Abstract
The multiple species concepts currently in use by the scientific community (e.g. Morphological, Biological, Phylogenetic) are united in that they all aim to capture the process of divergence between populations. For example, the Biological Species Concept (BSC) defines a species as a natural group of organisms that is reproductively isolated from other such groups. Here we synthesize nearly a century of research on the ciliate genus Paramecium that highlights the shortcomings of our prevailing notions on the nature of species. In this lineage, there is discordance between morphology, mating behavior, and genetics, features assumed to be correlated, at least after sufficient time has passed, under all species concepts. Intriguingly, epigenetic phenomena are well documented in ciliates where they influence features such as germline/soma differentiation and mating type determination. Consequently, we hypothesize that divergence within ciliate populations is due to a dynamic interaction between genetic and epigenetic factors. The growing list of examples of epigenetic phenomena that potentially impact speciation (i.e. by influencing the dynamics of sex chromosomes, fate of hybrids, zygotic drive and genomic conflicts) suggests that interactions between genetics and epigenetics may also drive divergence in other eukaryotic lineages.
Keywords: Species concepts, Paramecium, epigenetics, ciliates, molecular evolution
Introduction
Our conceptualization of species greatly influences our understanding of biodiversity and is crucial to dialogue within the life sciences. Currently, the scientific community uses a number of different species concepts (e.g. Morphological, Biological, Phylogenetic; reviewed in de Queiroz 2007; Donoghue 1985; Hey 2006; Mayden 2002). These concepts are united in that they all operate under the idea that a species is a natural unit of biodiversity and aim to capture the process of divergence between populations. Consequently, we expect species delineations under these various concepts to overlap and, given sufficient evolutionary time, to be concordant. However, as revealed by this synthesis of nearly a century of research on Paramecium and other genera, ciliates challenge the prevailing notions on the nature of species. This is because of the discordance between morphology, mating behavior, and genetics (e.g. Allen et al. 1973; Catania et al. 2009; Coleman 2005; Hori et al. 2006; Sonneborn 1937; Sonneborn 1938; Sonneborn 1957a; Sonneborn 1975; Sonneborn and Lynch 1932), which suggest that divergence between populations is more complex than our current species concepts envision.
The numerous species concepts used by biologists have been reviewed extensively (e.g. de Queiroz 2007; Donoghue 1985; Hey 2006; Mayden 2002; Schlegel and Meisterfeld 2003). We discuss just three here: the Morphological, Biological, and Phylogenetic species concepts. While they do not capture the complete breadth of species concepts, these examples arguably represent three of the major categories of species concepts (de Queiroz 2007).
According to the Morphological Species Concept (MSC), a species is a group of organisms with shared morphology (Donoghue 1985). Linnaeus and other early taxonomists used morphology to create taxonomies of living organisms and today the MSC remains pertinent in many instances. For example, morphology remains a useful tool for identifying organisms observed in the field and captures key aspects of biodiversity (McManus and Katz 2009; Mishler and Donoghue 1982). Furthermore, many organisms for which molecular and behavioral data do not currently exist are classified as species solely on the basis of morphology.
The Biological Species Concept (BSC) emerged in the mid-twentieth century when new theories about evolution were superimposed onto the existing Linnaean hierarchical system of classification (Dobzhansky 1940; Mayr 1942; Mayr 1996). At this time the concepts of Darwinian natural selection and Mendelian genetic inheritance were united under what has been called the Modern Synthesis (Mayr and Provine 1981). The BSC defines a species as a group of interbreeding organisms that is reproductively isolated from other such groups (Dobzhansky 1940; Mayr 1942). The BSC argues that divergence between populations at the molecular level ultimately results in reproductive incompatibility (Mayr 1996).
With the rise of molecular systematics in the latter half of the twentieth century, the phylogenetic species concept (PSC) emerged (de Queiroz 2007; Donoghue 1985). The PSC argues that evolution is pivotal to speciation and thus species can be recognized as distinct monophyletic clades (Donoghue 1985). The PSC is particularly useful for measuring diversity when other measurements are not possible, as with microorganisms that cannot be cultured in the lab.
Here, we evaluate the applicability of these various species concepts in light of observations on the Paramecium aurelia complex as well as insights from other ciliates. Ciliates are microbial eukaryotes that are characterized by the presence of cilia in at least one stage of the life cycle and by the presence of dimorphic nuclei (reviewed in Yao et al. 2002; McGrath et al. 2006; Juranek and Lipps 2007). Ciliates reproduce asexually via binary fission and separately go through meiosis and conjugation during the sexual life cycle phase (Paulin 1996). Within each cell there is a functional somatic macronucleus and a transcriptionally inactive germline micronucleus (reviewed in Yao et al. 2002; McGrath et al. 2006; Juranek and Lipps 2007). Research by T.M. Sonneborn on the Paramecium aurelia species complex brought ciliate biology to the attention of the scientific community in the early twentieth century (e.g. Sonneborn 1937; Sonneborn 1938; Sonneborn 1957a; Sonneborn 1975). Since then, molecular studies of the Paramecium complex (Fig. 1) as well as other ciliate lineages extensively have yielded conflicting data on the nature of ciliate species.
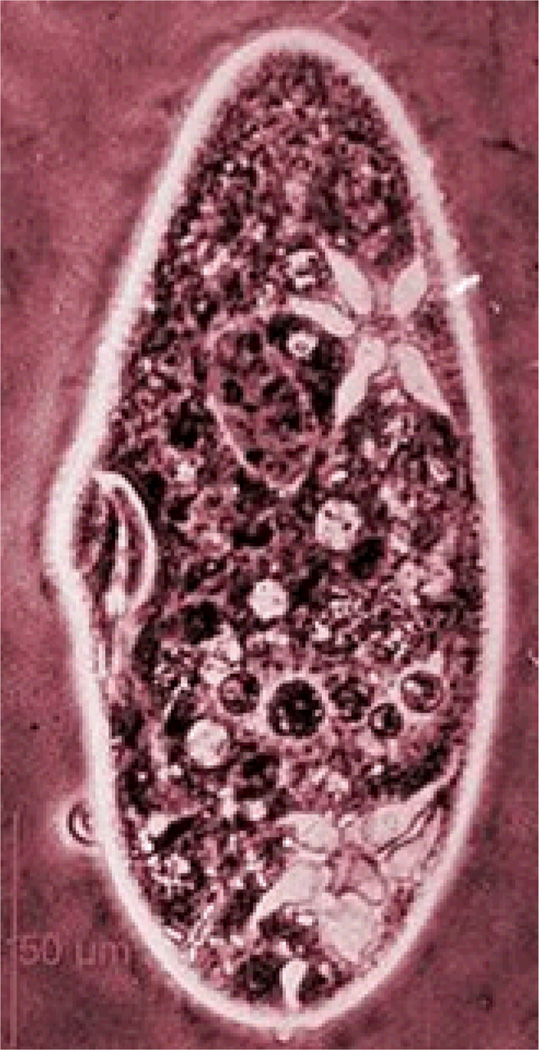
Figure 1.
Paramecium tetraurelia. Source: Brugerolle G. http://starcentral.mbl.edu/microscope/portal.php?pagetitle=assetfactsheet&imageid=23189
Sonneborn and the Paramecium aurelia complex
The geneticist T.M. Sonneborn was among the first to recognize that ciliates defy traditional species concepts. His skepticism emerged from his research on the ciliate morphospecies Paramecium aurelia (reviewed in Schlegel and Meisterfeld 2003; Schloegel 1999). Although P. aurelia was originally classified as a single morphological species in 1773 by O.F. Müller, Sonneborn’s observations suggested that its various strains did not constitute a single species (Pryzboś et al. 2007; Sonneborn 1938; Sonneborn 1957a). While the Paramecium aurelia cells are morphologically indistinguishable, Sonneborn found that the lineages belonged to one of fourteen subgroups. He referred to these subgroups as syngens, i.e. “generating together” (Sonneborn 1957a). Cells of the same syngen would readily form conjugant pairs with one another but were essentially reproductively isolated from the other syngens (Sonneborn 1957a).
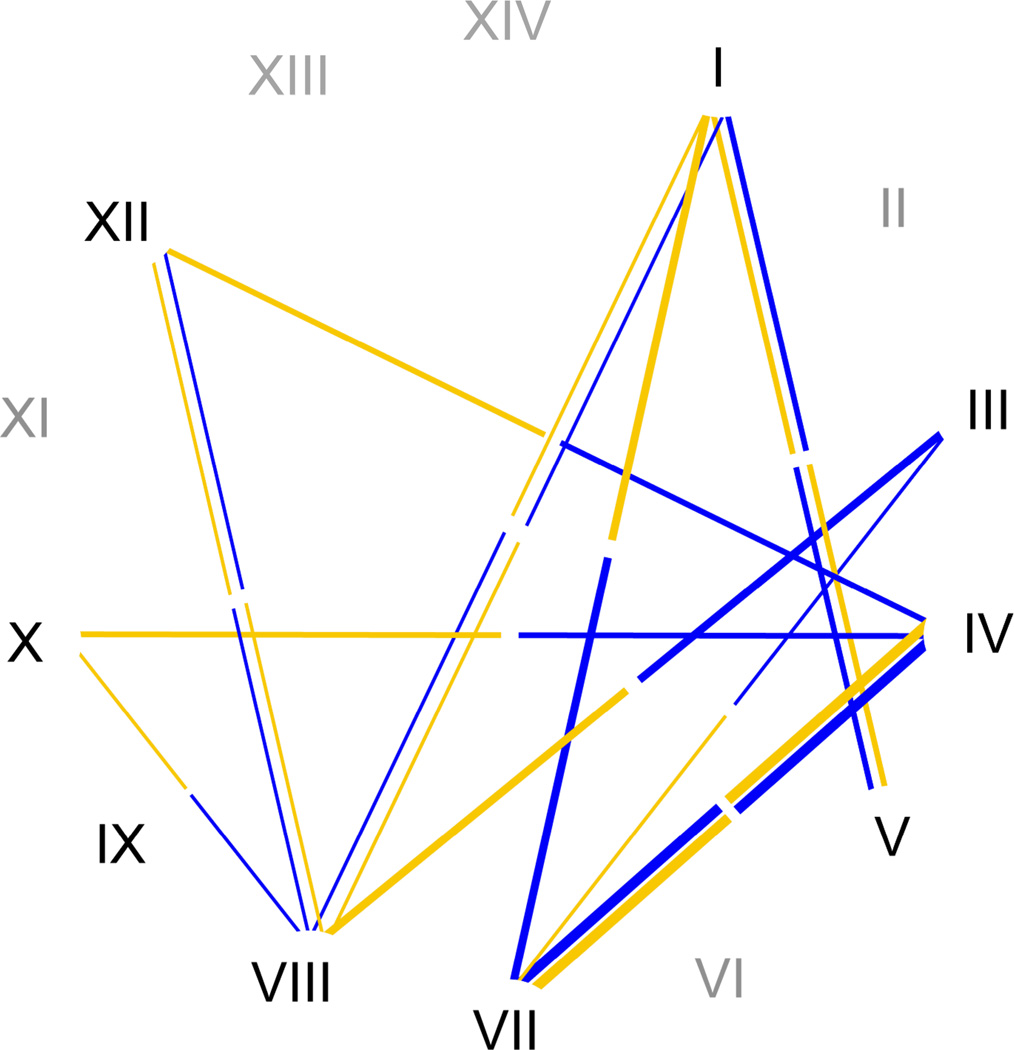
Sonneborn (1938) analyzed a total of twenty-six lines of the morphospecies Paramecium aurelia that were isolated from a number of sites in New York, Maryland, Massachusetts, and California. Sonneborn (1938) then conducted crosses between lines and observed variable rates of intersyngen clustering and conjugation. While most experimental crosses between syngens did not result in true conjugation, that is, involving the exchange of genetic material, Sonneborn observed that in a number of crosses the syngens would respond to one another by clustering together or even forming conjugant pairs (Fig. 2). In some instances, this involved the exchange of genetic material (Sonneborn 1975). The strongest intersyngen mating reactions were between syngen IV and syngen VIII (later, P. tetraurelia and P. octaurelia). Here, between 90 and 95% of the cells formed conjugant pairs when Sonneborn optimized the environmental conditions (Sonneborn 1975). Sonneborn estimated that 90% of the conjugant pairs in this cross were true, i.e. involved the exchange of genetic material (Sonneborn 1975).
Figure 2.
Mating relationships among members of the Paramecium aurelia complex. Roman numerals represent species Latin names (e.g. P. primaurelia = I). Light (mating type O) and dark (mating type E) bars indicate species pairings described in Sonneborn (1957a, 1975) as drawn in Coleman (2005). The thickness of a bar indicates the intensity of interspecies pairing, spanning from brief and weak contacts one pair at a time (e.g. Species III and VII) to 90–95% of cells forming conjugant pairs under optimal reactive conditions (Species IV and VIII; Sonneborn 1957a, 1975). Gray numerals indicate species not included in analysis.
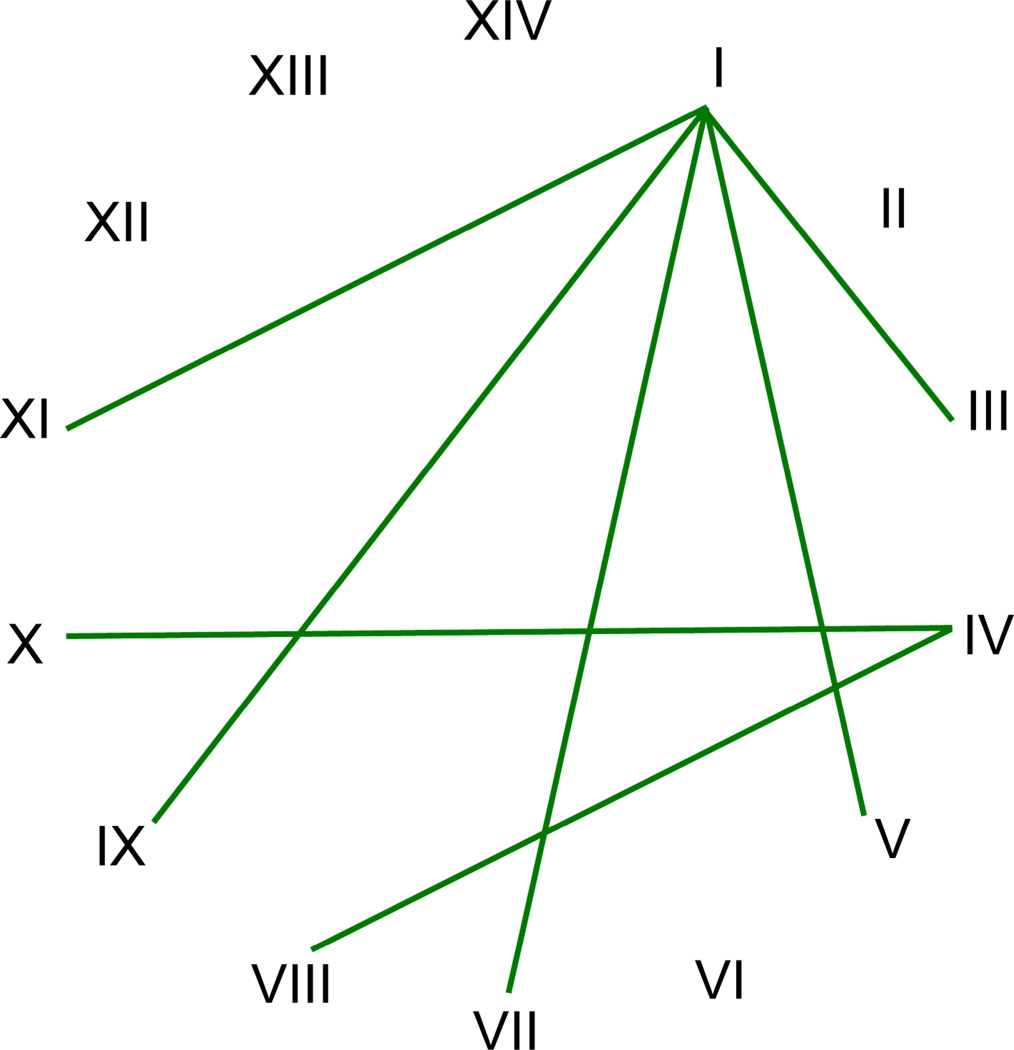
While there was evidence to suggest that the syngens were largely reproductively isolated, Sonneborn did not initially call the syngens individual species, in part because he believed that it would be impractical to identify the species on the basis of mating behavior (Sonneborn 1938). In the years that followed, Sonneborn continued to study the P. aurelia syngens (e.g. Sonneborn 1957a; Sonneborn 1975). Based on his research on the mating behavior, mode of mating type inheritance, serotype similarities and differences, symbiont distribution, strength of cytoplasmic influence, and geographic distribution, Sonneborn (1957a; 1975) argued that there was a deep evolutionary divide between the syngens I and IV and that their ancestors gave rise to syngens III, V, VII, IX and XI, and VIII and X respectively (Fig. 3). However, it was apparent that further research was needed in order to elucidate the relatedness of the P. aurelia syngens.
Figure 3.
Inferred evolutionary relationships between members of the Paramecium aurelia complex as proposed by Sonneborn (1957a) on the basis of mating type inheritance, intersyngen mating behavior, serotype profile, symbiont distribution, strength of cytoplasmic influence, and geographic distribution (bars). Abbreviations as in Figure 2.
Analysis of allozymes within the Paramecium aurelia complex
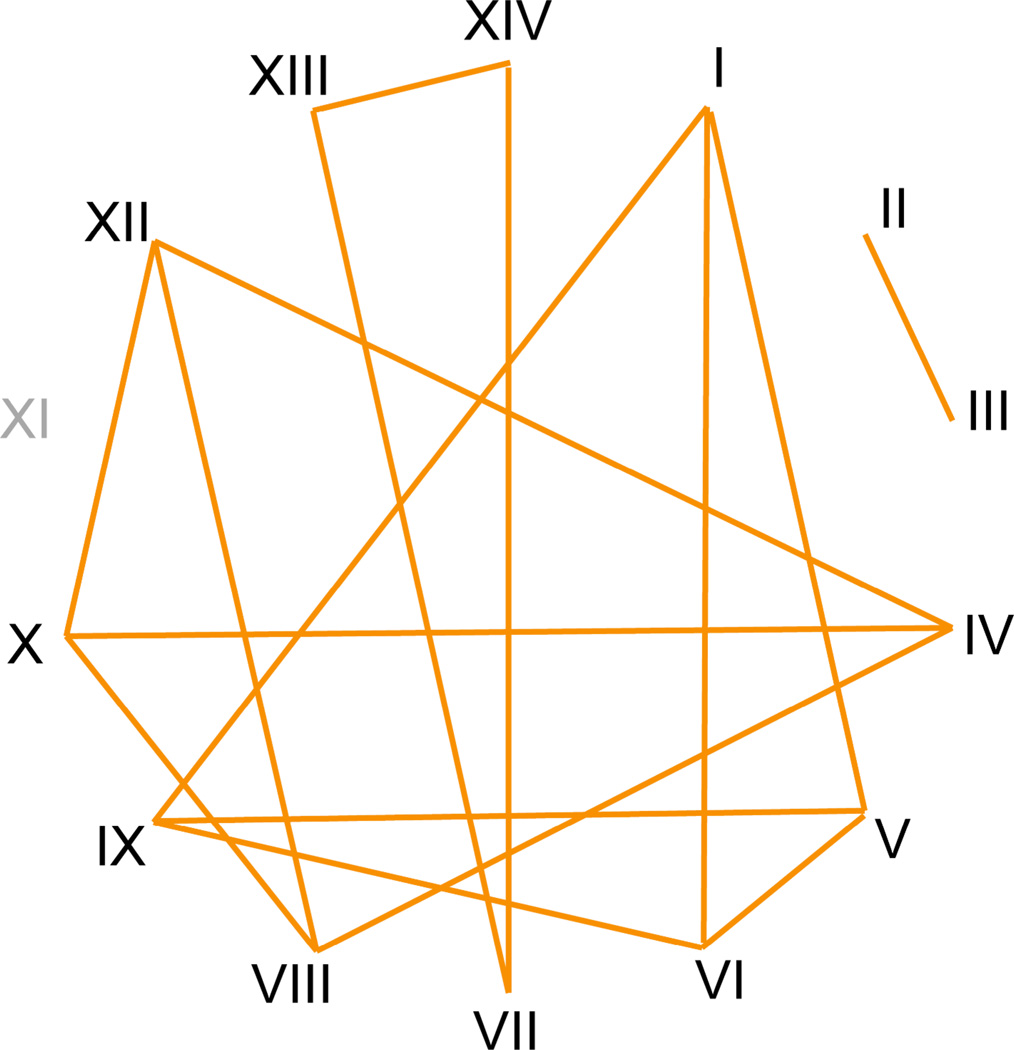
In the years following Sonneborn’s experiments, other researchers have sought to understand the P. aurelia complex through the use of molecular techniques (e.g. Allen et al. 1973; Catania et al. 2009; Coleman 2005; Tait 1970). One of the earliest molecular experiments on the P. aurelia complex was conducted using protein electrophoresis and revealed that the majority of the fourteen syngens could be distinguished on the basis of eight allozyme loci (Allen et al. 1973). Based on these data, Allen et al. (1983) theorized that there were four groups of closely related syngens within the P. aurelia complex (Fig. 4). This research suggested that the syngens were molecularly distinct from one another and could be considered separate species (Sonneborn 1975). Yet, there is only weak similarity between Sonneborn’s observations and the conclusions of Allen et al. (1973; 1983). For example, while syngens II and III shared a highly similar allozyme pattern and were deemed closely related by Allen et al. (1973), Sonneborn (1957a) did not find them forming conjugant pairs with one another. Syngen III, on the other hand, did form conjugant pairs with three other syngens (I, VII, and VIII), though all of these syngens fall into separate groups based upon allozyme analysis (Fig. 2, Fig. 3; Allen et al. 1973; Allen et al. 1983; Sonneborn 1957a). Furthermore, there were notable contradictions within the allozyme study. Two of the syngens (I and V) had identical allozyme patterns for all eight loci examined, which suggests that they might belong to a single species, at least under the PSC. Additionally, there were three instances where multiple strains of a single syngen contains allozyme variation though they readily mated with one another (Allen et al. 1973).
Figure 4.
Inferred evolutionary relationships between species of the Paramecium aurelia complex based on analysis of eight allozymes (bars; Allen et al. 1973; 1983). Abbreviations as in Figure 2. Gray numerals indicate species not included in analysis.
Despite these uncertainties, Sonneborn designated the fourteen members of the P. aurelia complex as distinct species (Sonneborn 1975). Species classification was based on Sonneborn’s observations of mating behavior (1938; 1957a) as well as the molecular data revealed in Allen et al. (1973). Each species was assigned a Latin binomial that corresponded with its original numeric syngen identifier (e.g. Syngen 1 became P. primaurelia; Table 1; Sonneborn 1975). Paramecium aurelia was henceforth referred to as a complex of sibling species, albeit with continued skepticism over the applicability of the BSC in this instance.
Table 1.
Initial syngen numeric indentifiers (Sonneborn 1938), subsequent designated species binomial (Sonneborn 1975), and mode of mating type inheritance of the members of the fourteen initial members of the Paramecium aurelia complex.
| Syngen | Species Name | Mating Type Inheritancea |
|
|---|---|---|---|
| 1 | I | Paramecium primaurelia | A |
| 2 | II | P. biaurelia | B |
| 3 | III | P. triaurelia | A |
| 4 | IV | P. tetraurelia | B |
| 5 | V | P. pentaurelia | A |
| 6 | VI | P. sexaurelia | B |
| 7 | VII | P. septaurelia | B |
| 8 | VIII | P. octaurelia | B |
| 9 | IX | P. novaurelia | A |
| 10 | X | P. decaurelia | B |
| 11 | XI | P. undecaurelia | A |
| 12 | XII | P. dodecaurelia | B |
| 13 | XIII | P. tridecaurelia | C |
| 14 | XIV | P. quadecaurelia | A |
Mating type inheritance: A: karyonidal inheritance = mating type determined independently in each macronucleus; B: cytoplasmic inheritance = mating type reflects that of the cytoplasmic parent; C: synclonal inheritance = inheritance in accordance to Mendelian genetics (Miyake 1996; Phadke and Zufall 2009).
Nuclear and mitochondrial DNA sequence analysis of the Paramecium aurelia complex
The development of methods for DNA sequence analysis brought new opportunities for additional studies on the Paramecium aurelia complex with the intent of reconstructing the evolutionary history of these organisms using molecular markers (e.g. Catania et al. 2009; Coleman 2005; Hori et al. 2006). However, just as the allozyme data of Allen et al. (1973) failed to correspond with Sonneborn’s observations of mating behavior, surveys of various molecular markers paint differing pictures of the nature of and relationships among the P. aurelia “species.”
Analyses of the internal transcribed spacer 2 (ITS2) in the P. aurelia complex (Coleman 2005) do not correspond strictly with studies of mating behavior or allozymes (Allen et al. 1973; Coleman 2005; Sonneborn 1957a; Sonneborn 1975). Based on a relatively limited sampling (i.e. one or two strains for each species), Coleman (2005) found that some species within the complex could be differentiated by ITS2 sequences, but that the relationships among strains were incongruent with other markers; for example, two reproductively isolated species with distinct patterns of allozymes, P. tetraurelia and P. novaurelia, shared identical ITS2 sequences (Coleman 2005; Sonneborn 1957a; Sonneborn 1975). However, the level of variation at this locus is relatively low and many P. aurelia species share identical or nearly identical sequences.
Analysis of the protein-coding gene heat shock protein 70 (hsp70; Hori et al. 2006) yielded results that differ from Coleman (2005). Hori et al. (2006) analyzed the heat shock protein 70 (hsp70) locus for one to three strains of each of the Paramecium aurelia species and found that multiple strains of the same species generally clustered together. However, in contrast to Coleman (2005), Hori et al (2006) found P. septaurelia had an hsp70 sequence that was nearly identical to the P. triaurelia strains while Coleman (2005) found this species to be most similar to P. primaurelia, P. triaurelia, and P. pentaurelia at the ITS2 locus.
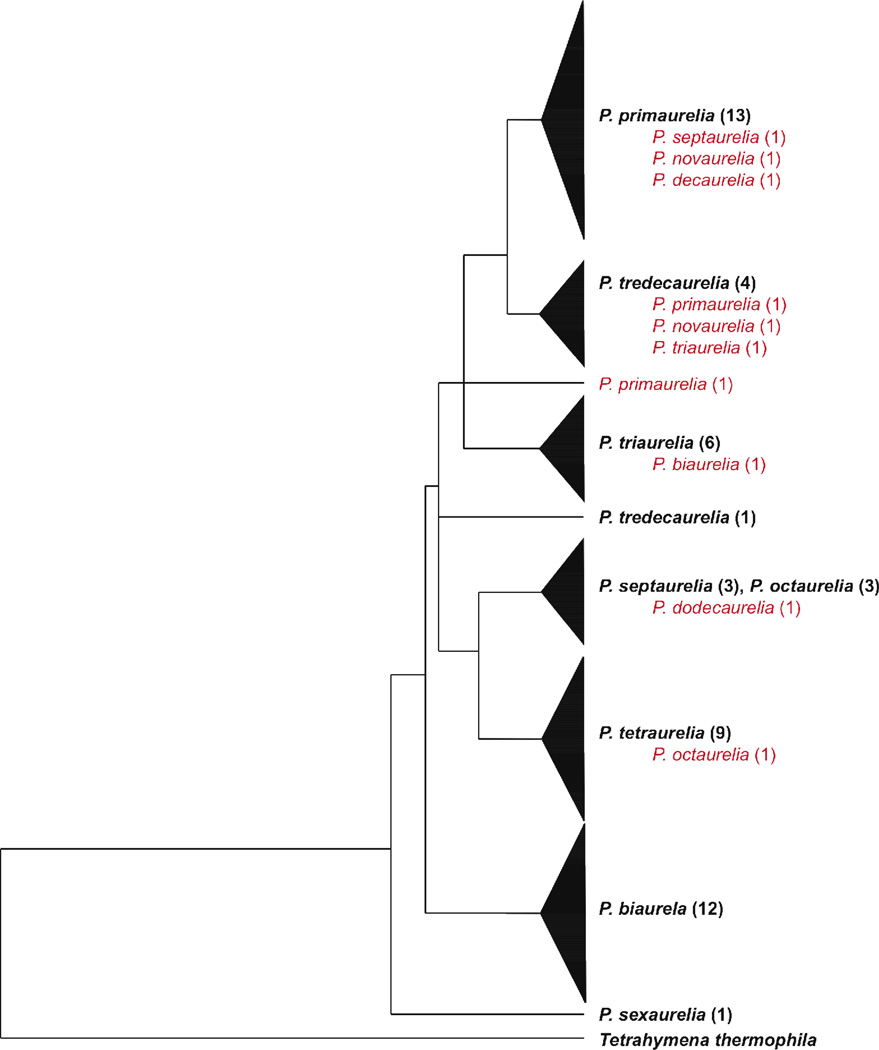
Studies of the Paramecium aurelia complex by Catania et al. (2009) further demonstrate that species delineations on the basis of molecular data (i.e. according to the PSC) do not correlate with species delineations on the basis of morphology or mating behavior (i.e. MSC or BSC). Catania et al. (2009) extended analysis of DNA sequence, analyzing 80 strains representing all 14 species of the P. aurelia complex at 10 nuclear and 5 mitochondrial protein-coding genes plus both nuclear and mitochondrial ribosomal RNA regions. Thirteen of the 80 strains were found to be discordant – that is, their species delineation according to mating tests did not match their phylogenetic position in molecular analyses where multiple strains of the same species failed to form a monophyletic group (Catania et al. 2009). For example, one strain of Paramecium octaurelia fell among the P. tetraurelia strains in a clade that is relatively distant from the other P. octaurelia sequence (Fig 5, redrawn from Fig. S1 in Catania et al. 2009). Furthermore, even at the exclusion of these discordant strains, the evolutionary relationships between species indicated by these analyses do not match those of previous studies. For example, according to Catania et al. (2009), P. septaurelia is sister to P. octaurelia (Fig. 5). However, Sonneborn (1957a) did not observe interspecies conjugation between the two (Fig. 2), Allen et al. (1973) found P. septaurelia to have allozyme patterns most similar to those of P. tredecaurelia and P. quadecaurelia (Fig. 4) and Hori et al. (2006) found P. septaurelia to be most similar to P. triaurelia.
Figure 5.
Phylogeny of 62 strains representing eleven species of the fourteen Paramecium aurelia complex adapted from figure S1 of Catania et al. (2009). The original tree was built by concatenating three mitochondrial protein-coding loci (Catania et al. 2009), though a similar tree was found for nuclear genes. Ten discordant strains, those that are designated as one species according to mating tests but another according to molecular analysis, appear in lighter-color font within the tree.
Ciliate evolution: towards a resolution?
Despite numerous attempts to sort out the nature of diversity within the Paramecium aurelia complex, species delineation remains a tangled mess. Further, evidence for ciliate genera with similar characteristics extends beyond the P. aurelia complex, including several additional examples within the genera Paramecium and Tetrahymena (Lynn 1996). In all cases, morphologically indistinguishable lineages possess complex features that confound the MSC, BSC, and PSC.
We hypothesize that epigenetics, particularly as it relates to the dynamics of mating type determination, conjugation, and macronuclear development, underlies the discordance between morphological, behavioral, and molecular characteristics in Paramecium and other ciliates. Although the impact of epigenetics has historically been omitted from discourse on evolution and speciation, it may in fact explain the conundrum of ciliate divergence as epigenetic phenomena can disrupt the predicted pattern of gene exchange central to all of the species concepts mentioned here. Evidence for epigenetics, which we define as inheritance that transcends what is encoded in the DNA sequence of genomes, is found across the ciliate tree of life and includes such examples as non-Mendelian inheritance of mating type and cortical structures (Meyer and Chalker 2007). As we will show, a number of these epigenetic features have implications for the processes of gene exchange and are consequently relevant to our understanding of ciliate evolution.
One of the first ciliate traits to be recognized as non-Mendelian was that of mating type inheritance in the Paramecium aurelia complex (Sonneborn 1937). In each of these species there are two mating types, Odd (O) and Even (E). Organisms of one mating type share physiological characters that prevent conjugation with one another (Miyake 1996; Sonneborn 1937). Rather, conjugation occurs between two cells of complementary mating types (reviewed in Miyake 1996; Phadke and Zufall 2009). While one species of the P. aurelia complex exhibits Mendelian inheritance of mating type (i.e. synclonal inheritance), the remaining species exhibit either cytoplasmic and karyonidal inheritance, both of which are epigenetic forms of mating type determination (Table 1; Miyake 1996; Phadke and Zufall 2009). In cytoplasmic mating type inheritance, mating type follows the phenotype of the cytoplasmic parent (Beale and Preer 2008a; Sonneborn and Lynch 1934). Consequently, in this system, all cells of a clonal line generally express the same mating type (Miyake 1996). Under the karyonidal model, the expression of mating type is determined during macronuclear development following sexual conjugation (Miyake 1996). In such instances, epigenetic genome-wide rearrangements during macronuclear development influence which mating type a cell will express. In karyonidal systems, determination of mating type is stochastic and cells of a clonal line may express different mating types.
Further work has revealed that systems of two or more mating types are a feature of all sexual ciliate lineages (Bleyman 1996; Jennings et al. 1932; Miyake 1996; Phadke and Zufall 2009). While karyonidal inheritance has been found in Tetrahymena thermophila and other ciliate lineages, cytoplasmic inheritance is only known to occur within the genus Paramecium (Phadke and Zufall 2009). Unfortunately, little research has been done in other ciliate lineages to reveal the genetic or epigenetic nature of mating type determination. However, there is evidence that an epigenetic system exists in Blepharisma japonica (Bleyman 1996). It therefore stands to reason that epigenetic mating type systems are widespread among many ciliate lineages.
In addition to the intracellular epigenetic factors inherent in cytoplasmic and karyonidal mating type determination, mating type is also influenced by environmental factors in many ciliate lineages. For example, in several species of Paramecium and Tetrahymena temperature and nutrient conditions influence mating type ratios (Bleyman 1996; Nanney 1960; Preer 2000). The precise mechanisms behind these observations are not fully understood. However, Orias (1981) theorized that in the case of Tetrahymena thermophila an intricate system involving genetic as well as intracellular epigenetic and extracellular environmental factors is at play. Cells inherit either the mat-1 or mat-2 allele. A cell with the mat-1 allele can exhibit mating types I, II, III, V, and VI whereas a cell with the mat-2 allele can exhibit any of the seven mating types except I. The mating type a cell expresses is determined during macronuclear development based on the parameters of its mating type allele, temperature and nutrient conditions (Nanney 1960; Orias 1981).
Similarly, mating type in P. multimicronucleatum is light-dependent (Sonneborn 1957b). While some P. multimicronucleatum cells express a static mating type, others share a trait where they experience a circadian cycling of mating type involving two switches from one mating type to the other within a 24-hour period (Barnett 1966). During periods of transition these cells are capable of mating with other cells of the same mating type (Beale and Preer 2008a). Due to these circumstances, there is a temporal fluidity to the reproductive compatibility of the individuals within this species. Hence, epigenetic and environmental dynamics likely play a role in creating prezygotic barriers between ciliate species.
Ciliates also exhibit the epigenetic phenomenon of genome-wide rearrangements during macronuclear development (Juranek and Lipps 2007; Meyer and Chalker 2007). Sexual conjugation in ciliates is characterized by the exchange of micronuclei and subsequent fusion of micronuclei from both parents. The resulting diploid zygotic nucleus then undergoes division and the resulting products then differentiate to form a new micronucleus and macronucleus. Unlike the micronuclear genome, the macronuclear genome undergoes DNA deletions and chromosome fragmentation prior to gene expression. Consequently, the genetic content of the somatic macronuclear can differ markedly from that of the germline micronucleus. The level of genome-wide rearrangement varies markedly between ciliate lineages. Some lineages fragment their chromosomes to such an extent that a single gene resides on each chromosome while others possess longer macronuclear chromosomes of 100 kb or more (reviewed in Juranek and Lipps 2007; McGrath et al. 2006; Yao et al. 2002; Zufall et al. 2006). These genome rearrangements are regulated epigenetically by both the content of the parental macronuclear genome and cytoplasmic factors (Coyne et al. 1996; Meyer and Chalker 2007). Hence, epigenetics impacts transmission of genes in the somatic macronucleus and may influence the pattern of divergence among strains.
Conclusions
Sonneborn’s discovery of unpredictable patterns within the Paramecium aurelia complex was unsettling at a time when the scientific community believed that, with the Modern Synthesis, the true nature of species had been discovered. The perplexing molecular and reproductive characteristics of P. aurelia, as well as those of a number of other ciliate lineages, challenge the species concepts currently in use (e.g. MSC, BSC, PSC). However, none of these concepts takes into account the potential of epigenetics to influence gene flow within and between populations. Epigenetic mechanisms, such as those highlighted above, suggest that differentiation of ciliate lineages is not solely the result of classical Mendelian processes. Epigenetic factors that affect mating behavior and genome processing are intricately involved in the transmission of genetic information and consequently may influence the process of speciation.
Though we have focused on the role of epigenetics in the evolution of ciliates, we argue that the complex evolutionary patterns of this clade are not exceptions to the rule but rather examples of the dynamics of evolution. It is now recognized that epigenetic mechanisms are found in organisms across the eukaryotic tree of life (e.g. Beale and Preer 2008b; Lolle et al. 2005; Meiklejohn and Tao 2009; ; Parfrey and Katz 2010; Rassoulzadegan et al. 2006). Epigenetic phenomena are argued to impact speciation by: altering the dynamics of sex chromosome evolution (Rice et al. 2008); contributing to genomic conflicts (Brown and O’Neil 2010; Rice et al. 2008), postmating isolation in fungi (Giraud et al. 2008) and post-hybridization outcomes in polyploid plants (Ainouche and Jenczewski 2010; Hegarty et al. 2011); and inducing genome remodeling through changing transposon dynamics in response to the environment (Rebello et al. 2010). In some instances these epigenetic phenomena directly influence sex determination (Meiklejohn and Tao 2009) and epigenetic alterations can be heritable (Lolle et al. 2005; Rassoulzadegan et al. 2006). Hence, it is possible that epigenetics has influenced the divergence of additional eukaryotic lineages and may help explain patterns of biodiversity on Earth.
Acknowledgements
L.A.K. is extremely grateful to R.G. Harrison for the excellent training and continued support. L.A.K. is also supported by grants from the National Science Foundation (DEB RUI:0919152, DEB 043115, DEB 0816828) and National Institutes of Health (1R15GM081865-01).
References
- Ainouche ML, Jenczewski E. Focus on polyploidy. New Phytol. 2010;186:1–4. doi: 10.1111/j.1469-8137.2010.03215.x. [DOI] [PubMed] [Google Scholar]
- Allen SL, Adams J, Rushford CL. Interspecies relationships in the Paramecium aurelia complex: Acid phosphatase variation. J Eukaryot Microbiol. 1983;30(1):143–147. [Google Scholar]
- Allen SL, Farrow SW, Golembiewski PA. Esterase variations between the 14 syngens of Paramecium aurelia under axenic growth. Genetics. 1973;73:561–573. doi: 10.1093/genetics/73.4.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett A. A circadian rhythm of mating type reversals in Paramecium multimicronucleatum syngen 2, and its genetic control. J Cell Physiol. 1966;67(2):239–270. doi: 10.1002/jcp.1040670206. [DOI] [PubMed] [Google Scholar]
- Beale GH, Preer JR., Jr . Paramecium: Genetics and Epigenetics. Boca Raton: CRC Press; 2008a. Chapter 5: The determination of mating types in Paramecium; pp. 51–62. [Google Scholar]
- Beale GH, Preer JR., Jr . Paramecium: Genetics and Epigenetics. Boca Raton: CRC Press; 2008b. Chapter 12: Epigenetics; pp. 175–182. [Google Scholar]
- Bleyman L. Ciliate genetics. In: Hausmann K, Bradbury PC, editors. Ciliates: Cells as Organisms. Stuttgart: Gustav Fischer; 1996. pp. 291–324. [Google Scholar]
- Brown JD, O’Neill RJ. Chromosomes, conflicts, and epigenetics: Chromosomal speciation revisited. Annu Rev Genom Human Genet. 2010;11:291–316. doi: 10.1146/annurev-genom-082509-141554. [DOI] [PubMed] [Google Scholar]
- Catania F, Wurmser F, Potehkin AA, Pryzboś E, Lynch M. Genetic diversity in the Paramecium aurelia species complex. Mol Biol Evol. 2009;26(2):421–431. doi: 10.1093/molbev/msn266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman AW. Paramecium aurelia revisited. J Eukaryot Microbiol. 2005;52(1):68–77. doi: 10.1111/j.1550-7408.2005.3327r.x. [DOI] [PubMed] [Google Scholar]
- Coyne RS, Chalker DL, Yao M-C. Genome downsizing during ciliate development: nuclear division of labor through chromosome restructuring. Annu Rev Genet. 1996;30:557–578. doi: 10.1146/annurev.genet.30.1.557. [DOI] [PubMed] [Google Scholar]
- de Queiroz K. Species concepts and species delimitation. Syst Biol. 2007;56(6):876–886. doi: 10.1080/10635150701701083. [DOI] [PubMed] [Google Scholar]
- Dobzhansky T. Speciation as a stage in evolutionary divergence. Am Nat. 1940;74:312–321. [Google Scholar]
- Donoghue MJ. A critique of the biological species concept and recommendations for a phylogenetic alternative. Bryologist. 1985;88(3):172–181. [Google Scholar]
- Giraud T, Refrégier G, Le Gac M, de Vienne DM, Hood ME. Speciation in fungi. Fungal Genet Biol. 2008;45:791–802. doi: 10.1016/j.fgb.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Hegarty MJ, Batstone T, Barker GL, Edwards KJ, Abbott RJ, Hiscock SJ. Nonadditive changes to cytosine methylation as a consequence of hybridization and genome duplication in Senecio (Asteraceae) Mol Ecol. 2011;20(1):105–113. doi: 10.1111/j.1365-294X.2010.04926.x. [DOI] [PubMed] [Google Scholar]
- Hey J. On the failure of modern species concepts. Trends Ecol Evol. 2006;21(8):447–450. doi: 10.1016/j.tree.2006.05.011. [DOI] [PubMed] [Google Scholar]
- Hori M, Tomikawa I, Pryzboś E, Fujishima M. Comparison of the evolutionary distances among syngens and sibling species of Paramecium. Mol Phylogenet Evol. 2006;38:697–704. doi: 10.1016/j.ympev.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Jennings HS, Raffel D, Lynch RS, Sonneborn TM. The diverse biotypes produced by conjugation within a clone of Paramecium aurelia. J Exper Zool. 1932;62(2):363–408. [Google Scholar]
- Juranek SA, Lipps HJ. New insights into the macronuclear development in ciliates. Int Rev Cytol. 2007;262:219–251. doi: 10.1016/S0074-7696(07)62005-1. [DOI] [PubMed] [Google Scholar]
- Lolle SJ, Victor JL, Young JM, Pruitt RE. Genome-wide non-mendelian inheritance of extra-genomic information in Arabidopsis. Nature. 2005;434:505–509. doi: 10.1038/nature03380. [DOI] [PubMed] [Google Scholar]
- Lynn DH. Systematics of ciliates. In: Hausmann K, Bradbury PC, editors. Ciliates: Cells as Organisms. Stuttgart: Gustav Fischer; 1996. pp. 51–72. [Google Scholar]
- Mayden RL. On biological species, species concepts and individuation in the natural world. Fish Fish. 2002;3:171–196. [Google Scholar]
- Mayr E. Systematics and the Origin of Species from the Viewpoint of a Zoologist. Cambridge: Harvard University Press; 1942. p. xxi. [Google Scholar]
- Mayr E. What is a species, and what is not? Philos Sci. 1996;63(2):262–277. [Google Scholar]
- Mayr E, Provine WB. The evolutionary synthesis. B Am Acad Arts Sci. 1981;34(8):17–32. [Google Scholar]
- McGrath CL, Zufall RA, Katz LA. Ciliate genome evolution. In: Katz LA, Bhattacharya D, editors. Genomics and Evolution in Microbial Eukaryotes. New York: Oxford University Press; 2006. pp. 64–77. [Google Scholar]
- McManus GB, Katz LA. Molecular and morphological methods for identifying plankton: What makes a successful marriage? J Plank Research. 2009;31:1119–1129. [Google Scholar]
- Meiklejohn CD, Tao Y. Genetic conflict and sex chromosome evolution. Trends Ecol Evol. 2009;25(4):215–223. doi: 10.1016/j.tree.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer E, Chalker DL. Epigenetics of ciliates. In: Allis CD, Jenuwein T, Reinberg D, Caperros M-L, editors. Epigenetics. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2007. pp. 127–150. [Google Scholar]
- Mishler BD, Donoghue MJ. Species concepts: A case for pluralism. Syst Zool. 1982;31(4):491–503. [Google Scholar]
- Miyake A. Fertilization and sexuality in ciliates. In: Hausmann K, Bradbury PC, editors. Ciliates: Cells as Organisms. Stuttgart: Gustav Fischer; 1996. pp. 243–290. [Google Scholar]
- Nanney DL. Temperature effects on nuclear differentiation in variety 1 of Tetrahymena pyriformis. Physiol Zool. 1960;33(2):146–151. [Google Scholar]
- Orias E. Probable somatic DNA rearrangements in mating type determination in Tetrahymena thermophila: A review and a model. Devel Genet. 1981;2:185–202. [Google Scholar]
- Parfrey LW, Katz LA. Dynamic genomes of eukaryotes and the maintenance of genomic integrity. Microbe. 2010;5(4):156–163. [Google Scholar]
- Paulin JJ. Morphology and cytology of ciliates. In: Hausmann K, Bradbury PC, editors. Ciliates: Cells as Organisms. Stuttgart: Gustav Fischer; 1996. pp. 1–40. [Google Scholar]
- Phadke SS, Zufall RA. Rapid diversification of mating systems in ciliates. Biol J Linn Soc. 2009;98:187–197. [Google Scholar]
- Preer JR., Jr Epigenetic mechanisms affecting macronuclear development in Paramecium and Tetrahymena. J Eukaryot Microbiol. 2000;47(6):515–524. doi: 10.1111/j.1550-7408.2000.tb00084.x. [DOI] [PubMed] [Google Scholar]
- Pryzboś E, Prajer M, Greczek-Stachura M, Skotarczak B, Maciejewska A, Tarcz S. Genetic analysis of the Paramecium aurelia species complex (Protozoa: Ciliophora) by classical and molecular methods. Syst Biodivers. 2007;5(4):417–434. [Google Scholar]
- Rassoulzadegan M, Grandjean V, Gounon P, Vincent S, Gillot I, Cuzin F. RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature. 2006;441:469–474. doi: 10.1038/nature04674. [DOI] [PubMed] [Google Scholar]
- Rebollo R, Horard B, Hubert B, Vieira C. Jumping genes and epigenetics: Towards new species. Gene. 2010;454:1–7. doi: 10.1016/j.gene.2010.01.003. [DOI] [PubMed] [Google Scholar]
- Rice WR, Gavrilets S, Friberg U. Sexually antagonistic “zygotic drive” of the sex chromosomes. PLOS Genet. 2008 doi: 10.1371/journal.pgen.1000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegel M, Meisterfeld R. The species problem in protozoa revisited. Europ J Protistol. 2003;39:349–355. [Google Scholar]
- Schloegel JJ. From anomaly to unification: Tracy Sonneborn and the species problem in protozoa 1954–1957. J Hist Biol. 1999;23(1):93–132. [Google Scholar]
- Sonneborn TM. Sex, sex inheritance and sex determination in Paramecium aurelia. Proc Nat Acad Sci. 1937;23(7):378–385. doi: 10.1073/pnas.23.7.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonneborn TM. Mating types in Paramecium aurelia: Diverse conditions for mating in different stocks; occurrence, number and interrelations of the types. P Am Philos Soc. 1938;79(3):411–434. [Google Scholar]
- Sonneborn TM. Breeding systems, reproductive methods, and species problems in protozoa. In: Mayr E, editor. The Species Problem. Washington DC: American Association for the Advancement of Science; 1957a. pp. 155–324. [Google Scholar]
- Sonneborn TM. Diurnal change of mating type in Paramecium. Anat Rec. 1957b;128:626. [Google Scholar]
- Sonneborn TM. The Paramecium aurelia complex of fourteen sibling species. T Am Microsc Soc. 1975;94(2):155–178. [Google Scholar]
- Sonneborn TM, Lynch RS. Racial differences in the early physiological effects of conjugation in Paramecium aurelia. Biol Bull. 1932;62(3):258–293. [Google Scholar]
- Sonneborn TM, Lynch RS. Hybridization and segregation in Paramecium aurelia. J Exper Zool. 1934;67(1):1–72. [Google Scholar]
- Tait A. Enzyme variation between syngens in Paramecium Aurelia. Biochem Genet. 1970;4(4):461–470. doi: 10.1007/BF00486596. [DOI] [PubMed] [Google Scholar]
- Yao M-C, Duharcourt S, Chalker DL. Genome-wide rearrangements of DNA in ciliates. In: Craig NL, Craigie R, Gellert M, Lambowitz A, editors. Mobile DNA II. Washington DC: ASM Press; 2002. pp. 730–758. [Google Scholar]
- Zufall RA, McGrath CL, Muse SV, Katz LA. Genome architecture drives protein evolution in ciliates. Mol Biol Evol. 2006;23(9):1681–1687. doi: 10.1093/molbev/msl032. [DOI] [PubMed] [Google Scholar]