Abstract
As for mammals, insect health is strongly influenced by the composition and activities of resident microorganisms. However, the microbiota of insects is generally less diverse than that of mammals, allowing microbial function in insects to be coupled to individual, identified microbial species. This trait of insect symbioses facilitates our understanding of the mechanisms that promote insect-microbial coexistence and the processes by which the microbiota affect insect wellbeing. As a result, insects are potentially ideal models to study various aspects of interactions between the host and its resident microorganisms that are impractical or unfeasible in mammals and to generate hypotheses for subsequent testing in mammalian models.
Keywords: coevolution, innate immunity, insect-microbial interaction, metabolite exchange, microbiota, symbiosis
Introduction
The common condition for animals is to be chronically infected by microorganisms, most of which are benign or beneficial. The influence of these resident microorganisms on their animal host is profound, and operates at two levels: in physiological time, such that the physiology and well-being of the animal is influenced by the composition, density and activities of colonizing microorganisms; and in evolutionary time, through selection on the magnitude and pattern of the animal response to the infecting microorganisms. We can only understand animal-microbial interactions by combining these physiological and evolutionary perspectives.
There is immense variation in the detail of the interactions between animals and their resident microbiota. A proper grasp of the nature of interactions between humans and their microbiota can only come from an awareness of the parallels and contrasts with other animals. Insects represent a superb system for comparison because, as a group, they display far greater diversity than mammals in their interactions, including some associations of remarkable morphological intimacy and molecular integration. Research on insect-microbial interactions has been invigorated in recent years by the technical advances that make it possible to identify and study unculturable microorganisms. The recent gains in mechanistic understanding of the relationships in molecular terms have immediate relevance to research on the impact of resident microbes on the physiology and health of humans.
This review summarizes these recent advances by focusing on four topics: the mechanisms that promote co-existence of insect host and microbes; the contribution of microbes to insect nutrition and defense; insect dependence on their resident microbiota; and coevolutionary interactions between the partners. The impact of insect research on our understanding of associations in humans is also briefly considered.
Diversity of insect-microbial symbioses
Insects and mammals share the common feature that they are inhabited by microorganisms. Even so, there are two important differences in the host-microbial relationship between the two animal groups: (1) the diversity of the microbiota tends to be an order of magnitude greater in mammals than in insects and (2) many insects, but no known mammals, have beneficial intracellular microorganisms. To address these differences, the gut microbiota and intracellular symbioses of insects are considered in turn.
The gut lumen is very densely colonized by microbes in most mammals and insects. The animal gut can be considered a portion of the external environment in which the conditions and resources are controlled largely by the animal. Microorganisms associated with food have unrestricted access to the gut; and those that can tolerate, modulate or evade the digestive processes and immune function of the animal gut gain access to a nutrient-rich environment and a vehicle for dispersal via the feces. Nevertheless, the habitats in the insect and mammalian guts cannot be equivalent because the patterns in the composition and diversity of the gut microbiota are markedly different. The identification of bacterial operational taxonomic units (OTU) by 97% sequence identity of 16S rRNA gene sequences revealed that the gut of most insects bears <20–30 taxa (e.g. Dillon and Dillon, 2004; Robinson et al., 2010; Wong et al., 2011), and that of mammals (or their feces) yield 500–1,000 taxa (Dethlefsen et al., 2007; Nemergut et al., 2011). The reasons for this difference are unclear. As with many other low diversity habitats (Connell, 1978), insect guts tend to be transient and have extreme disturbance regimes. Compounding the short lifespan of many insects, microbes associated with the foregut and hindgut are eliminated at every insect molt (when the cuticle lining these gut regions is shed), and all microbes are shed when the larval gut is broken down and the adult gut develops during metamorphosis of higher (`holometabolous') insects including the true flies, butterflies and beetles. It has also been suggested that the adaptive immune system of mammals may promote microbial diversity through its greater capacity than the innate immune system to discriminate among different microorganisms, enabling the mammalian host to maintain complex multispecies consortia (McFall-Ngai, 2007).
The greater microbial diversity in individual mammals than insects is overlain by a greater diversity across all insects relative to all mammals. Thus, the dominant microbes in all mammals are Bacteroidetes and Firmicutes, but the gut microbiota of insects vary widely among different taxa, including Proteobacteria, Firmicutes and Protists (Brugerolle and Radek, 2006; Dillon and Dillon, 2004; Morales-Jimenez et al., 2009; Robinson et al., 2010). This difference could be a consequence of the greater variation of diets and gut physiology in insects than mammals, which is linked to the greater phylogenetic diversity of insects. [The class Mammalia comprises 5,000 species, and the class Insecta has 900,000 known (and 2–30 million predicted) species.] For example, all mammals have an extremely acidic stomach, but an equivalent region is rare among insects, apparently restricted to the higher Diptera, including Drosophila (Shanbhag and Tripathi, 2009). The pH in many insects lies within the range of 6–8 units, and some, notably lepidopteran caterpillars, have a very basic midgut region, at 11–12 pH units (Wieczorek et al., 2009).
The second dominant habitat in insects utilized by microorganisms is cells. An estimated 10–20% of insect species bear intracellular symbionts that are localized to specialized cells, known as bacteriocytes, whose sole function appears to be to house and maintain their symbionts. These associations are widespread or universal in several groups, notably cockroaches, hemipterans, ten families of beetles, and lice, and they also occur in some flies and ants [see Table 1 in Douglas (2007)]. Although they have evolved independently multiple times, the microorganisms are invariably transmitted vertically (from mother to offspring) usually via the eggs in the female ovaries. The resultant perfect congruence between the phylogenies of the microbial symbiont and their insect hosts, in some insect groups over >100–200 million years (Dale and Moran, 2006; Moran et al., 2005), has no parallel in mammals. Correlated with the long history and intimacy of the association, both the insect and microbial partners are dependent on the relationship, such that insects experimentally deprived of their bacteria fail to grow and reproduce (Douglas, 2007), and the symbionts generally have much reduced genomes (<1 Mb) and are unculturable (Dale and Moran, 2006).
Bacteriocytes are not the only insect cell type that contains microorganisms. Across all insect groups, microorganisms have been reported in cells of various organs, including the fat body, gut epithelium and gonads. Some of these taxa (e.g. Wolbachia, Hamiltonella) can occupy multiple compartments, for example within and between the cells of insect organs and in the blood, although the location of the microorganisms may vary with host and symbiont genotype, as well as the age and physiological condition of the host (Oliver et al., 2010; Werren et al., 2008). This trait of broad and variable tissue distribution is facilitated by the open circulatory system of insects, meaning that blood is not restricted to closed vessels but is in direct contact with various organs and moves relatively sluggishly around the body.
Bacteriocyte symbioses are unknown among mammals, and virtually all microorganisms that adopt an intracellular phase in mammals mediate chronic or acute pathogenic infections. There is no entirely satisfactory explanation for this difference. It has been suggested repeatedly that the adaptive immune system of vertebrates poses a very high barrier to the evolution of intracellular microorganisms, which has been overcome predominantly by pathogens. Consistent with this argument, intracellular symbionts are common-place among many invertebrates but extremely rare across all vertebrates, with the alga Oophila in the embryos of a salamander (Kerney et al., 2011) as the sole documented example.
Mechanisms for Co-Existence
Much more is known about the patterns than the mechanisms that promote co-existence of insects and their resident microbes. The key feature of these patterns is a relative uniformity in the location and numbers of microorganisms, suggesting that the host exerts tight controls over its symbionts. A rich literature has demonstrated that symbionts of insects are restricted to specific anatomical sites or cell types, and their abundance varies predictably with developmental age and sex of the insect host and environmental conditions. For microbes that can be either mutualistic or deleterious depending on environmental circumstance or host genotype, a key feature of the deleterious phenotype is high proliferation rates and abundance, often accompanied by an expanded distribution within the insect body. For example, the negative effect of the popcorn strain of Wolbachia on the longevity of Drosophila is strongly correlated with high bacterial numbers (McGraw et al., 2002); and the γ-proteobacterium Hamiltonella is harmful to its aphid host only on certain rearing plants, correlated with a ten-fold increase in bacterial abundance (Chandler et al., 2008).
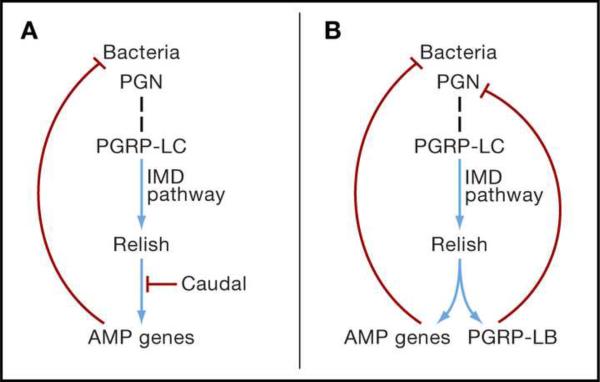
Several studies indicate that the immune system plays a central role in the persistence of the microbiota. One such study concerns the gut microbiota of Drosophila melanogaster, dominated by Acetobacter and relatives, and Lactobacillus. The epithelial cells of the insect midgut bear receptors for the IMD (Immune Deficiency) signaling pathway, which is also active in other tissues, including the fat body, where it mediates the expression of various antimicrobial peptides (AMPs) against bacteria (Lemaitre and Hoffmann, 2007). The resident gut bacteria induce IMD signaling in the gut epithelial cells, resulting in localization of the NF-κB transcription factor Relish to the epithelial cell nucleus but, contrary to expectation, the expression of AMPs is not induced (Ryu et al., 2008). The inhibition of AMP production is dependent on expression of the homeobox gene caudal (Figure 1A); when caudal expression is reduced by RNAi, AMP gene expression is upregulated. Importantly, the consequence of AMP production in the gut is not elimination of the gut microbiota but a change in the microbial composition. Specifically, the relative abundance of two bacteria is dramatically altered; in the untreated fly, a bacterium of the Acetobacteriaceae (strain A911, known as Commensalibacter intestini) is dominant, and the abundance of Gluconobacter morbifer (G707) is very low. When the AMPs are expressed, the ratio of A911:G707 shifts from nearly 200:1 (favoring A911) to 1:20 (favoring EW707) (Roh et al., 2008; Ryu et al., 2008). G. morbifer is deleterious to the host, resulting in severely depressed lifespan of the flies with a G. morbifer-dominated gut community.
Figure 1.
The Insect IMD Pathway and Persistence of Resident Microorganisms. The IMD (Immune Deficiency) pathway is triggered by binding of bacterial peptidoglycan (PGN) to the peptidoglycan recognition protein PGRP-LC (dashed vertical line). The resultant activation of the NF-κB transcription factor Relish leads to the upregulated expression of anti-microbial peptide (AMP) genes. Positive interactions (blue), negative interactions (red) (A) Resident microorganisms in the Drosophila gut activate the IMD pathway, but expression of AMPs is repressed by the transcription factor Caudal.
(B) The IMD pathway in bacteriocytes (insect cells bearing symbiotic bacteria) of the Glossina tsetse fly and weevil Sitophilus is activated, but the immunoreactivity of bacteriocytes is dampened by the very high expression of the IMD-responsive gene PGRP-LB. PGRP-LB is a PGN-amidase that degrades the PGN ligand, thereby down-regulating the IMD pathway.
The implications are two-fold. First, AMPs are not exclusively antagonistic to microorganisms, but can also act to manage and regulate the microbial community. This conclusion has been reached independently for AMPs in other invertebrate animals (Fraune et al., 2010), and there is also evidence that other immune-related molecules can function in immunological management. For example, Toll-like receptor-2 (TLR2), which contributes to pathogen recognition in mammals, also suppresses the host inflammatory response to a major resident gut microorganism, Bacteroides fragilis; specifically, a product of B. fragilis (polysaccharide A) signals through TLR2 expressed by regulatory T cells to suppress pro-inflammatory TH17 cells, thereby enabling B. fragilis to associate closely with the intestinal mucosa without inducing a host inflammatory response (Round et al., 2011). Second, although the great flexibility of the vertebrate adaptive immune system has been suggested to facilitate immunological management of the resident microbiota (McFall-Ngai, 2007), research on Drosophila demonstrates that the innate immune system can also function to regulate and manage the microbiota.
The study of Ryu et al. (2008) on the interaction between the gut microbiota and immune effectors of Drosophila focused exclusively on the immune responses localized to the gut. Data on Anopheles mosquitoes suggest that the gut microbiota induces a systemic immunological response that limits the abundance and distribution of the microorganisms. RNAi-mediated silencing of AMPs and immune signaling pathways has been shown to result in increased proliferation of the gut microbiota, including Pseudomonas and Novosphingobium species, and their localization to the hemolymph (Dong et al., 2009; Garver et al., 2008). The effect of the gut microbiota on the systemic immune response of other insects has not been investigated.
Further evidence that the innate immune system can serve to promote co-existence between insects and their resident microbiota comes from several studies on intracellular bacteria in bacteriocytes. Although they have much-reduced genomes, these bacteria are predicted to possess the molecular patterns that are recognized by the insect immune system. The host immune response is attenuated in the bacteriocytes of both the weevil Sitophilus and tsetse fly Glossina. In both cases, PGRP-LB (a peptidoglycan recognition protein with amidase activity) is strongly expressed in the bacteriocytes. Its expression is dependent on activation of the IMD pathway, and it functions to remove the peptidoglycan ligand that triggers the IMD pathway (Figure 1B). In this way, the responsiveness of the IMD pathway to symbiont peptidoglycan fragments is dampened, promoting persistence of the bacteria. The symbioses in Sitophilus and Glossina have separate evolutionary origins, indicating that the central role of PGRP-LB in these symbioses has arisen independently. PGRP-LB is not, however, the universal arbiter of symbiont persistence in bacteriocytes. Aphids, for example, lack both PGRPs and a functional IMD pathway, and it has been suggested that the selection pressure to accommodate its bacteriocyte symbionts (Buchnera) may have led to the evolutionary loss of this portion of the immune system (Gerardo et al., 2010). Taken together, these data illustrate how selection by resident microorganisms can influence the immune responsiveness of specific organs and cell types, and potentially of the entire animal. Although parasites have been invoked repeatedly as a selection pressure for diversification of the animal immune system (Rolff, 2007; Sackton et al., 2007), beneficial symbioses can also be important.
A recent study on Wolbachia in the mosquito Aedes aegypti has implicated insect miRNAs (18–25-mer nonprotein-coding RNAs) in the regulation of symbiont numbers (Hussain et al., 2011). Specifically, when either insects or cultured insect cells are infected by Wolbachia, the titer of one specific miRNA (aae-miR-2940) is elevated, resulting in the increased expression of an insect metalloprotease gene. When either the miRNA was inhibited or the metalloprotease gene expression silenced, Wolbachia numbers in the insect cells were reduced. Further research is required to establish the generality of miRNA-mediated promotion of insect-microbial co-existence, and how these mechanisms interact with the humoral immune system.
Overlaying these mechanisms acting in physiological time are selection pressures for co-existence operating in evolutionary time. In this regard, mode of transmission of the microorganisms plays a key role (Ewald, 1994). The selective interest of microorganisms that require a live host, and especially depend on a live host for transmission, overlaps with that of the host, and these microorganisms tend to be more benign that those that are transmitted efficiently from dead or dying hosts. Obligate vertical transmission generates an especially great overlap in selective interest between the microbial and host partners because the fitness of the microorganism is critically dependent on the fecundity of the host. Obligate vertical transmission is the norm in bacteriocyte symbioses of insects and is assured by behavioral mechanisms for various gut associations. The transmission of the γ-proteobacterium Ishikawaella in the distal midgut of various stinkbugs provides a particularly vivid example. The female insect deposits a fecal pellet containing Ishikawaella alongside an egg, the offspring feed on the pellet immediately after emerging from the egg, and the larval gut then undergoes dramatic changes involving the effective separation of the proximal blind-ended gut region for digestion and the distal symbiotic organ in the distal region (Fukatsu and Hosokawa, 2002; Hosokawa et al., 2007).
Benefits of the Resident Microbiota to the Host
The contribution of the resident microbiota to the wellbeing of an animal, and the underlying mechanisms, can most readily be investigated by comparing animals experimentally deprived of their microbiota with untreated animals bearing their natural microbial complement. Useful supplementary approaches to identify the contribution of specific microorganisms and the underlying mechanisms include the analysis of animals with modified microbial contents, e.g. associations with single taxa, microorganisms with known genetic mutations, or microbiota of different host genotypes or species. These manipulations come under the umbrella term of “gnotobiotics”, meaning the rearing of animals under germ-free conditions, either continuously through life or with experimental administration of specific microorganisms. Gnotobiotics are especially important for investigating potentially important functions of microbial taxa that are at low abundance in unmanipulated associations. Various insect associations are very amenable to gnotobiotics because the treatments can be administered easily and cheaply to hundreds-to-thousands of individual insects. It is generally straightforward to attribute function to individual microbial taxa because many insects bear a microbiota of low diversity (see above). The equivalent experiments using gnotobiotic mammals are technically demanding and, in some instances, extremely difficult to interpret because many microorganisms are members of complex, interdependent consortia with much functional redundancy, such that one host may bear multiple taxa with similar traits, and the dominant taxa mediating a function of interest may vary among different host individuals.
Because of their experimental tractability, insect symbioses can provide valuable lessons about the services that resident microbes provide to animal hosts. Analyses of both gut and intracellular symbioses in many insects have revealed two core microbial functions: nutrition and defense. In many instances, it has been straightforward to link function to individual microbial taxa.
Particular insights are being obtained from studying insects that feed through the life cycle on diets of extremely unbalanced composition or low overall nutritional value, e.g. vertebrate blood (deficient in B vitamins), plant sap (for which essential amino acids are in short supply), and sound wood ( grossly deficient in nitrogen and various essential nutrients). These insects all possess symbiotic microorganisms that provide specific nutrients in short dietary supply (Douglas, 2009). Strong indications that blood-feeding insects, including tsetse flies, lice and bedbugs, derive B vitamins from their symbionts come from experiments comparing the performance of insects bearing and lacking their symbionts on blood diets that are untreated or supplemented with B vitamins. The same experimental approach, together with complementary metabolic analysis, has been applied to plant sap feeders, revealing that these symbionts provide essential amino acids.
The most detailed information is available for the association between the plant phloem sapfeeding pea aphid Acyrthosiphon pisum and its bacteriocyte symbiont Buchnera, for which both genomes are sequenced and annotated (International Aphid Genomics Consortium, 2010; Shigenobu et al., 2000). Up to 50% of the essential amino acids synthesized by Buchnera cells are released to the surrounding host cell contents (Gunduz and Douglas, 2009). The genome of this bacterium has a dearth of recognizable regulatory sequences, its gene expression is remarkably unresponsive to dietary perturbation (Reymond et al., 2006; Wilson et al., 2006), and variation in aphid requirements for essential amino acids cannot generally be attributed to SNPs or other sequence variants in the Buchnera genome of the different aphids (Macdonald et al., 2011; Vogel and Moran, 2011). Taken together, these data suggest that the insect host plays a major role in shaping the composition and quantity of essential amino acids released from Buchnera. Analysis of both the transcriptome and proteome of bacteriocytes has demonstrated that the bacteriocytes are enriched in enzymes mediating the synthesis of precursors required for Buchnera-mediated essential amino acid synthesis (Hansen and Moran, 2011; Poliakov et al., 2011). Although the size and turnover of the precursor pools in the host cell remain to be established, these data raise the possibility that Buchnera metabolism is poised for maximal production of essential amino acids, and the realized synthesis rate is determined by precursor supply from the host cell.
Most research on the contribution of microorganisms resident in the gut to insect nutrition has focused on termites, which feed on various diets rich in plant fiber (wood, soil, humus). Historically – and erroneously - these associations were described as “miniature cows”, in which cellulose-rich plant material was degraded exclusively by the hindgut microbiota to short chain fatty acids (SCFAs) that were utilized by the termite. It is now realized that the termites are more complex and diverse than originally envisaged. The guts of termites (unlike mammals) have considerable intrinsic cellulase activity, which is partly or entirely responsible for cellulose degradation, varying among termite groups (Watanabe and Tokuda, 2010). Members of the gut microbiota additionally contribute to the nitrogen economy of the insect by recycling insect waste nitrogen or fixing atmospheric nitrogen, again varying among termite taxa (Burnum et al., 2011; Warnecke et al., 2007).
As the examples above illustrate, most research on the nutrition of insect-microbial symbioses has focused on animals with diets that are extremely nutrient-poor or nutritionally-unbalanced, and are not utilized generally by mammals. Even so, the nutritional significance of the resident gut microbiota in a few insects utilizing less extreme diets has been studied. Notably, elimination of the microbiota from Drosophila melanogaster has been reported to extend development time and shorten lifespan (Bakula, 1969; Brummel et al., 2004). The indication that the latter effect appears to be diet-dependent (Ren et al., 2007) suggests that the microbiota may have a nutritional role in this insect, but this has not been demonstrated definitively.
There is unambiguous evidence that specific resident microorganisms promote the resistance of insects to certain natural enemies, including viruses, bacteria, fungi, nematodes and parasitic wasps. Examples include: the α-proteobacterium Wolbachia, which protects Drosophila melanogaster against various viruses (Hedges et al., 2008); Spiroplasma bacteria that confer resistance in Drosophila neotestacea against the nematode parasite Howardula aoronymphium (Jaenike et al., 2010) and in Drosophila hydei against the parasitic wasp Leptopilina heterotoma (Xie et al., 2010); and the γ-proteobacterium Regiella insecticola, which reduces the mortality of pea aphids infected with entomopathogenic fungi (Scarborough et al., 2005). In these and many other interactions, the underlying mechanisms are not known, but for a few instances, an outline understanding of the mechanisms has been obtained. The following three examples illustrate the diversity of interactions mediated by defensive microbes.
The first example concerns actinobacteria with protective function. Beewolf digger wasps house cultures of actinobacteria `Candidatus Streptomyces philanthi' in antennal glands, and smear the bacteria onto cocoons deposited in their humid, microbe-rich burrows (Kaltenpoth et al., 2005). The complex mix of antibiotics synthesized by the bacteria confers a generalized protection against microbial attack (Kroiss et al., 2010). The thorax of leafcutting ants is enveloped in a multi-species actinobacterial mat (Mueller et al., 2008) that produces antibiotics with activity against fungi, including a major fungal pathogen, Escovopsis (Oh et al., 2009). Actinobacteria in a similar relationship with Dendronoctus beetles provide analogous protection (Scott et al., 2008). In each of these systems, the low diversity of the actinobacterial partners has facilitated the identification of the actinobacterial partner, its function and the chemical nature of its antimicrobials.
The second example relates to parasitic wasps, many of which exploit various insects by depositing an egg in the body cavity of their victim. The resultant wasp larva consumes tissues of the living insect over some days, and following the death of the insect, the adult wasp emerges from the dead insect “mummy”. The resistance of pea aphids to the parasitic wasp Aphidius ervi is heightened by the presence of the γ-proteobacterium Hamiltonella defensa, which occurs in the body cavity of some pea aphids (the prevalence of H. defensa varies widely among different pea aphid populations). Importantly, this defensive role is evident only for H. defensa isolates that contain the virus APSE bearing the genes for toxins, including homologs of cytolethal toxins (Oliver et al., 2009). The reasonable inference is that resistance against the parasitic wasp is mediated by these toxins, but neither this interpretation nor how the aphid tissues avoid toxin-incurred damage has been demonstrated.
The resident microbiota in Anopheles mosquitoes reduces infection of the mosquitoes by the malaria parasite Plasmodium ingested with the blood meal. Specifically, the invasion of the midgut epithelium by the Plasmodium ookinetes is inhibited. This effect is mediated by bacteria established in the gut, and co-feeding Anopheles with bacteria and Plasmodium or co-injecting bacteria into the insect body cavity with Plasmodium feeding mediates these effects, indicating that it is not caused by a direct interaction between the bacteria and Plasmodium (Dong et al., 2009). Instead, the insect immune system is involved, including immune effectors that are active against both bacteria and Plasmodium. The precise mechanism of protection is not fully established (Cirimotich et al., 2010). After feeding on blood, the mosquito may mount a vigorous immune response against its resident bacteria, which proliferate rapidly when blood is ingested, or against hemozoin, which is an immunogenic breakdown product of the blood, with an indirect effect on the Plasmodium. Alternatively or additionally, the mechanical disruption of the midgut epithelium by the invading Plasmodium ookinetes may introduce bacteria into the insect blood, triggering a systemic immune response. There is evidence for some specificity in this effect, with Gram-negative bacteria providing greater protection than Gram-positives (Cirimotich et al., 2010).
Dependence and Addiction
An important lesson from insect symbioses comes from the parasitic wasp Asobara tabida. In nature, every individual of this insect bears the bacterium Wolbachia, which is transmitted vertically via the eggs of the wasp. When Wolbachia is eliminated by antibiotic treatment, A. tabida is reproductively sterile with egg production halted by massive apoptosis of the nurse cells in the ovary. A limited incidence of apoptosis occurs during normal oogenesis in A. tabida bearing Wolbachia. A reasonable inference from these data is that Wolbachia inhibits apoptosis, and there is strong correlative evidence that Wolbachia perturbs iron metabolism in its host, resulting in increased oxidative stress that disrupts cellular physiology, including apoptosis (Kremer et al., 2009). This interaction has been argued to select for increased apoptotic signaling in the host to compensate for the otherwise harmful effects of Wolbachia on host physiology (Pannebakker et al., 2007). The putative compensatory response of A. tabida is constitutive, such that apoptosis is excessive in the absence of the inhibitory signal from Wolbachia. In this way, A. tabida has become dependent on Wolbachia without deriving any discernible benefit, a condition that is defined as addiction (Aanen and Hoekstra, 2007).
Dependence without benefit (i.e. addiction) has received little attention, partly because dependence is often interpreted as evidence for benefit, even in the absence of evidence for any service (e.g. nutrient provisioning, defensive role), but also because interactions between animal hosts and their resident microbiota are often investigated without considering their evolutionary context. Even so, there are candidate instances of dependence without benefit in various symbioses, including those between mammals and their resident microbiota. As one specific example, the capillary network in the small intestine of the mouse develops normally only in the presence of the gut microbiota (Stappenbeck et al., 2002). When mice are reared under aseptic conditions, the small intestine is poorly vascularized and the mice have reduced capacity to assimilate nutrients from the gut. Capillary growth is not induced by the bacteria directly, but by antibacterial peptides produced by the Paneth cells of the intestine in response to the presence of the microorganisms. In this way, the bacteria have become integrated into the animal signaling pathway required for normal development of the intestinal capillary bed, even though the capillaries function independently of the microbiota. There is no selection on the host to be independent of the microbiota because they are invariably infected by gut microorganisms soon after birth. Further candidate instances of dependence without benefit come from the central role of the gut microbiota in the development of the gut-associated lymph tissue and various populations of intestinal immune cells (Duan et al., 2010; Falk et al., 1998; Niess et al., 2008; Pollard and Sharon, 1970).
How can dependence without benefit evolve? One possibility is that these many instances are animal compensatory responses to microbial perturbations of the host immune system or metabolism. As in A. tabida, the host responses may be constitutive because there is no direct selection for function in the absence of the microbiota (because the microbiota are never absent under natural conditions). An alternative scenario is that certain signaling networks are truly symbiotic: that the resident microbiota in the ancestors of modern animals may have provided elements of the signaling networks regulating developmental processes, including angiogenesis and immune function, and that these elements have been retained in modern animals (Lee and Mazmanian, 2010; McFall-Ngai, 2002). Whatever the evolutionary origin of dependence, the consequent addiction of animals for their resident microbiota is evident only under the unnatural experimental conditions of axenic (germ-free) rearings, or large-scale perturbation of the microbiota, as in various immunological diseases or grossly unsuitable diets.
Coevolution between Insects and their Microbiota
Insect symbioses offer spectacular examples of coevolution with major consequences for the health and wellbeing of the animal host. For example, coevolutionary changes may include increased microbial production of nutrients valuable to the host and correlated changes in host metabolism, transporters etc. that promote the host processing of microbial nutrients.
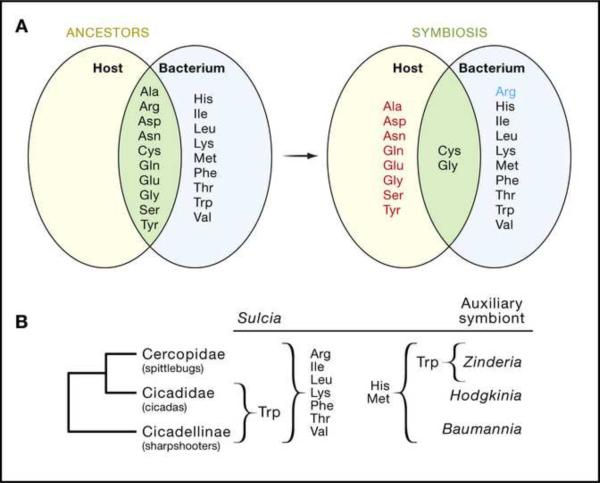
Coevolutionary interactions have been inferred from genome sequencing projects on plant sap-feeding insects and their microbial symbionts, including several instances of perfect complementarity in amino acid metabolism through differential gene loss in the partners in the association. For example, the genome of both the pea aphid Acyrthosiphon pisum and its Buchnera symbiont have been sequenced (International Aphid Genomics Consortium, 2010; Shigenobu et al., 2000), revealing a large-scale loss in the capacity of Buchnera to synthesize non-essential amino acids (which can be synthesized by the aphid host), and loss of the capacity to synthesize one amino acid, arginine, in the aphid host (Figure 2A). Unlike aphids which have a single obligate symbiont, plant sap-feeding insects of a related suborder (the Auchennorhyncha) possess dual symbioses, comprising a Bacteroidetes Sulcia mulleria and a second auxiliary bacterium, the identity of which varies with the insect group (Figure 2B). Remarkably, Sulcia has the capacity to synthesize 7 or 8 of the 10 essential amino acids (varying with insect group), and the auxiliary symbiont can synthesize the remaining essential amino acids (McCutcheon et al., 2009; McCutcheon and Moran, 2007, 2010). As a consequence, each symbiont supplies the required nutrients for both the other bacterium and the host (Figure 2B).
Figure 2.
Coevolution of Amino Acid Biosynthesis in Plant Sap-Feeding Insects and their Bacterial Symbionts (A) In the symbiosis between the pea aphid and Buchnera bacteria, the Buchnera (but not aphid) has lost the capacity to synthesize eight amino acids (red), and the aphid (but not Buchnera) has lost the capacity to synthesize arginine (blue) relative to the ancestral condition (E. coli, a relative of Buchnera, and a generalized insect, respectively). The one-reaction synthesis of glycine from serine is retained by both partners. Cysteine is synthesized by different pathways in the two partners (from methionine in the aphid, from serine in Buchnera).
(B) Spittlebugs, cicadas and sharpshooters bear two bacterial symbionts, Sulcia and an auxiliary symbiont with complementary amino acid biosynthetic capabilities. The amino acids produced by each bacterium are made available to both the insect host and the alternative bacterium. Both bacteria require the ten non-essential amino acids from the host.
[ala:alanine; arg: arginine; asn: asparagine; asp: aspartate; cys: cysteine; gln: glutamaine; glu: glutamate; gly: glycine; his: histidine; ile: isoleucine; leu: leucine; lys: lysine; met: methionine; phe: phenylalanine; pro: proline; ser: serine; thr: threonine; trp: tryptophan; tyr: tyrosine; val: valine].
The “perfect” metabolic complementarity in these insect symbioses is a product of the reciprocal coevolutionary changes in the host and symbiont(s), accompanied by genome reduction of the symbionts. Both of these latter traits are facilitated by the persistent obligate vertical transmission of the symbionts via the egg, for example, over an estimated 160 million years for Buchnera and 260 million years for Sulcia (Moran et al., 1993; Moran et al., 2005). The continuous interaction between individual lineages of host and symbiont provides the basis for both genome reduction of the symbiont (by relaxed selection and genome deterioration) and strict reciprocal coevolution.
These associations provide an important lesson in the power of coevolutionary interactions in shaping the functional traits of animals and their resident microbes. Nevertheless, we should not necessarily anticipate precisely equivalent relationships in mammals because no resident microorganisms of mammals are known to be obligately vertically transmitted and the coevolutionary interactions are more diffuse, i.e. likely to involve guilds of microbes (Oh et al., 2010).
Current and Future Lessons from Insect Symbioses
Insect symbioses offer various clear-cut exemplars of processes underlying interactions between animals and their resident microbiota. Most of these interactions are unlikely to be replicated precisely in mammalian systems, simply because symbioses in mammals and insects are different (Table 1). Their principal value to researchers investigating mammalian systems is conceptual, demonstrating, for example, how the persistence of microorganisms can be shaped by modulation of the innate immune system (Figure 1) or how the metabolic capabilities of hosts and multiple microorganisms can coevolve to perfect complementarity (Figure 2). The low diversity of microorganisms in most insects, compared to mammals, is important for the clarity of these exemplars. For insects, it is generally possible to couple microbial function to one (or several) well-defined microbial taxa, and functional redundancy is minimal.
The key traits of low diversity and minimal functional redundancy in the microbiota of many insects provide the opportunity to exploit insect systems to investigate fundamental problems in animal-microbial interactions that are impractical or unfeasible in mammalian systems. From insect-based research, specific hypotheses can be generated for subsequent analysis in biomedical rodent models or in human trials. The opportunities relate especially to the link between the gut microbiota and various human diseases, including metabolic syndrome (obesity, type 2 diabetes and cardiovascular disease), Crohn's disease and related immunological dysfunction (Lee and Mazmanian, 2010; Wang et al., 2011). The causal networks underpinning these diseases are difficult to disentangle because the mammalian microbiota is complex and variable. Various insect systems have potential value. Drosophila melanogaster offers a particularly strong model because the low diversity of its gut microbiota (Cox and Gilmore, 2007; Roh et al., 2008; Wong et al., 2011) can be harnessed with the unparalleled set of Drosophila genetic resources and tools. Insect symbioses can certainly provide many valuable lessons to be learnt, but their greatest value is not yet realized. We should anticipate a shift in the curriculum towards the explicit exploitation of insect symbioses as models for biomedical research on the role of the resident microbiota in health and disease.
ACKNOWLEDGEMENTS
This work was supported by NIH grant 1R01GM095372-01, NSF grant IOS-0919765, and the Sarkaria Institute for Insect Physiology and Toxicology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Aanen DK, Hoekstra RF. The evolution of obligate mutualism: if you can't beat 'em, join 'em. Trends Ecol Evol. 2007;22:506–509. doi: 10.1016/j.tree.2007.08.007. [DOI] [PubMed] [Google Scholar]
- Bakula M. The persistence of a microbial flora during postembryogenesis of Drosophila melanogaster. Journal of Invertebrate Pathology. 1969;14:365–374. doi: 10.1016/0022-2011(69)90163-3. [DOI] [PubMed] [Google Scholar]
- Brugerolle G, Radek R. Symbiotic protozoa of termites. Soil Biol. 2006;6:243–269. [Google Scholar]
- Brummel T, Ching A, Seroude L, Simon AF, Benzer S. Drosophila lifespan enhancement by exogenous bacteria. Proc Natl Acad Sci U S A. 2004;101:12974–12979. doi: 10.1073/pnas.0405207101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnum KE, Callister SJ, Nicora CD, Purvine SO, Hugenholtz P, Warnecke F, Scheffrahn RH, Smith RD, Lipton MS. Proteome insights into the symbiotic relationship between a captive colony of Nasutitermes corniger and its hindgut microbiome. ISME Journal. 2011;5:161–164. doi: 10.1038/ismej.2010.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler SM, Wilkinson TL, Douglas AE. Impact of plant nutrients on the relationship between a herbivorous insect and its symbiotic bacteria. Proceedings of the Royal Society B-Biological Sciences. 2008;275:565–570. doi: 10.1098/rspb.2007.1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirimotich CM, Dong Y, Garver LS, Sim S, Dimopoulos G. Mosquito immune defenses against Plasmodium infection. Dev Comp Immunol. 2010;34:387–395. doi: 10.1016/j.dci.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connell JH. Diversity in tropical rain forests and coral reefs. Science. 1978;199:1302–1310. doi: 10.1126/science.199.4335.1302. [DOI] [PubMed] [Google Scholar]
- Cox CR, Gilmore MS. Native microbial colonization of Drosophila melanogaster and its use as a model of Enterococcus faecalis pathogenesis. Infect Immun. 2007;75:1565–1576. doi: 10.1128/IAI.01496-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale C, Moran NA. Molecular interactions between bacterial symbionts and their hosts. Cell. 2006;126:453–465. doi: 10.1016/j.cell.2006.07.014. [DOI] [PubMed] [Google Scholar]
- Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449:811–818. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon RJ, Dillon VM. The gut bacteria of insects: nonpathogenic interactions. Annu Rev Entomol. 2004;49:71–92. doi: 10.1146/annurev.ento.49.061802.123416. [DOI] [PubMed] [Google Scholar]
- Dong Y, Manfredini F, Dimopoulos G. Implication of the mosquito midgut microbiota in the defense against malaria parasites. PLoS Pathog. 2009;5:e1000423. doi: 10.1371/journal.ppat.1000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas AE. Symbiotic microorganisms: untapped resources for insect pest control. Trends in Biotechnology. 2007;25:338–342. doi: 10.1016/j.tibtech.2007.06.003. [DOI] [PubMed] [Google Scholar]
- Douglas AE. The microbial dimension in insect nutritional ecology. Functional Ecology. 2009;23:38–47. [Google Scholar]
- Douglas AE, Bouvaine S, Russell RR. How the insect immune system interacts with an obligate symbiotic bacterium. Proc Biol Sci. 2010 doi: 10.1098/rspb.2010.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan J, Chung H, Troy E, Kasper DL. Microbial colonization drives expansion of IL-1 receptor 1-expressing and IL-17-producing gamma/delta T cells. Cell Host Microbe. 2010;7:140–150. doi: 10.1016/j.chom.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald PW. Oxford University Press. Oxford; UK: 1994. Evolution of Infectious Diseases. [Google Scholar]
- Falk PG, Hooper LV, Midtvedt T, Gordon JI. Creating and maintaining the gastrointestinal ecosystem: what we know and need to know from gnotobiology. Microbiol Mol Biol Rev. 1998;62:1157–1170. doi: 10.1128/mmbr.62.4.1157-1170.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraune S, Augustin R, Anton-Erxleben F, Wittlieb J, Gelhaus C, Klimovich VB, Samoilovich MP, Bosch TC. In an early branching metazoan, bacterial colonization of the embryo is controlled by maternal antimicrobial peptides. Proc Natl Acad Sci U S A. 2010;107:18067–18072. doi: 10.1073/pnas.1008573107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukatsu T, Hosokawa T. Capsule-transmitted gut symbiotic bacterium of the Japanese common plataspid stinkbug, Megacopta punctatissima. 2002;68:389–396. doi: 10.1128/AEM.68.1.389-396.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garver LS, Xi Z, Dimopoulos G. Immunoglobulin superfamily members play an important role in the mosquito immune system. Dev Comp Immunol. 2008;32:519–531. doi: 10.1016/j.dci.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerardo NM, Altincicek B, Anselme C, Atamian H, Barribeau SM, de Vos M, Duncan EJ, Evans JD, Gabaldon T, Ghanim M. Immunity and other defenses in pea aphids, Acyrthosiphon pisum. Genome Biol. 2010;11:R21. doi: 10.1186/gb-2010-11-2-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunduz EA, Douglas AE. Symbiotic bacteria enable insect to use a nutritionally inadequate diet. Proceedings of the Royal Society B-Biological Sciences. 2009;276:987–991. doi: 10.1098/rspb.2008.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen AK, Moran NA. Aphid genome expression reveals host-symbiont cooperation in the production of amino acids. Proc Natl Acad Sci U S A. 2011;108:2849–2854. doi: 10.1073/pnas.1013465108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges LM, Brownlie JC, O'Neill SL, Johnson KN. Wolbachia and virus protection in insects. Science. 2008;322:702. doi: 10.1126/science.1162418. [DOI] [PubMed] [Google Scholar]
- Hosokawa T, Kikuchi Y, Fukatsu T. How many symbionts are provided by mothers, acquired by offpsring, and needed for successful vertical transmission in an obligate insect-bacterium mutualism? Mol Ecol. 2007;16:5316–5325. doi: 10.1111/j.1365-294X.2007.03592.x. [DOI] [PubMed] [Google Scholar]
- Hussain M, Frentiu FD, Moreira LA, O'Neill SL, Asgari S. Wolbachia uses host microRNAs to manipulate host gene expression and facilitate colonization of the dengue vector Aedes aegypti. Proc Natl Acad Sci U S A. 2011;108:9250–9255. doi: 10.1073/pnas.1105469108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Aphid Genomics Consortium Genome sequence of the pea aphid Acyrthosiphon pisum. PLOS Biology. 2010;8:e1000313. doi: 10.1371/journal.pbio.1000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenike J, Unckless R, Cockburn SN, Boelio LM, Perlman SJ. Adaptation via symbiosis: recent spread of a Drosophila defensive symbiont. Science. 2010;329:212–215. doi: 10.1126/science.1188235. [DOI] [PubMed] [Google Scholar]
- Kaltenpoth M, Gottler W, Herzner G, Strohm E. Symbiotic bacteria protect wasp larvae from fungal infestation. Curr Biol. 2005;15:475–479. doi: 10.1016/j.cub.2004.12.084. [DOI] [PubMed] [Google Scholar]
- Kerney R, Kim E, Hangarter RP, Heiss AA, Bishop CD, Hall BK. Intracellular invasion of green algae in a salamander host. Proc Natl Acad Sci U S A. 2011;108:6497–6502. doi: 10.1073/pnas.1018259108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer N, Voronin D, Charif D, Mavingui P, Mollereau B, Vavre F. Wolbachia interferes with ferritin expression and iron metabolism in insects. PLoS Pathog. 2009;5:e1000630. doi: 10.1371/journal.ppat.1000630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroiss J, Kaltenpoth M, Schneider B, Schwinger MG, Hertweck C, Maddula RK, Strohm E, Svatos A. Symbiotic Streptomycetes provide antibiotic combination prophylaxis for wasp offspring. Nat Chem Biol. 2010;6:261–263. doi: 10.1038/nchembio.331. [DOI] [PubMed] [Google Scholar]
- Lee YK, Mazmanian SK. Has the microbiota played a critical role in the evolution of the adaptive immune system? Science. 2010;330:1768–1773. doi: 10.1126/science.1195568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annu Rev Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- Macdonald SJ, Thomas GH, Douglas AE. Genetic and metabolic determinants of nutritional phenotype in an insect-bacterial symbiosis. Mol Ecol. 2011;20:2073–2084. doi: 10.1111/j.1365-294X.2011.05031.x. [DOI] [PubMed] [Google Scholar]
- McCutcheon JP, McDonald BR, Moran NA. Convergent evolution of metabolic roles in bacterial co-symbionts of insects. Proc Natl Acad Sci U S A. 2009;106:15394–15399. doi: 10.1073/pnas.0906424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JP, Moran NA. Parallel genomic evolution and metabolic interdependence in an ancient symbiosis. Proc Natl Acad Sci U S A. 2007;104:19392–19397. doi: 10.1073/pnas.0708855104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JP, Moran NA. Functional convergence in reduced genomes of bacterial symbionts spanning 200 My of evolution. Genome Biol Evol. 2010;2:708–718. doi: 10.1093/gbe/evq055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall-Ngai M. Unseen forces: the influence of bacteria on animal development. Developmental Biology. 2002;242:1–14. doi: 10.1006/dbio.2001.0522. [DOI] [PubMed] [Google Scholar]
- McFall-Ngai M. Adaptive immunity: care for the community. Nature. 2007;445:153. doi: 10.1038/445153a. [DOI] [PubMed] [Google Scholar]
- McGraw EA, Merritt DJ, Droller JN, O'Neill SL. Wolbachia density and virulence attenuation after transfer into a novel host. Proc Natl Acad Sci U S A. 2002;99:2918–2923. doi: 10.1073/pnas.052466499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Jimenez J, Zuniga G, Villa-Tanaca L, Hernandez-Rodriguez C. Bacterial community and nitrogen fixation in the red turpentine beetle, Dendroctonus valens LeConte (Coleoptera: Curculionidae: Scolytinae) Microb Ecol. 2009;58:879–891. doi: 10.1007/s00248-009-9548-2. [DOI] [PubMed] [Google Scholar]
- Moran NA, Munson MA, Baumann P, Ishikawa H. A molecular clock in endosymbiotic bacteria is calibrated using the insect hosts. Proceedings of the Royal Society of London B. 1993;253:167–171. [Google Scholar]
- Moran NA, Tran P, Gerardo NM. Symbiosis and insect diversification: an ancient symbiont of sap-feeding insects from the bacterial phylum Bacteroidetes. Appl Environ Microbiol. 2005;71:8802–8810. doi: 10.1128/AEM.71.12.8802-8810.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller UG, Dash D, Rabeling C, Rodrigues A. Coevolution between attine ants and actinomycete bacteria: a reevaluation. Evolution. 2008;62:2894–2912. doi: 10.1111/j.1558-5646.2008.00501.x. [DOI] [PubMed] [Google Scholar]
- Nemergut DR, Costello EK, Hamady M, Lozupone C, Jiang L, Schmidt SK, Fierer N, Townsend AR, Cleveland CC, Stanish L. Global patterns in the biogeography of bacterial taxa. Environ Microbiol. 2011;13:135–144. doi: 10.1111/j.1462-2920.2010.02315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niess JH, Leithauser F, Adler G, Reimann J. Commensal gut flora drives the expansion of proinflammatory CD4 T cells in the colonic lamina propria under normal and inflammatory conditions. J Immunol. 2008;180:559–568. doi: 10.4049/jimmunol.180.1.559. [DOI] [PubMed] [Google Scholar]
- Oh DC, Poulsen M, Currie CR, Clardy J. Dentigerumycin: a bacterial mediator of an ant-fungus symbiosis. Nat Chem Biol. 2009;5:391–393. doi: 10.1038/nchembio.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh PL, Benson AK, Peterson DA, Patil PB, Moriyama EN, Roos S, Walter J. Diversification of the gut symbiont Lactobacillus reuteri as a result of host-driven evolution. ISME J. 2010;4:377–387. doi: 10.1038/ismej.2009.123. [DOI] [PubMed] [Google Scholar]
- Oliver KM, Degnan PH, Burke GR, Moran NA. Facultative symbionts in aphids and the horizontal transfer of ecologically important traits. Annu Rev Entomol. 2010;55:247–266. doi: 10.1146/annurev-ento-112408-085305. [DOI] [PubMed] [Google Scholar]
- Oliver KM, Degnan PH, Hunter MS, Moran NA. Bacteriophages encode factors required for protection in a symbiotic mutualism. Science. 2009;325:992–994. doi: 10.1126/science.1174463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannebakker BA, Loppin B, Elemans CP, Humblot L, Vavre F. Parasitic inhibition of cell death facilitates symbiosis. Proc Natl Acad Sci U S A. 2007;104:213–215. doi: 10.1073/pnas.0607845104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliakov A, Russell CW, Ponnala L, Hoops HJ, Sun Q, Douglas AE, van Wijk KJ. Large-scale label-free quantitative proteomics of the pea aphid-Buchnera symbiosis. Mol Cell Proteomics. 2011 doi: 10.1074/mcp.M110.007039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard M, Sharon N. Responses of the Peyer's Patches in Germ-Free Mice to Antigenic Stimulation. Infect Immun. 1970;2:96–100. doi: 10.1128/iai.2.1.96-100.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren C, Webster P, Finkel SE, Tower J. Increased internal and external bacterial load during Drosophila aging without life-span trade-off. Cell Metab. 2007;6:144–152. doi: 10.1016/j.cmet.2007.06.006. [DOI] [PubMed] [Google Scholar]
- Reymond N, Calevro F, Viñuelas J, Morin N, Rahbé Y, Febvay G, Laugier C, Douglas AE, Fayard J-M, Charles H. Different levels of transcriptional regulation due to trophic constraints in the reduced genome of Buchnera aphidicola APS. Applied and Environmental Microbiology. 2006;72:7760–7766. doi: 10.1128/AEM.01118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson CJ, Schloss P, Ramos Y, Raffa K, Handelsman J. Robustness of the bacterial community in the cabbage white butterfly larval midgut. Microb. Ecol. 2010;59:199–211. doi: 10.1007/s00248-009-9595-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh SW, Nam YD, Chang HW, Kim KH, Kim MS, Ryu JH, Kim SH, Lee WJ, Bae JW. Phylogenetic characterization of two novel commensal bacteria involved with innate immune homeostasis in Drosophila melanogaster. Appl Environ Microbiol. 2008;74:6171–6177. doi: 10.1128/AEM.00301-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round JL, Lee M, Li J, Tran G, Jabri B, Chatila TA, Mazmanian SK. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332:974–977. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolff J. Why did the acquired immune system of vertebrates evolve? Dev Comp Immunol. 2007;31:476–482. doi: 10.1016/j.dci.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Ryu JH, Kim SH, Lee HY, Bai JY, Nam YD, Bae JW, Lee DG, Shin SC, Ha EM, Lee WJ. Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science. 2008;319:777–782. doi: 10.1126/science.1149357. [DOI] [PubMed] [Google Scholar]
- Sackton TB, Lazzaro BP, Schlenke TA, Evans JD, Hultmark D, Clark AG. Dynamic evolution of the innate immune system in Drosophila. Nat Genet. 2007;39:1461–1468. doi: 10.1038/ng.2007.60. [DOI] [PubMed] [Google Scholar]
- Scarborough CL, Ferrari J, Godfray HC. Aphid protected from pathogen by endosymbiont. Science. 2005;310:1781. doi: 10.1126/science.1120180. [DOI] [PubMed] [Google Scholar]
- Scott JJ, Oh DC, Yuceer MC, Klepzig KD, Clardy J, Currie CR. Bacterial protection of beetle-fungus mutualism. Science. 2008;322:63. doi: 10.1126/science.1160423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanbhag S, Tripathi S. Epithelial ultrastructure and cellular mechanisms of acid and base transport in the Drosophila midgut. J Exp Biol. 2009;212:1731–1744. doi: 10.1242/jeb.029306. [DOI] [PubMed] [Google Scholar]
- Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature. 2000;407:81–86. doi: 10.1038/35024074. [DOI] [PubMed] [Google Scholar]
- Stappenbeck TS, Hooper LV, Gordon JI. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc Natl Acad Sci U S A. 2002;99:15451–15455. doi: 10.1073/pnas.202604299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel KJ, Moran NA. Sources of variation in dietary requirements in an obligate nutritional symbiosis. Proc Biol Sci. 2011;278:115–121. doi: 10.1098/rspb.2010.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, Feldstein AE, Britt EB, Fu X, Chung YM. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnecke F, Luginbuhl P, Ivanova N, Ghassemian M, Richardson TH, Stege JT, Cayouette M, McHardy AC, Djordjevic G, Aboushadi N. Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature. 2007;450:560–565. doi: 10.1038/nature06269. [DOI] [PubMed] [Google Scholar]
- Watanabe H, Tokuda G. Cellulolytic systems in insects. Annu Rev Entomol. 2010;55:609–632. doi: 10.1146/annurev-ento-112408-085319. [DOI] [PubMed] [Google Scholar]
- Werren JH, Baldo L, Clark ME. Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol. 2008;6:741–751. doi: 10.1038/nrmicro1969. [DOI] [PubMed] [Google Scholar]
- Wieczorek H, Beyenbach KW, Huss M, Vitavska O. Vacuolar-type proton pumps in insect epithelia. J Exp Biol. 2009;212:1611–1619. doi: 10.1242/jeb.030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson AC, Dunbar HE, Davis GK, Hunter WB, Stern DL, Moran NA. A dual-genome microarray for the pea aphid, Acyrthosiphon pisum, and its obligate bacterial symbiont, Buchnera aphidicola. BMC Genomics. 2006;7:50. doi: 10.1186/1471-2164-7-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CN, Ng P, Douglas AE. Low-diversity bacterial community in the gut of the fruitfly Drosophila melanogaster. Environ Microbiol. 2011 doi: 10.1111/j.1462-2920.2011.02511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Vilchez I, Mateos M. Spiroplasma bacteria enhance survival of Drosophila hydei attacked by the parasitic wasp Leptopilina heterotoma. PLoS One. 2010;5:e12149. doi: 10.1371/journal.pone.0012149. [DOI] [PMC free article] [PubMed] [Google Scholar]