Abstract
Adeno-associated virus type 2 (AAV) is a popular vector for human gene therapy, because of its safety record and ability to express genes long term. Yet large scale recombinant (r)AAV production remains problematic due to low particle yield. The adenovirus (Ad) and herpes (simplex) virus (HSV) helper genes for AAV have been widely used and studied, but the helper genes of human papillomavirus (HPV) have not. HPV-16 E1, E2 and E6 help wild type (wt) AAV productive infection in differentiating keratinocytes, however HEK293 cells are the standard cell line used for generating rAAV. Here we demonstrate that the three HPV genes were unable to stimulate significant rAAV replication in HEK293 cells when used alone. However, when used in conjunction (complementation) with the standard Ad5 helper gene set, E1, E2 and E6 were each capable of significantly boosting rAAV DNA replication and virus particle yield. Moreover, wt AAV DNA replication and virion yield were also significantly boosted by each HPV gene along with wt Ad5 virus co-infection. Mild to moderate changes in rep- and cap–encoded protein levels were evident in the presence of the E1, E2 and E6 genes. Higher wt AAV DNA replication was not matched by similar increases in the levels of rep-encoded protein. Moreover, while rep mRNA was up-regulated, cap mRNA was up-regulated more. Higher virus yields did correlate most consistently with increased Rep52, VP3 and VP-related 21/31 kDa species. The observed boost in wt and rAAV production by HPV genes was not unexpected, as the Ad and HPV helper gene sets do not seem to recapitulate each other. These results raise the possibility of generating improved helper gene sets derived from both the Ad and HPV helper gene sets.
Introduction
First used in 1984, adeno-associated virus (AAV) has become one of the most used vectors for gene delivery/gene therapy due to its safety and long term transgene delivery (1-4). AAV has a 4.7 kb single-stranded DNA genome containing two major open reading frames (ORF) (5). The left ORF, rep, and encodes the replication (Rep) proteins, while the right ORF, cap, encodes the capsid (VP) proteins (6,7), and the genome is flanked by 145bp inverted terminal repeats (ITR) which are cis required for DNA replication, packaging, chromosomal integration and rescue (8). Thus, only the ITRs are needed for rAAV production and all of the intervening sequences can be replaced by foreign DNA sequences.
While AAV will autonomously replicate in differentiating keratinocytes (9-11), taxonomically AAV is a Dependovirus, a genus of Parvoviridae. This categorization reflects AAV’s requirement for co-infection with a helper virus, such as adenovirus (Ad), herpes simplex virus (HSV), or human papillomavirus (HPV) in order for productive AAV infection to occur (10,12,13). Thus AAV receives important activities it lacks from a set of “helper genes”. The understanding and optimal use of these helper gene sets would be beneficial for both maximal recombinant (r) and wild type (wt) AAV production. The complexity of the helper gene sets plus the complexity of AAV’s own rep and cap gene expression make generating high titers of recombinant AAV a challenge for the field of gene therapy. Developing new methods for generating higher titers of recombinant AAV would be highly beneficial.
The adenovirus helper gene set is the most often used for generating recombinant (r)AAV. Studies on adenovirus have identified E1A, E1B, E2A, E4orf6, and the VA RNA as the critical helper genes. E1A, an oncogene, helps drive the cell into S phase for viral replication and transcriptionally activates the AAV p5 promoter (14-17). E1B and E4orf6, also oncogenes, interact as a heterodimer and, again, helps drive the cell into S phase (16,18-22). E4orf6 also modules components of DNA repair, which may also influence AAV DNA replication (23). The E2A is a SS DNA binding protein which is involved in up-regulating AAV gene expression, particularly the AAV capsid proteins (24-30). However, splicing of the AAV transcripts is greatly affected, lowered, without E2A. The VA RNA is also important in up-regulating AAV gene expression (23-29).
While the Ad (and HSV) helper genes have been well studied the HPV helper genes have not. Moreover, the HPV16 helper genes (E1, E2 and E6) have been studied in differentiating keratinocytes, cells very different than HEK293 cells normally used for generating rAAV (11). It is interesting that the HPV and Ad helper gene sets do not recapitulate one another, with the exception of the HPV16 E6 oncogene (30-32). The HPV16 E1 gene is an origin of HPV DNA replication binding protein, analogous to the AAV Rep78/Rep68 proteins, but without the endonuclease and covalent attachment activities of the large Rep proteins (30-33). Moreover HPV16 E1 is known to bind the AAV rep encoded replication protein Rep78 and modulate its replication-related biochemistries (34,35). HPV16 E2 is a DNA binding transcription factor (36-38). The limited overlap of functions between the two helper gene sets suggest the possibility of complementation. Here we demonstrate that the HPV16 E1, E2 or E6 helper genes individually lack the capability to stimulate rAAV DNA replication in HEK293 cells. However these same genes are capable of significantly enhancing rAAV DNA replication and virion production in collaboration with the full Ad helper gene set.
RESULTS
Generation of HEK293 cell lines carrying HPV-16 E1, E2 and E6
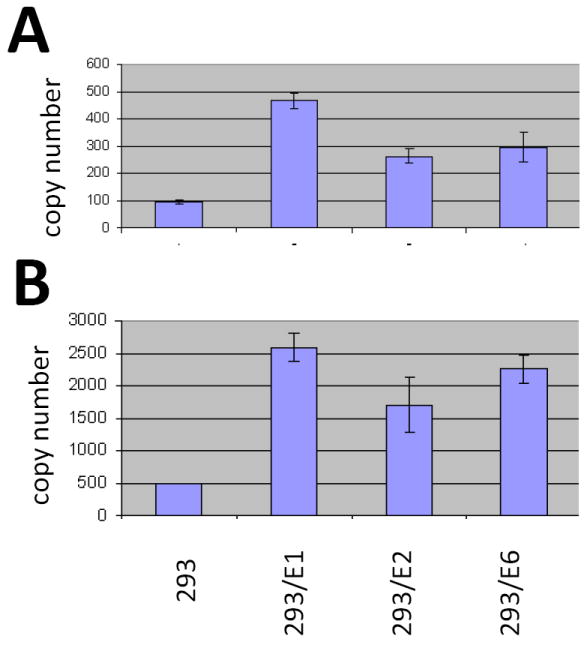
HEK293 cells were infected with AAV/E1/Neo, AAV/E2/Neo and AAV/E6/Neo, and G418 selected as described in the Material and Methods section. Each infection resulted in over 30 G418-resistant colonies and these cultures were grown as bulk cultures and used in all subsequent experiments as such and referred to as 293/E1, 293/E2 and 293/E6. These cells were generated in such a manner that many of the clones would rescue and replicate the AAV/helper gene provirus upon challenge with the Ad/AAV helper genes. To test this issue the cells were transfected with the Ad/AAV helper plasmid pSH3 (similar to pDG), Hirt DNA isolated at three days post-transfection and analyzed by PCR for the specific HPV helper gene DNA, as shown in Figures 1A-C. As seen, all three cell lines contained the appropriate HPV gene, as indicated by the appearance of the appropriately sized PCR product, and lacked the other HPV genes. Total cellular DNA of these cells were also analyzed by Q-PCR for Neo gene (transgene) DNA copy number and all contained relatively comparable levels of transgene DNA, as shown in Supplementary Figure 1.
Figure 1. Analysis of HPV sequences present in the 293-E1, 293-E2 and 293-E6 cell lines.
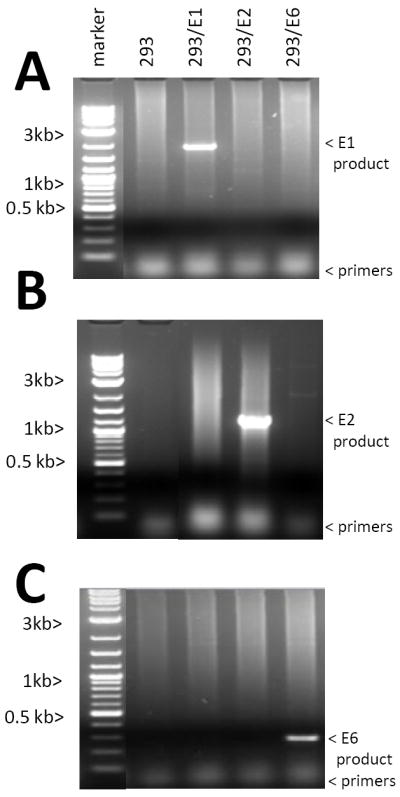
The three HPV+ cell lines were generated as described in the Materials and Methods. After pSH3 plasmid transfection Hirt DNA was extracted and analyzed by PCR for the HPV-16 E1, E2 and E6 sequences. Note that each cell line was positive for the expected HPV gene, but was negative for the others.
The individual E1, E2 and E6 genes don’t provide full helper function in HEK293 cells
We have reported that these three HPV genes serve as helper genes for wild type (wt) AAV replication in differentiating keratinocytes, AAV’s natural host tissue (11). However, their activity in HEK293 cells is unknown. To test this the three 293/HPV cell lines were transfected with a defective AAV vector plasmid (2μg)(pAAV/eGFP) plus a Rep78 expression plasmid (2μg)(pKEX-Rep78). The 293/E1 cell line was also transfected with pSH3 as a positive control. After five days low molecular weight Hirt DNA was isolated from the cells and analyzed for replication by Southern blot using 32P-eGFP DNA. As seen in Figure 2, none of the 293-HPV helper cell lines demonstrated significant rAAV production as determined by a monomer duplex or single stranded DNA-sized band. However, the 293/E2 and 293/E6 lanes show low levels of very high molecular weight species (possibly representing rolling circle replication).
Figure 2. Analysis of AAV helper function of 293-E1, 293-E2 and 293-E6 cell lines without the Ad5 helper gene set.
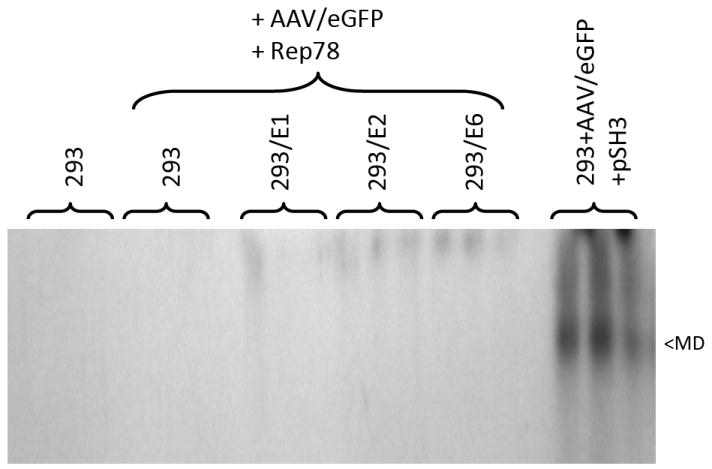
The three HPV+ cell lines, and control 293 cells, were transfected with AAV/eGFP plasmid as described in the Materials and Methods and the Rep78 expression plasmid pKEX-Rep78. Several days later Hirt DNA was extracted and analyzed by Southern blot, probed with 32P-eGFP sequences. Hirt DNA from 293 cells transfected AAV/eGFP plus pSH3 served as a positive control. Note that none of the HPV cell lines generated significant levels of AAV/eGFP replication. “MD” indicates the position of monomer duplex rAAV DNA. “SS” indicates the position of single stranded rAAV DNA. Each transfection was done in triplicate.
Higher rAAV DNA/virion production by E1, E2 and E6 with Ad/AAV helper plasmid
We next assayed if the three 293-HPV cell lines showed an advantage over control HEK293 cells in the generation of rAAV in conjunction with the Ad5 helper gene set, provided by pSH3. To test this, the three cell lines were transfected with pAAV/eGFP (2μg) plus pSH3 (2μg). After five days low molecular weight Hirt DNA was isolated from the cells and analyzed for replication levels by Southern blot using 32P-labeled eGFP DNA. As seen in Figure 3A and quantified in 3B, 293/E1, 293/E2 and 293/E6 all allowed for higher levels of rAAV/eGFP DNA replication than control HEK293 cells. Thus, E1, E2 and E6, each displayed helper activity for rAAV DNA replication. We also obtained similar results when pDG8 Ad/AAV helper plasmid was used in place of pSH3 (data not shown).
Figure 3. HPV16 E1, E2 and E6 serve as helper genes, enhancing rAAV DNA replication and virion production.
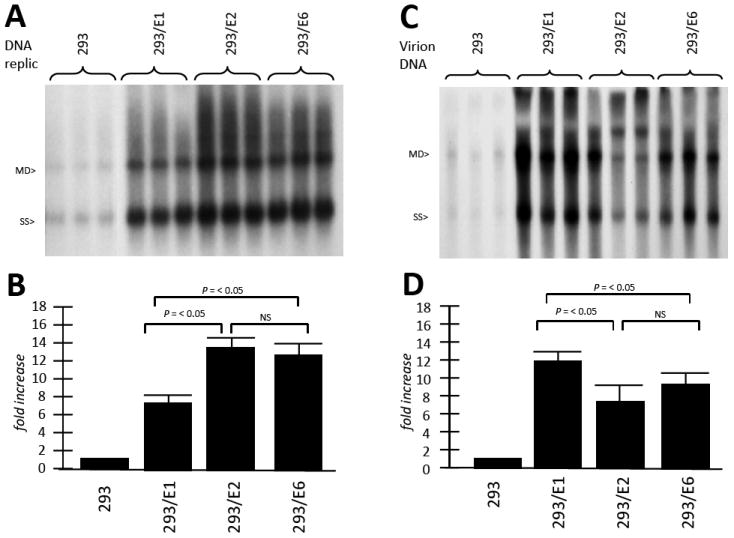
A. The three HPV+ cell lines, and control 293 cells, were transfected with AAV/eGFP plus pSH3 (Ad/AAV helper plasmid) as described in the Materials and Methods. Several days later Hirt DNA was extracted and analyzed by Southern blot, probed with 32P-eGFP sequences. Note that E1, E2 and E6 each enhanced rAAV/eGFP DNA replication. This enhancement was densitometrically quantitated in panel B. In C, DNA from the resulting virus stock was titered as described in the Material and Methods, again, by Southern blot, probed with 32P-eGFP sequences. Note that, again, E1, E2 and E6 each enhanced rAAV/eGFP virion production. The enhancement of rAAV was densitometrically quantitated in panel D. Each transfection was done in triplicate.
We next assayed if the three 293-HPV cell lines also exhibited increased rAAV virion production. Thus, an identical experiment was carried out as just above with the exception that the cells were freeze/thawed three times, cellular debris pelleted, then filtered, and the supernatant analyzed for the level of AAV/eGFP encapsidated genomes (eg) as described in the Materials and Methods section. As seen in Figure 3C and quantified in 3D, 293/E1, 293/E2 and 293/E6 all allowed for higher levels of rAAV/eGFP virion DNA than control HEK293 cells. The amount of rAAV virus produced by the four different cell lines, 293, 293/E1, 293/E2, 293/E6, were approximately 1.4×1010, 2×1011, 1.3×1011, and 1.6×1011, respectively, per 10 cm plate, transfected with 2 μgs each of vector and helper plasmid (Supplementary Figure 2). Thus, E1, E2 and E6, each displayed helper activity for rAAV virion production. Analysis of the relative infectivity of AAV/eGFP virus produced by each cell line, equalized for the same MOI and measured by fluorescence, showed that infectivity was similar (Supplementary Figure 3).
Higher wt AAV replication by E1, E2 and E6 during wt Ad5 coinfection
While rAAV generation is very important for gene therapy use, the study of the effects on natural wt AAV replication will give us more detailed information on how HPV E1, E2, and E6 regulate the AAV life cycle. To test this, the three cell lines were infected with wt AAV and Ad5 at a multiplicity of 10, with unaltered HEK293 cells serving as control. After two days low molecular weight Hirt DNA was isolated from the cells and analyzed for replication by Southern blot using 32P-rep DNA. As seen in Figure 4A and quantified in 4B, 293/E1, 293/E2 and 293/E6 all allowed for higher levels of wt AAV DNA replication than control HEK293 cells. Thus, E1, E2 and E6, each displayed helper activity for rAAV DNA replication. Wt AAV virion production was also analyzed, shown in Figure 4C and quantified in 4D, and also demonstrate significant helper function by the three HPV genes.
Figure 4. HPV16 E1, E2 and E6 serve as helper genes, enhancing wt AAV DNA replication and virion production.
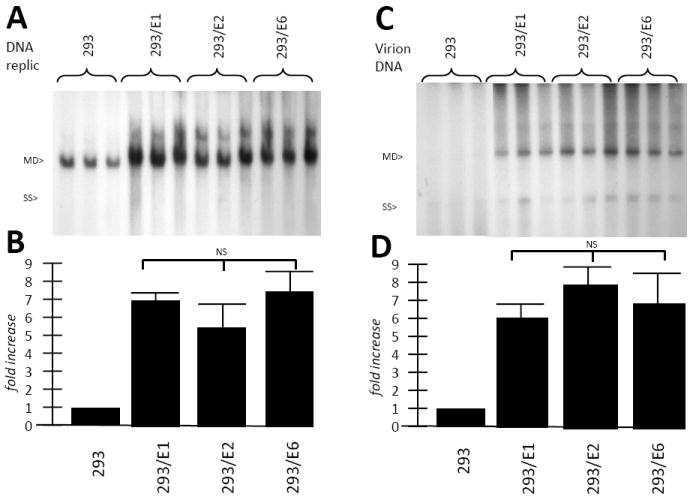
A. The three HPV+ cell lines, and control 293 cells, were infected with AAV2 and Ad5 at an moi of 10 as described in the Materials and Methods. Several days later Hirt DNA was extracted and analyzed by Southern blot, probed with 32P-rep sequences. Note that E1, E2 and E6 each enhanced wt AAV DNA replication. This enhancement was densitometrically quantitated in panel B. In C, DNA from the resulting virus stock was titered as described in the Material and Methods, again, by Southern blot, probed with 32P-rep sequences. Note that, again, E1, E2 and E6 each enhanced wt AAV virion production. The enhancement of rAAV was densitometrically quantitated in panel D. Each infection was done in triplicate.
E1, E2 and E6 are associated with altered wt AAV mRNA and protein levels
The effects of E1, E2 and E6 on wt AAV mRNA levels were also analyzed. RNA isolated from equivalently treated plates as in Figure 4, and analyzed by quantitative RNA as described in the Materials and Methods section, and the results are shown in Figure 5A for rep mRNA, and 5B for cap mRNA, compared to β-actin housekeeping control expression. As can be seen, all three HPV genes enhanced both rep and cap mRNA levels, with cap levels enhanced slightly more than rep.
Figure 5. HPV16 E1, E2 and E6 modulate wt AAV mRNA expression.
A. The three HPV+ cell lines, and control 293 cells, were transfected with AAV/eGFP plus pSH3 (Ad/AAV helper plasmid) as described in the Materials and Methods. Several days later mRNA was extracted and analyzed by Q-PCR for rep RNA expression. Note that E1, E2 and E6 enhanced rep mRNA levels 2.5-5 fold. B shows a similar Q-PCR analysis for cap mRNA levels, and show a corresponding enhancement of cap mRNA levels by E1, E2 and E6.
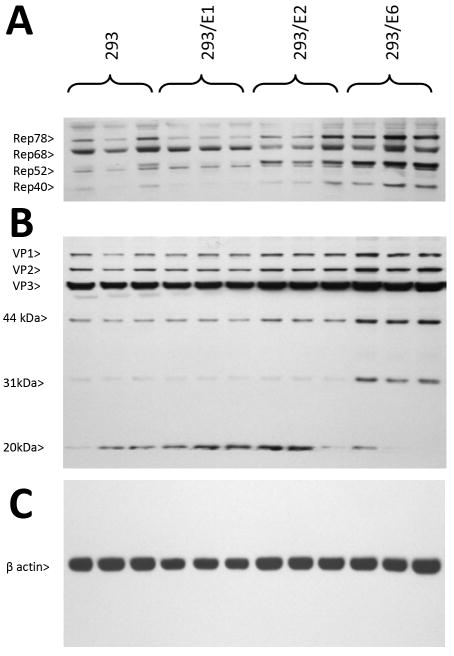
To study helper gene effects on wt AAV protein levels, again, equivalent plates (as in Figure 4 and 5) were analyzed by Western blot for Rep (rep) and VP (cap) proteins. The Western blot, Figure 6A, probed with anti-Rep antibodies, shows that Rep68 and Rep78 were present in all lanes. However differences were also noticeable. The predominant Rep in unaltered 293 and 293/E1 cells was Rep68, with low Rep78, Rep52 and Rep40. E2 helper activity resulted in a more balanced levels of these three proteins but largely lacked Rep40. 293/E6 cells displayed higher levels of all Rep proteins, including Rep40.
Figure 6. HPV16 E1, E2 and E6 modulate wt AAV protein expression.
A. The three HPV+ cell lines, and control 293 cells, were transfected with AAV/eGFP plus pSH3 (Ad/AAV helper plasmid) as described in the Materials and Methods. Several days later total protein was extracted and analyzed by Western blot rep- encoded protein expression using anti-Rep antibody. Note that E1, E2 and E6 had variable effects Rep protein expression. B shows a similar Western blot analysis for cap-encoded VP proteins. Note that VP protein expression was modestly affected. C shows a loading control probed with anti-β actin antibody. Each infection was done in triplicate.
The Western blot, Figure 6B, was probed with anti-VP antibodies, and shows that VP1-3 was lowest in control HEK293 cells, higher in 293/E1 cells, and highest in 293/E2 and 293/E6 cells. Moreover, smaller but previously uncharacterized VP proteins showed differential expression. VP-44kDa showed progressively higher expression levels from control HEK293 cells through 293-E1 and 293/E2, with 293/E6 expressing the highest level. VP-31kDa showed almost exclusive expression in 293/E6 cells. Finally, 293/E1 and 293/E2 cells showed higher levels of VP-20kDa protein over control HEK293 cells, but 293-E6 had little of this species. A β-actin control is shown in Figure 6C.
Discussion
Previously we have shown that HPV serves as a helper virus for AAV in differentiating keratinocytes and that the HPV E1, E2 and E6 genes are the helper genes (10,11). Here we demonstrate that these same HPV genes, while unable to provide full helper function individually in HEK293 cells, are able to complement the Ad helper gene set, resulting in significant enhancement of rAAV and wt AAV production (Figures 3 and 4). While this study is limited to HPV16 it is likely that, with the exception of E6, the analogous genes of the other HPV types can carry out similar helper activities. The E1 and E2 genes have largely been shown to have similar functions among the papillomaviruses, whereas E6 appears more pleomorphic in function. The generation of rAAV is very important, and all three HPV helper genes were able to enhance rAAV production. However, wt AAV replication with wt Ad5 in HEK293 cells is the most studied model of the natural life cycle of AAV. The analysis of E1, E2 and E6 phenotypes on wt AAV2 plus wt Ad5 co-infection gives us more meaningful and detailed data on the helper effects of each HPV gene. During Ad/AAV co-infections all three HPV helper genes upregulated rep and cap mRNA levels (Figure 5). Western blot analysis of AAV proteins identified multiple mild to moderate changes in the ratios and levels of certain rep- and cap–encoded proteins in the presence of the E1, E2 and E6 genes. It was interesting that higher wt AAV DNA replication was not matched by similar increases in the levels of rep-encoded proteins.
During wt AAV/Ad coinfection E1 helper activity correlated with higher rep and cap mRNA, however, a corresponding increase in levels of these proteins was not observed. This suggests that E1, while promoting the highest AAV transcription, may selectively inhibit translation of these same rep mRNAs. Moreover the ratio of the rep-encoded proteins was roughly the same as the HPV-negative control. Interestingly, a small truncated 20kDa VP protein (Figure 6B) appeared at higher levels in the presence of E1. E2 helper activity also correlated with higher rep and cap mRNA expression, but not as high as E1. The increases rep and cap mRNA did correlate more closely with higher protein levels, in particular for the cap-encoded VPs. It is unclear if any effect on translation is seen. The small 20kDa VP protein appeared at even higher levels in the presence of E2.
E6 helper activity also correlated with higher rep and cap mRNA expression, intermediate between E1 and E2. Among the HPV helper genes the E6-induced increase in AAV mRNA and proteins appeared to correlate best. Significant increases in both rep and cap-encoded proteins were observed. Significant levels of all four Rep proteins were seen, now including Rep40. Thus E6 appears to have no regulatory effect on translation. With the E6 gene the VP proteins are also observed to be at the highest levels of the study, with VP1, VP2, and VP3 all up-regulated. However, this up-regulation in VP proteins did not correspond with increased virion production compared to the other HPV helper genes. E6 helper activity also correlated with the highest level of the truncated 44kDa VP protein, the lowest level of the 20kDa VP protein, and the appearance of a new VP species of 31kDa. We can find no reference to these smaller VP proteins in the literature. The antibody used recognizes aa726-733 at the carboxy-terminus of all the VPs and there are internal (initiation) methiones within the cap sequence which will generate small VP proteins of approximately the observed sizes. However, they may be favored breakdown products of the larger VPs. E6 protein appears to have a negative effect on rep mRNA splicing as Rep78, from an unspliced rep transcript, appeared at the highest levels in of experimental situations. Yet it is confusing that Rep40, from a spliced rep transcript, also appeared at the highest levels of all experimental situations.
There are several interpretations which come from these data, beyond the documentation of overall helper activity by these three HPV helper genes. First is that E1, while having the most profound effect on AAV transcription, upregulating both rep and cap mRNA levels, also has a negative effect on the translation of their encoded AAV proteins. Second, E6 helper function shows the strongest effect on the AAV proteins, upregulating all (except VP20kDa), yet this increase did not correlate with significantly higher wt AAV virus production compared to E1 and E2 helper functions. Third, E1 appeared to produce the highest amount of wt AAV virus Figure per unit of Rep and VP protein. As both the large Rep and VP1-3 proteins are constituents of the mature AAV virus particle, this suggests that E1 may have a positive modulatory role on the maturation of the wt AAV virion. Regarding this role it is important to note Rep protein helicase activity has been speculated to be involved in AAV virion maturation (39), however E1 appears to modulate Rep78 helicase activity in a negative manner (35).
While this is an important descriptive study on HPV-AAV interaction and generation of rAAV and wt AAV, further analysis remains to be done, in particular on determining the specific mechanisms of action of the HPV helper functions. We have already shown that HPV E1 protein binds AAV Rep78 protein and results in an enhancement of Rep78’s replication-related biochemistries in vitro (34,35). Additionally E1 is known to bind/recruit the DNA polymerase alpha-primase complex, which may also be involved in E1 helper function (40). However, the helper effects of E2 and E6 remain to be determined. It should also be noted that E6 is an oncoprotein and may have some functional similarity to Ad E1B (41). HPV E2 is a DNA-binding transcription factor and HPV E1 is a replication-related helicase, and neither have homologues within the adenovirus genome (42). It is also unclear of possible complementations between the HPV genes themselves. In spite of the different individual effects of the HPV genes, taken together these data strongly suggest that the HPV helper genes will be useful in increasing rAAV yield for human gene therapy studies.
Materials and Methods
Plasmids
Plasmids AAV/E1/Neo, AAV/E2/Neo, AAV/E6/Neo, AAV/eGFP, pKEX-Rep78, pSH3, pDG and pSM620 have been described before (11,43-46).
Cells
HEK293 cells were obtained from the American Type Culture Collection. Virus stocks of AAV/E1/Neo, AAV/E2/Neo, AAV/E6/Neo were generated by transfecting 2μg each of the rAAV plasmids along with 2μg of pSH3 (Ad/AAV helper plasmid) into HEK293 cells. After 4 days the cells were freeze-thawed three times and filtered to generate virus stocks. HEK293 cells were plated at low confluency (104) and infected with 107 encapsidated genomes of virus stock and allowed to grow for several days before the addition of G418. G418 was initially added at 600μg/ml, then dropped to 400μg/ml after three days. Selection continued for approximately three weeks when G418 was dropped to 200μg/ml. At that time each plate had 30-50 G418-resistant colonies. These bulk G418-resistant cells were used for the HPV helper experiments. The cells lines will hereafter be referred to as 293-E1, 293-E2 and 293-E6. PCR analysis was carried out to confirm the presence of the HPV-16 E1, E2 and E6 genes in the appropriate cell lines.
Analysis of virus DNA replication by Southern blot
When studying rAAV replication, 10 cm plates of 70% confluent HEK293, 293-E1, 293-E2 and 293-E6 cells were transfected with to 2μg each of pAAV/eGFP and pSH3 using FuGENE-6 per manufacturer’s instructions. Five days after transfection low molecular weight Hirt DNA was isolated, agarose gel electrophoresed and Southern blotted as described previously (1) and probed with 32P-eGFP DNA. No Dpn I digestion is needed at this late time of harvest as all transfected input plasmid DNA has been degraded. When studying wt AAV replication, 10 cm plates of 80% confluent HEK293, 293-E1, 293-E2 and 293-E6 cells were infected with a multiplicity of infection (moi) of both wt AAV2 and Ad5. Sixty hours after transfection low molecular weight Hirt DNA was isolated, agarose gel electrophoresed, Southern blotted and probed with 32P-pSM620 DNA.
Analysis of virus titer
Plates of transfected HEK293 cells were freeze-thawed three times, cellular debris pelleted by centrifugation at 7,000 rpm for 25 minutes, and the supernatant pushed through a 0.22 μm filter. Three hundred μl of virus stock was treated with 20 units DNase for 30 minutes at 37°C. After heating the sample for 10 minutes at 100°C, the sample was digested with proteinase K (0.2 μg/ml) for 4 hrs, then phenol extracted and ethanol precipitated. The resulting DNA was then agarose gel electrophoresed, Southern blotted and probed with 32P-eGFP DNA when analyzing for rAAV production or with 32P-pSM620 DNA, when analyzing for wt AAV production.
Analysis of viral mRNA
Total RNA was isolated from cell using the RNeasy mini kit (Qiagen) and treated with DNase I. Reverse transcription to synthesize cDNA was performed using the Transcriptor First Strand cDNA Synthesis Kit (Roche) with anchored oligo(dT) primer. cDNA were then used for qRT-PCR analyses. The house-keeping gene beta-actin was subjected to qPCR as the internal controls for quantitation. The primer pairs specific to AAV genomic rep gene (forward: 5’- CCG TGG CCG AGA AGC TGC AG -3’, reverse: 5’- CCA CGA GCA CGT GCA TGT GG -3’) and cap gene (forward: 5’- GAC CGG CAG CTC GAC AGC GG -3’, reverse: 5’- GCC TGG AAG ACT GCT CGT CC -3’) and beta-actin (forward: 5’-ATC TGG CAC CAC ACC TTC TAC-3’, reverse: 5’-GAA GGT CTC AAA CAT GAT CTG G-3’ ) were synthesized by Integrated DNA Technologies, Inc. (Coralville, IA). Real-time PCRs were carried out on Applied Biosystems 7900HT Fast Real-Time PCR System using the SYBR® Green Master Mix kit (Applied Biosystems), with cycle parameters of initial denaturation at 95°C for 10 minutes, 40 denaturation cycles at 95°C for 15 seconds, and annealing at 60°C for 1 minute and a dissociation step. Data analysis was executed by 7900HT Fast System Sequence Detection Software.
Analysis of AAV proteins by Western blot
Whole cellular proteins were prepared as previously described (47), and were then separated by electrophoresis on 10% SDS-PAGE (Invitrogen) followed by being transferred to nitrocellulose membranes. After a blocking step with 5% nonfat milk (Fisher Scitific) for 1 h at room temperature, membranes were incubated overnight at 4°C with primary antibodies (American Research Products) specific to AAV-rep protein (1: 200 dilution) and AAV-capsid protein (1: 200 dilution). Membranes were then washed with 1×TBST buffer (10 mM Tris-Cl [pH 7.5], 150 mM NaCl, 0.1% Tween 20) and incubated with 1:2000 dilution HRP-conjugated secondary antibody at room temperature for 1 hour. Proteins were detected using the ECL system (Fisher-Scientific Pierce).
Supplementary Material
Acknowledgments
This work is supported by grant R01 CA104873 from the NIH, a VA Merit Review grant, and intramural funding support from the UAMS College of Medicine Research Council to P.L.H. The authors thank Drs Jurgen Kleinschmidt (Hiedelberg, Germany), James Trempe (Toledo, OH), Sergei Zolotukin (Gainesville, FL) and Nicholas Muzyczka (Gainesville, FL) for their kind donations of pKEX-Rep78, pSH3, pDG8, and pSM620, respectively. The authors thank Dr. Philip Palade (Little Rock, AR) for reviewing this manuscript.
Footnotes
Supplementary information is available at Gene Therapy’s website.
Conflict of Interest Statement: The authors declare no conflict of interest.
Literature Cited
- 1.Hermonat PL, Muzyczka N. Use of adeno-associated virus as a mammalian DNA cloning vector: transduction of neomycin resistance into mammalian tissue culture cells. Proc Natl Acad Sci USA. 1984;81:6466–6470. doi: 10.1073/pnas.81.20.6466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jiang H, Pierce GF, Ozelo MC, de Paula EV, Vargas JA, Smith P, Sommer J, Luk A, Manno CS, High KA, Arruda VR. Evidence of multiyear factor IX expression by AAV-mediated gene transfer to skeletal muscle in an individual with severe hemophilia B. Molec Ther. 2006;14:452–455. doi: 10.1016/j.ymthe.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 3.You CX, Liu Y, Shi M, Cao M, Luo R-C, Hermonat PL. Comparison of AAV/IL-7 autocrine (T cell) versus paracrine (DC) gene delivery for enhancing CTL stimulation and function. Can Imm Immunothery. 2010;59:779–787. doi: 10.1007/s00262-009-0798-0. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khan JA, Cao M, Kang BY, Liu Y, Mehta, Hermonat PL. Systemic hNetrin-1 gene delivery by AAV lowers monocyte/macrophage accumulation and atherogenesis in vivo. Gene Therapy. doi: 10.1038/gt.2010.155. In press. [DOI] [PubMed] [Google Scholar]
- 5.Srivastava A, Lusby EW, Berns KI. Nucleotide sequence and organization of the adeno-associated 2 genome. J Virol. 1983;45:555–564. doi: 10.1128/jvi.45.2.555-564.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hermonat PL, Labow MA, Wright R, Berns KI, Muzyczka N. Genetics of adeno-associated virus: isolation and preliminary characteriozation of adeno-associated virus type 2 mutants. J Virol. 1984;51:319–339. doi: 10.1128/jvi.51.2.329-339.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tratschin JD, Miller IL, Carter BJ. Genetic analysis of adeno-associated virus: properties of deletion mutants constructed in vitro and evidence for an adeno-associated virus replication function. J Virol. 198;51:611–619. doi: 10.1128/jvi.51.3.611-619.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Samulski RJ, Srivastava A, Berns KI, Muzyczka N. Rescue of adeno-associated virus into pBR322: rescue of intact virus from the recombinant plasmid in human cells. Proc Natl Acad Sci USA. 1982;79:2077–2081. doi: 10.1073/pnas.79.6.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meyers C, Mane M, Kokorina N, Alam S, Hermonat PL. Ubiquitous human adeno-associated virus type 2 autonomously replicates in differentiating keratinocytes of a normal skin model. Virology. 2000;272:338–346. doi: 10.1006/viro.2000.0385. [DOI] [PubMed] [Google Scholar]
- 10.Meyers C, Alam S, Mane M. Hermonat PL, Altered biology ofadeno-associated virus type 2 and human papillomavirus during dual infection of natural host tissue. Virology. 2001;287:30–39. doi: 10.1006/viro.2001.0968. [DOI] [PubMed] [Google Scholar]
- 11.You H, Liu Y, Prasad CP, Agrawal N, Zhang D, Bandyopadhyay S, Liu H, Kay HH, Hermonat PL. Multiple human papillomavirus genes affect the adeno-associated virus life cycle. Virology. 2006;344:532–40. doi: 10.1016/j.virol.2005.08.039. [DOI] [PubMed] [Google Scholar]
- 12.Casto BC, Atchinson RW, Hammon WMcD. Studies on the relationship between adenoassociated virus type 1 and adenovirues. I. Replication of AAV-1 in certain cell cultures and its effect on helper adenovirus. Virology. 1967;18:52–60. doi: 10.1016/0042-6822(67)90251-6. [DOI] [PubMed] [Google Scholar]
- 13.Buller RM, Janik JE, Subring ED, Rose JA. Herpes simplex virus type 1 and 2 completely help adeno-associated virus replication. J Virol. 1981;40:241–247. doi: 10.1128/jvi.40.1.241-247.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang L-S, Shenk T. Adeno-associated virus p5 promoter contains an adenovirus E1A inducible element and a binding site for the major late transcription factor. J Virol. 1989;63:3479–3488. doi: 10.1128/jvi.63.8.3479-3488.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tratschin JD, West MHP, Sandbank T, Carter BJ. A human parvovirus, adenoassociated virus as a eukaryotic vector: transient expression and encapsidation of the prokaryotic gene for chloramphenicil acetyltransferase. Mol Cell Biol. 1984;4:2072–2081. doi: 10.1128/mcb.4.10.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laughlin CA, Jones N, Carter BJ. Effects of deletions in adenovirus region 1 genes upon the replication of adeno-associated virus. J Virol. 1982;41:868–876. doi: 10.1128/jvi.41.3.868-876.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang L-S, Shenk T. The adenovirus DNA binding protein stimulates the rate of transcription directed by adenovirus and adeno-associated virus promoters. J Virol. 1990;64:2103–2109. doi: 10.1128/jvi.64.5.2103-2109.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carter BJ, Marcus-Sekura CJ, Laughlin CA, Ketner G. Properties of an adenovirus type 2 mutant, Ad2dl807, having a deletion near the right-hand genome terminus: failure to help AAV replication. Virology. 1983;126:505–516. doi: 10.1016/s0042-6822(83)80008-7. [DOI] [PubMed] [Google Scholar]
- 19.Richardson WD, Westphal WD. A cascade of adenovirus early functions are required for expression of adeno-associated virus. Cell. 1981;27:131–141. doi: 10.1016/0092-8674(81)90367-6. [DOI] [PubMed] [Google Scholar]
- 20.Samulski RJ, Shenk T. Adenovirus E1B 55-M, polypeptide facilitates timely cytoplasmic accumulation of adeno-associated virus mRNAs. J Virol. 1988;62:206–210. doi: 10.1128/jvi.62.1.206-210.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang M-M, Hearing P. Adenovirus early region 4 encodes two gene products with redundant effects in lytic infection. J Virol. 1989;63:2605–2615. doi: 10.1128/jvi.63.6.2605-2615.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hart LS, Yannone SM, Naczki C, Orlando JS, Waters SB, Akman SA, Chen DJ, Ornelles D, Koumenis C. The adenovirus E4orf6 protein inhibits DNA double strand break repair and radiosensitizes human tumor cells in an E1B-55K-independent manner. J Biol Chem. 2005;280:1474–1481. doi: 10.1074/jbc.M409934200. [DOI] [PubMed] [Google Scholar]
- 23.West MPH, Trempe JP, Tratschin JD, Carter BJ. Gene expression virus vectors: the effects of chimeric mRNA structure, helper virus, and adenovirus VA1 RNA. Virology. 1987;160:38–47. doi: 10.1016/0042-6822(87)90041-9. [DOI] [PubMed] [Google Scholar]
- 24.Janik JE, Huston MM, Cho K, Rose JA. Efficient synthesis of adeno-associated virus structural proteins requires both adenovirus DNA binding protein and VA1 RNA. Virology. 1989;168:320–329. doi: 10.1016/0042-6822(89)90272-9. [DOI] [PubMed] [Google Scholar]
- 25.McPherson RA, Ginsburg HS, Rose JA. Adeno-associated virus helper activity of adenovirus DNA binding protein. J Virol. 1982;46:523–529. doi: 10.1128/jvi.44.2.666-673.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strauss SE, Ginsburg HS, Rose JA. DNA-minus temperature sensitive mutants of adenovirus type 5 help-adeno-associated virus replication. J Virol. 1976;17:140–148. doi: 10.1128/jvi.17.1.140-148.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jay FT, Laughlin CA, Carter BJ. Eukaryotic translational control: adeno-associated virus protein synthesis is affected by a mutation in the adenovirus DNA-binding protein. Proc Natl Acad Sci USA. 1989;78:2827–2931. doi: 10.1073/pnas.78.5.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyers MW, Carter BJ. Assembly of adeno-associated virus. Virology. 1980;102:71–82. doi: 10.1016/0042-6822(80)90071-9. [DOI] [PubMed] [Google Scholar]
- 29.Richardson WD, Westphal WD. Requirement for either early region la or early region lb adenovirus gene products in the helper effect of adeno-associated virus. J Virol. 1984;10:1–8. doi: 10.1128/jvi.51.2.404-410.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duan D, Sharma P, Dudus L, Zhang Y, Sanlioglu S, Yan Z, Yue Y, Ye Y, Lester R, Yang J, Fisher KJ, Engelhardt JF. Formation of adeno-associated virus circular genomes is differentially regulated by adenovirus E40RF6 and E2a gene expression. J Virol. 1999;73:161–1699. doi: 10.1128/jvi.73.1.161-169.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Androphy EJ, Hubbert NL, Schiller JT, Lowy DR. Identification of the I-IPV-16 E6 protein from transformed mouse cells and human cervical carcinoma cell lines. EMBO J. 1987;6:989–992. doi: 10.1002/j.1460-2075.1987.tb04849.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bedell MA, Jones KH, Grossman SG, Laimins LA. Identification of human papillomavirus type 18 transformation genes in immortalized and primary cells. J Virol. 1989;63:1247–1255. doi: 10.1128/jvi.63.3.1247-1255.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Del Vecchio AM, Romanczuk H, Howley PM, Baker CC. Transient replication of human papillomavirus DNAs. J Virol. 1992;66:5949–5958. doi: 10.1128/jvi.66.10.5949-5958.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bandyopadhyay S, Raney KD, Liu Y, Hermonat PL. AAV-2 Rep78 and HPV-16 E1 interact in vitro, modulating their ATPase activity. Biochemistry. 2008;47:845–56. doi: 10.1021/bi701579v. [DOI] [PubMed] [Google Scholar]
- 35.Bandyopadhyay S, Cao M, Liu Y, Hermonat PL. HPV E1 up-regulates replication-related biochemistries of AAV Rep78. Virology. 2010;402:94–101. doi: 10.1016/j.virol.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ustav M, Stenlund A. Transient replication of BPV- 1 requires two viral polypeptides encoded by the E1 and E2 open reading frames. EMBO J. 1991;10:449–457. doi: 10.1002/j.1460-2075.1991.tb07967.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sandier AB, Vande Pol SB, Spalholz BA. Repression of bovine papillomavirus type 1 transcription by the E1 replication protein. J Virol. 1993;67:5079–5087. doi: 10.1128/jvi.67.9.5079-5087.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scheffner M, Romanczuk H, Monger K, Huibregtse JM, Mietz JA, Howley PM. Functions of human papillomavirus proteins. In: zur Hausen H, editor. Human Pathogenic Papillomaviruses. Heidelberg: Springer-Verlag; 1994. pp. 83–100. [DOI] [PubMed] [Google Scholar]
- 39.King JA, Dubielzig R, Grimm D, Kleinschmidt JA. DNA helicase-mediated packaging of adeno-associated virus type 2 genomes into preformed capsids. EMBO J. 2001;20:3282–3291. doi: 10.1093/emboj/20.12.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bonne-Andrea C, Santucci S, Clertant P, Tillier F. Bovine papillomavirus E1 protein binds specifically DNA polymerase alpha but not replication protein A. J Virol. 1995;69:2341–2350. doi: 10.1128/jvi.69.4.2341-2350.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moody CA, Laimins LA. Human papillomavirus oncoproteins: pathways to transformation. Nat Rev Can. 2010;10:550–560. doi: 10.1038/nrc2886. [DOI] [PubMed] [Google Scholar]
- 42.Lambert PF. Papillomavirus DNA replication. J Virol. 1991;65:3417–3420. doi: 10.1128/jvi.65.7.3417-3420.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Horer M, Weger S, Butz K, Hoppe-Seyler F, Geisen C, Kleinschmidt JA. Mutational analysis of adeno-associated virus Rep protein-mediated inhibition of homologous promoters. J Virol. 1995;69:5485–5496. doi: 10.1128/jvi.69.9.5485-5496.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Collaco RF, Cao X, Trempe JP. A helper virus-free packaging system for recombinant adeno-associated virus. Gene. 1999;238:397–405. doi: 10.1016/s0378-1119(99)00347-9. [DOI] [PubMed] [Google Scholar]
- 45.Beattie SG, Goetzman E, Conlon T, Germain S, Walter G, Campbell-Thompson M, Matern D, Vockley J, Flotte TR. Biochemical correction of short-chain acyl-coenzyme A dehydrogenase after portal vein injection of rAAV8-SCAD. Hum Gene Ther. 2008;19:579–588. doi: 10.1089/hum.2007.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Samulski RJ, Berns KI, Tan M, Muzyczka N. Cloning of adeno-associated virus into pBR322: rescue of intact virus from the recombinant plasmid in human cells. Proc Natl Acad Sci USA. 1982;79:2077–2081. doi: 10.1073/pnas.79.6.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dandapat A, Hu CP, Chen J, Liu Y, Khan JA, Remeo F, Carey RM, Hermonat PL, Mehta JL. Over-expression of angiotensin II type 2 receptor (agtr2) decreases collagen accumulation in atherosclerotic plaque. Biochem Biophys Res Commun. 2008;366:871–877. doi: 10.1016/j.bbrc.2007.11.061. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.