedia in biochemistry and molecular biology education. Int Union Biochem Mol Biol. 2006;34(3):232–4. doi: 10.1002/bmb.2006.49403403232. [DOI] [PubMed] [Google Scholar]
- 3.Goble CA, Bhagat J, Aleksejevs S, et al. My experiment: a repository and social network for the sharing of bioinformatics workflows. Nucleic Acids Res. 2010;38(Suppl. 2):W677–82. doi: 10.1093/nar/gkq429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferragina P, Scaiella U. Proc. 19th ACM International Conference on Information and Knowledge Management CIKM 2010, 26–30 October 2001. Toronto, Canada: ACM New York, NY, USA; TAGME: on-the-fly annotation of short text fragments (by Wikipedia entities) pp. 1625–8. [Google Scholar]
- 5.Fitzpatrick B, Recordon D. Thoughts on the Social Graph. http://bradfitz.com/social-graph-problem (25 August 2011, date last accessed) [Google Scholar]
- 6.Kang T, Kagal L. Enabling Privacy-Awareness in Social Networks, AAAI Spring Symposium Series. Menlo Park, California, USA: 2010. [Google Scholar]
- 7.Yeung CA, Liccardi I, Lu K, et al. Decentralization: The Future of Online Social Networking, W3C Workshop on the Future of Social Networking. Position Papers, 2009. Barcelona, Spain. [Google Scholar]
- 8.Ahmadi N, Jazayeri M, Lelli F, et al. 23rd IEEE/ACM International Conference on Automated Software Engineering 2008, 15-16 September, 2008. L'Aquila, Italy: A Survey of Social Software Engineering. [Google Scholar]
- 9.Whitehead J, Mistrík I, Grundy J, et al., editors. Collaborative Software Engineering. Berlin Heidelberg: Springer; 2010. [Google Scholar]
- 10.Brickley D, Miller L. FOAF Vocabulary Specification. http://xmlns.com/foaf/spec/ (25 August 2011, date last accessed) [Google Scholar]
- 11.Facchiano A, Romano P. Network Tools and Applications in Biology NETTAB–BBCC 2010 Biological Wikis, November 29–December 1, 2010, Napoli, Italy. Roma, Italy: Aracne editrice S.r.l.; 2010. [Google Scholar]
- 12.Huss JW, Lindenbaum P, Martone M, et al. The Gene Wiki: community intelligence applied to human gene annotation. Nucleic Acids Res. 2010;38(Database issue):D633–9. doi: 10.1093/nar/gkp760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huss JW, III, Orozco C, Goodale J, et al. A Gene Wiki for community annotation of gene function. PLoS Biol. 2008;6(7):e175. doi: 10.1371/journal.pbio.0060175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffmann R. A wiki for the life sciences where authorship matters. Nat Genet. 2008;40:1047–51. doi: 10.1038/ng.f.217. [DOI] [PubMed] [Google Scholar]
- 15.Pico AR, Kelder T, Van Iersel MP, et al. WikiPathways: pathway editing for the people. PLoS Biol. 2008;6(7):e184. doi: 10.1371/journal.pbio.0060184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mons B, Ashburner M, Chichester C, et al. Calling on a million minds for community annotation in WikiProteins. Genome Biol. 2008;9:R89. doi: 10.1186/gb-2008-9-5-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Studer R, Benjamins R, Fensel D. Knowledge engineering: principles and methods. Data Knowl Eng. 1998;25(1–2):161–198. [Google Scholar]
- 18.Noy NF, Chugh A, Alani H. The CKC Challenge: exploring tools for collaborative knowledge construction. Intell Syst IEEE. 2008;23(1):64–8. doi: 10.1109/MIS.2008.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rubin DL, Noy NF, Musen MA. Protégé: a tool for managing and using terminology in radiology applications. J Digit Imaging. 2007;20(Suppl. 1):34–46. doi: 10.1007/s10278-007-9065-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tudorache T, Noy NF, Tu SW, et al. Seventh International Semantic Web Conference, ISWC 2008. Karlsruhe, Germany: Springer; 2008. Supporting collaborative ontology development in Protégé. [Google Scholar]
- 21.Tudorache T, Vendetti J, Noy NF. Fifth International Workshop, Karlsruhe, Germany, 26–27 October 2008 co-located with ISWC 2008. Germany: Springer, Heidelberg; 2008. Web-Protégé: A Lightweight OWL Ontology Editor for the Web. In OWL: Experiences and Directions (OWLED 2008) [Google Scholar]
- 22.Storey MA-D, Cubranic D, German DM. Proceedings of the 2005 ACM Symposium on Software Visualization. St Louis, MO: 2005. On the use of visualization to support awareness of human activities in software development: a survey and a framework; pp. 193–202. [Google Scholar]
- 23.Gutwin C, Greenberg S, Roseman M. Proc. of the International Conference on Human–Computer Interaction: People and Computers XI. London: 1996. Workspace awareness in real-time distributed groupware: framework, widgets, and evaluation; pp. 281–298. [Google Scholar]
- 24.Sarma A. A survey of collaborative tools in software development. UCI ISR Technical Report, UCI-ISR-05-3, 2005. [Google Scholar]
- 25.Dewan P. Proceedings of the 2008 International Workshop on Recommendation Systems for Software Engineering. Atlanta, GA: 2008. Dimensions of tools for detecting software conflicts; pp. 21–25. [Google Scholar]
- 26.Jimenez M, Piattini M, Vizcaìno A. Challenges and improvements in distributed software development: a systematic review. Advances in Software Engineering. New York, USA: Hindawi Publishing Corporation; 2009. [Google Scholar]
- 27.Lanubile F, Ebert C, Prikladnicki R, et al. Collaboration tools for global software engineering. IEEE Software. 2010;27(2):52–55. [Google Scholar]
- 28.Dourish P, Bellotti V. Proc. of the ACM Conference on Computer Supported Cooperative Work (CSCW) Toronto: 1992. Awareness sand coordination in shared workspaces; pp. 107–14. [Google Scholar]
- 29.Omoronyia I, Ferguson J, Roper M, et al. A review of awareness in distributed collaborative software engineering. Softw-Pract Exp. 2010;40:1107–33. [Google Scholar]
- 30.Kantor M, Redmiles D. Creating an Infrastructure for Ubiquitous Awareness. Eighth IFIP TC 13 Conference on Human-Computer Interaction (INTERACT 2001) Tokyo, Japan: 2001. [Google Scholar]
- 31.Saul ZM, Filkov V, Devanbu P, et al. Recommending Random Walks. ACM New York, NY, USA: ACM SIGSOFT; 2007. [Google Scholar]
- 32.Robillard MP. Automatic Generation of Suggestions for Program Investigation. ACM New York, NY, USA: ACM SIGSOFT; 2005. [Google Scholar]
- 33.Kleinberg J. Authoritative Sources in a Hyperlinked Environment. ACM-SIAM Symposium on Discrete Algorithms; 1998. Extended version in Journal of the ACM 1999;46:604–632. [Google Scholar]
- 34.de Souza CRB, Quirk S, Trainer E, et al. Supporting Collaborative Software Development through the Visualization of Socio-Technical Dependencies. ACM Conference on Supporting Group Work. Sanibel Island, FL: ACM Press; 2007. [Google Scholar]
- 35.Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Margaria T, Kubczak C, Njoku M, et al. Proc. of the 11th International Conference on Engineering of Complex Computer Systems (ICECCS), 15-17 August 2006. Stanford, CA, USA: IEEE Computer Society and Press; 2006. Model-based design of distributed collaborative bioinformatics processes in the jABC; pp. 169–176. [Google Scholar]
- 38.Margaria T, Kubczak C, Steffen B. Bio-jETI: a service integration, design, and provisioning platform for orchestrated bioinformatics processes. BMC Bioinformatics. 2008;9(Suppl. 4):S12. doi: 10.1186/1471-2105-9-S4-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lamprecht AL, Margaria T, Steffen B. Bio-jETI: a framework for semantics-based service composition. BMC Bioinformatics. 2009;10(Suppl. 10):S8. doi: 10.1186/1471-2105-10-S10-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang J, Kuc D, Lu S. Proceedings of IEEE International Conference on Web Services (ICWS) Miami, FL, USA: 2010. Confucius: a scientific collaboration system using collaborative scientific workflows; pp. 567–75. [Google Scholar]
- 41.Zhang J. Proceedings of SCC. Miami, FL, USA: 2010. Co-Taverna: a tool supporting collaborative scientific workflows. [Google Scholar]
- 42.Chine K. IEEE International Workshop on Open Source Software for Scientific Computation (OSSC) Piscataway, NJ, USA: IEEE Press; 2009. Scientific Computing Environments in the age of Virtualization, toward a universal platform for the Cloud; pp. 44–48. [Google Scholar]
- 43.Rosemberg MJ. Beyond E-Learning: Approaches and Technologies to Enhance Organizational Knowledge, Learning, and Performance. San Francisco, CA, USA: Pfeiffer; 2005. [Google Scholar]
- 44.Chermack TJ, van der Merwe L. The role of constructivist learning in scenario planning. Futures. 2003;35(5):445–60. [Google Scholar]
- 45.Grabinger RS, Dunlap JC. Rich environments for active learning in the higher education classroom. In: Wilson BG, editor. Constructivist Learning Environments: Case Studies in Instructional Design. Englewood Cliffs. New Jersey: Educational Technology Publications; 1996. [Google Scholar]
- 46.Cole J, Foster H. Using Moodle: Teaching with the Popular Open Source Course Management System. Sebastopol, CA, USA: O'Reilly Media; 2007. [Google Scholar]
- 47.Anderson WL, Mitchell SM, Osgood MP. Comparison of student performance in cooperative learning and traditional lecture-based biochemistry classes. Int Union Biochem Mol Biol. 2005;33(6):387–93. doi: 10.1002/bmb.2005.49403306387. [DOI] [PubMed] [Google Scholar]
- 48.Weisman D. Incorporating a collaborative web-based virtual laboratory in an undergraduate bioinformatics course. Biochem Mol Biol Educ. 2010;38(1):4–9. doi: 10.1002/bmb.20368. [DOI] [PubMed] [Google Scholar]
- 49.Dougiamas M, Taylor PC. Proceedings of the EDMEDIA 2003 Conference. Honolulu, Hawaii: 2003. Moodle: Using Learning Communities to Create an Open Source Course Management System. [Google Scholar]
- 50.Dougiamas M. Developing tools to foster online educational dialogue. In: Martin K, Stanley N, Davison N, editors. Teaching in the Disciplines/Learning in Context, 119–123. Proc. of the 8th Annual Teaching Learning Forum, The University of Western Australia, Perth, UWA, 1999. [Google Scholar]
- 51.Mercer D. Create and Operate any Type of Website Quickly and Efficiently. Birmingham, UK and Mumbai, India: Packt Publishing; 2010. Drupal 7. [Google Scholar]
- 52.Fitzgerald B. Drupal for Education and E-learning, Teaching and Learning in the Classroom using Drupal CMS. Birmingham, UK and Mumbai, India: Packt Publishing; 2009. [Google Scholar]