SUMMARY
Human glycolipid transfer protein (GLTP) fold represents a novel structural motif for lipid binding/transfer and reversible membrane translocation. GLTPs transfer glycosphingolipids (GSLs) which are key regulators of cell growth, division, surface adhesion, and neurodevelopment. Herein, we report structure-guided engineering of the lipid binding features of GLTP. New crystal structures of wild-type GLTP and two mutants (D48V and A47D||D48V), each containing bound N-nervonoyl-sulfatide, reveal the molecular basis for selective anchoring of sulfatide (3-O-sulfo-galactosylceramide) by D48V-GLTP. Directed point mutations of ‘portal entrance’ residues, A47 and D48, reversibly regulate sphingosine access to the hydrophobic pocket via a mechanism that could involve homo-dimerization. ‘Door-opening’ conformational changes by phenylalanines within the hydrophobic pocket are revealed during lipid encapsulation by new crystal structures of bona fide apo-GLTP and GLTP complexed with N-oleoyl-glucosylceramide. The development of ‘engineered GLTPs’ with enhanced specificity for select GSLs provides a potential new therapeutic approach for targeting GSL-mediated pathologies.
Keywords: GLTP-fold, sulfatide-binding pocket, structure-based protein design, lipid chain re-arrangement, dimer interface of ligand-bound human GLTP
INTRODUCTION
3-O-sulfogalactosylceramide or sulfatide (SF) is a major component of the myelin sheath in the central and peripheral nervous system in mammals. SF makes up 4–6% of myelin lipids and, together with its precursor galactosylceramide (GalCer), accounts for more than half (50–60 wt%) of the outer surface lipids of the myelin sheath (Eckhardt, 2008). Accumulation of SF has been implicated in neurological diseases such as metachromatic leukodystrophy (Eckhardt, 2008; Halder et al, 2007; Jeon et al, 2008; Zeng et al, 2008), which often are characterized by progressive demyelination. Elevation of sulfatide by 30–40% has also been found in the superior frontal and cerebellar gray matter in Parkinson’s disease patients (Eckhardt, 2008). By contrast, substantial decrease in SF has been observed in a case of progressive epilepsy with mental retardation (Hermansson et al, 2005) and in the brain and cerebrospinal fluid of Alzheimer’s disease patients (Eckhardt, 2008; Han, 2007; 2010). Not surprisingly, sulfatide homeostasis is critical for the normal function of central and peripheral nerves. Nonetheless, the metabolic alterations underlying the disease-linked changes in sulfatide homeostasis remain poorly understood, especially with respect to alterations in sulfatide inter- and intracellular trafficking (Han, 2007). Mammalian glycolipid transfer proteins (GLTPs) accelerate the intermembrane trafficking of various glycosphingolipids (GSLs) including sulfatide (Sasaki, 1990; Brown and Mattjus, 2007). In contrast, a fungal GLTP in Podospora anserina has a narrower specificity for GSLs and transfers sulfatide very poorly (Kenoth et al, 2010). Development of human ‘engineered GLTPs’ with altered lipid binding features, such as enhanced selectivity for sulfatide, could provide a potential new avenue of biomedical therapy for targeting GSL-mediated pathologies.
Previously, we determined the structure of human GLTP in GSL-free form as well as complexed with various galactosyl- and lactosylceramide species (GalCers and LacCers) (Malinina et al, 2004; 2006). The studies showed the human GLTP-fold to be unique among lipid binding/transfer proteins and serve as the structural prototype that defines the GLTP superfamily. To acquire GSL, GLTP utilizes: (i) a GSL recognition center to selectively bind the sugar-amide headgroup via a specific network of hydrogen-bond interactions, (ii) a cleft-like gating mechanism to facilitate lipid chain entry/exit into/out of the hydrophobic pocket in the protein interior; (iii) two modes of GSL binding. In the GSL ‘sphingosine-in’ mode, the acyl and sphingosine lipid chains are both encapsulated within the same hydrophobic pocket of GLTP. In the GSL ‘sphingosine-out’ mode, only the acyl chain occupies the hydrophobic pocket while the sphingosine chain remains outside and cross-bridges to a partner GLTP molecule, containing a similarly complexed glycolipid, to form a dimer. Structure determination of GLTP complexed with different GSLs has revealed that pocket occupancy, optimal fit by the ceramide chains, and monomer/dimer stability of the GLTP/GSL complex all play roles in determining whether the GSL lipid-chain encapsulation mode is sphingosine-in or sphingosine-out (Malinina et al, 2004; 2006). However, the intrinsic molecular events most likely to function as primary regulators of the GSL binding mode by GLTP, i.e. control the opening/closing of the entrance portal to the hydrophobic pocket, as well as its expansion/contraction during lipid chain encapsulation remain poorly understood, further complicating the engineering of GLTPs with altered lipid binding features.
In the present study, we show that point mutation of ‘portal entrance’ residues in the human GLTP-fold can switch the binding mode for lipid-chain encapsulation in a manner that can be reversed by introduction of a second adjacent point mutation. More remarkably, the GSL selectivity of the point-mutated GLTP is enhanced by virtue of continued binding/transfer of sulfatide, but not of simple neutral GSLs (e.g. GalCer) that typically are most highly preferred by wt-GLTP. The molecular basis for alteration of transfer selectivity in favor of sulfatide, switching of the GLTP-GSL binding mode, and optimization of hydrophobic pocket expansion/contraction are elucidated by five new high resolution crystal structures of GSL-GLTP complexes presented herein.
RESULTS AND DISCUSSION
Phe ‘cluster’ within apo-GLTP and its reshaping on GSL-binding
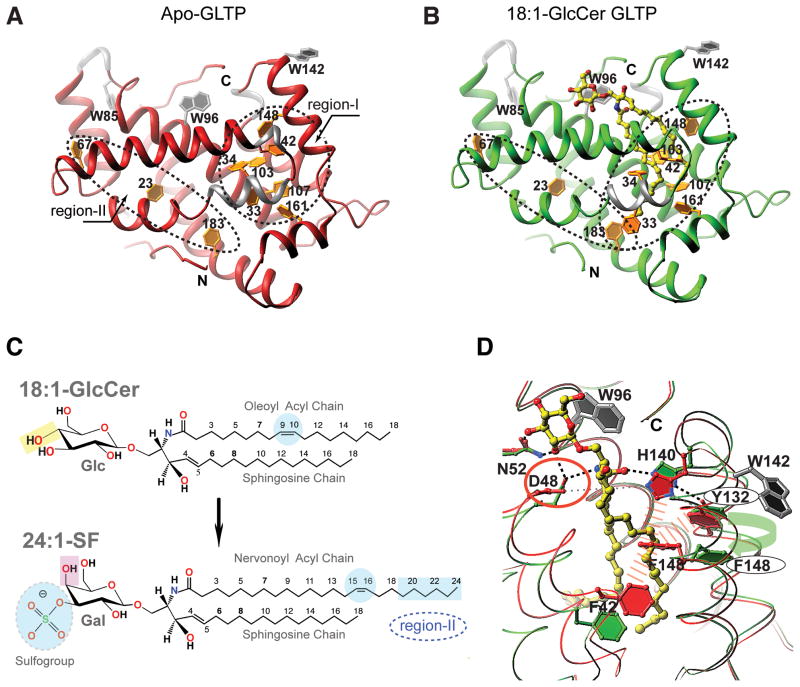
The hydrophobic pocket that encapsulates the GSL hydrocarbon chains is conforma-tionally adaptable and highly nonpolar (Malinina et al, 2004; 2006; Airenne et al, 2006). In GLTP, seven of ten phenylalanine residues (33, 34, 42, 103, 107, 148 and 161) are grouped closely and line the hydrophobic pocket, providing an endogenous marker for the main lipid chain encapsulation region (region-I; circled area in Figure 1A). Three other phenylalanine residues (183, 23 and 67) are more distant and located in a separate, extended region (region-II; long narrow ellipse in Figure 1A). Both region-I and region-II are fully collapsed and contain no trace of bound hydrocarbon chain, verifying this to be the first bona fide structure (1.5 Å) of human apo-GLTP (Table 1). In contrast, previously reported ‘glycolipid-free’ GLTP structures all contain non-glycolipid hydrocarbons near the junction of region-I and region-II (Malinina et al, 2004; 2006; Airenne et al, 2006), possibly obscuring some conformational changes that occur when the lipid chains are encapsulated by the hydrophobic pocket (Figure S1).
Figure 1. Role of Phenylalanine Residues in human GLTP-fold Functionality.
(A, B) A comparison of the collapsed hydrophobic pocket of bona fide apo-GLTP versus the occupied and expanded hydrophobic pocket of GLTP complexed with 18:1-GlcCer. Two differently ‘shaped’ regions (I and II) containing phenylalanine ‘clusters’ are outlined by dashed-lines. The protein chain is colored in red for apo-GLTP (panel A) whereas in green in the complex with 18:1-GlcCer (panel B). 310 helices are colored silver. Glycolipid atoms are colored yellow, red and blue for carbon, oxygen and nitrogen, respectively. Tryptophan and phenylalanine side chains are colored silver and gold.
(C) Glycolipid structures for N-oleoyl (18:1) glucosylceramide (top panel) and for N-nervonyl (24:1) 3-O-sulfo-galactosylceramide (bottom panel). Highlights emphasize distinguishing features that affect accommodation mode by GLTP.
(D) ‘Door opening’ mechanism for Phe148 shown by superimposition of apo-GLTP (colored red) and 18:1-GlcCer/GLTP complex (colored green), with glycolipid colored as in panel B. Two tryptophan residues are colored in silver; the wide, red-dashed lines symbolize the hydrophobic contacts between Phe148 and the three neighboring side chains of Phe42, Tyr132, and His140 in apo-GLTP, while the green arrow emphasizes the Phe 148/Tyr132 stacking interaction that occurs when the hydrophobic pocket is occupied by the ceramide chains of 18:1-GlcCer. Dashed lines show hydrogen-bonds; Asp48, the residue to be mutated, is highlighted by a red circle. See also Figure S1.
Table 1.
X-ray data collection and refinement statistics for human GLTP structures.
| Protein Ligand | apo-GLTP | WT-GLTP 18:1 GlcCer |
WT-GLTP 24:1 SF |
D48V-GLTP 24:1 SF |
D48V||A47D-GLTP 24:1 SF |
|---|---|---|---|---|---|
| Data collection | |||||
| Space group | P43212 | P212121 | C2 | C2 | C2 |
| a, (Å) | 82.21 | 50.66 | 74.97 | 77.97 | 76.87 |
| b, (Å) | 82.21 | 61.65 | 50.6 | 49.04 | 48.13 |
| c, (Å) | 148.28 | 67.21 | 65.43 | 62.63 | 69.08 |
| β, (°) | 90.0 | 90.0 | 118.5 | 125.0 | 123.8 |
| α= γ = 90° | |||||
| Resolution (Å) | 50-1.5 (1.55-1.50)a | 50-1.4 (1.45-1.40) | 50-1.1 (1.15-1.1) | 50-1.5 (1.55-1.50) | 50-2.1 (2.15-2.1) |
| Rmerge (%) | 7.2(24.9) | 5.2(39.2) | 8.0(29.3) | 6.1(44.4) | 5.0(55.0) |
| I/σI | 15.6(4.6) | 15.0(2.6) | 19.0(3.67) | 8.8(2.0) | 14.2(2.9) |
| Completeness (%) | 99.5(98.1) | 98.2(90.4) | 99.8(99.5) | 99.9(99.9) | 96.0(93.7) |
| Redundancy | 4.6(3.1) | 4.9(3.6) | 4.5(2.6) | 5.1(3.2) | 4.6(4.6) |
| Refinement | |||||
| Resolution (Å) | 15-1.5 | 15-1.4 | 15-1.1 | 15-1.5 | 15-2.1 |
| No. reflections | 76833 | 39019 | 96462 | 28741 | 10513 |
| Rwork/Rfree (%) | 17.8/20.7 | 15.98/19.36 | 13.84/16.40 | 17.9/21.0 | 18.6/21.8 |
| No. atoms | |||||
| Protein | 3329 | 1708 | 1679 | 1636 | 1660 |
| GSL | - | 51 | 61 | 61 | 61 |
| Water | 877 | 294 | 415 | 180 | 42 |
| B-factors | |||||
| Protein | 20.23 | 17.31 | 17.37 | 33.47 | 47.4 |
| GSL | - | 24.68 | 24.21 | 34.3 | 44.62 |
| Water | 48.64 | 30.94 | 34.5 | 44.18 | 47.8 |
| Rmsd | |||||
| Bond lengths (Å) | 0.015 | 0.012 | 0.013 | 0.019 | 0.016 |
| Bond angles (°) | 1.46 | 1.54 | 1.55 | 1.62 | 1.57 |
| PDB entry | 3RWV | 3S0K | 3RZN | 3S0I | 3RIC |
Rmsd, root-mean-square deviation from ideal values.
Values in parentheses are for the highest-resolution shell.
Rfree is calculated for 5% of randomly selected reflections excluded from refinement.
Region-I is the major zone of GSL ceramide accommodation where chains of various length and unsaturation can show two binding modes: ‘sphingosine-in’ or ‘sphingosine-out’ (Malinina et al, 2004; 2006). In Figure 1B, the ‘sphingosine-in’ mode is clearly evident in the crystal structure (1.4 Å; Table I) of GLTP complexed with N-oleoyl-glucosylceramide (18:1-GlcCer; Figure 1C, top panel) verifying the ability of region-I to simultaneously accommodate both ceramide chains in side-by-side alignment as originally reported for the GLTP/GSL structure involving N-oleoyl-lactosylceramide (Malinina et al, 2004). The fully extended 18:1 acyl chain approaches the Phe183 phenyl ring but is too short to insert into region-II while the Phe148 and Phe33 side chains in region-I assume ‘open door’ conformations compared to apo-GLTP. In general, this repositioning of the phenylalanine side chains in region-I has the net effect of transforming the circular ‘Phe cluster’ of apo-GLTP (Figure 1A) to an elliptical ‘Phe cluster’ (Figure 1B) for GLTP containing bound N-oleoyl-glucosylceramide.
Hypotheses for switching lipid chain binding within GLTP
Despite structural evidence indicating connectivity between Region-I and Region-II (Figures 1A and 1B), the role of region II in lipid chain accommodation is unclear. Previously, we observed that a naturally occurring GalCer with a nervonoyl acyl chain (24:1), which is sufficiently long to reach region II, did not enter into it (Malinina et al, 2006). Instead, the long acyl chain assumed a serpentine conformation within region-I and, by so doing, obstructed entry of the sphingosine chain, resulting in a ‘sphingosine-out’ binding mode (Figure S3A). We speculated that the cis-double-bond positioning between carbons 15 and 16 in the 24:1 acyl chain (Figure 1C, bottom panel) might interfere with chain entry into the narrow bottom compartment (region-II), much like difficulties encountered when attempting to ‘thread the eye of a needle’. In such situations, chances for success often are improved by slight loosening of the hold on the thread and adjusting the ‘push direction’. Accordingly, to switch the binding mode to ‘sphingosine-in’, we hypothesized that weakened anchoring of the ceramide amide would be needed at the ‘portal entrance’ of the hydrophobic pocket while preserving (or strengthening) both portal opening and GSL headgroup anchoring. Development of this hypothesis was facilitated by the new data for apo-GLTP (1.5 Å) and by superposition analysis against the new high-resolution structure (1.4 Å) of the GLTP/18:1-GlcCer complex (Figure 1D) that reveal the side chain interactions controlling the dramatic changes in positioning of specific Phe residues, i.e. ‘open door’ conformations needed for unobstructed entry of lipid hydrocarbon chains. In apo-GLTP (Figure 1D; red), the Phe148 ‘closed’ conformation is aided by hydrophobic contacts with side chains of Tyr132, Phe42, and His140. Opening of this Phe148 ‘door’ (Figure 1D; green) to accommodate GSL hydrocarbon chain(s) requires headgroup anchoring of GSL and disruption of hydrophobic contacts involving Phe148-His140 and Phe148-Phe42. To attain hydrogen-bonding with the ceramide amide group, His140 moves ‘upward’ and away from Phe148 (Figure 1D). To compensate the energy loss associated with disruption of the hydrophobic contacts, Phe148 undergoes stacking with the Tyr132 phenol ring (Figure 1D) to achieve the ‘open door’ conformation. This movement also facilitates Phe42 ‘liberation’, creating additional space for sphingosine chain encapsulation. Thus, hydrogen-bonding of the ceramide amide group with His140 is likely to be a critical initial step for triggering the mechanistic process that promotes accommodation of sphingosine within GLTP hydrophobic pocket. The essential role of His140 in GLTP action is supported by the complete inactivation observed in H140L-GLTP (Malinina et al, 2004).
Enhancing GLTP selectivity for sulfatide
In wt-GLTP, Asp48 and His140 interact via hydrogen bonding through a water molecule and regulate the opening of the portal entrance to the hydrophobic pocket. We considered mutations that might weaken the anchoring of the ceramide amide at the ‘portal entrance’ of the hydrophobic pocket while preserving (or strengthening) portal opening and GSL headgroup anchoring. We hypothesized that such mutational changes also might increase GLTP selectivity for certain glycolipids. Given the inactivation encountered by H140 mutation, Asp48 and adjacent residues emerged as leading candidates for directed mutation-based engineering. From the lipid standpoint, we focused on 3-O-sulfo-galactosylceramide (sulfatide) because wt-GLTP was known to transfer sulfatide and other neutral or negatively-charged GSLs (Sasaki, 1990). Although D48V-GLTP mutant was known to transfer GalCer poorly (Malinina et al, 2004; Malakhova et al., 2005), its ability to transfer sulfatide was untested. The focus on sulfatide containing nervonoyl (24:1) acyl chains (24:1-SF) was based on the hypothesized need to have an acyl chain sufficiently long to reach Region-II, as well as to have sulfate present to maintain strong interaction with the GLTP recognition center. Figure 1C (bottom panel) illustrates the chemical structure of 3-O-sulfo-GalCer including the location of the negatively-charged, sulfate group on the galactose moiety.
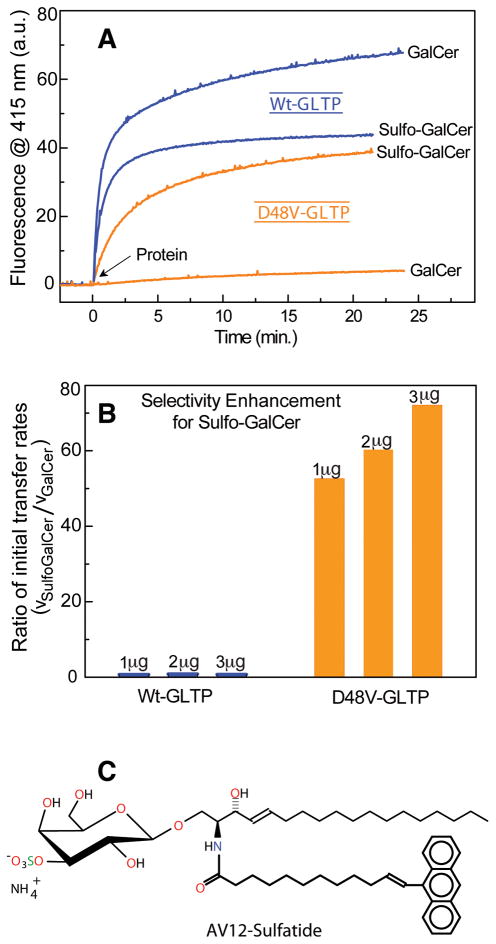
To initially test our hypotheses, we compared the transfer activity of wild-type GLTP with that of the D48V-mutant. Real-time intermembrane transfer kinetics of sulfatide and of GalCer by D48V-GLTP and wt-GLTP were measured by Förster resonance energy transfer (Figure 2A-B) using GSLs with acyl chains containing omega-linked anthrylvinyl (AV) fluorophores (Figure 2C). Whether the cleft-like gating mechanism of glycolipid uptake by GLTP (Malinina et al., 2004) enables entry of the bulky, nonpolar AV group into the hydrophobic pocket remains unclear. Regardless, the approach is well established for assessing GLTP transfer activity rates and lipid specificity (Brown and Mattjus, 2007). The outcomes are supported by competition analyses involving the selective slowdown in AV-glycolipid transfer rate by glycolipids with naturally-occurring, unaltered acyl chains (Figure S2A-D), as well as by transfer assays involving radioactive sulfatide and GalCer (Figure S2E-F). Remarkably, sulfatide transfer by wt-GLTP or by D48V-mutant proceeds moderately well (Figures 2B, S2A, S2E), in contrast to the deleterious effect of D48V mutation on GalCer transfer and the highly efficient transfer of GalCer by wt-GLTP. Ratiometric comparison of the initial kinetic transfer rates of sulfatide/GalCer by D48V-GLTP and wt-GLTP indicates a >50-fold enhancement in the transfer selectivity of D48V-GLTP for sulfatide over GalCer (Figure 2B).
Figure 2. Transfer Activity of wild-type GLTP and D48V-GLTP for Sulfatide and Galactosylceramide.
(A) Transfer of AV-glycolipid by wt-GLTP or D48V-GLTP (2 μg) as a function of time. The increase in fluorescence emission at 415 nm (AV emission) occurs because of decreased Förster resonance energy transfer that occurs as AV-glycolipid is removed from donor vesicles containing 3-perylenoyl PC and is transported to POPC acceptor vesicles, as described in the Methods.
(B) Selectivity enhancement of D48V-GLTP for sulfatide compared to wt-GLTP. The ratio of the initial rates of sulfatide and GalCer transfer by D48V-GLTP is increased 50- to 70-fold compared to wt-GLTP. Determinations are shown for three different protein levels (1, 2, and 3 μg of protein).
(C) Structure of 3-O-Sulfo-D-galactosyl-β1-1′-N-[12-(9-anthryl)-11E-dodecenoyl]-D-erythro-sphingosine (AV-sulfatide). See also Figure S2.
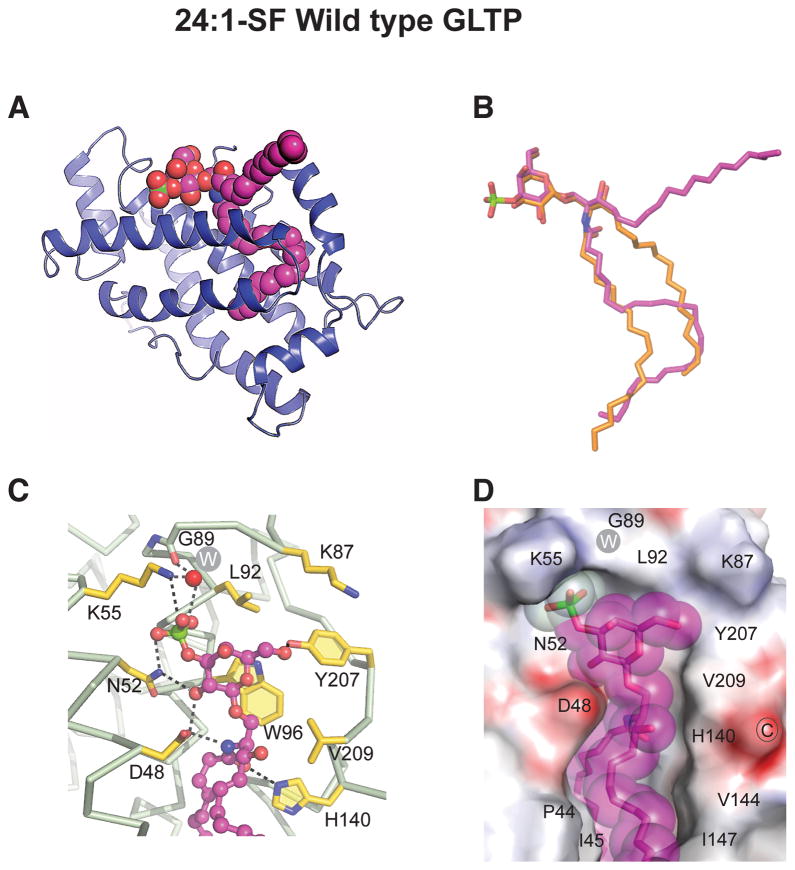
Structure of 24:1 Sulfatide bound to wild-type GLTP
To elucidate the molecular basis for the enhanced specificity of D48V-GLTP for sulfatide, we solved the crystal structures of both wt-GLTP/24:1-sulfatide complex (1.1 Å) and D48V-GLTP/24:1-sulfatide complex (1.5 Å). The crystallographic parameters are summarized in Table 1. With wt-GLTP (Figure 3A), the 24:1-sulfatide binding mode remains ‘sphingosine-out’, as also was the case for wt-GLTP/24:1-GalCer (Malinina et al, 2006). The conformations of bound 24:1-sulfatide (Figure 3A) and bound 24:1-GalCer in wt-GLTP (Figure S3A) are nearly identical and differ significantly from the ‘sphingosine-in’ conformation of 18:1 GlcCer bound to wt-GLTP (Figure 3B).
Figure 3. Crystal Structure of human Wild-Type GLTP in Complex with N-nervonoyl (24:1) Sulfatide.
(A) Ribbon representation of human wt-GLTP showing 24:1-sulfatide bound in the sphingosine-out mode.
(B) Superpositioned conformations 24:1-sulfatide (magenta) and 18:1-GlcCer (orange) as bound by wt-GLTP.
(C) Anchoring of 3-O-sulfo-galactose headgroup of sulfatide to wt-GLTP sugar recognition centre residues. Hydrogen bonds are indicated by dashed lines. The protein Cα-backbone is silver with gold side chains. Glycolipid carbon atoms are magenta colored. Oxygen atoms are red, nitrogen atoms are blue, and the sulfur atom is green. The white W inside the gray circle represents a water molecule.
(D) Electrostatic surface view (blue, positive; red, negative; grey, neutral) of the wt-GLTP sugar headgroup recognition center occupied by 24:1-sulfatide (space-filled and magenta colored). See also Figures S3 and S6.
Anchoring of the 3-O-sulfated-Gal headgroup to the headgroup recognition center of wt-GLTP occurs via a network of intermolecular hydrogen-bonds (Figure 3C) involving the same residues (Asp48, Asn52, Lys55, His140, Tyr207, Trp96) as for the Gal headgroup of GalCer (Figure S3B). The carboxy group of Asp48 forms two bifurcated hydrogen-bonds (Figure 3C) that are also observed in other GLTP complexes with non-sulfated GSLs (Malinina et al, 2004; 2006). However, there are differences in the way that residues Asn52 and Lys55 interact with sulfatide compared to GalCer. For instance, Asn52 undergoes bidentate hydrogen bonding with the 2-OH group of galactose and the OS1-atom of the sulfo-group in sulfatide (Figure 3C), rather than with the 2-OH and 3-OH groups of galactose in GalCer (Figure S3B). The ε-amino group of Lys55 interacts with OS2-atom of the sulfo-group rather than with the 3-OH group of galactose in GalCer. Also, the sulfate-OS3-atom undergoes hydrogen-bond bridging with the main chain oxygen of Gly89 through a highly conserved water molecule (Figure 3C). Thus, all three OS-atoms contribute to the hydrogen-bond network that anchors the sulfo-group of sulfatide to the GLTP surface (Figure 3D).
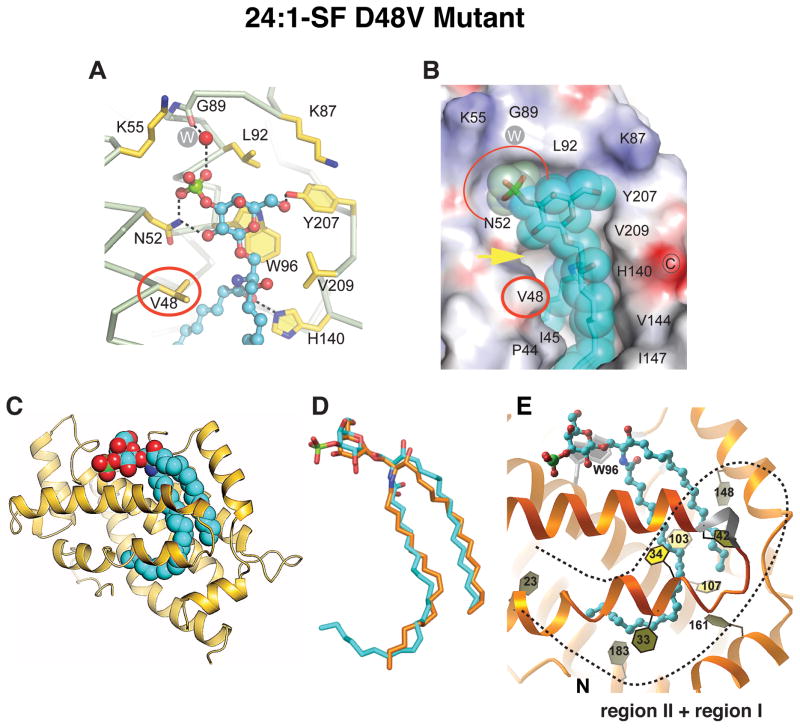
Structure of 24:1 Sulfatide bound to D48V-GLTP
As observed in the wt-GLTP/24:1-sulfatide complex (Figure 3C), Asn52 forms bidentate hydrogen bonds with the 2-OH group of galactose and the OS1-atom of the sulfo-group in the D48V-GLTP/24:1-sulfatide complex (Figure 4A). However, the recognition center of D48V-GLTP (Figure 4A) reveals a missing hydrogen-bond between the ε-amino group of Lys55 and the sulfate OS2-atom, as well as a lack of the two bifurcated hydrogen-bonds originating from Asp48 replacement with Val. Regardless, 3-O-sulfated GalCer still is transferred well by D48V-GLTP even though GalCer is not (Figures 2B, S2). Hence, the 3-O-sulfo-group makes an essential contribution to sulfatide anchoring in the recognition center of D48V-GLTP. This conclusion is further supported by the severely weakened interaction known for GalCer and D48V-GLTP (Malinina et al., 2004; Malakhova et al., 2005), which also involves the loss of the two bifurcated hydrogen bonds provided by D48. Without sulfate to compensate and secure sugar headgroup anchoring, GalCer binding becomes weak.
Figure 4. Crystal Structure of D48V-GLTP in Complex with N-nervonoyl (24:1) sulfatide.
(A) Anchoring of 3-O-sulfo-galactose headgroup of sulfatide to D48V-GLTP sugar recognition centre residues. Hydrogen bonds are indicated by dashed lines. The protein Cα-backbone is silver with gold side chains. Glycolipid carbon atoms are cyan colored. Oxygen atoms are red, nitrogen atoms are blue, and the sulfur atom is green. The white W inside the gray circle represents a water molecule.
(B) Electrostatic surface view (blue, positive; red, negative; grey, neutral) of the D48V-GLTP sugar headgroup recognition center occupied by 24:1-sulfatide (space-filled and cyan colored). Replacement residue Val48 and complementary cavity for the sulfate group are highlighted by red oval and red semi-oval, respectively. The yellow arrow emphasizes the affected region of inter-molecular contact associated with the D48V mutation.
(C) Ribbon representation (gold) of human D48V-GLTP showing 24:1-sulfatide (cyan) bound in the sphingosine-in mode.
(D) Superpositioned conformations 24:1-sulfatide (cyan) as bound by D48V-GLTP and 18:1-GlcCer (orange) as when bound by wt-GLTP.
(E) Sphingosine-in binding mode of 24:1-SF by D48V-GLTP. The ‘open door’ conformations of both Phe148 and Phe33 enable occupancy of Regions I and II (dashed line) by the long nervonoyl chain. See also Figure S4 and S6.
To elucidate the intermolecular contacts that account for the favorable accommodation of sulfatide headgroup at the protein surface, we compared surface topologies. The surface view of D48V-GLTP (Figure 4B) reveals that the diminished hydrogen bonding with sulfate by Lys55 results in repositioning and uncovering of a cavity (highlighted by a red semicircle) that surrounds the bulky sulfate and appears to aid in its anchoring to GLTP. The cavity also is present in wt-GLTP/sulfatide complexes but the increased H-bonding to sulfate by Lys55 results in partial obstruction (Figure 3D). When GalCer lacks the 3-O-sulfo group, Lys55 completely obstructs the cavity because of positioning for hydrogen bonding with the 3-OH group of galactose (Malinina et al, 2006). These findings show that Lys55 interaction with sulfatide in GLTP is more complex than a simple charge-charge interaction among chemical groups.
The changes to GLTP surface topology brought about by D48V mutation, which eliminates the bifurcated hydrogen-bonds to the 2-OH group of galactose and the amide group of ceramide in sulfatide, are illustrated in Figures 3D and 4B. Comparison of the van-der-Waals contacts between sulfatide and wt-GLTP (Figure 3D) or D48V-GLTP (Figure 4B) reveals significantly more free space around the V48 residue (Figure 4B, yellow arrows) because of the smaller volume of the valine side chain compared to aspartic acid. In fact, water molecules occupy the liberated space. However, their positions and number (two or three) differ in various crystals of D48V-GLTP complexed with sulfatide, indicating that the D48V mutation diminishes the clamp-like hold of D48/H140 observed for GSLs bound to GLTP at the ‘portal entrance’ of the hydrophobic pocket (Malinina et al., 2004; 2006) and liberates the ceramide region of sulfatide.
Switching of lipid chain encapsulation within GLTP
As hypothesized, introduction of the D48V mutation into human GLTP resulted in switching of the 24:1-sulfatide binding mode to ‘sphingosine-in’ (Figure 4C). Overall, the conformation resembles that of 18:1 GlcCer in wt-GLTP (Figure 4D). The initial segment of the long nervonoyl acyl chain is straight in the D48V mutant (Figures 4C and 4D), rather than S-shaped as in wt-GLTP (Figures 3A and 3B), enabling the sphingosine chain to be accommodated within the hydrophobic pocket rather than being excluded. Moreover, the distal portion of the 24:1 acyl chain occupies region-II of D48V-GLTP (Figure 4E and Figure S4A-B). It is noteworthy that the sharp bend that enables the distal portion of the 24:1 acyl chain to enter region-II corresponds to the location of the cis double bond. The ‘open door’ conformation of phenyl ring of Phe33 allows clear passage of the distal portion of the 24:1 acyl chain into region-II (Figure 4E) and simultaneously shields this portion of acyl chain from outside hindrance (Figure S4A and S4C).
A ‘hot spot’ for homodimerization in D48V-GLTP
Although the sulfate-accommodating surface cavity of D48V-GLTP explains how sulfatide can bind when GalCer cannot, this mutant must display additional attributes to enable accommodation of 24:1-sulfatide by a ‘sphingosine-in’ binding mode compared to wt-GLTP, which uses ‘sphingosine-out’ binding. The dimerization interfaces of the D48V-GLTP/sulfatide and wt-GLTP/sulfatide complexes reveal subtle but potentially important differences. The dimerization contact regions of GSL/GLTP complexes generally coincide with their membrane interaction domains (Malinina et al, 2006; Kamlekar et al, 2010). In the absence of membranes, binding of 24:1-sulfatide by wt-GLTP or D48V-GLTP does promote homodimerization in solution as determined by dynamic light scattering analyses (Table S1).
Role for Pro44 in hydrophobic interfacial contacts
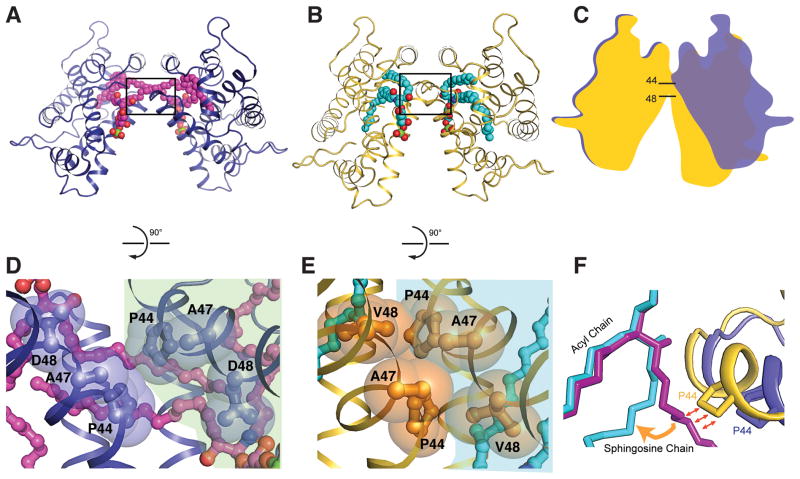
Figures 5A and 5B show the dimeric structures of wt-GLTP and D48V-GLTP complexed with 24:1-sulfatide. The two structures differ both in their lipid chain conformations and in the inclinations of the monomers forming their dimerization interfaces. The end result is a more ‘open’ dimer conformation for the wt-GLTP complex compared to D48V-GLTP (depicted schematically in Figure 5C).
Figure 5. D48V Mutation Generates GLTP Dimeric ‘Hotspot’ near Portal Entrance of Hydrophobic Pocket.
(A, B) Dimeric arrangements of 24:1-sulfatide/wt-GLTP (A) and 24:1-sulfatide/D48V-GLTP (B) with proteins shown in ribbon representation and glycolipids shown in space-filling representation. WT-GLTP and D48V-mutant are colored blue and gold, respectively. Glycolipid carbon atoms are colored magenta for wt-GLTP and cyan for D48V-GLTP.
(C) Schematic superimposition of two dimeric arrangements, highlighting the different mutual monomer inclination in D48V-GLTP complex compared to wt-GLTP.
(D, E) The boxed regions of panels A, B rotated by 90 degrees and amplified in panels D, E with residues Pro44, Ala47, Asp48 and Val48 shown in space-filling and ball-and-stick representations. Two molecules of the dimeric structures are sub-colored differently to emphasize a weak protein-protein contact between two Pro44 residues in the wt-GLTP dimer. The intermolecular contact region increases in D48V dimer as a result of formation of a larger hydrophobic contact surface.
(F) Superimpositions of two dimeric arrangements, highlighting the ‘interaction-point’ of a protein partner in a dimer with 24:1-sulfatide sphingosine chain with lipids and proteins colored as in panels A and B.
The dimer interface involves helix-loop-helix contacts that are associated primarily with α-helix 2 and its adjacent α1–2 loop. Within this contact region, Pro44 is an especially important residue that undergoes hydrophobic contact with its partner Pro44 in the dimeric complex (Figures 5D and 5E). However, it is noteworthy that the Pro44 side chain forms a ‘single-residue’ protein-protein contact in the wild-type dimer (Figure 5D), whereas the dimerization interface of D48V-GLTP contains a larger multi-residue hydrophobic contact area formed by Pro44, Ala47 and Val48 (Figure 5E). Since only one of the interface residues differs in D48V-GLTP, we conclude that replacement of Asp48 by Val48 facilitates the close proximity and interlocking along the dimer interface.
Pro44 also forms van der Waals contacts with the nonpolar sphingosine chains of both partner lipid molecules in the dimeric structure in the ‘sphingosine-out’ binding mode (Malinina et al, 2006). Figure 5F shows the contacts of Pro44 with a partner sphingosine chain in the wt-GLTP complex compared to the D48V-GLTP complex to emphasize that, if the sphingosine chain remained in the ‘out’ conformation in D48V-GLTP, Pro44 contacts would be impossibly close in the D48V dimer. Thus, changes in dimer conformation appear to facilitate the ‘sphingosine-in’ binding mode of 24:1-sulfatide in D48V-GLTP.
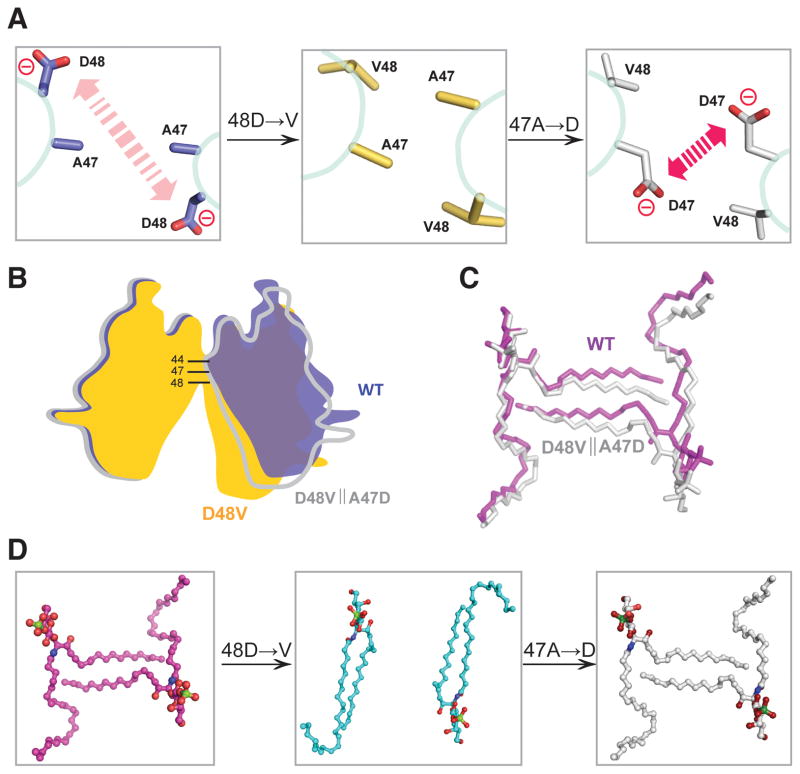
Redesign of lipid binding mode in A47D||D48V double-mutant
To further test our idea, we introduced an adjacent negatively-charged aspartic acid into D48V-GLTP through mutation of alanine-47 for aspartate, i.e. A47D||D48V double-mutant GLTP. We reasoned that the electrostatic repulsion originally provided by Asp48 in wtGLTP (Figure 6A, left panel), would be restored by insertion of A47D into D48V along with the open-dimer conformation that promotes the ‘sphingosine-out’ binding mode of 24:1 sulfatide. Figure 6A (middle panel) shows how the lost electrostatic repulsion in D48V-GLTP is replaced by the ‘van der Waals attraction’ of the hydrophobic residues. In contrast, in the A47D||D48V double-mutant, the repulsion provided by the newly created aspartic acid, D47, (Figure 6A, right panel) restores the open-dimer conformation. Figure 6B illustrates the dimer transitions from open (wtGLTP; yellow) to closed (D48V-GLTP; blue) and back to open (A47D||D48V-GLTP; white) in schematic fashion; whereas Figure 6C displays the similar (but not identical) mutual orientations of interacting sulfatide molecules in the ‘open’ dimers of wtGLTP and A47D||D48V-GLTP. Figure 6D shows the spatial arrangement of the three ‘sulfatide•sulfatide pairs’ and the engineered transitions induced in the complexes of wtGLTP (left panel; ‘sphingosine-out’), D48V-GLTP (middle panel; ‘sphingosine-in’), and A47D||D48V-GLTP (right panel, ‘sphingosine-out’). The overall structure of the A47D||D48V double-mutant GLTP with its ‘restored’ sphingosine-out 24:1-sulfatide is illustrated in Figure S5.
Figure 6. Comparison of the ‘Open’ Conformation and the ‘Sphingosine-out’ Binding Mode of 24:1-SF Found in A47D||D48V Dimer with those of WT-GLTP.
(A) Schematic highlighting the distinctions in intermolecular interactions observed in dimers of wt-GLTP (left panel), D48V-mutant (middle panel) and double-mutant A47D||D48V (right panel): the ‘red-arrowed’ repulsion between two negatively-charged residues Asp48 in wt-GLTP is replaced for the attraction of hydrophobic residues in D48V-mutant and ‘restored’ in double-mutant A47D||D48V by means of an additional mutation A47D.
(B) The ‘open’ arrangement of the dimer in WT-GLTP and A47D||D48V mutant with conserved local negative charge of dimeric ‘hotspot’ versus ‘close’ conformation of ‘neutral’ D48V mutant.
(C) Similarity of the ‘sphingosine-out’ binding mode in dimeric WT-GLTP and A47D||D48V mutant.
(D) Lipid dimers ‘derived’ from the dimeric arrangments of WT-GLTP (left panel), D48V-mutant (middle panel) and double-mutant A47D||D48V (right panel).
Glycolipids and amino acids are shown in stick representations, proteins are indicated schematically. Coloring is blue, gold and white for WT-GLTP, D48V mutant and A47D||D48V mutant, respectively, and magenta, cyan and white for lipid derived from WT-GLTP, D48V mutant and A47D||D48V mutant, respectively, with red, blue and green colors indicating atoms of oxygen, nitrogen and sulfur. See also Figure S5.
Recognition center adaptability for sulfated-GSL headgroups
Nearly all residues known to comprise the sugar headgroup recognition center of GLTP play essential roles in binding of sulfatide, including Asp48, Asn52, His140 and Trp96. The only exception is Lys55, which would be expected to use its ε-amino group to interact strongly with the 3-O-sulfo moiety of sulfatide in both wt-GLTP and D48V-GLTP. However, this clearly is not the case, especially for D48V-GLTP (Figure 4A), where a special surface cavity with a complementary contact surface for the 3-O-sulfo-group (Figures 4B) becomes visible when Lys55 repositions. The complementary cavity seems to play a key role in anchoring the negatively-charged, sulfate group to the GLTP surface. A conserved water molecule was discovered that hydrogen bonds with the oxygen atom of Gly89 and undergoes tight van-der-Waals contacts with the NH-group of Gly89 and the Cβ-atom of Leu92 (Figures 3C and 4A). The water molecule favorably shapes the surface cavity for complementary contact with the 3-O-sulfo-group (Figures 3D and 4B). The water molecule is present in all apo-and holo-forms of human GLTP resolved to date.
Intriguingly, a water molecule is also observed at the same location in a GLTP-like protein (PDB 2Q52) from thermoacidophilic unicellular red algae, Galdieria sulphuraria (Figure S6). The conserved nature of this water molecule is illustrated in Figure S6B, which shows the G. sulphuraria GLTP-fold superpositioned against human wt-GLTP complexed with sulfatide. Although the lipid composition of G. sulphuraria does not include 3-O-sulfo-GalCer, a ‘virtual’ sulfatide is well accommodated, suggesting binding of an analogous sulfoglycolipid. Surface views of G. sulphuraria GLTP-fold (Figure S6C) and human GLTP (Figure S6A) show similar outward projecting conformations for Lys66/55, respectively, further emphasizing the similarity in shape and size of the potential sulfo-group interaction site.
Uniqueness of GLTP Among Sulfatide-binding Proteins
It is noteworthy that our high-resolution structures of sulfatide complexed with wtGLTP, D48V-GLTP, and A47D||D48V-GLTP (Table 1) differ substantially from the structures of two other proteins known to bind sulfatide, sulfatide/saposin B (2.2 Å) and sulfatide–CD1a (2.15 Å). The hydrogen-bond interaction network responsible for the recognition specificity of the 3-O-sulfo-galactose and ceramide moieties distinguishes the GLTP complexes (Figures 3C and 4A) from the corresponding CD1a complex, which uses fewer hydrogen bond recognition contacts (Zajonc et al, 2003; 2005; Zajonc and Kroneberg, 2007). In the case of saposin B, uncertainties in the specific features of the lipid conformation prevented detailed analysis of the protein–lipid interactions in this complex (Ahn et al, 2003). In the CD1a complex, the sphingoid-base and N-acyl chains insert into separate interconnecting hydrophobic pockets; whereas a single hydrophobic pocket accommodates one or both lipid chains in the GLTP complex. For saposin B, protein dimerization is needed for formation of the hydrophobic cavity that accommodates both hydrocarbon chains of sulfatide. These unique glycolipid–protein interactions appear to be associated with distinct functional events, namely glycolipid transfer by the GLTP, in contrast to glycolipid presentation by the CD1 major histocompatibility proteins, as well as by saposin B.
CONCLUDING REMARKS
The engineering of proteins with enhanced stabilities, ligand specificities, and functionalities is a rapidly emerging field with potential applications in biotechnology and biomedical therapeutics (Hancock et al, 2009; Gaj et al, 2011; Picollo et al, 2009; Wörsdörfer et al, 2011). Our structure-guided, point mutational engineering of human GLTP illustrates how a detailed understanding of structure-function relationships can enable the development of ‘designer GLTPs’ with enhanced selectivity for certain GSLs. The five new high-resolution crystal structures of human GLTP presented herein provide unprecedented insights into the workings of the hydrophobic pocket of the human GLTP-fold. Most notably, our data reveal the molecular basis for the enhanced selectivity of D48V-GLTP for sulfatide but not its non-sulfated precursor, which is very well accommodated by wild-type GLTP. The protein surface cavity that accommodates the sulfo-group is formed at the junction of two alpha-helices and involves a conserved water molecule. Our data establish the functionality of a narrow bottom compartment of the GLTP hydrophobic pocket and, for the first time, demonstrate that structure-guided, point mutational redesign of GLTP can switch the binding mode of lipid chains. Taken together, the five new high-resolution GLTP structures provide unprecedented insights into the molecular changes that optimize and regulate the expansion/contraction of the hydrophobic pocket during lipid accommodation.
Gaining an understanding of the structural principles that enable human GLTP to selectively transfer certain GSLs provides a new avenue for creating ‘designer GLTPs’ with highly targeted specificity, thus facilitating the development of potential new therapeutic applications for the treatment of GSL-related neurodegenerative diseases.
EXPERIMENTAL PROCEDURES
Plasmid Construction and Mutagenesis
The ORF encoding human GLTP (NCBI GenBank Acc.# AF209704) was subcloned into the pET-30 Xa/LIC expression vector (Novagen) by ligation independent cloning, enabling cleavage of the N-terminal His6-S-tag to yield protein identical in sequence to native GLTP. The site-directed D48V-GLTP mutant was obtained using QuikChange Site-directed Mutagenesis Kit (Stratagene) and verified by sequencing.
Protein expression and purification
Transformed BL21 (DE3) cells (Novagen) were grown in Luria-Bertani medium at 37° C, induced with 0.1 mM IPTG, and grown an additional 16–20 h at 15° C. Wild-type GLTP and D48V-GLTP were purified from soluble lysate by Ni-affinity chromatography as detailed previously (Airenne et al, 2006). His- and S-tag sequences were removed using Factor-Xa and GLTP was repurified by FPLC size exclusion chromatography using a HiLoad 16/60 Superdex-75 prep grade column (Amersham Biosciences).
Crystallization and X-ray data collection
Crystals of wild-type-GLTP and D48V-GLTP complexed with 24:1 SF, as well as apo-GLTP and GLTP complexed with 18:1 GlcCer, were grown by the hanging-drop vapor-diffusion method using PEG 3350 or 8000 (15–20%) as precipitant and 100 mM MES (pH 5–7) containing 150 mM NaCl as buffer (Malinina et al, 2004;2006). All lipids were obtained from Avanti Polar Lipids (Alabaster AL). Protein-lipid complexes were prepared by mixing protein and lipid in 1:1 molar ratio using lipids dissolved in ethanol (final concentration ~20% EtOH). Crystals were transferred into well solution containing 20% glycerol as cryoprotectant, then mounted in a fiber loop, and flash-frozen in a cold nitrogen stream. X-ray data were collected at 100°K using synchrotron radiation at ESRF (Grenoble, France) on beamlines ID 14-4 and ID 23-1, except for double mutant A47D||D48V-GLTP complexed with 24:1-sulfatide which was collected at CICbioGUNE on X8 Proteum System (Bruker) at CuKα wavelength 1.54 Å using a CCD detector. All data were processed and scaled using the program suite HKL2000.
Structure determination and refinement
Crystals of apo-GLTP belonged to the P43212 space group, containing two molecules in the asymmetric unit. The 18:1-GlcCer/GLTP complex crystallized as a monomer with P212121 space group symmetry. Three other crystals belonged to the C2 space group and had one molecule of protein-lipid complex in the asymmetric unit. All structures were determined by the Molecular Replacement (MR) method, using the AMoRe program (Navaza, 1997) along with previously refined complexes as models (Malinina et al, 2004; 2006). Refinement was performed with REFMAC (Murshudov et al, 1997), alternating with manual model-rebuilding using Turbo-Frodo and Coot. The ARP/wARP automatic procedure was used to add solvent molecules (Lamzin and Wilson, 1993). Final structures were validated by PROCHECK. The coordinates have been deposited in the Protein Data Bank. Data collection and refinement statistics, along with unit cell dimensions, are summarized in Table 1.
Fluorescent lipids
N-[(11E)-12-(9-anthryl)-11-dodecenoyl]-1-O-β-galactosylsphingosine (AV-GalCer) and 1-acyl-2-[9-(3-perylenoyl)nonanoyl]-sn-glycero-3-phosphocholine (Per-PC)] were synthesized as described previously(Molotkovsky et al, 1982; 1991). 3-O-sulfo-D-galactosyl-β1-1′-N-[(11E)-12-(9-anthryl)-11-dodecenoyl]-D-erythro-sphingosine (AV-sulfatide) synthesis relied on a similar approach to be detailed elsewhere. 1-Palmitoyl-2-oleoylphosphatidylcholine (POPC) was purchased from Avanti Polar Lipids. The chemical structure of AV- sulfatide is shown in Figure 2C.
Fluorescence Lipid Transfer between Membranes
Real-time intermembrane transfer rates of fluorescent glycolipids by wt-GLTP and D48V-GLTP were obtained by Förster resonance energy transfer using a SPEX FluoroMax spectrofluorimeter (Horiba Scientific), with excitation and emission bandpasses of 5 nm, and a stirred (~100 rpm), temperature-controlled (25 ±0.1°C) sample cuvette holder (Brown, 1992). Both fluorescent lipids were localized initially to the donor vesicles, formed by rapid ethanol injection and comprised of POPC plus 1 mole% AV-glycolipid and 1.5 mole% Per-PC. Minimal emission by AV-glycolipid occurred upon excitation (370 nm) because of resonance energy transfer to nearby Per-PC. In competition assays, donor vesicles also contained ‘competitor’ lipids with naturally-occurring hydrocarbon chains at 1, 2, and 4 mole%. Addition of ~10-fold excess of sonicated POPC acceptor vesicles produced minimal change in fluorescence signal because of the very slow spontaneous transfer of lipids with long acyl chains (Mattjus et al, 1999; Brown, 1992). Addition of catalytic GLTP amounts triggered a sudden, exponential increase in AV emission intensity (415 nm) as the protein transported AV-labeled glycolipids from donor vesicles to acceptor vesicles, creating separation from ‘nontransferable’ Per-PC. Addition of detergent after extended incubation provided a measure of maximum AV intensity achievable by ‘infinite’ separation from Per-PC. Nonlinear regression analyses using ORIGIN 7.0 software (Origin Lab) provide the initial lipid transfer rate, νo, for the first-order exponential transfer process. Standard deviations were calculated at 95% confidence interval. R2 values for all of the estimates were>0.96.
Preparation of Donor and Acceptor Vesicles
Donor vesicles composed of POPC (97.5 mole%), AV-glycolipid (1 mole%) and Per-PC (1.5 mole%) were prepared by rapid ethanol injection into buffer being rapidly stirred in the cuvette at 25°C as described previously (Mattjus et al, 1999). Prior to injection, all three lipids were mixed together in hexane, dried under nitrogen, and then re-dissolved in ethanol (HPLC grade). Each ethanol injection (5 μl) contained ~175 pmol of AV-glycolipid. After dilution, the final ethanol concentration was less than 0.2%. The acceptor vesicles were prepared in the following way. POPC was dried onto a glass round bottom flask in vacuo before hydrating in sodium phosphate buffer (pH 6.6) at 50 mM concentration and suspending by vortexing. The suspension was probe sonicated intermittently under nitrogen until opalescent and then centrifuged for 90 min at 100,000 g to remove probe particles and multilamellar vesicles. The size of the acceptor vesicle populations averaged ~25 nm in diameter. The final acceptor vesicle concentration used in the FRET lipid transfer assay was ~85 μM, which was 10–15 fold higher than the donor concentration.
Dynamic light scattering
Dynamic light scattering (DLS) measurements were performed using a DynaPro Titan instrument, Wyatt Technology Corporation (with laser of ~830 nm wavelength). Studies were carried out in the protein concentration range of 1–4 mg/ml in 150 mM NaCl, 20 mM Tris-Cl buffer (pH 8.0) at 18°C. Complexes with lipid were prepared immediately prior to measurement using protein:lipid ratios in complexes of 1:1. Prior to DLS, all buffers were centrifuged at 27000g for 30 min and filtered through 0.2 mm membrane filters (Whatman). Data were collected and analyzed using the DYNAMICS software for the DynaPro Titan instrument (Wyatt Technology Corporation).
Supplementary Material
Acknowledgments
This research was supported by Spanish Ministerio de Ciencia e Innovacion (MICINN BFU2010-17711), Russian Foundation for Basic Research (#09-04-00313), USPHS (NIGMS GM45928 & NCI CA121493), the Abby Rockefeller Mauzé Trust, and Maloris and Hormel Foundations. We thank Margarita Malakhova for helpful advice, the personnel at ESRF beamline ID 23-1 and ID 14-4 for assistance with X-ray data collection, our colleagues Sandra Delgado, Jevgenia Tamjar, Oscar Millet and Inma Gómez García from CICbioGUNE for help with fluorescence and DLS measurements, and Helen Pike from the UMN-Hormel Institute for expressing and purifying protein used for transfer activity measurements.
Footnotes
Supplemental information includes six figures, one table, ‘Experimental Procedures’ paragraph and two references. Correspondence and requests for materials should be addressed to L.M. or R.E.B
ACCESSION NUMBERS
Atomic coordinates and structures factors have been deposited in the Protein Data Bank. PDB entries are stated in the Table 1.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahn VE, Faull KF, Whitelegge JP, Fluharty AL, Privé GG. Crystal structure of saposin B reveals a dimeric shell for lipid binding. Proc Natl Acad Sci USA. 2003;100:38–43. doi: 10.1073/pnas.0136947100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Airenne TT, Kidron H, Nymalm Y, Nylund M, West G, Mattjus P, Salminen TA. Structural evidence for adaptive ligand binding of glycolipid transfer protein. J Mol Biol. 2006;355:224–236. doi: 10.1016/j.jmb.2005.10.031. [DOI] [PubMed] [Google Scholar]
- Brown RE. Spontaneous lipid transfer between organized lipid assemblies. Biochim Biophys Acta. 1992;1113:375–389. doi: 10.1016/0304-4157(92)90007-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RE, Mattjus P. Glycolipid transfer proteins. Biochim Biophys Acta. 2007;1771:746–760. doi: 10.1016/j.bbalip.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhardt M. The role and metabolism of sulfatide in the nervous system. Mol Neurobiol. 2008;37:93–103. doi: 10.1007/s12035-008-8022-3. [DOI] [PubMed] [Google Scholar]
- Gaj T, Mercer AC, Gersbach CA, Gordley RM, Barbas CF., III Structure-guided reprogramming of serine recombinase DNA sequence specificity. Proc Natl Acad Sci USA. 2011;108:498–503. doi: 10.1073/pnas.1014214108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halder RC, Jahng A, Maricic I, Kumar V. Mini Review: Immune response to myelin-derived sulfatide and CNS-demyelination. Neurochem Res. 2007;32:257–262. doi: 10.1007/s11064-006-9145-4. [DOI] [PubMed] [Google Scholar]
- Han X. Potential mechanisms contributing to sulfatide depletion at the earliest clinically recognizable stage of Alzheimer’s disease: a tale of shotgun lipidomics. J Neurochem. 2007;103 (Suppl 1):171–179. doi: 10.1111/j.1471-4159.2007.04708.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X. Multi-dimensional mass spectrometry-based shotgun lipidomics and the altered lipids at the mild cognitive impairment stage of Alzheimer’s disease. Biochim Biophys Acta. 2010;1801:774–783. doi: 10.1016/j.bbalip.2010.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermansson M, Käkelä R, Berghäll M, Lehesjoki AE, Somerharju P, Lahtinen U. Mass spectrometric analysis reveals changes in phospholipid, neutral sphingolipid and sulfatide molecular species in progressive epilepsy with mental retardation, EPMR, brain: a case study. J Neurochem. 2005;95:609–617. doi: 10.1111/j.1471-4159.2005.03376.x. [DOI] [PubMed] [Google Scholar]
- Hancock SM, Rich JR, Caines MEC, Strynadka NCJ, Withers SG. Designer enzymes for glycosphingolipid synthesis by directed evolution. Nature Chem Biol. 2009;5:508–514. doi: 10.1038/nchembio.191. [DOI] [PubMed] [Google Scholar]
- Jeon SB, Yoon HL, Park SH, Kim IH, Park EJ. Sulfatide, a major lipid component of myelin sheath activates inflammatory responses as an endogenous stimulator in brain-resident immune cells. J Immunol. 2008;181:8077–8087. doi: 10.4049/jimmunol.181.11.8077. [DOI] [PubMed] [Google Scholar]
- Kamlekar RK, Gao Y, Kenoth R, Molotkovsky JG, Prendergast FG, Malinina L, Patel DJ, Wessels WS, Venyaminov SY, Brown RE. Human GLTP: Three distinct functions for three tryptophans in a novel peripheral amphitropic fold. Biophys J. 2010;99:2626–2635. doi: 10.1016/j.bpj.2010.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenoth R, Simanshu DK, Kamlekar RK, Pike HM, Molotkovsky JG, Benson LM, Bergen HR, 3rd, Prendergast FG, Malinina L, Venyaminov SY, Patel DJ, Brown RE. Structural determination and tryptophan fluorescence of heterokaryon incompatibility C2 protein (HET-C2), a fungal glycolipid transfer protein (GLTP), provide novel insights into glycolipid specificity and membrane interaction by the GLTP-fold. J Biol Chem. 2010;285:13066–13078. doi: 10.1074/jbc.M109.093203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamzin VS, Wilson KS. Automated refinement of protein models. Acta Cryst. 1993;D49:129–149. doi: 10.1107/S0907444992008886. [DOI] [PubMed] [Google Scholar]
- Malakhova ML, Malinina L, Pike HM, Kanack AT, Patel DJ, Brown RE. Point mutational analysis of the liganding site in human glycolipid transfer protein. Functionality of the complex. J Biol Chem. 2005;280:26312–26320. doi: 10.1074/jbc.M500481200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinina L, Malakhova M, Teplov A, Brown RE, Patel DJ. Structural basis for glycosphingolipid transfer specificity. Nature. 2004;430:1048–1053. doi: 10.1038/nature02856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinina L, Malakhova ML, Kanack AT, Lu M, Abagyan R, Brown RE, Patel DJ. The liganding of glycolipid transfer protein is controlled by glycolipid acyl structure. PLoS Biol. 2006;4:e362. doi: 10.1371/journal.pbio.0040362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattjus P, Molotkovsky JG, Smaby JM, Brown RE. A fluorescence resonance energy transfer approach for monitoring protein-mediated glycolipid transfer between vesicles. Anal Biochem. 1999;268:297–304. doi: 10.1006/abio.1998.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molotkovsky JG, Bergelson LD. Perylenoyl-labeled lipid-specific fluorescent probes. Bioorgan Khim. 1982;8:1256–1262. [Google Scholar]
- Molotkovsky JG, Mikhalyov II, Imbs AB, Bergelson LD. Synthesis and characterization of new fluorescent glycolipid probes. Molecular organisation of glyco-sphingolipids in mixed-composition lipid bilayers. Chem Phys Lipids. 1991;58:199–212. [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Cryst. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Navaza J. AMoRe: an automated package for molecular replacement. Acta Cryst. 1997;A50:157–163. [Google Scholar]
- Picollo A, Malvezzi M, Houtman JCD, Accardi A. Basis of substrate binding and conservation of selectivity in the CLC family of channels and transporters. Nature Struct Mol Biol. 2009;16:1294–1301. doi: 10.1038/nsmb.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T. Glycolipid transfer protein and intracellular traffic of glucosylceramide. Experientia. 1990;46:611–616. doi: 10.1007/BF01939700. [DOI] [PubMed] [Google Scholar]
- Wörsdörfer B, Woycechowsky KJ, Hilvert D. Directed evolution of a protein container. Science. 2011;331:589–592. doi: 10.1126/science.1199081. [DOI] [PubMed] [Google Scholar]
- Zajonc DM, Elsliger MA, Teyton L, Wilson IA. Crystal structure of CD1a in complex with a sulfatide self-antigen at a resolution of 2.15 Å. Nat Immunol. 2003;4:808–815. doi: 10.1038/ni948. [DOI] [PubMed] [Google Scholar]
- Zajonc DM, Maricic I, Wu D, Halder R, Roy K, Wong CH, Kumar V, Wilson IA. Structural basis for CD1d presentation of a sulfatide derived from myelin and its implications for autoimmunity. J Exp Med. 2005;202:1517–1526. doi: 10.1084/jem.20051625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zajonc DM, Kronenberg M. CD1 mediated T cell recognition of glycolipids. Curr Opin Struct Biol. 2007;17:521–529. doi: 10.1016/j.sbi.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng J, Cheng H, Jiang X, Han X. Endosomes and lysosomes play distinct roles in sulfatide-induced neuroblastoma apoptosis: potential mechanisms contributing to abnormal sulfatide metabolism in related neuronal diseases. Biochem J. 2008;410:81–92. doi: 10.1042/BJ20070976. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.