Abstract
Sinapic acid is an intermediate in syringyl lignin biosynthesis in angiosperms, and in some taxa serves as a precursor for soluble secondary metabolites. The biosynthesis and accumulation of the sinapate esters sinapoylglucose, sinapoylmalate, and sinapoylcholine are developmentally regulated in Arabidopsis and other members of the Brassicaceae. The FAH1 locus of Arabidopsis encodes the enzyme ferulate-5-hydroxylase (F5H), which catalyzes the rate-limiting step in syringyl lignin biosynthesis and is required for the production of sinapate esters. Here we show that F5H expression parallels sinapate ester accumulation in developing siliques and seedlings, but is not rate limiting for their biosynthesis. RNA gel-blot analysis indicated that the tissue-specific and developmentally regulated expression of F5H mRNA is distinct from that of other phenylpropanoid genes. Efforts to identify constructs capable of complementing the sinapate ester-deficient phenotype of fah1 mutants demonstrated that F5H expression in leaves is dependent on sequences 3′ of the F5H coding region. In contrast, the positive regulatory function of the downstream region is not required for F5H transcript or sinapoylcholine accumulation in embryos.
Many investigations of plant metabolic pathways, gene regulation, and DNA transposition have exploited the dispensable nature of phenylpropanoid compounds. Most of these efforts have focused on phlobaphenes and anthocyanins because these conspicuous pathway end products have greatly facilitated genetic analyses. These investigations have resulted in the isolation and characterization of genes encoding enzymes and transcription factors required for the accumulation of these secondary metabolites (for review, see Dooner et al., 1991). In Arabidopsis phenylpropanoid metabolism gives rise to flavonoids, lignin, and sinapic acid esters. Mutants of Arabidopsis that are altered in flavonoid biosynthesis are collectively known as transparent testa mutants because these mutations decrease or eliminate the flavonoid-based condensed tannins that pigment the seed coat. Some of these loci have been shown to encode biosynthetic enzymes and others encode regulatory proteins (Koornneef, 1990; Shirley et al., 1995). Although flavonoid biosynthesis in Arabidopsis has been studied extensively at the genetic and molecular levels, much less is known about the genes involved in the biosynthesis of sinapic acid esters. Because these compounds are dispensable under laboratory conditions (Chapple et al., 1992), they provide additional targets for the genetic analysis of phenylpropanoid metabolism.
Arabidopsis and other members of the Brassicaceae accumulate three major sinapic acid esters, sinapoylglucose, sinapoylcholine, and sinapoylmalate (Fig. 1) (Bouchereau et al., 1991; Chapple et al., 1992), and the relative abundance of each of these compounds is regulated developmentally during the plant's life cycle (Strack, 1977; Mock et al., 1992; Lorenzen et al., 1996). Leaves contain only sinapoylmalate, whereas seeds accumulate primarily sinapoylcholine and smaller amounts of sinapoylglucose. During seed development de novo synthesis of sinapic acid leads to the production of sinapoylcholine. Through a series of interconversion reactions that are initiated upon imbibition, seed sinapoylcholine reserves provide the phenylpropanoid moiety for the synthesis of sinapoylmalate in expanding cotyledons. As seeds germinate, sinapoylcholine is hydrolyzed to yield sinapic acid, which is then re-esterified by sinapic acid:UDPG sinapoyltransferase to form sinapoylglucose. Sinapoylglucose is subsequently converted to sinapoylmalate by the activity of sinapoylglucose:malate sinapoyltransferase (Strack, 1982; Lorenzen et al., 1996). These interconversions are complete at approximately d 6 of seedling development, when de novo synthesis of sinapic acid contributes to the accumulation of sinapoylmalate in developing leaves.
Figure 1.
The phenylpropanoid pathway and the pathways leading to sinapate esters in Arabidopsis. CCoA OMT, Caffeoyl CoA O-methyltransferase; C3H, p-coumarate-3-hydroxylase; pCCoA 3H, p-coumaroyl CoA-3-hydroxylase; SCE, sinapoylcholinesterase; SCT, sinapoylglucose:choline sinapoyltransferase; SGT, sinapic acid:UDPG sinapoyltransferase; SMT, sinapoylglucose:malate sinapoyltransferase. The enzyme catalyzing the step from sinapic acid to sinapoyl CoA is shown as “4CL?” to reflect the uncertainty surrounding the identity of the protein involved.
Sinapate esters can be visualized by their blue fluorescence under UV light both in vivo and after TLC separation. The ease with which these compounds can be detected has facilitated the isolation of mutants defective in sinapate ester synthesis. These mutants have provided insights into the biological function of sinapate esters, and have enabled the isolation of genes involved in their synthesis. The best studied of these mutants is the sinapoylmalate-deficient fah1 mutant (Chapple et al., 1992). Experiments with fah1 demonstrated that sinapoylmalate is an important UV-B sunscreen in Arabidopsis (Landry et al., 1995), and cloning of the FAH1 gene revealed that it encodes F5H, a Cyt P450-dependent monooxygenase required for the synthesis of sinapate esters and sinapic acid-derived syringyl lignin (Meyer et al., 1996). It has since been shown that F5H catalyzes the rate-limiting step in syringyl lignin biosynthesis, and that its expression determines the monomer composition of the lignin in xylem and sclerified parenchyma (Meyer et al., 1998). Arabidopsis xylem cell walls contain only ferulic acid-derived guaiacyl lignin, whereas the interfascicular parenchyma of the rachis deposits syringyl lignin. When transformed with F5H ectopic-overexpression constructs, plants deposit syringyl-rich lignin in all cells that normally lignify, indicating that F5H is an important regulatory site for hydroxycinnamic acid production, at least with respect to lignin biosynthesis.
We investigated F5H expression in Arabidopsis in the context of sinapate ester biosynthesis. These experiments indicate that F5H transcript accumulation is regulated in a manner distinct from that of other phenylpropanoid genes. Furthermore, F5H expression in leaves is dependent on a regulatory domain that is located 3′ of the F5H stop codon, whereas its expression in embryos is independent of this downstream element. Although the pattern of F5H expression is consistent with a role for F5H in the determination of sinapate ester content, overexpression of F5H does not alter the temporal or tissue-specific regulation of sinapate ester accumulation. Thus, although F5H catalyzes the rate-limiting step in syringyl lignin biosynthesis, these findings cannot be extrapolated to imply a regulatory role for F5H in the biosynthesis of all sinapic acid-derived metabolites.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis plants were grown under a 16-h light/8-h dark photoperiod in potting mix (ProMix, Premier Horticulture, Red Hill, PA) at 22°C. For the growth of seedlings under axenic conditions, seeds were surface-sterilized in 30% (v/v) commercial bleach containing 0.07% (v/v) Triton X-100, rinsed, and sown on plates containing modified Murashige-Skoog medium lacking ammonium nitrate, as described previously (Lorenzen et al., 1996).
RNA Analysis
For the isolation of RNA, plant tissues were harvested, frozen in liquid nitrogen, and stored at −70°C until ready for extraction. Total RNA was isolated as described previously (Goldsbrough and Cullis, 1981). Samples were electrophoretically separated, transferred to membranes (Hybond N+, Amersham), hybridized at 65°C, washed, and exposed to film. DNA probes for the Arabidopsis genes encoding 4CL (Lee et al., 1995), C4H (Bell-Lelong et al., 1997), F5H (Meyer et al., 1996), OMT (Arabidopsis expressed sequence tag no. 12052; clone no. 154J19T7), and PAL (PAL1, PAL2, and PAL3) (Wanner et al., 1995) were made using the DECAprime II system (Ambion, Austin, TX).
DNA Analysis and Sequencing
Genomic Arabidopsis DNA carrying the F5H coding sequence and 5′ and 3′ regulatory regions was subcloned into pGEM-7Zf(+) (Promega) or pMBL18 (Nakano et al., 1995) for manual (Sequenase kit version 2.0 [United States Biochemical]) or automated (model 373A sequencer [Applied Biosystems] or model ALF Express sequencer [Pharmacia]) sequencing. The position of the T-DNA insertion in the fah1-9 allele was determined by cloning and sequencing a 0.5-kb PCR product made by amplification of the junction between the T-DNA right border and the F5H genomic DNA using the PCR primers GCCAACCACGCGCCTCATCT (F5H) and GTCACCTTAGGCGACTTTTGA (T-DNA right border). The F5H transcription start site was determined by primer extension as described previously (Bell-Lelong et al., 1997) using the primer GAGACGTCGTGGGATCTGATAG.
Construction of Transgenic Lines
Standard techniques were used for DNA manipulations (Sambrook et al., 1989). The isolation of cosmid pBIC20-F5H, the construction of plasmids 35S-F5H and C4H-F5H, and the introduction of these three constructs into the fah1-2 mutant line was described previously (Meyer et al., 1996, 1998). The F5H(HX) construct was made by ligating a 5.15-kb HindIII-XhoI fragment, derived from pBIC20-F5H and containing the F5H promotor and coding region, into the binary vector pGA482 (An, 1987). This plasmid was transformed into the fah1-2 mutant by vacuum infiltration (Bent et al., 1994), as described previously (Bell-Lelong et al., 1997). Two representative lines containing a single T-DNA insertion were made homozygous and used for subsequent experiments.
Analysis of Sinapate Esters
Sinapate esters were extracted from plant tissues in 50% methanol containing 0.75% (v/v) phosphoric acid. A 20-μL sample of each extract was analyzed by HPLC on a C18 column (Microsorb-MV, Rainin Instruments, Woburn, MA) using a gradient from 1.5% phosphoric acid to 35% acetonitrile in 1.5% phosphoric acid for elution and UV detection at 335 nm. Sinapate esters were quantitated using the extinction coefficient of sinapic acid. For TLC analyses, seed sinapate esters were extracted in 50% methanol and separated on silica gel K6 TLC plates (Whatman) using a solvent mixture of n-butanol, acetic acid, and water (5:2:3, v/v), and visualized under UV light.
RESULTS
F5H Transcript Accumulation Is Distinct from That of Other Phenylpropanoid Pathway Genes
To evaluate the tissue specificity of F5H expression, the abundance of F5H transcript in various organs of wild-type Arabidopsis was examined (Fig. 2). RNA-blot analysis using the F5H cDNA (Meyer et al., 1996) as a probe indicated that F5H mRNA accumulated in all tissues examined. As a fraction of total RNA, the highest level of F5H message was found in the rachi, which is consistent with the role of F5H in lignin biosynthesis. F5H expression in mature leaves was substantially lower than that in older or younger leaves.
Figure 2.
F5H transcript accumulation in tissues of wild-type Arabidopsis. Total RNA was extracted from the tissues indicated and probed with the F5H cDNA in RNA-blot analysis. The bottom panel illustrates ethidium-bromide staining of rRNA as a loading control.
To compare the expression of F5H with that of other phenylpropanoid genes, we determined their mRNA levels in developing light- and dark-grown seedlings and in seedlings grown in the dark for 4 d before transfer to the light. Except for a weak signal in 1-d-old seedlings, F5H transcript was nearly undetectable in dark-grown seedlings and in light-grown plants before d 3 (Fig. 3A). In light-grown seedlings F5H mRNA increased slowly over the remainder of the 10-d experimental period. When dark-grown seedlings were shifted to light conditions, F5H transcript levels increased during the next 6 d at approximately the same rate as in light-grown seedlings. This pattern of expression differed from that of the other genes examined. The mRNA of PAL1, PAL2, C4H, OMT, and 4CL accumulated more rapidly than that of F5H in light-grown seedlings, reaching maximal levels within 4 d of planting (Fig. 3B). In etiolated seedlings mRNAs corresponding to all of these genes were readily detectable, but were generally lower than in light-grown plants. These transcripts still reached maximal levels in the dark within 4 d of planting, but then gradually decreased. In the shift experiments transcripts reached maximal levels within 2 d after transfer to the light. Consistent with a previous report (Wanner et al., 1995), PAL3 mRNA accumulation was undetectable at all time points tested (data not shown).
Figure 3.
Transcript accumulation of phenylpropanoid genes during seedling development. Arabidopsis seedlings were germinated and grown under aseptic conditions on modified Murashige-Skoog agar plates (Lorenzen et al., 1996) in light (L; 16-h light/8-h dark photoperiod), in darkness (D), or in darkness for 4 d before being shifted to light conditions (D→L). Total RNA was extracted on the days indicated and RNA analysis was performed using probes from cDNAs of the indicated genes. A, Total RNA from wild-type seedlings probed with the F5H cDNA. The lower panel illustrates ethidium-bromide staining of rRNA as a loading control. B, Total RNA from wild-type seedlings probed with cDNAs corresponding to the phenylpropanoid genes indicated. C, Total RNA from a fah1-2: F5H(HX) transgenic line probed with the F5H cDNA. All blots were exposed to film for 24 h except PAL2, which was exposed for 48 h.
Sinapoylmalate Accumulation in Seedlings Does Not Change in Response to Constitutive Expression of F5H
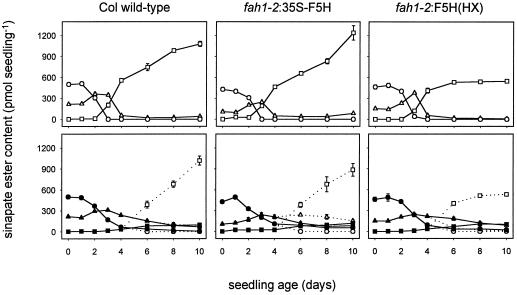
To correlate the results of the previous expression analysis with phenylpropanoid metabolism, the interconversion and biosynthesis of sinapate esters was evaluated in germinating light- and dark-grown wild-type seedlings (Fig. 4). In light-grown wild-type seedlings, the levels of sinapoylcholine decreased to undetectable levels by d 3. Coincident with this decrease in sinapoylcholine was a transient increase in sinapoylglucose, which peaked between d 2 and 3. By d 4 sinapoylglucose levels had decreased to nearly undetectable levels. Sinapoylmalate, which is undetectable in seeds, began to accumulate by d 2 and increased until at d 6 it was the dominant sinapate ester. By this time its levels approximated the total sinapate ester content of seeds. Sinapoylmalate continued to increase in abundance each day thereafter, indicating that de novo synthesis begins to contribute to the sinapate ester content of wild-type seedlings at approximately d 6. In dark-grown wild-type seedlings, the levels of sinapoylcholine and sinapoylglucose were found to be nearly the same as in light-grown seedlings for the first 2 d after imbibition.
Figure 4.
Accumulation of sinapate esters in wild-type and transgenic fah1 Arabidopsis seedlings. Wild-type seedlings and transgenic fah1-2 seedlings were grown on modified Murashige-Skoog agar plates as described for Figure 3. Sinapate esters were fractionated by HPLC, detected at 335 nm, and quantitated using the extinction coefficient of sinapic acid. Each point represents the average of three replicates of 10 seedlings each ± se. Top row, Seedlings grown in the light (open symbols). Bottom row, Seedlings grown in the dark (solid lines, solid symbols) or shifted from dark to light on d 4 (dotted lines, open symbols). Circles, Sinapoylcholine; triangles, sinapoylglucose; squares, sinapoylmalate.
During the next 8 d the levels of sinapoylglucose and, to a lesser extent, sinapoylcholine remained elevated, and sinapoylmalate failed to accumulate. The total sinapate ester content of dark-grown seedlings never exceeded the levels found in seeds, suggesting that de novo synthesis was not initiated under these conditions. When dark-grown seedlings were transferred to the light on d 4, sinapoylglucose levels decreased and sinapoylmalate levels increased gradually, indicating that transfer to the light permitted the completion of the interconversion phase and the onset of the de novo phase of sinapate ester biosynthesis. In both light- and dark-grown seedlings transferred to the light, the onset of de novo sinapate ester biosynthesis coincided with the initiation of F5H mRNA accumulation, which is consistent with the hypothesis that F5H expression regulates the biosynthesis of these metabolites in developing Arabidopsis seedlings.
To test the hypothesis that de novo accumulation of sinapoylmalate in seedlings is determined at the level of F5H expression, sinapate ester content was compared in light- and dark-grown transgenic seedlings constitutively expressing F5H (Fig. 4). If F5H expression were rate limiting for de novo sinapoylmalate biosynthesis, it would be expected that its constitutive expression would cause a precocious accumulation of sinapoylmalate in light-grown seedlings and possibly permit its accumulation in etiolated seedlings. Instead, constitutive expression of F5H under the direction of the CaMV 35S promoter in the fah1-2:35S-F5H transgenic line (Fig. 5) had virtually no effect on the onset of sinapoylmalate biosynthesis (Fig. 4). This result argues that F5H expression is not a control point for the temporal regulation of sinapoylmalate accumulation. Furthermore, the absence of sinapoylmalate in dark-grown fah1-2:35S-F5H seedlings suggests that additional light-dependent factors are required for sinapoylmalate accumulation in etiolated seedlings.
Figure 5.
Diagrammatic representation of the F5H gene and F5H transgenes used in this study. Exons are represented by open rectangles. A, F5H genomic region. An inverted triangle indicates the location of a T-DNA insertion in the fah1-9 mutant. B, Constructs used for F5H expression analysis in the fah1-2 mutant background. 35S, CaMV 35S promoter; E, EcoRI; H, HindIII; K, KpnI; S, SacI; X, XhoI.
F5H Expression Does Not Determine Sinapoylmalate Distribution in Mature Leaves
The distribution of sinapoylmalate in Arabidopsis leaves can be visualized by examining plants under UV light. Sinapoylmalate leads to a blue-green fluorescence in the adaxial epidermis, whereas the abaxial epidermis fluoresces red. The adaxial surfaces of rosette leaves of wild type, fah1-2, and the fah1-2 transgenic lines are indistinguishable when observed under white light (Fig. 6). Under UV light sinapoylmalate in leaves of the wild-type and the fah1-2:35S-F5H and fah1-2:C4H-F5H transgenic plants (Fig. 5) causes them to appear blue-green. In contrast, the leaves of the fah1 mutant appear red because they lack sinapoylmalate (Chapple et al., 1992). These data are consistent with previous biochemical analyses showing that the 35S-F5H and C4H-F5H constructs are capable of complementing the fah1-2 mutant phenotype (Meyer et al., 1996, 1998). Under UV illumination the abaxial surfaces of the wild-type, mutant, and transgenic leaves appear red (Fig. 6), indicating the absence of sinapoylmalate. Thus, ectopic overexpression of F5H did not lead to the accumulation of sinapoylmalate in the abaxial leaf epidermis. Although F5H expression determines the spatial deposition of syringyl units in lignifying tissues (Meyer et al., 1998), this result demonstrates that ectopic F5H expression is not sufficient to lead to sinapoylmalate accumulation in the abaxial leaf epidermis.
Figure 6.
Leaves of wild type, fah1-2, and transgenic lines as viewed under visible and UV light. Top panel, Adaxial leaf surfaces illuminated by visible light; middle panel, adaxial leaf surfaces illuminated by UV light; bottom panel, abaxial leaf surfaces illuminated by UV light (peak wavelength = 302 nm).
Sinapoylcholine Accumulation in Embryos Is Not Regulated at the Level of F5H Transcription
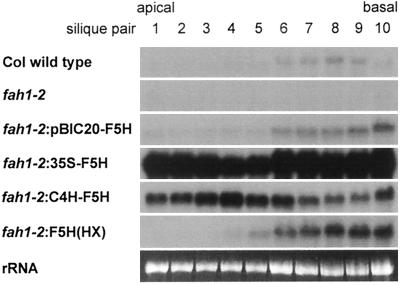
We examined F5H transcript accumulation in developing siliques of the wild type and various transgenic lines to investigate whether F5H expression could be the control point for sinapoylcholine biosynthesis in developing embryos (Fig. 7). For the collection of these data total RNA was prepared from siliques of the primary inflorescence that had been sampled in pairs, beginning with the first expanding silique. F5H mRNA in transgenic fah1-2 plants can only be the result of transgene expression, because the mutant lacks endogenous F5H transcript (Meyer et al., 1996). In the wild type F5H mRNA was nearly undetectable in the first five silique pairs and increased gradually thereafter. The pattern of F5H expression was similar in the fah1-2:pBIC20-F5H line. In contrast, a high level of F5H transcript was detected in the youngest siliques of both the fah1-2:35S-F5H and the fah1-2:C4H-F5H lines and this level remained relatively constant over the course of silique development. To determine whether the alteration of F5H expression had an effect on sinapate ester biosynthesis, the accumulation of sinapoylcholine was examined in comparable siliques (Fig. 8). As expected from earlier studies (Chapple et al., 1992), this compound was nearly undetectable in the fah1-2 mutant. In the wild type, sinapoylcholine was first detected in the fifth to seventh silique pairs, and its accumulation paralleled the increase in F5H expression. Although these data were consistent with a regulatory role for F5H expression, overexpression of F5H had no effect on the developmental onset of sinapoylcholine accumulation (Fig. 8). It seems unlikely that transcriptional regulation of F5H is a control point for sinapoylcholine biosynthesis in embryos.
Figure 7.
Accumulation of F5H transcript in developing siliques. Siliques were collected in pairs from 5-week-old plants of the lines indicated, beginning with the first expanding silique. Total RNA was extracted and probed with the F5H cDNA in RNA analysis. Blots were exposed to film for 24 h (fah1-2:35S-F5H and fah1-2:C4H-F5H) or 48 h. The bottom panel illustrates ethidium-bromide staining of rRNA as a loading control. Col, Arabidopsis ecotype Columbia.
Figure 8.
Accumulation of sinapoylcholine in developing siliques. Siliques were collected in pairs as described for Figure 7 and extracted in acidic 50% methanol. Sinapoylcholine was quantified by HPLC as described for Figure 4. Each point represents the average of three replicates of three silique pairs ± se. Col, Arabidopsis ecotype Columbia.
Sequences Downstream of the F5H Gene Are Required to Complement the fah1 Mutant Phenotype
The pBIC20-F5H cosmid contains 2.5 kb of DNA upstream and approximately 12.5 kb of DNA downstream of the F5H coding region (Fig. 5) and complements both the sinapoylmalate and syringyl lignin deficiencies of the fah1-2 mutant (Meyer et al., 1996). Similarly, transformation of the mutant with constructs in which expression of the F5H gene is driven by the CaMV 35S promoter (Odell et al., 1985) or the Arabidopsis C4H promoter (Bell-Lelong et al., 1997) restores all of the wild-type phenotypes (Meyer et al., 1996, 1998; this study). To further delimit the region of DNA sufficient to direct F5H gene expression, the plasmid F5H(HX) (Fig. 5) was introduced into fah1-2 plants. Despite having the same upstream DNA as pBIC20-F5H and the same amount of downstream DNA as the 35S-F5H and C4H-F5H constructs, observation under UV light of more than 50 independent transgenic lines indicated that F5H(HX) failed to complement the fah1-2 mutant phenotype (Fig. 6; data not shown). To explore this observation further, F5H mRNA abundance was examined in homozygous fah1-2:F5H(HX) transgenic seedlings. These analyses indicated that F5H transcript in fah1-2:F5H(HX) seedlings on d 1 (Fig. 3C) was similar to that in the wild type (Fig. 3A), but that there was no subsequent increase in F5H mRNA.
Although sinapoylmalate is absent in mature fah1-2:F5H(HX) plants, initial experiments demonstrated its presence in young seedlings (data not shown). To further explore these preliminary observations, the levels of sinapate esters were measured in developing fah1-2:F5H(HX) seedlings (Fig. 4). The sinapate ester profile of these seedlings was nearly identical to that of the wild type during the first 4 d of development. Thereafter, sinapoylmalate levels remained constant, which is consistent with the absence of F5H mRNA accumulation in seedlings of this transgenic line (Fig. 3C) and the requirement of F5H expression for de novo sinapoylmalate biosynthesis. fah1-2:F5H(HX) seeds (d 0) contained wild-type levels of sinapoylcholine. These data indicate that F5H must have been expressed during the development of the embryos within these seeds. As predicted by the presence of sinapoylcholine in fah1-2:F5H(HX) seeds (Fig. 4), developing siliques also accumulated sinapoylcholine (Fig. 8) and F5H transcript (Fig. 7). The high level of F5H expression in this line is likely the result of a transgene positional effect, because a second fah1-2:F5H(HX) line accumulated approximately wild-type levels of F5H transcript in maturing siliques (data not shown).
Our results indicate that the 630 bp of 3′ DNA in the 35S-F5H, C4H-F5H, and F5H(HX) constructs was sufficient for expression and mRNA stability in leaves when F5H was driven with a heterologous promoter, but in vegetative tissue F5H expression required additional downstream DNA in the context of its own promoter. From these data we conclude that an element in this downstream DNA functions as a positive regulator of gene expression. In contrast, the presence of F5H transcript (Fig. 7) and sinapoylcholine (Fig. 8) in fah1-2:F5H(HX) embryos indicates that the F5H 3′-flanking DNA is not required for F5H expression in developing embryos.
A T-DNA Insertion in the Downstream DNA of the fah1-9 Allele Results in a Phenotype That Is Similar to That of the fah1-2:F5H(HX) Transgenic Line
To determine how the T-DNA insertion in the fah1-9 allele leads to the mutant phenotype (Meyer et al., 1996), its position was determined by the cloning and sequencing of a PCR product made from the T-DNA right border-F5H junction. The T-DNA right border was found to be 283 bp 3′ of the F5H stop codon and 38 bp downstream of the F5H polyadenylation site (Fig. 5). This observation suggested that the T-DNA may interfere with the regulatory functions of the F5H 3′ DNA. Because the 3′-flanking DNA is not required for F5H expression in embryos, this hypothesis would predict that the F5H gene would be transcribed in fah1-9 embryos and would permit sinapate ester accumulation. Indeed, sinapoylcholine is present at approximately wild-type levels in fah1-9 seeds (Fig. 9). Together with the observation that fah1-9 homozygotes fail to accumulate sinapoylmalate in leaves, these results provide independent genetic evidence for the regulatory role of the 3′ region of the F5H gene and its tissue-specific function.
Figure 9.
Sinapate ester accumulation in seeds of the fah1-9 mutant. Seed extracts were separated by TLC and photographed under UV light. Col, Arabidopsis ecotype Columbia; WS, Arabidopsis ecotype Wassilewskija.
DISCUSSION
Tracheophytes accumulate myriad phenylpropanoid derivatives that range from taxon-specific flavonoids and simple phenylpropanoid glucosides and esters to the more complex and ubiquitous phenylpropanoid polymer lignin. The sinapic acid esters sinapoylmalate and sinapoylcholine are most commonly found in members of the Brassicaceae. In Arabidopsis these compounds provide convenient endogenous fluorescent reporters of the activity of the phenylpropanoid pathway and the enzymes specific to sinapate ester biosynthesis. Using these markers in combination with conventional molecular approaches, we have demonstrated that F5H expression is modulated independently of other phenylpropanoid pathway genes in Arabidopsis, but does not regulate sinapate ester accumulation. These studies have also shown that F5H expression involves a regulatory element located 3′ of its stop codon, a feature not previously associated with phenylpropanoid pathway gene regulation.
Comparison of F5H Expression with That of Other Phenylpropanoid Genes
The core phenylpropanoid biosynthetic genes (PAL, C4H, and 4CL) are expressed at relatively high basal levels even in dark-grown plants, and these transcripts increase in response to light and wounding (Hahlbrock and Scheel, 1989; Ohl et al., 1990; Logemann et al., 1995; Bell-Lelong et al., 1997; Mizutani et al., 1997). In contrast, the basal level of F5H expression in etiolated seedlings is very low compared with that of light-grown seedlings. In this respect F5H mRNA accumulation resembles that of the Arabidopsis flavonoid biosynthetic genes encoding chalcone synthase, chalcone isomerase, and dihydroflavonol reductase in developing dark-grown seedlings (Kubasek et al., 1992). The regulation of F5H is distinct, however, because in light-grown seedlings transcript levels for these three flavonoid genes peak at d 3 to 4 of growth and then decrease to nearly undetectable levels. The expression of PAL, C4H, OMT, and 4CL shows a similar increase at d 4, although their transcripts persist at higher levels than those of the flavonoid genes during the next week of growth (Kubasek et al., 1992; this study). In contrast, F5H mRNA accumulates continuously for the first 10 d of seedling development.
In mature plants F5H mRNA was found in all organs examined, but was most abundant in the rachis, which is consistent with the role of F5H in the lignification of the sclerified interfascicular parenchyma. F5H transcript was also abundant in young leaves, where it is required for the synthesis of UV-protective sinapoylmalate during leaf expansion. The relatively low levels of F5H mRNA in mature leaves may suggest that the sinapoylmalate synthesized in young leaves is relatively stable and can serve as a UV protectant during the lifetime of the leaf. Alternatively, it may indicate that the F5H protein has a long half-life and can support continued synthesis of sinapoylmalate.
The high levels of F5H mRNA in senescent leaves are more difficult to explain, because sinapoylmalate levels are known to decline in older rosettes (Chapple et al., 1992). High levels of transcript in these leaves may reflect continued synthesis of sinapoylmalate coupled with rapid turnover or fortuitous expression that is not correlated with secondary metabolite production caused by a limitation imposed by another step in the biosynthetic pathway. The high level of expression in roots is also surprising, because we have not been able to detect substantial levels of sinapate-derived metabolites in this tissue (M. Ruegger and C. Chapple, unpublished results). C4H is also highly expressed in Arabidopsis roots (Bell-Lelong et al., 1997), suggesting that phenylpropanoid gene expression is activated for the production of compounds that have not yet been identified, perhaps because of secretion or transport to the aerial portion of the plant. Alternatively, the levels of F5H expression in roots and senescent leaves may represent stress induction of phenylpropanoid gene expression. We believe that this is unlikely because, although many experiments have demonstrated the wound inducibility of the core phenylpropanoid genes (Ohl et al., 1990; Bell-Lelong et al., 1997; Mizutani et al., 1997), we have been unable to detect wound induction of F5H transcript (data not shown).
F5H Is Not a Regulatory Site for Sinapate Ester Biosynthesis
By using alternative promoters to overexpress F5H, we have demonstrated that its transcription regulates the flux of phenylpropanoid precursors through sinapic acid and toward syringyl lignin in lignifying cells of Arabidopsis (Meyer et al., 1998). The same study also showed that xylem-targeted expression of F5H was sufficient to permit the deposition of syringyl lignin in cells that normally accumulate only guaiacyl lignin. In contrast, although F5H expression parallels the accumulation of sinapate esters in developing seedlings and siliques, overexpression of F5H has no effect on the temporal regulation of their appearance. Similarly, although sinapoylmalate is distributed in a tissue-specific fashion, ectopic expression of F5H is not sufficient to permit its accumulation in the abaxial epidermis. It could be argued that, as in previous lignin-modification studies, F5H expression must be targeted to the specific cells in which sinapate esters are made and that our overexpression constructs failed to do so. On the other hand, we believe that sinapate ester synthesis is a cell-autonomous trait, and because C4H activity is required for sinapate ester synthesis, the C4H promoter should effectively target F5H expression to the correct cells. Thus, although F5H expression does control the biosynthesis of syringyl lignin, we conclude that it does not have a general regulatory role in the production of sinapic acid-derived metabolites.
These results leave open the question of how sinapate ester synthesis is regulated in Arabidopsis. It seems unlikely that sinapoylmalate synthesis is regulated by the activity of sinapoylglucose:malate sinapoyltransferase, because sinapoylglucose does not accumulate in wild-type leaves. Sinapoylglucose:choline sinapoyltransferase may be a regulatory point in sinapoylcholine synthesis, because low levels of sinapoylglucose are found in seeds of the Columbia ecotype of Arabidopsis. Free sinapic acid is found in neither seeds nor leaves, suggesting that sinapic acid:UDPG sinapoyltransferase does not have a regulatory role. Thus, steps earlier in the pathway probably regulate the synthesis of sinapate esters. The light independence and rapid accumulation of PAL, C4H, and OMT mRNAs in developing seedlings make it unlikely that sinapate ester synthesis is transcriptionally regulated within the phenylpropanoid pathway. The expression pattern of these genes is very different from the gradual and light-dependent increase in seedling sinapoylmalate content. The inability of F5H overexpression to alter sinapoylcholine accumulation in siliques leads to a similar conclusion.
These data suggest that if sinapate ester biosynthesis is controlled upstream of F5H, its regulation may be posttranscriptional in nature. Alternatively, transcriptional regulation may occur within the shikimate pathway, which supplies Phe for phenylpropanoid biosynthesis. Finally, with regard to the adaxial specificity of sinapoylmalate accumulation, the possibility that sinapate esters or their precursors are synthesized and then transported to the upper epidermis has not been excluded. Ectopic expression of F5H would not be expected to alter a transport or source/sink mechanism involved in such a movement of metabolites.
The Role of Downstream DNA in Gene Expression
The fah1-9 mutant and the fah1-2:F5H(HX) transgenic lines failed to accumulate sinapoylmalate in their leaves. In contrast, these same lines accumulated F5H transcript and sinapoylcholine in developing siliques and seeds, indicating that the 3′-flanking DNA necessary for F5H expression in adult tissues is not required for expression in embryos. A number of reports have demonstrated the involvement of 3′-flanking DNA in plant gene expression (Thornburg et al., 1987; Dean et al., 1989; Elliott et al., 1989; Dietrich et al., 1992; Larkin et al., 1993; Viret et al., 1994; Fu et al., 1995a, 1995b; Chinn et al., 1996; Marshall et al., 1997). Downstream sequences have been shown to be required for gene expression in response to wounding (Thornburg et al., 1987), light (Viret et al., 1994), and Suc (Fu et al., 1995a). They have also been shown to be necessary for correct spatial expression either by activating (Dietrich et al., 1992; Larkin et al., 1993) or repressing (Viret et al., 1994) gene expression. Certain 3′-flanking sequences are thought to act at the level of transcriptional regulation (Dean et al., 1989; Larkin et al., 1993; Viret et al., 1994), whereas others are thought to affect mRNA stability (Elliott et al., 1989) or chromatin structure (Chinn et al., 1996).
The requirement for 3′-flanking sequences in F5H expression further differentiates the regulation of F5H from that of other phenylpropanoid genes. Thus, the regulatory factors that control F5H expression in Arabidopsis may be independent of those that control upstream genes. This observation may be related to the fact that sinapate ester accumulation is taxonomically restricted, and that sinapate-derived secondary metabolites are less common than derivatives of hydroxycinnamic acids such as caffeic and ferulic acids. A recombination or transposition event that positioned a regulatory element downstream of the F5H-coding sequence early in the evolutionary history of the Brassicaceae may have been critical to the acquisition of the ability to express F5H and to accumulate sinapic acid derivatives in leaf tissue.
The differential requirements for F5H expression in seedlings and leaves versus embryos indicate that the 3′-flanking DNA may be a determining factor for tissue specificity. Although the mechanism underlying this activity remains to be characterized, our preliminary results suggest that it is unlikely to involve mRNA stabilization. The 35S-F5H and C4H-F5H constructs, both of which lack the 3′ DNA, lead to high levels of F5H mRNA in transformed plants (Meyer et al., 1996, 1998). These data suggest that the 3′ region is not required for F5H transcript stability. On the other hand, these relatively strong promoters may compensate for the absence of the stabilization provided by the 3′ sequence, which might be important in the context of the relatively weak F5H promoter. Preliminary results indicate that the 3′ DNA is required for expression of an F5H promoter-driven GUS reporter gene in leaves and stems of adult plants, but is not required for expression in embryos (M. Ruegger and C. Chapple, unpublished results). These results also argue against a role for the 3′ sequences in transcript stability. By the addition of downstream restriction fragments to the F5H(HX) construct and complementation of the fah1 leaf phenotype, we have recently shown that sequences that constitute this downstream regulatory element are contained within a 3326-bp region 3′ of the F5H stop codon (M. Ruegger and C. Chapple, unpublished results).
The Utility of Sinapate Ester Accumulation as a Tool to Understand Plant Gene Regulation
The ease of detection and dispensable nature of sinapate esters make Arabidopsis an attractive model for the study of phenylpropanoid gene regulation. Although the requirement for 3′-flanking sequences has been demonstrated for a number of plant genes, we are unaware of any reports in which downstream cis-acting elements have been defined in detail or shown to be involved in phenylpropanoid gene expression. We believe that the study of secondary metabolism in Arabidopsis, particularly with respect to F5H expression and sinapate ester accumulation, can provide critical insights into these and other factors that regulate gene expression. The use of F5H-containing transgenes for mutant complementation avoids issues associated with most reporter-gene experiments, such as stability of the transgene product and the elimination or altered spatial organization of cis-acting elements. Unlike many reporter constructs, F5H transgenes contain a full complement of cis-acting elements in their native context. When introduced into the fah1 mutant, indirect but quantitative assays of F5H activity and expression can be made by the determination of sinapate ester content in embryos and leaves, and syringyl lignin content in stems. Finally, the isolation of novel sinapate ester-deficient mutants may lead to the identification of trans-acting factors that regulate F5H and/or other phenylpropanoid biosynthetic genes.
Abbreviations:
- 4CL
4-hydroxycinnamoyl CoA ligase
- CaMV
cauliflower mosaic virus
- C4H
cinnamate-4-hydroxylase
- F5H
ferulate-5-hydroxylase
- OMT
caffeic acid/5-hydroxyferulic acid O-methyltransferase
- PAL
Phe ammonia-lyase
- UDPG
UDP-Glc
The accession number for the sequence reported in this article is AF068574.
Footnotes
This research was supported by the Division of Energy Biosciences, U.S. Department of Energy (grant no. DE-FG02-94ER20138 to C.C.), and by postdoctoral fellowships from the Swiss National Science Foundation and the Alexander von Humboldt Foundation (Feodor Lynen Fellowship) to K.M. This is journal paper no. 15, 853 of the Purdue University Agricultural Experiment Station.
LITERATURE CITED
- An G. Binary Ti vectors for plant transformation and promoter analysis. Methods Enzymol. 1987;153:292–305. [Google Scholar]
- Bell-Lelong DA, Cusumano JC, Meyer K, Chapple C. Cinnamate-4-hydroxylase expression in Arabidopsis. Regulation in response to development and the environment. Plant Physiol. 1997;113:729–738. doi: 10.1104/pp.113.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent AF, Kunkel BN, Dahlbeck D, Brown KL, Schmidt R, Giraudat J, Leung J, Staskawicz BJ. RPS2 of Arabidopsis thaliana: a leucine-rich repeat class of plant disease resistance genes. Science. 1994;265:1856–1860. doi: 10.1126/science.8091210. [DOI] [PubMed] [Google Scholar]
- Bouchereau A, Hamelin J, Lamour I, Renard M, Larher F. Distribution of sinapine and related compounds in seeds of Brassica and allied genera. Phytochemistry. 1991;30:1873–1881. [Google Scholar]
- Chapple CCS, Vogt T, Ellis BE, Somerville CR. An Arabidopsis mutant defective in the general phenylpropanoid pathway. Plant Cell. 1992;4:1413–1424. doi: 10.1105/tpc.4.11.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinn AM, Payne SR, Comai L. Variegation and silencing of the Heat Shock Cognate 80 gene are relieved by a bipartite downstream regulatory element. Plant J. 1996;9:325–339. doi: 10.1046/j.1365-313x.1996.09030325.x. [DOI] [PubMed] [Google Scholar]
- Dean C, Favreau M, Bond-Nutter D, Bedbrook J, Dunsmuir P. Sequences downstream of translation start regulate quantitative expression of two petunia rbcS genes. Plant Cell. 1989;1:201–208. doi: 10.1105/tpc.1.2.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich RA, Radke SE, Harada JJ. Downstream DNA sequences are required to activate a gene expressed in the root cortex of embryos and seedlings. Plant Cell. 1992;4:1371–1382. doi: 10.1105/tpc.4.11.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner HK, Robbins TP, Jorgensen RA. Genetic and developmental control of anthocyanin biosynthesis. Annu Rev Genet. 1991;25:173–199. doi: 10.1146/annurev.ge.25.120191.001133. [DOI] [PubMed] [Google Scholar]
- Elliott RC, Dickey LF, White MJ, Thompson WF. cis-Acting elements for light regulation of pea ferredoxin I gene expression are located within transcribed sequences. Plant Cell. 1989;1:691–698. doi: 10.1105/tpc.1.7.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Kim SY, Park WD. High-level tuber expression and sucrose inducibility of a potato Sus4 sucrose synthase gene require 5′ and 3′ flanking sequences and the leader intron. Plant Cell. 1995a;7:1387–1394. doi: 10.1105/tpc.7.9.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Kim SY, Park WD. A potato Sus3 sucrose synthase gene contains a context-dependent 3′ element and a leader intron with both positive and negative tissue-specific effects. Plant Cell. 1995b;7:1395–1403. doi: 10.1105/tpc.7.9.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsbrough PB, Cullis CA. Characterization of the genes for ribosomal RNA in flax. Nucleic Acids Res. 1981;9:1301–1309. doi: 10.1093/nar/9.6.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahlbrock K, Scheel D. Physiology and molecular biology of phenylpropanoid metabolism. Annu Rev Plant Physiol Plant Mol Biol. 1989;40:347–369. [Google Scholar]
- Koornneef M. Mutations affecting the testa color in Arabidopsis. Arabidopsis Inf Serv. 1990;28:1–4. [Google Scholar]
- Kubasek WL, Shirley BW, McKillop A, Goodman HM, Briggs W, Ausubel FM. Regulation of flavonoid biosynthetic genes in germinating Arabidopsis seedlings. Plant Cell. 1992;4:1229–1236. doi: 10.1105/tpc.4.10.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry LG, Chapple CCS, Last R. Arabidopsis mutants lacking phenolic sunscreens exhibit enhanced ultraviolet-B injury and oxidative damage. Plant Physiol. 1995;109:1159–1166. doi: 10.1104/pp.109.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin JC, Oppenheimer DG, Pollock S, Marks MD. Arabidopsis GLABROUS1 gene requires downstream sequences for function. Plant Cell. 1993;5:1739–1748. doi: 10.1105/tpc.5.12.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Ellard M, Wanner LA, Davis KR, Douglas CJ. The Arabidopsis thaliana 4-coumarate:CoA ligase (4CL) gene: stress and developmentally regulated expression and nucleotide sequence of its cDNA. Plant Mol Biol. 1995;28:871–884. doi: 10.1007/BF00042072. [DOI] [PubMed] [Google Scholar]
- Logemann E, Parniske M, Hahlbrock K. Modes of expression and common structural features of the complete phenylalanine ammonia-lyase gene family in parsley. Proc Natl Acad Sci USA. 1995;92:5905–5909. doi: 10.1073/pnas.92.13.5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzen M, Racicot V, Strack D, Chapple C. Sinapic acid ester metabolism in wild type and a sinapoylglucose-accumulating mutant of Arabidopsis. Plant Physiol. 1996;112:1625–1630. doi: 10.1104/pp.112.4.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall JS, Stubbs JD, Chitty JA, Surin B, Taylor WC. Expression of the C4 Me1 gene from Flaveria bidentis requires an interaction between 5′ and 3′ sequences. Plant Cell. 1997;9:1515–1525. doi: 10.1105/tpc.9.9.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K, Cusumano JC, Somerville C, Chapple CCS. Ferulate-5-hydroxylase from Arabidopsis thaliana defines a new family of cytochrome P450-dependent monooxygenases. Proc Natl Acad Sci USA. 1996;93:6869–6874. doi: 10.1073/pnas.93.14.6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K, Shirley AM, Cusumano JC, Bell-Lelong DA, Chapple C. Lignin monomer composition is determined by the expression of a cytochrome P450-dependent monooxygenase in Arabidopsis. Proc Natl Acad Sci USA. 1998;95:6619–6623. doi: 10.1073/pnas.95.12.6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani M, Ohta D, Sato R. Isolation of a cDNA and a genomic clone encoding cinnamate 4-hydroxylase from Arabidopsis and its expression manner in planta. Plant Physiol. 1997;113:755–763. doi: 10.1104/pp.113.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mock H-P, Vogt T, Strack D. Sinapoylglucose:malate sinapoyltransferase activity in Arabidopsis thaliana and Brassica rapa. Z Naturforsch. 1992;47c:680–682. [Google Scholar]
- Nakano Y, Yoshida Y, Yamashita Y, Koga T. Construction of a series of pACYC-derived plasmid vectors. Gene. 1995;162:157–158. doi: 10.1016/0378-1119(95)00320-6. [DOI] [PubMed] [Google Scholar]
- Odell JT, Nagy F, Chua N-H. Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature. 1985;313:810–812. doi: 10.1038/313810a0. [DOI] [PubMed] [Google Scholar]
- Ohl S, Hedrick SA, Chory J, Lamb CJ. Functional properties of a phenylalanine ammonia-lyase promoter from Arabidopsis. Plant Cell. 1990;2:837–848. doi: 10.1105/tpc.2.9.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Shirley BW, Kubasek WL, Storz G, Bruggemann E, Koorneef M, Ausubel FM, Goodman HM. Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J. 1995;8:659–671. doi: 10.1046/j.1365-313x.1995.08050659.x. [DOI] [PubMed] [Google Scholar]
- Strack D. Sinapic acid ester fluctuations in cotyledons of Raphanus sativus. Z Pflanzenphysiol. 1977;84:139–145. [Google Scholar]
- Strack D. Development of 1-O-sinapoyl-β-d-glucose:l-malate sinapoyltransferase activity in cotyledons of red radish (Raphanus sativus L. var. sativus) Planta. 1982;155:31–36. doi: 10.1007/BF00402928. [DOI] [PubMed] [Google Scholar]
- Thornburg RW, An G, Cleveland TE, Johnson R, Ryan CA. Wound-inducible expression of a potato inhibitor II-chloramphenicol acetyltransferase gene fusion in transgenic tobacco plants. Proc Natl Acad Sci USA. 1987;84:744–748. doi: 10.1073/pnas.84.3.744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viret J-F, Mabrouk Y, Bogarad L. Transcriptional photoregulation of cell-type-preferred expression of maize rbcS-m3: 3′and 5′ sequences are involved. Proc Natl Acad Sci USA. 1994;91:8577–8581. doi: 10.1073/pnas.91.18.8577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner LA, Li G, Ware D, Somssich IE, Davis KR. The phenylalanine ammonia-lyase gene family in Arabidopsis thaliana. Plant Mol Biol. 1995;27:327–338. doi: 10.1007/BF00020187. [DOI] [PubMed] [Google Scholar]