SUMMARY
Therapy-related myelodysplasia or acute myeloid leukemia (t-MDS/AML) is a major complication of cancer treatment. We compared gene expression in CD34+ cells from patients who developed t-MDS/AML after autologous hematopoietic cell transplantation (aHCT) for lymphoma with controls who did not develop t-MDS/AML. We observed altered gene expression related to mitochondrial function, metabolism, and hematopoietic regulation in pre-aHCT samples from patients who subsequently developed t-MDS/AML. Progression to overt t-MDS/AML was associated with additional alterations in cell-cycle regulatory genes. An optimal 38-gene PBSC classifier accurately distinguished patients who did or did not develop t-MDS/AML in an independent group of patients. We conclude that genetic programs associated with t-MDS/AML are perturbed long before disease onset, and accurately identify patients at risk for this complication.
INTRODUCTION
Therapy-related myelodysplasia or acute myeloid leukemia (t-MDS/AML) is a lethal complication of cytotoxic cancer therapy. Among patients undergoing autologous hematopoietic cell transplantation (aHCT) for Hodgkin lymphoma (HL) or non-Hodgkin lymphoma (NHL), t-MDS/AML is a leading cause of non-relapse mortality (Bhatia et al., 2005; Bhatia et al., 1996; Miller et al., 1994; Pedersen-Bjergaard et al., 2000; Stone et al., 1994). Pre-aHCT therapeutic exposures, transplant conditioning, autograft collection and hematopoietic regeneration contribute to development of t-MDS/AML (Bhatia et al., 1996; Kalaycio et al., 2006; Krishnan et al., 2000). The overwhelming majority of patients develop t-MDS/AML within 6 years after aHCT. However the timing and sequence of acquisition of molecular abnormalities leading to t-MDS/AML is unknown. t-MDS/AML accounts for 15% of all AML and MDS cases and shares morphologic and cytogenetic characteristics with primary MDS and AML in the elderly. Study of t-MDS/AML offers a unique opportunity to understand leukemogenesis since known genotoxic exposures can be temporally and causally related to genetic changes associated with subsequent development of leukemia (Mason, 2003; Pedersen-Bjergaard, 2005).
To better understand the pathogenetic mechanisms underlying t-MDS/AML, we have constructed a prospective cohort of patients undergoing aHCT for HL or NHL in order to improve our understanding of the pathogenesis of t-MDS/AML. Patients are followed longitudinally with collection of peripheral blood stem cells (PBSC) and bone marrow (BM) samples prior to a-HCT, and serial BM samples till 5-years post-aHCT. This design allows use of a nested case-control approach to compare gene expression profiles in CD34+ hematopoietic stem and progenitor cells (HSC) from “cases” that developed t-MDS/AML after aHCT with “controls” who did not develop t-MDS/AML. In the current report, PBSC procured pre-aHCT and BM samples obtained at time of t-MDS/AML post-aHCT were studied. This approach facilitated identification of gene expression changes pre-aHCT in patients who subsequently developed t-MDS/AML after aHCT. This approach also allowed a comparison of gene expression pre-aHCT with that seen at development of overt t-MDS/AML. Finally, using an independent sample set (test set), we investigated whether gene expression in pre-aHCT samples could accurately identify patients at risk for development of post-aHCT t-MDS/AML.
RESULTS
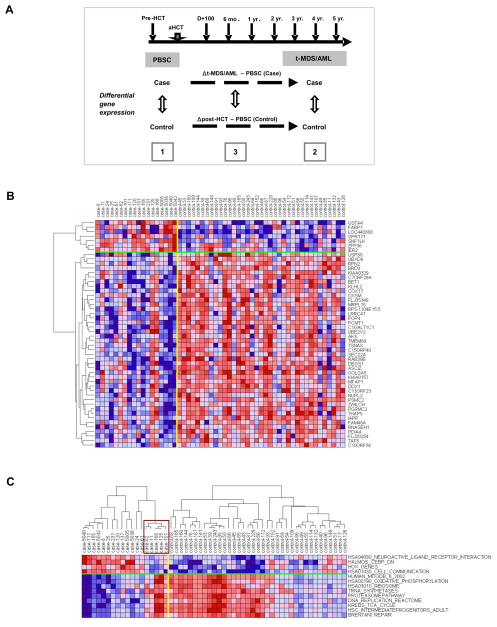
We compared gene expression in CD34+ cells from the training set consisting of 18 cases that developed t-MDS/AML and 37 matched controls that did not develop t-MDS/AML after aHCT for HL or NHL. One to three randomly selected controls were individually matched to each case for primary diagnosis [HL/NHL], age at aHCT [±10 years], and race/ethnicity [Caucasians, African-Americans, Hispanics, other]. The median time to t-MDS/AML post-aHCT was 2.7 years (range, 0.5 to 5.2 years). For each case, controls were selected that had been followed for a length of time that exceeded the latency from aHCT to t-MDS for the index case, to ensure that the probability of the controls developing t-MDS subsequently was minimized. The length of follow-up from aHCT for cases is 33.4 months (range: 5.9 to 63.7 months) and for controls is 116 months (range: 75.8 to 136 months). The clinical and demographic characteristics of the cases and controls are shown in Table S1. Comparison of cases with controls revealed no significant differences in primary diagnosis, sex, race/ethnicity, age at primary diagnosis and aHCT, stem cell source and mobilization regimens, number of PBSC collections, CD34+ cell dose, and conditioning regimens. Detailed analysis of pre-aHCT therapeutic exposures (including cumulative doses), HCT-related conditioning, and post-aHCT therapeutic exposure (in the event of relapse) did not reveal any statistically significant difference in the intensity or nature of therapeutic exposures between case and controls (Table S1). The clinical and pathological characteristics of the 18 patients with t-MDS/AML are shown in Table S2. We studied PBSC samples from the 18 cases and 37 matched controls, and BM samples obtained at time of t-MDS/AML from a sub-cohort of 12 cases and 21 matched controls (Figure 1A). This subset of 12 cases did not differ significantly in clinical or demographic characteristics from the parent group. Gene expression profiles in CD34+ cells from t-MDS/AML cases and matched controls were compared using conditional logistical model (CLM) (Figure 1A). The following comparisons were made: (1) pre-aHCT PBSC from cases versus controls; (2) BM from cases at t-MDS/AML versus BM from controls at a comparable time point post-aHCT; and (3) changes in gene expression from pre-aHCT to development of t-MDS/AML in cases versus controls over a comparable time period (Δ t-MDS/AML – PBSC). The results of the training set were validated in an independent group of 36 patients (test set) consisting of 16 cases that developed t-MDS/AML post-aHCT and 20 matched controls.
Figure 1. Altered gene expression in PBSC CD34+ cells from t-MDS/AML cases compared with controls.
(A) The study design is shown. Gene expression was compared between [1] pre-aHCT PBSC samples obtained from cases (n=18) versus controls (n=37); [2] BM samples obtained from cases at time of t-MDS/AML (n=12) versus BM samples obtained from controls at comparable time points post-aHCT (n=21); and [3] changes in gene expression over time from PBSC collection to development of t-MDS/AML in cases compared with gene expression changes over a comparable time period in controls (Δ t-MDS/AML – PBSC). (B) Differences in gene expression in PBSC CD34+ cells from cases and controls are shown for top 50 genes [with the smallest P-values and 4-fold change in OR (>4 or <0.25)]. (C) Expression of representative gene sets within individual PBSC CD34+ cells from individual cases and controls is shown. The expression was averaged over the genes in each gene set and hierarchical clustering was performed for samples within the case and control groups. The box indicates the five cases with gene expression resembling that of controls. See also Figure S1 and Tables S1, S2 and S4.
Gene expression in PBSC CD34+ cells preceding onset of t-MDS/AML
This analysis was directed towards identifying genetic changes in pre-aHCT samples. Unsupervised clustering of cases and controls using all genes (filtered to remove those that were not expressed or with minimal differences across the cohort) showed that samples clustered into two major groups, with 5 cases clustering with controls and 11 controls clustering with cases (Figure S1A). PBSC from patients who developed t-MDS/AML demonstrated significant differences in gene expression compared to controls (Figure 1B). 779 genes were upregulated and 2220 genes were downregulated in t-MDS/AML cases compared to controls, based on the criteria of absolute odds ratio >4 and P-value<0.05. If using absolute odds ratio > 4 and P-value <0.01, 44 genes were upregulated and 301 genes were downregulated in t-MDS/AML cases compared to controls.
GSEA was performed to determine concordant differences between differentially expressed genes and curated gene sets (Subramanian et al., 2005) (Table 1, S3). PBSC from patients who subsequently developed t-MDS/AML (cases) showed significant downregulation of gene sets related to mitochondria and oxidative phosphorylation (Figure S1B), citrate cycle, ribosomal proteins (Figure S1B), aminoacyl-tRNA biosynthesis, amino acid metabolism, cell cycle regulation, DNA repair, and hematopoietic differentiation. G-protein coupled receptors, cell communication (Figure S1B), hematopoietic regulation, and cell adhesion related genes were upregulated in PBSC from cases. There was reduced expression of genes with binding motifs for NRF2 and GABP (regulators of mitochondrial enzymes)(Lenka et al., 1998), and E2F1 (regulator of cell cycle)(Chen et al., 2009), and increased expression of genes with binding motifs for GFI1 and FOXA1 (regulators of hematopoiesis)(Tothova et al., 2007) and OCT1 (regulator of DNA damage response) (Kang et al., 2009) (Table S3). In agreement with GSEA results, Ingenuity Pathway Analysis (IPA) indicated that pathways related to mitochondrial function, oxidative phosphorylation, protein ubiquitination, aminoacyl-tRNA synthesis, cell cycle regulation, citrate cycle, and amino acid metabolism, were significantly altered in cases compared to controls (Table S3). Gene Ontology (GO) analysis also indicated significant reduction in mitochondrial function and oxidative phosphorylation, cell metabolism, protein synthesis, and cell cycle regulation in cases; and enrichment of genes related to GTPase activity and transcription factors, tissue/organ development and cell communication (Table S3). Thus, multiple analytical approaches consistently demonstrated abnormalities in gene expression related to mitochondrial function, oxidative phosphorylation, and cellular response to oxidative and genotoxic stresses in PBSC samples from patients who subsequently develop t-MDS/AML.
Table 1.
Gene sets enriched in PBSC CD34+ cells from patients who later develop t-MDS/AML
| Gene set | Training set | Test Set | ||
|---|---|---|---|---|
| NES | FDR q-val | NES | FDR q-val | |
| Enriched in Cases | ||||
| G-Protein coupled receptors | ||||
| HSA04080_NEUROACTIVE_LIGAND_RECEPTOR_INTERACTION | 2.83 | <0.001 | 3.89 | <0.001 |
| GPCRDB_CLASS_A_RHODOPSIN_LIKE | 2.60 | <0.001 | 3.35 | <0.001 |
| GPCRS_CLASS_A_RHODOPSIN_LIKE | 2.52 | <0.001 | 2.83 | <0.001 |
| MONOAMINE_GPCRS | 2.32 | 0.003 | 2.62 | <0.001 |
| PEPTIDE_GPCRS | 2.21 | 0.005 | 2.86 | <0.001 |
| Hematopoietic regulation | ||||
| HALMOS_CEBP_DN | 2.33 | 0.003 | 1.73 | 0.079 |
| HOX_GENES | 1.89 | 0.036 | 1.06 | 0.738 |
| HSA04340_HEDGEHOG_SIGNALING_PATHWAY | 1.76 | 0.058 | 2.15 | 0.005 |
| Cell adhesion/communication | ||||
| HSA01430_CELL_COMMUNICATION | 2.05 | 0.009 | 2.86 | <0.001 |
| CELL_ADHESION | 1.63 | 0.108 | 1.82 | 0.061 |
| Xenobiotic metabolism | ||||
| GAMMA_HEXACHLOROCYCLOHEXANE_DEGRADATION | 2.04 | 0.009 | 2.01 | 0.018 |
| Enriched in controls (reduced in cases) | ||||
| Mitochondria and oxidative phosphorylation | ||||
| HUMAN_MITODB_6_2002 | −2.92 | <0.001 | −3.93 | <0.001 |
| HSA00190_OXIDATIVE_PHOSPHORYLATION | −2.88 | <0.001 | −3.01 | <0.001 |
| ELECTRON_TRANSPORT_CHAIN | −2.85 | <0.001 | −3.17 | <0.001 |
| MITOCHONDRIA | −2.84 | <0.001 | −3.85 | <0.001 |
| MOOTHA_VOXPHOS | −2.74 | <0.001 | −3.08 | <0.001 |
| OXIDATIVE_PHOSPHORYLATION | −2.58 | <0.001 | −2.67 | <0.001 |
| Ribosomes | ||||
| HSA03010_RIBOSOME | −3.02 | <0.001 | −4.06 | <0.001 |
| RIBOSOMAL_PROTEINS | −3.01 | <0.001 | −3.77 | <0.001 |
| tRNA biosynthesis | ||||
| TRNA_SYNTHETASES | −2.68 | <0.001 | −2.71 | <0.001 |
| AMINOACYL_TRNA_BIOSYNTHESIS | −2.41 | <0.001 | −2.74 | <0.001 |
| HSA00970_AMINOACYL_TRNA_BIOSYNTHESIS | −2.27 | <0.001 | −2.76 | <0.001 |
| Proteasomal pathway | ||||
| PROTEASOMEPATHWAY | −2.47 | <0.001 | −3.14 | <0.001 |
| HSA03050_PROTEASOME | −2.47 | <0.001 | −2.69 | <0.001 |
| PROTEASOME_DEGRADATION | −2.32 | <0.001 | −2.52 | <0.001 |
| Cell cycle regulation | ||||
| DNA_REPLICATION_REACTOME | −2.37 | <0.001 | −2.64 | <0.001 |
| P21_P53_MIDDLE_DN | −2.26 | <0.001 | −1.45 | 0.107 |
| P21_P53_ANY_DN | −2.16 | 6.79E-04 | −2.33 | <0.001 |
| SERUM_FIBROBLAST_CELLCYCLE | −2.14 | 0.014456 | −1.74 | 0.029 |
| CELL_CYCLE_KEGG | −1.91 | 0.005942 | −1.97 | 0.008 |
| Metabolism | ||||
| KREBS_TCA_CYCLE | −2.46 | <0.001 | −2.46 | <0.001 |
| HSA00020_CITRATE_CYCLE | −2.23 | 2.31E-04 | −2.55 | <0.001 |
| PENG_LEUCINE_DN | −2.22 | 4.46E-04 | −3.65 | <0.001 |
| HSA00252_ALANINE_AND_ASPARTATE_METABOLISM | −2.05 | 0.001727 | −1.00 | 0.535 |
| PROPANOATE_METABOLISM | −1.98 | 0.003869 | −2.82 | <0.001 |
| Hematopoietic stem cells | ||||
| HSC_INTERMEDIATEPROGENITORS_ADULT | −2.42 | <0.001 | −3.15 | <0.001 |
| HSC_LATEPROGENITORS_ADULT | −2.36 | <0.001 | −3.73 | <0.001 |
| HSC_INTERMEDIATEPROGENITORS_SHARED | −2.35 | <0.001 | −3.19 | <0.001 |
| HSC_LATEPROGENITORS_FETAL | −2.31 | <0.001 | −3.61 | <0.001 |
| HSC_LATEPROGENITORS_SHARED | −2.28 | <0.001 | −3.87 | <0.001 |
| DNA repair | ||||
| BRENTANI_REPAIR | −2.23 | <0.001 | −1.68 | 0.039 |
See also Table S3
Expression of enriched genes from representative gene sets in individual PBSC samples is shown in Figure 1C, S1C. Five of 18 PBSC samples from the cases demonstrated gene expression resembling that of controls. These 5 cases (#11, #62, #101, #125, and #168,) were the same as those identified on hierarchical clustering analysis using all genes. For the 5 patients that were misclassified, one patient (#11) had transient myelodysplasia with del 7 which subsequently resolved; one patient (#168) developed transient del(20q); 2 patients developed persistent del(20q) or del(13q) abnormalities with mild dysplasia (#62 and #125); and one patient developed AML with 11q23 translocation (#101). (Gupta et al., 2007; Han and Theil, 2007). The remaining 13 cases showing clearly altered gene expression in PBSC samples compared to controls, later presented with overt t-MDS/AML. Reanalysis of data after removing the two transient cases and their controls showed enrichment of gene sets in cases and controls that were highly similar to those seen in the original analysis (Figure S1D). The major categories of gene sets upregulated in cases [GPCRs, hematopoietic transcription factors (CEBP), cell communication, xenobiotic metabolism] and downregulated in cases (upregulated in controls) [mitochondrial oxidative phosphorylation, ribosomes, aminoacyl-tRNA synthetases, proteasomal degradation, citric acid cycle, cell cycle, DNA repair and hematopoietic stem cells] were maintained in the new analysis (Table S4).
Gene expression at time of t-MDS/AML
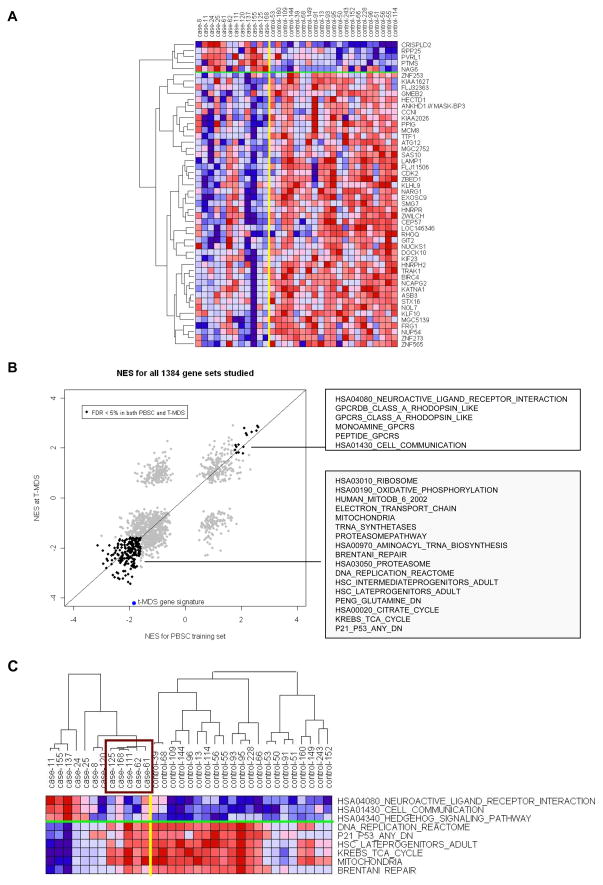
To evaluate genetic changes at the time of clinically overt t-MDS/AML, we compared gene expression in BM cells at t-MDS/AML from cases with BM samples obtained from controls at comparable time points post-aHCT (Figure 2A). We did not observe significant differences in distribution of subsets of CD34+ cells (HSC, CMP, GMP, and MEP) in a subset of t-MDS/AML cases (n=5) and controls (n=6) (Figure S2A), indicating that the gene expression differences identified in CD34+ cells were not simply a reflection of differences in cellular composition and were related to t-MDS/AML. GSEA analysis (Table 2, S5) showed significantly downregulated gene sets related to cell cycle regulation, DNA replication (Figure S2B), DNA repair (Figure S2B), hematopoietic differentiation, mitochondria, proteasome, citrate cycle, and amino acid metabolism in t-MDS/AML cases. Genes related to G-protein coupled receptors, cell communication (Figure S2B), and hematopoietic regulatory factors were upregulated in cases. There was increased expression of genes with binding motifs for hematopoietic regulatory TF including GFI1, GATA1, and OCT1, and reduced expression of genes with binding motifs for E2F1 in cases (Table S5). IPA and GO analysis confirmed significantly downregulated DNA damage response, DNA repair and cell cycle regulation, and upregulated cell communication and adhesion in cases compared with controls (Tables S5, S5). Of note, several gene sets enriched in BM CD34+ cells at time of t-MDS/AML were also enriched in PBSC CD34+ cells obtained pre-aHCT, long before the onset of clinical disease. This is illustrated in Figure 2B where gene sets significantly upregulated (n=10) or downregulated (n=185) in both PBSC and t-MDS/AML samples are highlighted. A 50-gene t-MDS/AML signature derived from differential gene expression at time of t-MDS/AML was significantly enriched in PBSC from cases (downregulated) (NES= −1.84, P-value <0.001, FDR=1.0%).
Figure 2. Altered gene expression in BM CD34+ cells at time of t-MDS/AML compared with controls.
(A) Differences in gene expression between cases and controls at time of t-MDS/AML are shown for top 50 genes with smallest P-values and 4-fold change in OR (OR>4 or OR<0.25). (B) Normalized enrichment scores (NES) from GSEA analyses for altered gene sets in PBSC CD34+ and BM CD34+ cells at time of t-MDS/AML are plotted. Gene sets significantly upregulated (n=15) or downregulated (n=203) in both PBSC and t-MDS/AML samples with FDR <5% are highlighted and representative gene sets listed in the boxes. A 50-gene t-MDS/AML signature set representing gene differentially expressed at time of development of t-MDS/AML is also shown. (C) Expression of representative gene sets in BM CD34+ cells from individual cases at time of t-MDS/AML and controls is shown. The expression was averaged over the genes in each gene set and hierarchical clustering performed for samples within the case and control groups. The box indicates the five cases with gene expression resembling that of controls. See also Figure S2 and Table S6.
Table 2.
Gene sets enriched in BM CD34+ cells from patients at time of development of t-MDS/AML
| NES | FDR q-val | |
|---|---|---|
| Enriched in Cases | ||
| G-Protein coupled receptors | ||
| HSA04080_NEUROACTIVE_LIGAND_RECEPTOR_INTERACTION | 3.11 | <0.001 |
| GPCRDB_CLASS_A_RHODOPSIN_LIKE | 2.70 | <0.001 |
| GPCRS_CLASS_A_RHODOPSIN_LIKE | 2.69 | <0.001 |
| MONOAMINE_GPCRS | 2.50 | <0.001 |
| PEPTIDE_GPCRS | 1.80 | 0.143 |
| Cell adhesion/communication | ||
| HSA01430_CELL_COMMUNICATION | 2.59 | <0.001 |
| HSA04512_ECM_RECEPTOR_INTERACTION | 2.22 | 0.001 |
| HSA04514_CELL_ADHESION_MOLECULES | 1.94 | 0.016 |
| Hematopoietic regulation | ||
| HSA04340_HEDGEHOG_SIGNALING_PATHWAY | 2.04 | 0.012 |
| Enriched in controls (reduced in cases) | ||
| Cell cycle regulation/checkpoints | ||
| SERUM_FIBROBLAST_CELLCYCLE | −3.34 | <0.001 |
| DNA_REPLICATION_REACTOME | −3.15 | <0.001 |
| P21_P53_ANY_DN | −3.12 | <0.001 |
| P21_P53_MIDDLE_DN | −2.87 | <0.001 |
| CELL_CYCLE | −2.77 | <0.001 |
| CELL_CYCLE_KEGG | −2.70 | <0.001 |
| HSA04110_CELL_CYCLE | −2.63 | <0.001 |
| DNA_DAMAGE_SIGNALING | −2.45 | <0.001 |
| GOLDRATH_CELLCYCLE | −2.42 | 0.002 |
| Hematopoietic stem cells | ||
| HSC_LATEPROGENITORS_ADULT | −2.60 | <0.001 |
| HSC_LATEPROGENITORS_SHARED | −2.58 | <0.001 |
| HSC_LATEPROGENITORS_FETAL | −2.57 | <0.001 |
| HSC_INTERMEDIATEPROGENITORS_ADULT | −2.51 | <0.001 |
| HSC_EARLYPROGENITORS_SHARED | −2.47 | <0.001 |
| HSC_INTERMEDIATEPROGENITORS_SHARED | −2.39 | <0.001 |
| Metabolism | ||
| PENG_LEUCINE_DN | −2.56 | <0.001 |
| KREBS_TCA_CYCLE | −2.55 | <0.001 |
| PENG_GLUTAMINE_DN | −2.43 | <0.001 |
| HSA00020_CITRATE_CYCLE | −2.26 | <0.001 |
| Mitochondria and oxidative phosphorylation | ||
| MITOCHONDRIA | −2.66 | <0.001 |
| HUMAN_MITODB_6_2002 | −2.58 | <0.001 |
| ELECTRON_TRANSPORT_CHAIN | −2.42 | <0.001 |
| Proteasome pathway | ||
| PROTEASOMEPATHWAY | −2.30 | <0.001 |
| PROTEASOME | −2.16 | <0.001 |
| DNA repair | ||
| BRENTANI_REPAIR | −2.91 | <0.001 |
See also Table S5
Expression of enriched genes from representative gene sets in individual subjects is shown in Figure 2C. At time of t-MDS/AML, BM from five cases showed gene expression resembling that of controls. Four of these cases presented with transient del (20q) or del (13q) abnormalities, three of whom had also shown gene expression resembling that of controls in the corresponding PBSC samples. The fifth patient presented with AML with del (7q). The remaining seven cases with clearly altered gene sets compared to controls presented with overt t-MDS/AML.
We also compared gene expression CD34+ cells from t-MDS/AML patients with normal BM CD34+ cells. A heat map showing up- and down-regulated genes in t-MDS/AML compared with normal CD34+ cells (FC>2 and FDR<0.05, 133 genes) is shown in Figure S2C. We observed difference in gene sets enriched in this analysis with those enriched when comparing t-MDS/AML samples with samples from control NHL/HL patients who did not develop t-MDS/AML (Table S6) We also compared gene expression in normal BM CD34+ cells with control NHL/HL patients who did not develop t-MDS/AML (Figure S2D, Table S6). There was considerable overlap in gene sets enriched in BM CD34+ cells from t-MDS/AML and control NHL/HL patients compared to normal CD34+ cells with 87 of 219 significant gene sets (40%) common to both analysis, as shown in Figure S2E. These results suggest a significant contribution of underlying disease, therapeutic exposures and hematopoietic regeneration post-aHCT to gene expression changes observed in t-MDS/AML CD34+ cells, and support the experimental design of using controls with similar disease and therapeutic exposure as t-MDS/AML cases to identify changes in gene expression specific to t-MDS/AML. On the other hand comparison with normal BM helps place changes in CD34+ cells from t-MDS/AML patients in the context of changes in controls that do not develop t-MDS/AML post-aHCT. For example expression of cell cycle and oxidative phosphorylation-related genes is increased in HL/NHL controls compared to normal CD34+ cells, suggesting that the reduction in these pathways in t-MDS/AML samples compared to controls may reflect a failure to upregulate these pathways post-aHCT.
Changes in gene expression from pre-aHCT to development of t-MDS/AML
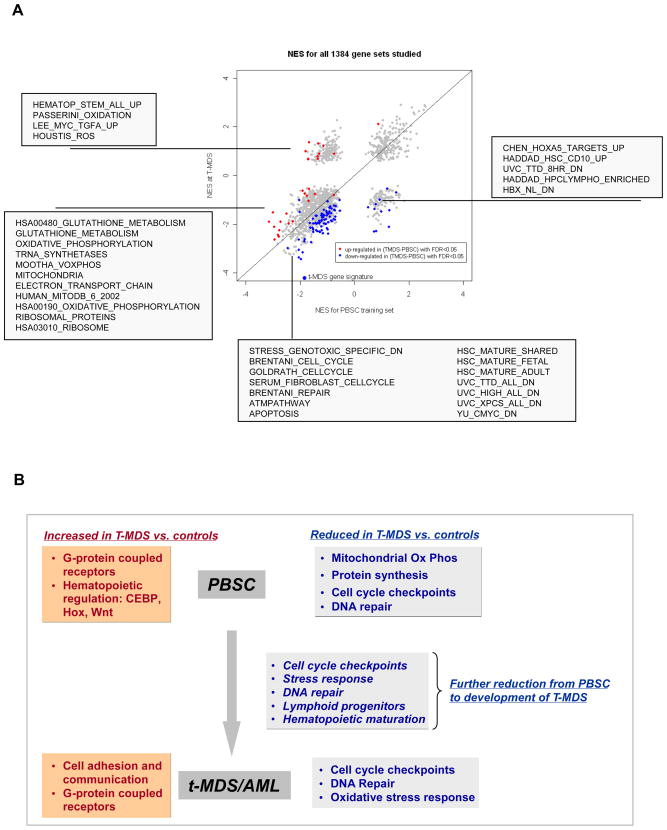
To assess the evolution of genetic changes from pre-aHCT to development of clinically overt disease, we compared changes in individual gene expression in cases from PBSC collection pre-aHCT to time of t-MDS/AML post-aHCT, with controls over a similar time period (Δ t-MDS/AML – PBSC). GSEA analysis of genes showing increased or decreased expression over time was performed (Table S7). Significant gene sets (FDR <5%) are illustrated in Figure 3A. Gene sets downregulated at time of t-MDS/AML but not in PBSC reflect changes acquired during development of t-MDS/AML. Genes related to early lymphoid progenitors were present in this group. In addition several gene sets downregulated in both pre-aHCT and t-MDS/AML samples showed significantly enhanced downregulation at time of t-MDS/AML compared to pre-aHCT samples, indicating progression of these alterations during t-MDS/AML development. These included genes related to cell cycle regulation, DNA repair, genotoxic stress response and hematopoietic maturation. Other gene sets were downregulated in PBSC but not at t-MDS/AML, or showed enhanced downregulation in PBSC compared to t-MDS/AML, likely representing alterations important early in the course of t-MDS/AML development. These included genes related to anti-oxidant response, mitochondrial oxidative phosphorylation and ribosomes.
Figure 3. Changes in gene expression in CD34+ cells in the course of development of t-MDS/AML.
(A) Changes in gene expression from PBSC to development of t-MDS/AML. Normalized enrichment scores (NES) from GSEA analyses for altered gene sets in PBSC CD34+ and BM CD34+ cells at time of t-MDS/AML are plotted. Gene sets significantly upregulated (red) or downregulated (blue) in cases compared to controls from time of PBSC collection to time of t-MDS/AML with FDR <5% are highlighted and representative gene sets are listed in the boxes. (B) Gene expression signatures that are upregulated and downregulated in cases compared to controls in PBSC and at time of development of t-MDS/AML, as well as changes in gene expression from time of PBSC to development of t-MDS/AML are summarized. See also Table S7.
Gene expression changes at different stages of development of t-MDS/AML are summarized in Figure 3B. PBSC from patients who develop t-MDS/AML demonstrate downregulation of mitochondrial, oxidative phosphorylation, protein synthesis, cell cycle checkpoint, and DNA repair, and upregulation of GPCR, cell adhesion, and certain hematopoietic transcription factors. These gene alterations continue to be present at the time of development of t-MDS/AML. However progression to t-MDS/AML is associated with further downregulation of cell cycle checkpoint, DNA repair, genotoxic response and hematopoietic differentiation genes.
Outcome prediction
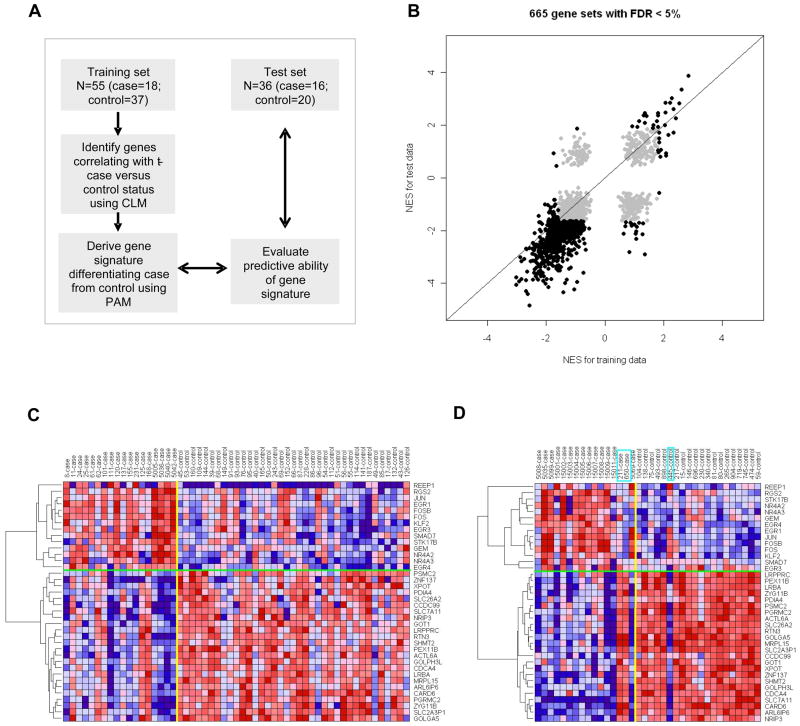
Since gene expression in PBSC samples from the training set was associated with later development of t-MDS/AML we sought to identify a PBSC gene signature that could identify NHL and HL patients pre-aHCT at high risk for developing t-MDS/AML following aHCT. PBSC samples from the training set were used to derive the gene signature which was then applied to an independent test set of 16 patients who subsequently developed t-MDS/AML after aHCT for NHL or HL, and 20 matched controls that did not develop t-MDS/AML (Figure 4A, Table S2). The length of follow up for cases was 44 months (range: 4.9 to 101 months) and for controls was 76 months (range: 34 to 138 months). The clinical and demographic characteristics of t-MDS/AML patients in the training and test sets revealed differences in primary diagnosis, time to t-MDS/AML, and cytogenetic abnormalities (Table S8, S8). However, patients in the two sets did not differ by age at diagnosis or HCT, number of stem cell collections, stem cell dose infused, stem cell mobilization techniques, conditioning regimens, and pre-aHCT exposure to radiation or topoisomerase II inhibitors. Independent analysis of differential gene expression between cases and controls in the test set revealed extensive overlap of up and down-regulated gene sets in t-MDS/AML cases between training and test sets (Figure 4B, Table S9). Gene expression changes related to mitochondria, metabolism, cell cycle regulation and hematopoietic progenitors that were observed in the training set were validated in the test set (Table 1). A cross-validated 38-gene classifier was derived from the training set using Prediction analysis of microarray (PAM, Table S10). Expression of the 38-gene signature in both training and test sets is shown in Figure 4C, D. Hierarchical clustering of the 38 genes revealed two major clusters with significant correlation for up- and down-regulated expression of genes in cases or controls in training and test sets (P <0.001). Application of the 38-gene signature to the test set correctly classified 19 of the 20 subjects who did not subsequently develop t-MDS/AML, and 14 of the 16 subjects who did develop t-MDS/AML (Figure S3A). There was significant correlation between predicted and true disease status [19/20 controls (95%) and 14/16 cases (87.5%), P<0.001]. The 38-gene classifier had an accuracy of 33/36=91.7% and a precision (positive predictive value) of 14/15=93.3%. The specificity of the test was 19/20=95% and the sensitivity was 14/16=87.5%. It is noteworthy that gene expression signatures derived from the training set were predictive of case versus control status in the test set despite differences in clinical characteristics between the two sets, suggesting that the gene expression signature is robust across different subsets of t-MDS/AML. The two t-MDS/AML patients who were misclassified presented with features typical of t-MDS/AML; and did not have any identifiable characteristics that distinguished them from other patients in the set. The control patient who was misclassified did not differ from other controls in terms of clinical features but developed relapse of lymphoma three years after aHCT and underwent allogeneic transplantation, and could not be followed further for development of t-MDS/AML. The misclassification of these samples may reflect a degree of heterogeneity in gene expression of CD34+ cells obtained from patients at time points prior to development of t-MDS. Repetition of the analysis after removing the two training cases with transient t-MDS/AML (#11 and #168) and their respective controls yielded a 31-gene signature which misclassified 4/16 cases in the test set as controls and 1/20 controls in the test set as cases (Figure S3B), and did not improve the error rate compared with the original gene signature. These results indicate that the gene expression profile of hematopoietic cells pre-aHCT can reliably and accurately identify patients at risk for t-MDS/AML post-aHCT.
Figure 4. Validation of altered gene expression in PBSC from t-MDS/AML cases and outcome prediction.
(A) Strategy for development and validation of a gene signature to differentiate PBSC from cases and controls. PAM represents prediction analysis of microarrays. (B) Normalized enrichment scores (NES) from GSEA analyses for 665 gene sets with FDR <5% in PBSC CD34+ from either training set or test set are highlighted. Six hundred and thirty six (95.6%) sets were in agreement between the two studies, with 34 gene sets upregulated and 602 gene sets downregulated in both training and test sets. (C) Expression for the 38-gene signature derived using PAM in the training PBSC set. (D) Expression for the 38-gene signature in the test PBSC set. The boxes indicate the three subjects that were misclassified by the gene signature. See also Figure S3 and Tables S8, S9, S10 and S11.
It was previously reported that same cytogenetic abnormality observed at the time of t-MDS diagnosis could be detected in pre-aHCT specimens by FISH.(Abruzzese et al., 1999) Cytogenetic analysis performed on pre-aHCT bone marrow (BM) samples for all subjects in this study did not show evidence of clonal chromosomal abnormalities characteristic of t-MDS/AML (Table S11). FISH analysis performed on a subset of CD34+ cells from PBSC samples (n=9), which were representative of the spectrum of cytogenetic abnormalities seen, and for which sufficient samples were available, did not show evidence of the t-MDS/AML clone in PBSC CD34+ cells (Table S11). Therefore cells bearing clonal cytogenetic abnormalities do not contribute significantly to the altered gene expression profile in PBSC CD34+ cells from t-MDS cases. The difference between our results and previous reports in the literature may reflect the practice of routinely performing cytogenetic analyses on pre-HCT BM samples from patients undergoing aHCT at our center.
Mitochondrial dysfunction in PBSC from patients who later developed t-MDS/AML
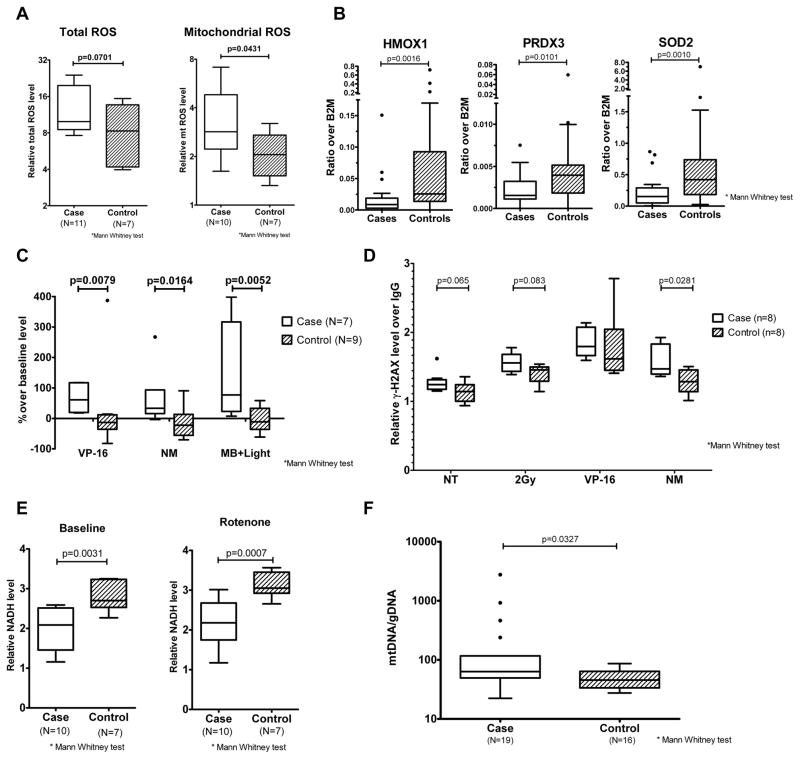
Gene signatures related to mitochondrial oxidative phosphorylation were prominently downregulated in PBSC CD34+ cells from cases. The mitochondrial electron transport chain, in addition to its role in energy production, is a major site of reactive oxygen species (ROS) generation(Kowaltowski et al., 2009). Impaired electron transfer could lead to increased ROS generation in CD34+ cells from patients who develop t-MDS/AML (Wallace, 2005). We conducted additional studies of mitochondrial function in PBSC cells from cases and controls. PBSC CD34+ cells from cases showed increased baseline levels of mitochondrial ROS (P=0.04) and total ROS (P=0.07) compared to controls (Figure 5A). PBSC CD34+ cells from cases also demonstrated reduced expression of anti-oxidant genes, which may further increase ROS levels. Q-PCR analysis showed significant downregulation of the anti-oxidant genes HMOX1 (p=0.0016), PRDX3 (p=0.01), and SOD2 (p=0.001), in PBSC from cases compared to controls (Osburn and Kensler, 2008) (Figure 5B). PBSC from cases exhibited sustained elevation of ROS following exposure to oxidative stressors including etoposide, nitrogen mustard and methylene blue with visible light compared to controls, consistent with reduced ROS detoxification (Figure 5C). We observed significantly increased γ-H2AX levels in PBSC from cases after exposure to mechlorethamine (nitrogen mustard, p=0.0281), and a trend toward increased γ-H2AX levels after radiation and etoposide treatment (p=0.065 and 0.083 respectively), indicating increased DNA damage (Kinner et al., 2008) (Figure 5D). PBSC from cases also demonstrated significantly reduced NADH levels compared to controls, both with and without treatment with the electron transport chain inhibitor Rotenone, suggesting impaired NADH production (Grivennikova and Vinogradov, 2006) (Figure 5E). This observation is consistent with reduced TCA cycle activity and a broad defect in mitochondrial energy metabolism. Mitochondrial mass, estimated through measurement of mitochondrial DNA content, was increased in PBSC from cases compared to controls, possibly representing a compensatory response to mitochondrial dysfunction or reduced autophagy of damaged mitochondria (Nugent et al., 2007; Tolkovsky, 2009) (Figure 5F). We did not observe significant differences in ROS levels both at baseline (n=6) and after exposure to oxidative stress (n=5) in CD34+ subsets (LT-HSC, CMP, GMP, MEP) selected by flow cytometry from normal PBSC samples (Figure S4). Therefore differences in ROS levels between PBSC CD34+ cells from t-MDS/AML cases and controls cannot be explained by differences in their cellular composition. These results indicate that PBSC CD34+ cells from cases that develop t-MDS/AML have altered mitochondrial function, increased ROS generation, reduced ROS detoxification, and enhanced DNA damage after therapeutic exposure, and support the results of gene expression analysis.
Figure 5. Mitochondrial dysfunction and metabolic abnormalities in PBSC from patients developing t-MDS/AML.
(A) Mitochondrial ROS and total ROS levels in PBSC CD34+ cells from cases (red) and controls (blue) detected with C-H2DCFDA and MitoSox RED respectively. Results represent median, interquartile range and range of values. (B) Expression of Nrf2 regulated anti-oxidant genes heme oxygenase 1 (HMOX1), peroxiredoxin 3 (PRDX3) and superoxide dismutase 2 (SOD2) in PBSC CD34+ cells from cases (red, n=20) and controls (blue, n=39) quantified using Q-PCR. Results represent median, interquartile range and range of values. (C) ROS levels in PBSC CD34+ cells from cases (red) and controls (blue), measured 2 hours after exposure to etoposide (VP-16), nitrogen mustard (NM) and methylene blue with visible light (MB+Light) by C-H2DCFDA labeling. Results represent median, interquartile range and range of values. (D) DNA damage in PBSC CD34+ cells from cases (red) and controls (blue), measured 4 hours after exposure to radiation (2Gy), VP-16 and NM as well as no treatment controls (NT) by measuring γ-H2AX levels. Results represent median, interquartile range and range of values. (E) NADH levels in PBSC CD34+ cells from cases (red) and controls (blue) measured at baseline and after rotenone treatment. Results represent median, interquartile range and range of values. (F) Mitochondrial mass was estimated in PBSC MNC isolated from cases (n=19) and controls (n=16) through measurement of mitochondrial DNA content. Results represent median, interquartile range and range of values. See also Figure S4.
DISCUSSION
Changes in gene expression associated with development of t-MDS/AML after aHCT for lymphoma were identified in CD34+ cells from PBSC obtained pre-aHCT, several months to years before development of clinically overt disease. These changes may represent factors predisposing to risk of t-MDS/AML and/or effects of pre-aHCT therapeutic exposures. We identified a 38-gene signature in PBSC that could distinguish patients who developed t-MDS/AML post-aHCT from those who did not. This gene signature was validated in an independent set of PBSC samples, supporting the potential prognostic utility of this approach to identify patients at high risk for developing t-MDS/AML.
PBSC CD34+ cells from patients who develop t-MDS/AML demonstrate altered expression of genes related to mitochondria, oxidative phosphorylation, oxidative stress response, ribosomes, and DNA repair. In additional studies we directly show that PBSC CD34+ cells from cases that develop t-MDS/AML demonstrate altered mitochondrial function, increased ROS generation, reduced ROS detoxification, and enhanced DNA damage after therapeutic exposure, validating and extending the results of gene expression analysis. It is hypothesized that mitochondrial defects are central to cancer cell biology, through enhanced ROS generation leading to mutation of critical genes that regulate cell proliferation. Mitochondrial oxidative phosphorylation, in addition to its role in energy production, is a major source of ROS generation (Kowaltowski et al., 2009). Impaired electron transfer could result in increased ROS generation in PBSC from patients at risk for t-MDS/AML (Wallace, 2005). Reduced anti-oxidant gene expression may further increase ROS levels. Tight regulation of ROS is essential for normal hematopoietic function (Ito et al., 2004). We have previously shown that development of t-MDS/AML post-aHCT is preceded by impaired hematopoietic function evidenced by impaired PBSC mobilization; reduced progenitor regeneration, and accelerated telomere loss (Bhatia et al., 2005; Chakraborty et al., 2009). Our findings support a model of t-MDS/AML where therapeutic exposure results in increased ROS levels related to mitochondrial dysfunction, increased DNA damage, mutagenesis, and impaired hematopoiesis (Figure 6). Impaired ribosomal function may also contribute to hematopoietic impairment in patients developing t-MDS/AML. Ribosomal gene mutations and impaired ribosome biogenesis occur in congenital BM failure syndromes and the 5q– syndrome, and may impair hematopoiesis through altered protein translation, or through p53-dependent apoptosis and senescence (Ebert et al., 2008; Faber et al., 2006; Fumagalli et al., 2009).
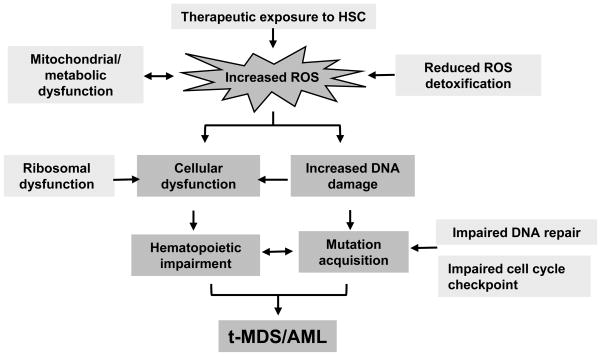
Figure 6. Proposed model for pathogenesis of t-MDS/AML.
Therapeutic exposure to HSC results in elevated ROS levels related to mitochondrial dysfunction. Excessive ROS generation results in increased DNA damage, cellular dysfunction and impaired hematopoiesis. Impaired DNA repair and cell cycle regulation leads to accumulation of mutations which potentially contributes to the development of t-MDS/AML.
Changes in gene expression from PBSC to development of t-MDS/AML likely represent additional abnormalities associated with transformation from the pre-leukemic to leukemic state. Progression to t-MDS/AML was associated with reduced expression of DNA repair and cell cycle regulatory genes, indicating loss of genome protective mechanisms, potentially allowing acquisition of additional mutations and disease evolution (Harper and Elledge, 2007; Kastan and Bartek, 2004). Such changes may result from acquisition of additional mutations or epigenetic changes in pre-malignant cells. Mutations in HRR genes are reported in t-MDS/AML patients (Rassool et al., 2007). Increased expression of genes associated with cell cycle progression was seen in a previous analysis of gene expression in t-MDS/AML CD34+ cells (Qian et al., 2002), and mutations in p53 are relatively frequent in t-MDS/AML (Ben-Yehuda et al., 1996). Loss of p53-related cell cycle regulation may contribute to genetic instability as well as survival and expansion of altered hematopoietic cells in t-MDS/AML (Feldser and Greider, 2007; Fumagalli et al., 2009).
The observed heterogeneity in gene expression with t-MDS/AML cases could be related in part to alternative genetic pathways to t-MDS/AML development, as have been defined based on characteristic chromosome abnormalities (Pedersen-Bjergaard et al., 2007). A previous analysis performed at time of development of t-MDS/AML revealed that certain gene expression patterns were common whereas others differed between different cytogenetic subgroups (Qian et al., 2002). Indeed in the current study variability of gene expression was associated with certain types of chromosomal abnormalities (20q-; 13q-). However, formal analysis of difference in gene expression in samples amongst different cytogenetic subgroups at early time points preceding the development of t-MDS/AML requires assembly of an even larger sample set.
The 38-gene predictor compares well with other recently reported multi-gene signatures for various cancers. Although a perfect test should have 100%, sensitivity and specificity, this is not achieved by any currently available biomarker (Wagner et al., 2004). For example, the best biomarker for prostate cancer, PSA, has a sensitivity of 90% and specificity of 25%. A 29 microRNA-gene signature for non-small cell lung cancer in peripheral blood mononuclear cells had 76% sensitivity and 82% specificity of prediction in an independent set of 38 cases and 17 controls (Raponi et al., 2009). A 75-probe signature in CD34+ cells predicted drug response to imatinib in CML patients with 88%sensitivity and 83% specificity in an independent test set of 17 responders and 6 non-responders (Oehler et al., 2009). Thus, taking into account the previously published predictors, the 38-gene signature for t-MDS/AML performed favorably in classifying cases and controls. This is especially notable since the signature was obtained from and applied to samples procured several years prior to development of overt disease. However, these findings are based on relatively small numbers of patients and additional larger studies are required as part of the stepwise development of this classifier to further validate its predictive value prior to its application as a clinical test. Furthermore, detection of this signature is currently limited to PBSC obtained prior to aHCT, at a time when patients have already been exposed to pre-aHCT therapeutic exposures. It will be of interest to determine whether this signature can be detected in samples obtained at diagnosis of NHL or HL, prior to initiation of treatment.
Despite these limitations, the current study has potential clinical significance, since early detection of patients at high risk for t-MDS/AML using a gene expression signature could facilitate application of interventions to prevent development of this lethal malignancy. Detection of the high risk profile could guide therapeutic decision-making including the use of alternative treatment approaches such as allogeneic transplantation (Litzow et al.), and application of targeted interventions for those at high risk. Although interventions that reduce the risk of progression to t-MDS/AML do not currently exist, insights into critical molecular mechanisms contributing to susceptibility to and emergence of t-MDS/AML could provide potential targets for development of preventive or therapeutic interventional strategies. For example, enhanced mitochondrial ROS levels can be explored as a potential target for interventions to prevent t-MDS/AML in patients receiving genotoxic cancer therapy (Colburn and Kensler, 2008). These translational implications of our results merit further investigation in future studies.
EXPERIMENTAL PROCEDURES
Patients and Samples
The study was approved by the institutional review board of City of Hope (COH) in accordance with an assurance filed with and approved by the Department of Health and Human Services, and met all requirements of the Declaration of Helsinki. Informed consent was obtained from all subjects. Patients receiving aHCT for HL or NHL at COH constituted the sampling frame for selection of cases and controls in this nested case-control study. PBSC samples obtained pre-aHCT and BM samples at the time of development of t-MDS/AML post-HCT were studied. The training set consisted of 18 patients who developed t-MDS/AML (“cases”) after aHCT, matched with 37 controls who underwent aHCT, but did not develop t-MDS/AML. Up to three controls were selected per case, matched for primary diagnosis (HL/NHL), age at aHCT (±10years), and ethnicity (Caucasians, African-Americans, Hispanics, other). Length of follow-up after aHCT for controls was longer than the time from aHCT to t-MDS/AML in the corresponding case. The results of the training set were validated in an independent group of 36 patients (test set) consisting of 16 cases that developed t-MDS/AML post-aHCT for HL or NHL, and 20 matched controls. Relevant demographic and clinical data were obtained from medical records and included age at diagnosis and aHCT, gender, race/ ethnicity, disease characteristics, pre-aHCT cumulative therapeutic exposures, conditioning regimens, priming with growth factors and/or chemotherapy for PBSC mobilization and collection, number of PBSC collections, dose of CD34+ cells infused, recovery of WBC counts, vital status, and disease status after aHCT.
Gene expression analysis
In the training set, 55 PBSC samples from 18 cases and 37 matched controls were studied. BM samples from time of development of t-MDS/AML were available for 12 cases and 21 matched controls obtained at a comparable time from aHCT. For validation, 36 PBSC samples from a test set consisting of 16 cases and 20 matched controls were studied. All samples had been cryopreserved as mononuclear cells in liquid nitrogen. Frozen cells were thawed and incubated in IMDM supplemented with 20% FBS and DNAse I (Sigma) for 3 hour incubation at 37°C. Samples were labeled with anti-CD34-APC and anti-CD45-FITC (BD biosciences) and CD34+CD45dim cells selected using flow cytometry (Beckman-Coulter, Miami, FL). Total RNA was extracted using the RNeasy kit (Qiagen). RNA from 1000 cells was amplified and labeled using GeneChip® Two-Cycle Target Labeling and Control Reagents from Affymetrix (Santa Clara, CA). 15 μg of cRNA each was hybridized to Affymetrix HG U133 plus 2.0 Arrays.
Statistical analysis
Microarray data were analyzed using R (version 2.9) with genomic analysis packages from Bioconductor (version 2.4). Details of analysis are provided in the supplemental experimental procedures. Following quality control, data for PBSC and BM samples were normalized separately. Probesets with low expression or variability were filtered. Using conditional logistic model (CLM) to retain matching between cases and controls, we analyzed the magnitude of association [expressed as odds ratio (OR)] between t-MDS/AML and 1) gene expression levels in PBSC at the pre-aHCT time point; 2) gene expression levels in BM at time of t-MDS/AML; and 3) change in expression of individual genes from PBSC to development of t-MDS/AML. False discovery rate (FDR) was applied to adjust for multiple testing. Gene set enrichment analysis (GSEA) was performed on ranked lists of genes differentially expressed between cases and controls generated using CLM. Where multiple significant gene sets were related to each other, analysis was performed to identify a subset of common enriched genes. Hierarchical clustering was performed within each of the case and control group. Gene Ontology (GO) and pathway analysis was performed using DAVID 2008 and Ingenuity IPA 7.5 respectively.
The association between gene expression in the PBSC product and subsequent development of t-MDS/AML identified in the training set was validated in the test set. Differential expression between cases and controls was analyzed using CLM. GSEA analysis was performed on the ranked list of differentially expressed genes. PAM was used to derive a prognostic gene signature from the training set to classify patients as case or control. Based on the misclassification error in cross-validation, a 38-gene signature was selected for prediction and applied to the test set.
Reactive oxygen species detection
To detect ROS, PBSC MNCs were incubated with carboxy-H2DCFDA (10 μM) and MitoSOX™ Red (3 μM) (Invitrogen, Calsbad, CA) at 37°C for 30 min to detect total ROS and mitochondrial ROS respectively after exposing to etoposide (VP-16, 34 nM), mechlorethamine (NM, 2μg/ml) or methylene blue with visible light. Cells were then labeled with CD34-PE-Cy7, CD45-APC-Cy7 (Ebioscience, San Diego, CA) and AnnexinV-Cy5 (BD Biosciences, San Jose, CA) on ice for 30 min, washed and immediately analyzed by flow cytometry using a LSRII flow cytometer (BD Biosciences, San Jose, CA).
Anti-oxidant gene expression
For anti-oxidant gene expression measurement, 10ng total RNA from FACS-sorted PBSC CD34+ cells was used to generate cDNA using SuperScript® III First-Strand Synthesis System (Invitrogen, Carlsbad, CA). Quantitative RT-PCR for expression of heme oxygenase 1 (HMOX1), peroxiredoxin 3 (PRDX3), superoxide dismutase 2 (SOD2) was performed on an 7900HT Fast Real-Time PCR System using Taqman® gene expression assays (Applied Biosystems, Foster City, CA). Results were normalized to endogenous control β2-microglobulin (B2M) expression.
DNA damage analysis
DNA damage was evaluated based on γ-H2AX level using flow cytometry. PBSC MNCs were treated with irradiation (2Gy), etoposide (VP-16, 34 nM) or mechlorethamine (nitrogen mustard, 2μg/ml) and γ-H2AX levels were detected in CD34+ fractions 4 hours after removal of DNA damage inducers by staining with Anti-phospho-Histone H2A.X (Tyr142) (Millipore, Temecula, CA) following the protocol from the manufacture.
NADH detection
NADH levels in PBSC CD34+ cells were assessed by endogenous cellular fluorescence measured at an excitation of 350 nm and an emission of 440nm using LSRII flow cytometer. MFI (median fluorescence intensity) was normalized to control unlabeled beads (BD Biosciences, San Jose, CA) for each sample.
Supplementary Material
SIGNIFICANCE.
Therapy-related myelodysplasia or acute myeloid leukemia (t-MDS/AML) is a lethal complication of cancer treatment. However, the pathogenesis of t-MDS/AML is poorly understood and methods to predict risk of t-MDS/AML development in individual patients are not available. Using gene expression analysis we detected abnormalities in mitochondrial function, metabolism, and hematopoietic regulation in hematopoietic cells early in t-MDS/AML pathogenesis, long before development of cytogenetic and pathological abnormalities. A t-MDS/AML gene expression signature applied to an independent group of patients accurately distinguished patients who did or did not subsequently develop t-MDS/AML. These results indicate that perturbations in genetic programs associated with t-MDS/AML are present long before disease onset, and can accurately identify patients at risk for developing this complication in the future.
Acknowledgments
This work was supported by NIH grants R01 HL083050 and P50 CA107399 and General Clinical Research Center Grant #5M01 RR00043. We thank Marilyn Slovak for cytogenetic analysis, Paul Hengen and Xiwei Wu for assistance with data analysis, and the City of Hope Analytical Cytometry, Cytogenetics, Functional Genomics and Bioinformatics shared resources. We are indebted to the physicians and nurses of the Department of Hematology/HCT, and to all the patients who participated in this study.
Footnotes
Accession number
Microarray data has been deposited in the Gene Expression Omnibus database (Accession number GSE23025).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abruzzese E, Radford JE, Miller JS, Vredenburgh JJ, Rao PN, Pettenati MJ, Cruz JM, Perry JJ, Amadori S, Hurd DD. Detection of abnormal pretransplant clones in progenitor cells of patients who developed myelodysplasia after autologous transplantation. Blood. 1999;94:1814–1819. [PubMed] [Google Scholar]
- Ben-Yehuda D, Krichevsky S, Caspi O, Rund D, Polliack A, Abeliovich D, Zelig O, Yahalom V, Paltiel O, Or R, et al. Microsatellite instability and p53 mutations in therapy-related leukemia suggest mutator phenotype. Blood. 1996;88:4296–4303. [PubMed] [Google Scholar]
- Bhatia R, Van Heijzen K, Palmer A, Komiya A, Slovak ML, Chang KL, Fung H, Krishnan A, Molina A, Nademanee A, et al. Longitudinal assessment of hematopoietic abnormalities after autologous hematopoietic cell transplantation for lymphoma. J Clin Oncol. 2005;23:6699–6711. doi: 10.1200/JCO.2005.10.330. [DOI] [PubMed] [Google Scholar]
- Bhatia S, Ramsay NK, Steinbuch M, Dusenbery KE, Shapiro RS, Weisdorf DJ, Robison LL, Miller JS, Neglia JP. Malignant neoplasms following bone marrow transplantation. Blood. 1996;87:3633–3639. [PubMed] [Google Scholar]
- Chakraborty S, Sun CL, Francisco L, Sabado M, Li L, Chang KL, Forman S, Bhatia S, Bhatia R. Accelerated telomere shortening precedes development of therapy-related myelodysplasia or acute myelogenous leukemia after autologous transplantation for lymphoma. J Clin Oncol. 2009;27:791–798. doi: 10.1200/JCO.2008.17.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HZ, Tsai SY, Leone G. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat Rev Cancer. 2009;9:785–797. doi: 10.1038/nrc2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colburn NH, Kensler TW. Targeting transcription factors for cancer prevention--the case of Nrf2. Cancer Prev Res (Phila) 2008;1:153–155. doi: 10.1158/1940-6207.CAPR-08-0025. [DOI] [PubMed] [Google Scholar]
- Ebert BL, Pretz J, Bosco J, Chang CY, Tamayo P, Galili N, Raza A, Root DE, Attar E, Ellis SR, Golub TR. Identification of RPS14 as a 5q- syndrome gene by RNA interference screen. Nature. 2008;451:335–339. doi: 10.1038/nature06494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber P, Fisch P, Waterhouse M, Schmitt-Graff A, Bertz H, Finke J, Spyridonidis A. Frequent genomic alterations in epithelium measured by microsatellite instability following allogeneic hematopoietic cell transplantation in humans. Blood. 2006;107:3389–3396. doi: 10.1182/blood-2005-08-3431. [DOI] [PubMed] [Google Scholar]
- Feldser DM, Greider CW. Short telomeres limit tumor progression in vivo by inducing senescence. Cancer Cell. 2007;11:461–469. doi: 10.1016/j.ccr.2007.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli S, Di Cara A, Neb-Gulati A, Natt F, Schwemberger S, Hall J, Babcock GF, Bernardi R, Pandolfi PP, Thomas G. Absence of nucleolar disruption after impairment of 40S ribosome biogenesis reveals an rpL11-translation-dependent mechanism of p53 induction. Nat Cell Biol. 2009;11:501–508. doi: 10.1038/ncb1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikova VG, Vinogradov AD. Generation of superoxide by the mitochondrial Complex I. Biochim Biophys Acta. 2006;1757:553–561. doi: 10.1016/j.bbabio.2006.03.013. [DOI] [PubMed] [Google Scholar]
- Gupta R, Soupir CP, Johari V, Hasserjian RP. Myelodysplastic syndrome with isolated deletion of chromosome 20q: an indolent disease with minimal morphological dysplasia and frequent thrombocytopenic presentation. Br J Haematol. 2007;139:265–268. doi: 10.1111/j.1365-2141.2007.06776.x. [DOI] [PubMed] [Google Scholar]
- Han JY, Theil KS. Karyotypic identification of abnormal clones preceding morphological changes or occurring with no definite morphological features of myelodysplastic syndrome: a preliminary study. Lab Hematol. 2007;13:17–21. doi: 10.1532/LH96.06047. [DOI] [PubMed] [Google Scholar]
- Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- Ito K, Hirao A, Arai F, Matsuoka S, Takubo K, Hamaguchi I, Nomiyama K, Hosokawa K, Sakurada K, Nakagata N, et al. Regulation of oxidative stress by ATM is required for self-renewal of haematopoietic stem cells. Nature. 2004;431:997–1002. doi: 10.1038/nature02989. [DOI] [PubMed] [Google Scholar]
- Kalaycio M, Rybicki L, Pohlman B, Sobecks R, Andresen S, Kuczkowski E, Bolwell B. Risk factors before autologous stem-cell transplantation for lymphoma predict for secondary myelodysplasia and acute myelogenous leukemia. J Clin Oncol. 2006;24:3604–3610. doi: 10.1200/JCO.2006.06.0673. [DOI] [PubMed] [Google Scholar]
- Kang J, Shakya A, Tantin D. Stem cells, stress, metabolism and cancer: a drama in two Octs. Trends Biochem Sci. 2009;34:491–499. doi: 10.1016/j.tibs.2009.06.003. [DOI] [PubMed] [Google Scholar]
- Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- Kinner A, Wu W, Staudt C, Iliakis G. Gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res. 2008;36:5678–5694. doi: 10.1093/nar/gkn550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowaltowski AJ, de Souza-Pinto NC, Castilho RF, Vercesi AE. Mitochondria and reactive oxygen species. Free Radic Biol Med. 2009;47:333–343. doi: 10.1016/j.freeradbiomed.2009.05.004. [DOI] [PubMed] [Google Scholar]
- Krishnan A, Bhatia S, Slovak ML, Arber DA, Niland JC, Nademanee A, Fung H, Bhatia R, Kashyap A, Molina A, et al. Predictors of therapy-related leukemia and myelodysplasia following autologous transplantation for lymphoma: an assessment of risk factors. Blood. 2000;95:1588–1593. [PubMed] [Google Scholar]
- Lenka N, Vijayasarathy C, Mullick J, Avadhani NG. Structural organization and transcription regulation of nuclear genes encoding the mammalian cytochrome c oxidase complex. Prog Nucleic Acid Res Mol Biol. 1998;61:309–344. doi: 10.1016/s0079-6603(08)60830-2. [DOI] [PubMed] [Google Scholar]
- Litzow MR, Tarima S, Perez WS, Bolwell BJ, Cairo MS, Camitta BM, Cutler CS, de Lima M, Dipersio JF, Gale RP, et al. Allogeneic transplantation for therapy-related myelodysplastic syndrome and acute myeloid leukemia. Blood. 2010;115:1850–1857. doi: 10.1182/blood-2009-10-249128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason PJ. Stem cells, telomerase and dyskeratosis congenita. Bioessays. 2003;25:126–133. doi: 10.1002/bies.10229. [DOI] [PubMed] [Google Scholar]
- Miller JS, Arthur DC, Litz CE, Neglia JP, Miller WJ, Weisdorf DJ. Myelodysplastic syndrome after autologous bone marrow transplantation: an additional late complication of curative cancer therapy. Blood. 1994;83:3780–3786. [PubMed] [Google Scholar]
- Nugent SM, Mothersill CE, Seymour C, McClean B, Lyng FM, Murphy JE. Increased mitochondrial mass in cells with functionally compromised mitochondria after exposure to both direct gamma radiation and bystander factors. Radiat Res. 2007;168:134–142. doi: 10.1667/RR0769.1. [DOI] [PubMed] [Google Scholar]
- Oehler VG, Yeung KY, Choi YE, Bumgarner RE, Raftery AE, Radich JP. The derivation of diagnostic markers of chronic myeloid leukemia progression from microarray data. Blood. 2009;114:3292–3298. doi: 10.1182/blood-2009-03-212969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osburn WO, Kensler TW. Nrf2 signaling: an adaptive response pathway for protection against environmental toxic insults. Mutation research. 2008;659:31–39. doi: 10.1016/j.mrrev.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen-Bjergaard J. Insights into leukemogenesis from therapy-related leukemia. N Engl J Med. 2005;352:1591–1594. doi: 10.1056/NEJMe048336. [DOI] [PubMed] [Google Scholar]
- Pedersen-Bjergaard J, Andersen MK, Christiansen DH. Therapy-related acute myeloid leukemia and myelodysplasia after high-dose chemotherapy and autologous stem cell transplantation. Blood. 2000;95:3273–3279. [PubMed] [Google Scholar]
- Pedersen-Bjergaard J, Andersen MT, Andersen MK. Genetic pathways in the pathogenesis of therapy-related myelodysplasia and acute myeloid leukemia. Hematology Am Soc Hematol Educ Program. 2007:392–397. doi: 10.1182/asheducation-2007.1.392. [DOI] [PubMed] [Google Scholar]
- Qian Z, Fernald AA, Godley LA, Larson RA, Le Beau MM. Expression profiling of CD34+ hematopoietic stem/ progenitor cells reveals distinct subtypes of therapy-related acute myeloid leukemia. Proc Natl Acad Sci U S A. 2002;99:14925–14930. doi: 10.1073/pnas.222491799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raponi M, Dossey L, Jatkoe T, Wu X, Chen G, Fan H, Beer DG. MicroRNA classifiers for predicting prognosis of squamous cell lung cancer. Cancer Res. 2009;69:5776–5783. doi: 10.1158/0008-5472.CAN-09-0587. [DOI] [PubMed] [Google Scholar]
- Rassool FV, Gaymes TJ, Omidvar N, Brady N, Beurlet S, Pla M, Reboul M, Lea N, Chomienne C, Thomas NS, et al. Reactive oxygen species, DNA damage, and error-prone repair: a model for genomic instability with progression in myeloid leukemia? Cancer Res. 2007;67:8762–8771. doi: 10.1158/0008-5472.CAN-06-4807. [DOI] [PubMed] [Google Scholar]
- Stone RM, Neuberg D, Soiffer R, Takvorian T, Whelan M, Rabinowe SN, Aster JC, Leavitt P, Mauch P, Freedman AS, et al. Myelodysplastic syndrome as a late complication following autologous bone marrow transplantation for non-Hodgkin’s lymphoma. J Clin Oncol. 1994;12:2535–2542. doi: 10.1200/JCO.1994.12.12.2535. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolkovsky AM. Mitophagy. Biochim Biophys Acta. 2009;1793:1508–1515. doi: 10.1016/j.bbamcr.2009.03.002. [DOI] [PubMed] [Google Scholar]
- Tothova Z, Kollipara R, Huntly BJ, Lee BH, Castrillon DH, Cullen DE, McDowell EP, Lazo-Kallanian S, Williams IR, Sears C, et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 2007;128:325–339. doi: 10.1016/j.cell.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Wagner PD, Verma M, Srivastava S. Challenges for biomarkers in cancer detection. Ann N Y Acad Sci. 2004;1022:9–16. doi: 10.1196/annals.1318.003. [DOI] [PubMed] [Google Scholar]
- Wallace DC. Mitochondria and cancer: Warburg addressed. Cold Spring Harb Symp Quant Biol. 2005;70:363–374. doi: 10.1101/sqb.2005.70.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.