Abstract
While trypsin remains the most commonly used protease in mass spectrometry, other proteases may be employed for increasing peptide-coverage or generating overlapping peptides. Knowledge of the accurate specificity rules of these proteases is helpful for database search tools to detect peptides, and becomes crucial when label-free mass spectrometry is used to discover in vivo proteolytic cleavages. Since in vivo cleavages are inferred by subtracting digestion-induced cleavages from all observed cleavages, it is important to ensure that the specificity rule used to identify digestion-induced cleavages are broad enough to capture even minor cleavages produced in digestion, to avoid erroneously identifying them as in vivo cleavages. In this study, we describe MS-Proteolysis, a software tool for identifying putative sites of in vivo proteolytic cleavage using label-free mass spectrometry. The tool is used in conjunction with digestion by trypsin and three other proteases, whose specificity rules are revised and extended before inferring proteolytic cleavages. Finally, we show that comparative analysis of multiple proteases can be used to detect putative in vivo proteolytic sites on a proteome-wide scale.
Keywords: mass spectrometry, label-free, protease specificity, trypsin, V8 protease, CNBr, chymotrypsin, proteolysis
1 Introduction
Proteases are molecular scissors that play a critical role in the regulatory processes inside the cell as well as molecular tools in the laboratory. The defining characteristic of a protease is its specificity, i.e. the rule that determines the selection of its cleavage-substrates. Knowledge of specificity is important for understanding the function and mechanism of proteases and for their laboratory applications. One such application of proteases is in the form of digestive enzymes in mass spectrometry-based proteomics [1], where they are used to cleave proteins into smaller peptides that are easier to analyze than intact proteins. Trypsin is the most commonly used protease for this purpose, partly because of its well-defined and robust specificity rules [2]. As we argued in [3], having precise knowledge of specificity of the protease is important not only for peptide identification (many peptide identification tools incorporate specificity rules into their search algorithms), but is also critical in some emerging applications of mass spectrometry such as label-free analysis of regulatory proteolysis [4, 5, 6]. In such studies, the sample is digested with a protease with known specificity (e.g., trypsin, V8 protease, etc.) and a regulatory protease (e.g., a caspase) with the goal to discover the (unknown) specificity of the regulatory protease. Tandem mass spectrometry (MS/MS) is then employed to determine all cleavages in the resulting sample. Afterwards, one has to “subtract” expected in vitro cleavages (e.g., trypsin-induced cleavages) from all found cleavages to identify the in vivo cleavages caused by the regulatory proteases. However, if the model of the protease specificity is even slightly inaccurate, these label-free studies are likely to fail. For example, while the rule “trypsin cuts after R and K but not before P” is a reasonable description of trypsin specificity for most applications, it becomes inaccurate if one attempts to find in vivo proteolytic sites (since trypsin actually cuts before P albeit with reduced e ciency [3]). As a result, if one uses an inaccurate rule for trypsin specificity, the cuts before P will not be subtracted resulting in a surprising “discovery” of many in vivo cleavage sites before P. In reality, this “discovery” reveals limitations of the common rule describing trypsin specificity rather than a new protease activity. Therefore, it is important that the specificity rule used to identify digestion-induced cleavages are broad enough to capture even minor cleavages produced in digestion, to avoid erroneously identifying them as in vivo cleavages.
Another area that requires detailed knowledge of protease specificity is the proteome-wide analysis of in vivo proteolytic events in the sample subjected to trypsin with the goal to infer the natural proteolytic cleavages induced by various proteases (without attempting to infer the specificities of individual proteases). In the past, informationabout proteolysis has been mainly gained by performing in vitro experiments with individual proteins and proteases that may not represent true in vivo scenarios at the proteome-wide scale. Recently, Manes et al., 2007 [7], and Shen et al., 2008 [8] addressed the challenge of proteome-wide proteolysis analysis in the studies of native (short) peptides in Salmonella enterica and Saccharomyces cerevisiae. However, longer native peptides require digestion with trypsin or other proteases, and there is still no software tool that can identify in vivo proteolytic sites from such digests.
Determining specificity of proteases has traditionally been a strenuous experimental process, and consequently, often limited to analysis of a small number of substrates [9]. Combinatorial library approaches address this short-coming by employing large libraries of substrates treated by the protease [10, 11, 12], although analyzing the cleaved products from these libraries may require use of laborious fluorescence or sequencing technology. Mass spectrometry presents a rapid approach for sequencing a large number of substrates from a peptide library. Recently, Schilling and Overall, 2008 [13] described peptide libraries derived from human proteome that could be easily analyzed by mass spectrometry through standard database-search methods. This approach, however, required the use of biotin-labeling to separate the N-terminal and C-terminal side of the cleavage sites.
In Rodriguez et al. 2008 [3], we demonstrated that it is possible to determine accurate specificity rules for the enzyme used for digestion in a standard mass spectrometry experiment when analyzing large spectral datasets. This approach can be easily implemented (even if the data were generated for a di erent purpose), without the requirement of expensive labeling methods. In contrast to most proteomics approaches (that typically identify peptides with FDR 1% and higher), this approach requires extremely accurate peptide identifications (typically FDR 0.1% and lower) since even a small fraction of incorrect assignments may contribute many (pseudo) cleavages that distort the analysis of protease specificity. Rodriguez et al. 2008 [3] used doubly-confirmed cuts (start- or end-points shared by two or more peptides) to arrive at a reliable set of identified peptides.
This study extends the above label-free approach to analyze the specificity of three other proteases used for digestion in mass spectrometry. Using multiple enzymes for digestion can be helpful for increasing the peptide-coverage of proteins, or in applications where overlapping peptides are desirable, such as in the construction of spectral networks and de novo protein sequencing [14, 15]. Staphylococcus aureus V8 protease (also known as Glu-C), chymotrypsin and CNBr are popular alternatives to trypsin. Here, we empirically derive the known specificity rules for these proteases/reagents and present evidence for some notable deviations from these rules, suggesting thatthe reaction specificity is not as simple as previously assumed. We extend the specificity rules to capture even the low-propensity cleavages detected in mass spectrometry, to minimize the possibility of digestion-induced cleavages being identified as in vivo cleavages. This “upper-bound” approach on specificity rules favors accuracy over sensitivity in the identification of proteolytic sites in the proteome. We allow for the possibility that some low-propensity cleavages could arise from non-specific activity of the protease, or due to contamination in the protease sample. By estimating the “effective” specificity of the actual protease sample in the given experimental conditions, instead of the hypothetical 100% pure sample under ideal conditions, we try to minimize the possibility of false discoveries. It must be noted that our pragmatic goal is to use the derived specificity for label-free analysis of regulatory proteolysis, rather than to derive the specificity of “perfectly” purified proteases in a “perfect” experiment. These specificities may vary slightly across multiple vendors and conditions (e.g., CNBr can be used in conjunction with TFA, formic acid etc.). The levels of in-source fragmentation (which might affect the observed cleavages) may also vary across instruments and conditions. The approach presented here attempts to offset these variations in specificity within the given experimental samples, for follow-up analyses of regulatory proteases using data generated under the same conditions.
While Rodriguez et al. 2008 [3] introduced the doubly-confirmed cleavages to infer reliable cleavage sites, we illustrate that it is possible to determine equally reliable but significantly larger list of cleavage sites using MS-GeneratingFunction [16]. We show that comparative analysis of multiple digests allows one to reliably identify N-terminal methionine excisions, signal peptide cleavages and other putative proteolytic events using our MS-Proteolysis software tool. Multiple protease digests provide independent evidence to confirm in vivo proteolytic sites and differentiate them from computational or experimental artifacts. MS-Proteolysis can be used to analyze any MS/MS dataset (including ones that were not generated to study proteolysis) to discover in vivo proteolytic events.
2 Methods
2.1 Culture Conditions and Cell Lysis
The following chemicals used, unless otherwise noted, were obtained from the Sigma-Aldrich Company (St. Louis, MO) and were of analytical grade. Wild type Shewanella oneidensis strain MR-1 was cultured and grown on M1 media without Ca2+, with FeNTA + Se. The media also contained 30 mM PIPES, 30 mM lactate, and 30 mM fumarate. The cells were grown aerobically (100 mL each) in 1 liter flasks at 30°c and at 120 RPM for 20 hours. The OD600 for the cell culture at the time of harvest was 0.765 ( 4 × C cells/mL). Cells were harvested via centrifugation at 6000 × g for 5 minutes. The supernatant was decanted and the cells were washed with 25 mL wash buffer (20 mM HEPES, pH 7.4, + 150 NaCl) and re-centrifuged at 6000 × g for 5 minutes. The supernatant again was decanted and 1 mL of wash buffer was added to the pellets. The pellets were again centrifuged at 6000 × g for 5 minutes and the supernatant was aspirated and the cells were frozen in liquid nitrogen and stored at ×80 °c until ready for lysis. Lysis was achieved by beadbeating the cells suspended in a volume of nanopure water which was 2 x the volume of the cell pellet. The cells were vortexed into a suspension and were beadbeated with 0.1 mm zirconia/silica beads in a mini-beadbeater (Biospec, Bartlesville OK) for 90 s at 4500 rpm. Lysates were collected and placed immediately on ice to inhibit proteolysis.
2.2 Protein Mass Determination and Addition of an Internal Protein Standard
The protein concentration of each whole cell lysate were measured using a Coomassie Plus protein assay (Pierce, Rockford, IL) using a bovine serum albumin standard. A measured volume of each sample was transferred to a new 1.5 mL microfuge tube and thus the total protein mass was known for each tube. 1 mg of apomyoglobin (equine), 0.363 mg of cytochrome c (bovine) and 0.210 mg of G-2-PHD (rabbit) was added to 1 mL total volume of nanopure water. This was the internal protein reference standard used in the samples to ensure that replicate analysis and instrument performance in time was consistent. To each whole protein samples 10 μL (15.73 μg) of the internal protein reference was added to every 984 μg of sample. Each lysate was then divided into 5 aliquots of equal and known protein mass (700 μg) and each aliquot was placed into a new 1.5 mL microfuge tube and all samples were dried down completely using centrifugation under vacuum.
2.3 Chymotrypsin Digestion
An aliquot from each cell lysate was digested with sequencing grade chymotrypsin (Promega, Madison WI) in accordance with manufacturer’s guidelines except as noted. First, 150 μL of freshly made 8 M urea and 1.5 μL of Neutralized TCEP (Bond Breaker, Pierce, Rockford IL) was added to the sample. TCEP has been successfully demonstrated for use as a disulfide reducing agent in this concentration in many prior proteolytic experiements [17]. The samples were sonicated for about a minute in a sonicating water bath to solubilize the samples. The samples were diluted 10-fold with freshly made 50 mM ammonium bicarbonate, pH 7.8 (manufacturer guidelines suggested the pH range of 7.0-9.0 for optimal chymotrypsin activity) and CaCl2 was added to be at a final concentration of 1 mM. Chymotrypsin was added in a 1:75 protease to sample protein ratio. Proteins were digested for 5 hours at 37 °c (also previously used [18]). Alkylation was performed in the dark with the addition of iodoacetamide at a final concentration of 20 mM for 1 hr at room temperature. The peptides were then desalted immediately after alkylation as described below.
2.4 Staphylococcus aureus V8 protease Digestion
An aliquot from each cell lysate was digested with Staphylococcus aureus V8 protease (ThermoScientific-Pierce, Rockford IL) following manufacturer’s guidelines except as noted. 150 μL of freshly made 8 M urea and 1.5 μL of Neutralized TCEP (Bond Breaker, Pierce, Rockford IL) was added to each sample (as described above for chymotrypsin). The samples were sonicated for about a minute in a sonicating water bath to solubilize the samples. The samples were diluted 10 fold with freshly made 100 mM sodium phosphate, pH 7.8 (recommended by the manufacturer to target proteolytic cleavage at both glutamic and aspartic acid residues) and CaCl2 was added to be at a final concentration of 1 mM. V8 protease was added in a 1:20 protease to sample protein ratio. Proteins were digested for 5 hours at 37 °c , as recommended by the manufacturer. After digestion, the peptides from the V8 protease digest were alkylated with iodoacetamide at a concentration of 20 mM rotating in the dark at room temperature for 30 minutes.
2.5 CNBr Digestion
An aliquot from each cell lysate was digested with cyanogen bromide. Each lyophilized sample (700 μg) was resuspended in 120 μL of 4% CHAPS in nanopure water and 1.5 μL of Neutralized TCEP (Bond Breaker, Pierce, Rockford IL) was added. CHAPS was previously found to be a useful detergent which is easily removed prior to mass spectrometric analysis, and was, therefore, selected to aid in the solubilization of the proteins [19]. The samples were incubated at 60 °c for 30 minutes. 240 μL of formic acid was added to each sample and 40 μL of a CNBr solution (1 mg/mL CNBr/formic acid) was added. The samples were vortexed and incubated in the dark at room temperature for 5 hours. The digests were lyophilized in a centrifugal vacuum to dryness. Then 200 μL of freshly made 50 mM ammonium bicarbonate was added along with 1.5 μL of TCEP and 10 μL of 36 mg/mL iodoacetamide in water. The samples were alkylated with the iodoacetamide in the dark for 30 minutes at room temperature.
2.6 Peptide Concentration and Cleanup
Immediately desaltation following alkylation allowed controlling the reaction time of alkylation [19, 20]. The chymotrypsin and V8 protease digests were desalted using Supelco (St. Louis, MO) Supelclean C-18 tubes as described elsewhere [21]. Since CHAPS used in CNBr digests binds to C-18 [22], this method was not applied to CNBr digests. Instead, Supelco Supelclean SCX tubes were used to desalt the CNBr digests, as described here. The digestion mixture pH was adjusted to 3.5 by the addition of dilute acetic acid and an equal volume of 10 mM ammonium acetate, pH 3.5 with 25% acetonitrile. The resin was conditioned with one column volume of acetonitrile followed by one column volume of 25% acetonitrile in 10 mM ammonium acetate, pH 3.5. After the peptide mixtures were loaded onto the resin, the peptides were washed with six column volumes of the same 25% acetonitrile in 10 mM ammonium acetate, pH 3.5. Peptide elution was accomplished with one column volume of 35% acetonitrile in 500 mM ammonium acetate, pH 8.5, followed by 100% acetonitrile. All eluted peptides were concentrated via speedvac (ThermoSavant, San Jose CA) until protein concentrations were 1.0 mg/mL. Peptide concentrations were determined by BCA assay (Pierce, Rockford IL) with a bovine serum albumin standard.
2.7 SCX Fractionation, Capillary LC Separation and Data Acquisition
300 μg of each desalted digest were separated with a strong cation exchange (SCX) fractionation as described elsewhere [23] with the exception that 50 fractions were collected for each sample. All SCX fractions were then separated by an automated in-house designed HPLC system as summarized elsewhere [21, 24] and were eluted directly into an ion trap for MS/MS as described below. The eluate from the HPLC was directly electrosprayed into an ion trap MS (LCQ, ThermoFinnigan, San Jose, CA) using electrospray ionization (ESI). For the MS/MS detection, the peptide material obtained from each SCX fraction was resuspended in 10 μL of nanopure water and was then loaded onto the reversed phase column for each analysis. The mass spectrometer operated in a data-dependent MS/MS mode. The peptide fractions produced from the SCX peptide separations were analyzed with one full m/z range (400-2000) each. The details of the data acquisition are described elsewhere [25].
3 Results
3.1 Peptide Identification
High-throughput LC-MS/MS experiments (see Methods) generated 1.51 million, 1.24 million and 1.54 million spectra for Shewanella oneidensis MR-1 sample digested with V8 protease, chymotrypsin and CNBr respectively. These spectra were analyzed with InsPecT [26], as previously described in Rodriguez et al., 2008 [3], using the default settings (fragment ion tolerance of 0.5Da and parent mass tolerance of 2.5Da, fixed modification of +57 Da on cysteine). We used the scoring function model that was not specific to any particular digestion enzyme, so that the peptide identifications are not biased by prior knowledge of their cleavage specificities. Shewanella oneidensis MR-1 protein sequences obtained from TIGR Comprehensive Microbial Resource, were used as the protein database (total size ≈1.5MB). A decoy database of the same size (containing shuffled protein sequences [27]) was used to estimate the peptide-level False Discovery Rate (FDR) and limit it to 5% (the spectrum-level FDR is less than 2%). 31630, 9390 and 5317 peptides were identified in V8 protease, chymotrypsin and CNBr digests respectively.
The use of delta scores (difference between the highest and the second-highest scoring peptides for a given spectrum) in scoring functions makes the scoring dependent on the size of the sequence database. This might be a concern when using small bacterial databases with search tools like Sequest and InsPecT, but not with database-independent scoring functions such as MS-GeneratingFunction [16]. Using MS-GeneratingFunction at the stringent 0.1% FDR, 19868, 6388 and 3442 peptides were identified in V8 protease, chymotrypsin and CNBr digests respectively. We also analyzed the previously published Shewanella samples digested with trypsin [28] with MS-GeneratingFunction and identified 32531 peptides at 0.1% FDR.
MS/MS spectra from trypsin digests of Saccharomyces cerevisiae proteome were obtained from the PeptideAtlas repository [29], and analyzed using InsPecT and MS-GeneratingFunction as in the case of Shewanella. 7488 peptides were identified at 0.1% FDR.
3.2 Reliable cleavage sites
Each identified peptide reveals two cleavage sites through its end-points. A doubly-confirmed cleavage site is defined as a position in the proteome which is an end-point for two or more identified peptides [3]. 8635, 2146 and 866 such sites were identified for V8 protease, chymotrypsin and CNBr digests respectively. To ensure that the peptides considered in this analysis are produced by the protease and not by post-digestion breakup, a filtering step is applied before constructing the final list of doubly-confirmed cleavages [3]. In Rodriguez et al., 2008 [3], the error rate for doubly-confirmed sites was found to be only 0.1% when the peptide level error rate was 5%, as in this study. Therefore, less than 9 among all doubly-confirmed cleavages in V8 protease, 2 in chymotrypsin and 1 in CNBr are expected to be false positive identifications.
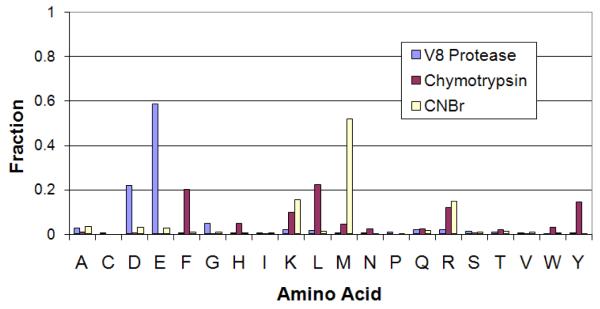
A protease substrate is conventionally labeled as ..P5, P4, P3, P2, P1, P1′, P2′, P3′, P4′, P5′.., where the cleavage is between P1 and P1′ positions. The commonly used specificity rules for the three proteases studied here are based on the amino acid at P1 position. V8 protease is known to cleave after acidic residues D and E [30, 31], CNBr is known to cleave after M [32] and chymotrypsin is known to cleave after aromatic amino acids Y, F and W and partially after L [33]. To compare these rules with the cleavages observed in our dataset, we analyze the fraction of different amino acids at the P1 position (Table 1). Figure 1 illustrates that the amino acids expected at the P1 position by known specificity rules are indeed highly over-represented at that position in our identified cleavages, thus supporting the rules as well as showing that our mass spectrometry-based approach can independently derive the specificity rules without prior knowledge. We now focus on the disagreements between the two to see if the observed cleavages can be used to extend the known specificity rules for these proteases to include even the low-propensity cleavages.
Table 1.
Frequency of different amino acids at P1 position in the double-confirmed cleavage sites observed for the three protease digests. The last column indicates the background frequency (count) of each amino acid in the entire Shewanella proteome. The amino acids defining the commonly accepted specificity for V8, chymotrypsin, and CNBr are shown in bold. The amino acids that do not contribute to known specificity rules but have surprisingly large counts in P1 positions of the cleavage sites are shown in italics.
| Amino Acid | V8 protease | Chymotrypsin | CNBr | Background |
|---|---|---|---|---|
| A | 215 | 22 | 30 | 136659 |
| C | 20 | 0 | 0 | 16251 |
| D | 1894 | 10 | 26 | 77030 |
| E | 5044 | 4 | 24 | 83281 |
| F | 47 | 431 | 6 | 57812 |
| G | 393 | 4 | 8 | 98698 |
| H | 23 | 101 | 3 | 34257 |
| I | 29 | 6 | 3 | 88040 |
| K | 146 | 209 | 135 | 75790 |
| L | 135 | 476 | 10 | 159360 |
| M | 39 | 96 | 448 | 37920 |
| N | 45 | 50 | 2 | 60337 |
| P | 53 | 0 | 1 | 59308 |
| Q | 164 | 48 | 13 | 71641 |
| R | 158 | 255 | 127 | 68627 |
| S | 90 | 10 | 8 | 95067 |
| T | 72 | 42 | 11 | 78882 |
| V | 32 | 6 | 6 | 98094 |
| W | 3 | 65 | 3 | 18888 |
| Y | 33 | 311 | 2 | 44781 |
|
| ||||
| Total | 8635 | 2146 | 866 | 1460723 |
Figure 1.
Fraction of different amino acids at P1 position in the doubly-confirmed cleavage sites, plotted for each of the three protease digests.
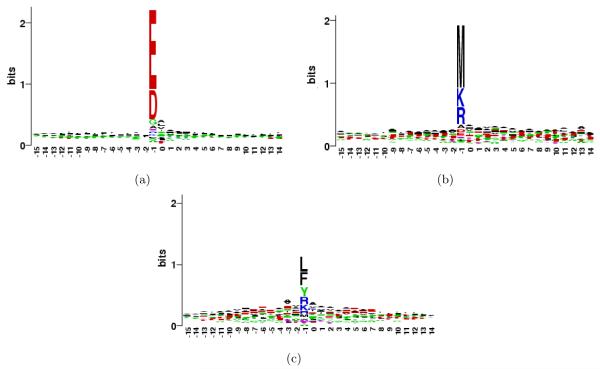
To extend the analysis from just P1 position to a longer motif around the cleavage site, we constructed the sequence logos [34] for regions containing P15 to P15′ positions, shown in Figure 2. The figure indicates that P1 position indeed plays the dominant role in determining the specificity of all three proteases. P1′ position reveals a small signal, which might represent a secondary preference contingent upon P1 position. To analyze this, we categorize each cleavage site by the pair of amino acids (di-AA) between which the site is located (P1 and P1’ positions). The observed frequency distribution for di-AAs flanking the cleavage sites can be used to better infer the specificity of the protease used for digestion [3]. Since not all di-AAs are equally likely to occur in the proteome, we normalize their observed frequencies by their background amino acid frequencies in Shewanella proteome. Supplementary Table 1 provides the list of the 400 di-AAs for each protease, sorted in the decreasing order of their normalized frequencies. In the following three sections, we use this data to analyze specificity rules for each protease in detail. We will use the notation X.Y to represent a di-AA, where X is the amino acid at P1 position and Y is the amino acid at P1′ position (use of * for X or Y indicates that any amino acid can be present at that position).
Figure 2.
Sequence logo for the observed cleavage sites in (a) V8 protease (b) CNBr and (c) chymotrypsin. The P1 position is numbered −1 in the logos.
3.3 V8 protease specificity
V8 protease is expected to cleave after D and E [30, 31], as is also observed in our data (Figure 1). The figure also shows that cleavages after E are more likely than cleavages after D, in agreement with previous observation [35]. Austen et al., 1976 [35] claimed that the protease does not cleave between E and P, while such cleavages were supported by Houmard et al., 1972 [30]. We find that E.P di-AA has rank 33 among all di-AAs, well ahead of many D.* cleavages like D.Q and D.R. In fact, the relative frequency of E.P cut is similar to the relative frequency of E.Q cut (rank 27). This suggests that V8 protease does cleave between E and P, although the propensity of such cleavages is lower than other E.* sites. We also notice very low propensity of E.E, D.D, and D.E cleavages suggesting that in such cases V8 cleaves after the second amino acid. In contrast, however, E.D cleavage is frequent.
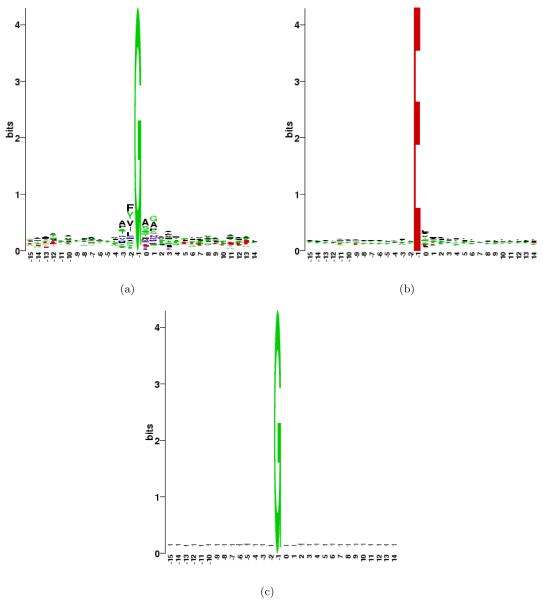
While the standard rule suggests that the top 40 di-AA cleavage sites should be D.* and E.*, followed by a random mix of other di-AAs, we surprisingly find 7 G.* cleavages (G.A, G.S, G.M, G.H, G.T, G.G, G.N) among the top 50. This leads to a new hypothesis that V8 protease also cleaves after G, although less efficiently than after D and E. Cleavages after G have not been previously reported for this protease. Since these cleavages are observed only in the V8 protease digests and not other proteases (see Table 1), it is unlikely that all of them (393) could represent in vivo proteolytic cleavages present in the proteome before digestion. We constructed the sequence logo to look at the sequence patterns around cleavage sites that have G at P1 position. Figure 3 shows the sequences logo for these sites, and for comparison, the logo for sites that have E at the P1 position. While the sites with E at P1 show only modest preferences at P1′ position and none at other positions, the sites with G at P1 position show a larger motif involving P2, P3, P1′ and P2′ positions. For example, F and Y are over-represented at P2 position while A is over-represented at P3, P1′ and P2′. Relatively lower preference for G, as compared to D or E, at P1 position may indicate that a longer sequence motif is needed for these sites to be recognized by the protease. To ensure that these trends observed for G are specific to cleavages and do not reflect a general preference of G to co-occur with certain amino acids, we constructed the sequence logo for all positions containing G in whole Shewanella proteome. Figure 3c shows that there is no such bias in the proteome; therefore, the patterns observed here are specific to the cleavages.
Figure 3.
(a) Sequence logo for the observed cleavages in V8 protease digests at sites that have G at P1 position. (b) Similar logo for sites with E at P1 position. (c) Logo for all the 98,698 sites in Shewanella proteome that contain G (placed at −1 position in the logo).
3.4 CNBr specificity
CNBr is known to cleave after M [32], and this is also clearly visible in the observed frequency table (Table 1). Note that although CNBr is a chemical, unlike other proteases analyzed, we will continue to use the notation of P1, P1′ etc. for positions around cleavage sites for convenience. It appears that cleavages are less likely when the amino acid at P1′ position is Q or T. No cleavages are observed between M and M, which indicates that in such cases of adjacent possible cleavage sites, CNBr cleaves at the second site. We also do not see any cleavages between M and W, and between M and C; however, this may be because of the low frequency of these di-AAs in the proteome.
While cleavages with M at P1 position are predominant in the observed list of di-AA pairs, we find that CNBr also shows a minor preference for R and K at P1 position (Figure 1). In fact, among the ranks 15 to 55 of top di-AA pairs for this protease (Supplementary Table 1), 16 have K at the P1 position while 17 have R at the P1 position (while only 2 of each type were expected by chance). This suggests that besides its primary specificity, CNBr may also have a small propensity to cleave after the basic amino acids. Table 1 shows that while R and K have some preference to be at P1 position in all proteases, the trend is particularly strong for CNBr indicating the role of protease in these cleavages. Even if these low-specificity cleavages are specific to sample processing in mass spectrometry (including the possibility of trypsin contamination), it is useful to include them in the “effective” specificity rule of CNBr in this context so that they are later not identified erroneously as in vivo proteolytic sites.
3.5 Chymotrypsin specificity
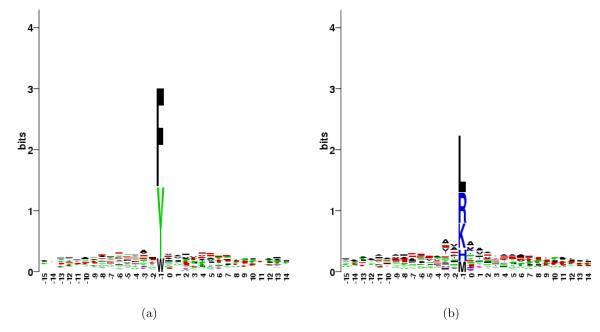
Figure 1 indicates that chymotrypsin cuts after a number of different amino acids. Chymotrypsin is usually expected to cleave after F, Y and W [33]. However, this rule is not unanimous, and some studies also include L in this list [13]. While F and Y stand out at P1 position, we observe that the preference for H, K, L, M and R at P1 is comparable to the preference for W (after adjusting for background distribution of amino acids). Constructing a sequence logo for these unexpected sites (Figure 4(b)) indicates that positions P3, P2, P1′ and P2′ are relatively more important for specificity at these sites than for the expected sites with F, Y or W (Figure 4(a)). Alanine is found to be the most commonly present amino acid at these positions among the unexpected sites, indicating its possible role in determining the specificity. While we cannot totally discard the possibility of trypsin contamination in these samples, we argue that if such contamination is commonplace, it is practical to include R and K in the empirical specificity rules of chymotrypsin (and subtract the cleavages with R/K at P1), especially when using mass spectrometry for discovery of in vivo proteolytic events.
Figure 4.
(a) Sequence logo for the observed cleavages in chymotrypsin digests at sites that have F, W or Y at P1 position. (b) Similar logo for sites with H, K, L, M or R at P1 position.
3.6 Using MS-GeneratingFunction for identifying cleavage sites
When using mass spectrometry-derived peptides to infer the specificity of proteases, it is critical to have extremely low error rates at the peptide level. This becomes even more important when the goal is to analyze the secondary (lower frequency) cleavage preferences, to minimize the possibility that erroneous peptide identifications are mis-attributed as secondary cleavage sites. The notion of doubly-confirmed cleavage sites allowed us to limit the analysis to very reliable cleavage sites [3]. However, this approach may be too restrictive, since we do not always expect multiple peptides to begin or end at all real cleavage sites in the proteome (particularly for low-abundance proteins).
Below we suggest that the same level of stringency can be achieved with even higher sensitivity, if one can control the False Positive Rate (FPR) of individual Peptide-Spectrum matches, without restricting to doubly-confirmed cuts. Recently, Kim et al., 2008 [16] described MS-GeneratingFunction approach that computes the FPR of individual peptide identifications, as opposed to the False Discovery Rate of all peptide identifications computed using the standard target-decoy approaches. MS-GeneratingFunction, therefore, not only controls the overall error rate but also ensures that every individual peptide identification selected above the threshold is reliable. In particular, it identifies a much larger number of peptides with virtually 0% FDR (i.e, no peptides identified in decoy database for the same score threshold) than other popular tools (see [16]).
We used MS-GeneratingFunction to calculate the FPR of peptide identifications for each of the three proteases, and thresholds were chosen to limit the FDR to 0.1% (at par with doubly-confirmed cuts). From each identified peptide, a cleavage was inferred at its N-terminus. We noticed that cleavages at C-termini show increased frequency of basic amino acids (R/K) at P1 position (perhaps due to ionization bias and/or detection preferences of existing MS/MS database search tools) and, therefore, were not included in the current analysis1. Note that similar over-representation of carboxy-terminal R and K residues in peptide identifications has been previously reported in native peptides even in absence of trypsin digestions [7]. For each of the Supplementary Tables 2A, 3A and 4A (described below) generated using N-terminal cleavages of peptides, additional Supplementary Tables 2B, 3B and 4B are also provided showing the corresponding data using both N-terminal and C-terminal cleavages.
Possible post-digestion breakup products of intact peptides were filtered off as described earlier [3]. 13116, 5116 and 2698 cleavage sites were detected in V8 protease, chymotrypsin and CNBr digests respectively, a significant increase compared to the doubly-confirmed cleavage approach. Supplementary Table 2A shows the distribution of amino acids at the P1 position in these cleavages for each of the 3 proteases, and confirms the specificity trends obtained with doubly-confirmed sites in Table 1.
Thus, MS-GeneratingFunction can be used to detect reliable cleavage sites with the same stringency and accuracy as doubly-confirmed cleavages. The larger list of cleavage sites obtained through this approach, however, can be particularly valuable for detecting in vivo proteolytic events, as discussed in the next section.
3.7 Detection of putative in vivo regulatory proteolytic sites
While most of the cleavages detected in the proteome (after discarding the post-digestion breakup products) are generally expected to be produced by the protease used for digestion, biological samples may also contain some in vivo cleavages representing N-terminal methionine excisions, removal of signal peptides, and other regulatory proteolytic events.By subtracting the cleavage sites explained by the specificity of the protease (e.g., cleavages after R and K for trypsin), one can filter the list of all cleavage sites to find candidates for such in vivo proteolytic events [28]. However, extra evidence is usually required to confirm these candidate sites as regulatory proteolytic sites. For example, Gupta et al., 2008 [6] compared candidate sites from three Shewanella species to find a set of evolutionary conserved putative proteolytic sites. Here, we argue that detection of a cleavage site across different digests of the same proteome can also provide evidence to confirm in vivo proteolytic sites.
From the list of cleavage sites obtained by MS-GeneratingFunction at 0.1% error rate, the sites explained by the protease specificity were excluded. The specificity rules were kept broad (based on the results obtained above) to minimize the possibility of any in vitro cleavage being considerd as an in vivo cleavage. We excluded all cleavages with the following amino acids at P1 position: D/E/G for V8 protease, M/R/K for CNBr, and F/Y/W/L/R/K/M/H for chymotrypsin. Besides the three proteases analyzed in this study, we also used trypsin as the fourth protease for increasing the coverage, using data from Gupta et al., 2007 [28] (sites with R/K at P1 position were excluded [2]). 6226, 3728, 627 and 561 candidate proteolytic sites were detected in trypsin, V8 protease, chymotrypsin and CNBr digests respectively (using only N-terminal cleavages, as discussed in the previous section). 513 of these cleavage sites were found in two or more protease digests, including 28 that were found in three, and 3 that were present in all digests (Supplementary Table 3A). The complete lists of candidate proteolytic sites identified from each digest are provided in Supplementary Table 4A.
One of the 3 sites found in all digests is between A23-A24 in protein SO1164 (dacA-1). This site represents signal peptide cleavage site that was also predicted by SignalP and PrediSi [28, 36]. Another site detected in all digests is between A52-K53 in protein SO4509 (formate dehydrogenase, alpha subunit), which was previously detected in orthologous positions in two Shewanella species [6]. The third site is between T106-A107 in SO0417 which is annotated as putative pilin, and no prior knowledge is available for this protein. It is noteworthy that among the 513 sites present in two or more digests, 111 have A at P1 position, indicating the presence of many signal peptides, which are known to have a strong preference for A at P1 position in bacteria [28, 36].2 Similarly, while one would expect false sites to be distributed uniformly across the lengths of the proteins, the sites with A at P1 position tend to appear in the first 40 positions (as expected for signal peptides). Given an average length of ≈ 300 residues for Shewanella proteins, we expect only 2 of the 513 sites to start at the second position of the proteins by chance. However, we find 55 cleavage sites at this position indicating the presence of many N-terminal methionine excisions (NME) [28]. Comparative analysis of multiple digests, therefore, is a promising approach for reliable identification of regulatory proteolytic sites.
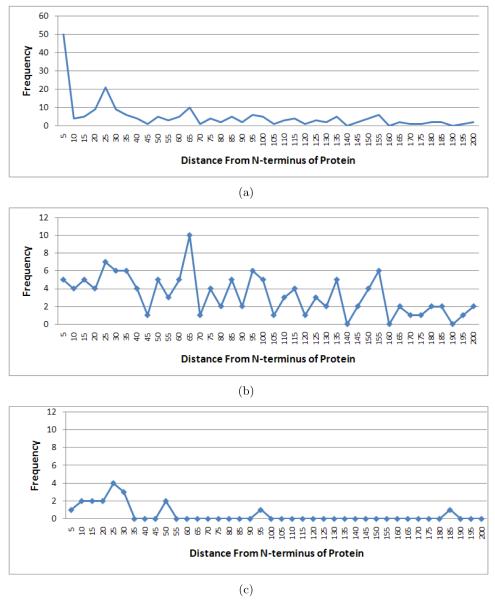
One can observe that many cleavage sites detected by MS-Proteolysis belong to highly expressed proteins. For example, the translation elongation factor Tu (tufB) has so many identified peptides (230) that they result in appearance of 21 putative cleavage sites in tufB generated by MS-Proteolysis. However, while tufB is known to undergo proteolysis in bacteria [37], most of these sites are likely to represent artifacts rather than real proteolytic events. For example, various degradation variants of a highly expressed protein may be detectable via MS/MS thus resulting in an artificial appearance of a cleavage site [8]. Since in vivo proteolytic events in such proteins are difficult to distinguish from artifacts, MS-Proteolysis generates an additional table that excludes all highly expressed proteins3 and reports only the remaining peptides. This filtering results in a set of 242 putative sites shown in Supplementary Table 5A. Figure 5(a) shows the distribution of the starting positions of the detected cleavage sites and reveals pronounced peaks at the beginning of the protein (NME) and around position 25 (signal peptides). We further removed from consideration NME sites expected from NME specificity rules [28] and signal peptide cleavage sites predicted by SignalP (Supplementary Table 5 lists the resulting 175 putative cleavage sites) and generated Figure 5(b) similar to Figure 5(a). Figure 5(b) still shows (a smaller) peak around position 25 indicating that SignalP failed to correctly predict some signal peptides. The peak becomes more pronounced, as shown in Figure 5(c), if we look at only the most upstream sites in proteins, indicating N-terminal proteolytic events like NME and signal peptide cleavage.4 Therefore, MS-Proteolysis is a useful tool for detecting the proteolytic events that software tools like SignalP miss. Since little is known about regulatory proteolysis in Shewanella apart from NME and signal peptides (CutDB database [38] does not report any proteolytic events in Shewanella), it remains to be verified which of these 175 putative cleavage sites represent in vivo proteolytic events. However, analysis of these sites reveals surprising biases that may warrant further studies. 33 out of these 175 sites have form A.* (19% of all sites) with surprisingly many A.A (11) and A.S (6) cleavages. While some of these sites may correspond to signal peptides missed by SignalP, others do not fit the profile of typical signal peptides and may reflect a still unknown proteolytic activity. Other surprisingly frequent cleavages are represented by Q.A and T.A (while these cleavages are expected to appear less than once by chance, they appear 6 and 7 times, respectively).
Figure 5.
(a) The histogram of positions in the corresponding protein sequence of the proteolytic sites in Supplementary Table 5. A bin size of 5 is used in the construction of histogram, and the plot is truncated at position 200 for brevity. (b) Similar plot as in (a), after removing the sites at second positions of proteins (NME) and those predicted as signal peptide cleavage sites by SignalP. (c) Similar plot as in (b), but keeping only the most upstream peptide detected in a protein (to infer N-terminal proteolytic events like NME or signal peptide cleavage [28]).
Supplementary Table 3A does not contain any site with K or R at P1 position, since these residues were included in the specificity rules for trypsin, CNBr and chymotrypsin digests, leaving only V8 protease that could identify such sites as candidates for in vivo proteolysis. Since we require a candidate proteolytic site to be identified in at least two digests, these K/R sites could not make it to the final list. However, some K/R sites may represent in vivo proteolytic sites, and we provide the list of all sites with K/R at P1 position that were identified in at least two of CNBr, V8 protease and chymotrypsin digests in Supplementary Table 6.
We also analyzed yeast (S. cerevisiae) proteome with MS-Proteolysis using trypsin digests. 7488 yeast peptides (identified with 0.1% FDR) yielded 11851 cleavage sites (Supplementary Table 7A), of which 1047 were not explained by trypsin specificity (Supplementary Table 7B) and represented candidates for in vivo proteolytic sites. One of the tryptic cleavage sites is listed in CutDB database for S. cerevisiae, corresponding to the cleavage by protease Kexin between positions R40-Y41 in the protein Exg1p [39]. Having only a small overlap between annotated proteolytic sites in CutDB and sites revealed by MS/MS in yeast may be indicative of (i) limited coverage of proteolytic sites in CutDB, (ii) the fact that some proteolytic events are not represented in MS/MS sample since they appear only under specific conditions, and (iii) peptide detectability limitations in MS/MS analysis [40].
4 Discussion
Mass spectrometry is a reliable technology to determine the specificity of enzymes. We had previously demonstrated its application for trypsin [3], which was known to be very specific [2]. Here, we studied the specificity of V8 protease, chymotrypsin and CNBr, validated the known specificity rules, and found some interesting deviations from these known rules for the conditions used. We are not claiming that the observed deviations from the canonical specificity rules represent the native specificity of the perfectly purified samples of these reagents, and we acknowledge that the observed deviations could vary across vendors or conditions. However, a typical mass spectrometry laboratory works with “real” rather than “perfect” reagents and thus it is important to have a pragmatic rather than idealistic method of deducing specificity. We argue that these pragmatic, rather than idealistic, rules for specificity may be more useful for downstream applications. Knowledge of these deviations is important for setting up parameters in database searches and for analysis of regulatory proteolysis using mass spectrometry. It is important to include all possible low-propensity cleavages produced by digestion-enzymes during mass spectrometry in the effective specificity rules of the enzyme, so that they are “subtracted” out from the list of possible cleavages when identifying in vivo proteolytic sites. Using comparative analysis of multiple proteases, we identified a set of putative in vivo proteolytic cleavage sites in Shewanella, which represent strong candidates for verification by future experiments. While some of these sites may represent various experimental and computational artifacts rather than than proteolytic cleavages, MS-Proteolysis represents the first step towards utilization of vast MS/MS datasets for studies of proteolysis.
Reliable peptide identifications are important for accurate determination of protease specificity from mass spectrometry. While we used ion-trap mass spectrometers to generate data for this study, we were able to keep the error rate extremely low by using doubly-confirmed cleavages or MS-GeneratingFunction. For future studies, using high precision instruments will be of additional help in detecting reliable cleavage sites. In the present study, we used In-sPecT database search without allowing optional modifications, and this approach proved capable of high-throughput detection of proteolytic cleavages. Future work could also include examination of modified peptides, especially for detecting proteolytic cleavages that are often accompanied by post-translational modifications.
Supplementary Material
5 Acknowledgments
This work was supported by NIH 5R01RR016522-05 and 1-P41-RR024851-01 grants and Howard Hughes Medical Institute Professor Award to PP. Portions of this research were supported by the NIH Center of Proteomics Research Resource for Integrative Biology RR018522 (to R.D.S.). The proteomics measurements were performed in the Environmental Molecular Sciences Laboratory, a U.S. Department of Energy (DOE) national scientific user facility on the PNNL campus. PNNL is multi-program national laboratory operated by Battelle for the DOE under Contract No. DE-AC05-76RLO 1830.
Abbreviations
- FDR
False Discovery Rate
- MS/MS
Tandem Mass Spectrometry
- NME
N-terminal Methionine Excision
Footnotes
Note that trypsin contamination alone does not explain this bias, since in that case, even the cleavages at N-termini of the peptides are expected to show similar preference for R/K at their P1 position, even if those R/K residues are not present in the detected peptides.
One would expect only ≈ 30 cleavage sites containing A at P1 by chance.
E.g., proteins that have over threshold = 50 identified peptides (threshold can be set to a different value by the users depending on the levels of protein degradation in their samples)
If the most upstream peptide identified in a protein (in a trypsin-digested sample, for example) starts at a non-tryptic position, it provides evidence for an N-terminal proteolytic event in the protein [28].
References
- [1].Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- [2].Olsen JV, Ong S, Mann M. Trypsin cleaves exclusively C-terminal to arginine and lysine residues. Mol Cell Proteomics. 2004;3:608–614. doi: 10.1074/mcp.T400003-MCP200. [DOI] [PubMed] [Google Scholar]
- [3].Rodriguez J, Gupta N, Smith RD, Pevzner PA. Does trypsin cut before proline? J Proteome Res. 2008;7:300–305. doi: 10.1021/pr0705035. [DOI] [PubMed] [Google Scholar]
- [4].Timmer JC, Enoksson M, Wildfang E, Zhu W, Igarashi Y, Denault JB, Ma Y, Dummitt B, Chang YH, Mast AE, Eroshkin A, Smith J, Tao WA, Salvesen GS. Profiling constitutive proteolytic events in vivo. Biochem. J. 2007;407:41–48. doi: 10.1042/BJ20070775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Enoksson M, Li J, Ivancic MM, Timmer JC, Wildfang E, Eroshkin A, Salvesen GS, Tao WA. Identification of Proteolytic Cleavage Sites by Quantitative Proteomics. J. Proteome Res. 2007;6:2850–2858. doi: 10.1021/pr0701052. [DOI] [PubMed] [Google Scholar]
- [6].Gupta N, Benhamida J, Bhargava V, Goodman D, Kain E, Nguyen N, Ollikainen N, Rodriguez J, Wang J, Lipton MS, Romine M, Bafna V, Smith RD, Pevzner PA. Comparative Proteogenomics: Combining Mass Spectrometry and Comparative Genomics to Analyze Multiple Genomes. Genome Res. 2008;18:1133–1142. doi: 10.1101/gr.074344.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Manes NP, Gustin JK, Rue J, Mottaz HM, Purvine SO, Norbeck AD, Monroe ME, Zimmer JSD, Metz TO, Adkins JN, et al. Targeted Protein Degradation by Salmonella under Phagosome-mimicking Culture Conditions Investigated Using Comparative Peptidomics. Molecular and Cellular Proteomics. 2007;6:717–727. doi: 10.1074/mcp.M600282-MCP200. [DOI] [PubMed] [Google Scholar]
- [8].Shen Y, Hixson KK, Tolic N, Camp DG, Purvine SO, Moore RJ, Smith RD. Mass Spectrometry Analysis of Proteome-Wide Proteolytic Post-Translational Degradation of Proteins. Analytical Chemistry. 2008;80:5819–5828. doi: 10.1021/ac800077w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Keil B. Specificity of Proteolysis. Springer-Verlag Berlin; Germany: 1992. [Google Scholar]
- [10].Rano TA, Timkey T, Peterson EP, Rotonda J, Nicholson DW, Becker JW, Chapman KT, Thornberry NA. A combinatorial approach for determining protease specificities: application to interleukin-1 converting enzyme (ICE) Chem. Biol. 1997;4:149–155. doi: 10.1016/s1074-5521(97)90258-1. [DOI] [PubMed] [Google Scholar]
- [11].Harris JL, Backes BJ, Leonetti F, Mahrus S, Ellman JA, Craik CS. Rapid and general profiling of protease specificity by using combinatorial fluorogenic substrate libraries. Proc Natl Acad Sci US A. 2000;97:7754–9. doi: 10.1073/pnas.140132697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Turk BE, Huang LL, Piro ET, Cantley LC. Determination of protease cleavage site motifs using mixture-based oriented peptide libraries. Nature Biotechnology. 2001;19:661–667. doi: 10.1038/90273. [DOI] [PubMed] [Google Scholar]
- [13].Schilling O, Overall CM. Proteome-derived, database-searchable peptide libraries for identifying protease cleavage sites. Nature Biotechnology. 2008;26:685–694. doi: 10.1038/nbt1408. [DOI] [PubMed] [Google Scholar]
- [14].Bandeira N, Tsur D, Frank A, Pevzner PA. Protein identification by spectral networks analysis. PNAS. 2007;104:6140–6145. doi: 10.1073/pnas.0701130104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Bandeira N, Pham V, Pevzner P, Arnott D, Lill JR. Automated de novo protein sequencing of monoclonal antibodies. Nature Biotechnology. 2008;26:1336–1338. doi: 10.1038/nbt1208-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kim S, Gupta N, Pevzner PA. Spectral Probabilities and Generating Functions of Tandem Mass Spectra: a Strike Against Decoy Databases. J. Proteome Res. 2008;7:3354–3363. doi: 10.1021/pr8001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lopez-Ferrer D, Hixson KK, Smallwood H, Squier TC, Petritis K, Smith RD. Evaluation of a high-intensity focused ultrasound-immobilized trypsin digestion and 18O-labeling method for quantitative proteomics. Anal. Chem. 2009;81:6272–6277. doi: 10.1021/ac802540s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Salplachta J, Marchetti M, Chmelik J, Allmaier G. A new approach in proteomics of wheat gluten: combining chymotrypsin cleavage and matrix-assisted laser desorption/ionization quadrupole ion trap reflectron tandem mass spectrometry. Rapid communications in mass spectrometry. 2005;19:2725–2728. doi: 10.1002/rcm.2092. [DOI] [PubMed] [Google Scholar]
- [19].Hixson KK, Adkins JN, Baker SE, Moore RJ, Chromy BA, Smith RD, McCutchen-Maloney SL, Lipton MS. Biomarker candidate identification in Yersinia pestis using organism-wide semiquantitative pro-teomics. J. Proteome Res. 2006;5:3008–3017. doi: 10.1021/pr060179y. [DOI] [PubMed] [Google Scholar]
- [20].Shen Y, Tolic N, Hixson KK, Purvine SO, Pasa-Tolic L, Qian WJ, Adkins JN, Moore RJ, Smith RD. Proteome-wide identification of proteins and their modifications with decreased ambiguities and improved false discovery rates using unique sequence tags. Anal. Chem. 2008;80:1871–1882. doi: 10.1021/ac702328x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Masselon C, Pasa-Tolic L, Tolic N, Anderson GA, Bogdanov B, Vilkov AN, Shen Y, Zhao R, Qian WJ, Lipton MS, et al. Targeted comparative proteomics by liquid chromatography-tandem Fourier ion cyclotron resonance mass spectrometry. Anal. Chem. 2005;77:400–406. doi: 10.1021/ac049043e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Hixson KK, Rodriguez N, Camp DG, II, Strittmatter EF, Lipton MS, Smith RD. Evaluation of enzymatic digestion and liquid chromatography-mass spectrometry peptide mapping of the integral membrane protein bacteriorhodopsin. Electrophoresis. 2002;23:3224–3232. doi: 10.1002/1522-2683(200209)23:18<3224::AID-ELPS3224>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- [23].Qian WJ, Liu T, Monroe ME, Strittmatter EF, Jacobs JM, Kangas LJ, Petritis K, Camp DG, Smith RD. Probability-based evaluation of peptide and protein identifications from tandem mass spectrometry and SEQUEST analysis: the human proteome. J. Proteome Res. 2005;4:53–62. doi: 10.1021/pr0498638. [DOI] [PubMed] [Google Scholar]
- [24].Shen Y, Tolic N, Zhao R, Pasa-Tolic L, Li L, Berger SJ, Harkewicz R, Anderson GA, Belov ME, Smith RD. High-throughput proteomics using high-efficiency multiple-capillary liquid chromatography with on-line high-performance ESI FTICR mass spectrometry. Anal. Chem. 2001;73:3011–3021. doi: 10.1021/ac001393n. [DOI] [PubMed] [Google Scholar]
- [25].Smith RD, Anderson GA, Lipton MS, Pasa-Tolic L, Shen Y, Conrads TP, Veenstra TD, Udseth HR. An accurate mass tag strategy for quantitative and high-throughput proteome measurements. Proteomics. 2002;2:513–523. doi: 10.1002/1615-9861(200205)2:5<513::AID-PROT513>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- [26].Tanner S, Shu H, Frank A, Wang L, Zandi E, Mumby M, Pevzner PA, Bafna V. Inspect: Fast and accurate identification of post-translationally modified peptides from tandem mass spectra. Anal. Chem. 2005;77:4626–4639. doi: 10.1021/ac050102d. [DOI] [PubMed] [Google Scholar]
- [27].Stephan C, Reidegeld KA, Hamacher M, van Hall A, Marcus K, Taylor C, Jones P, Muller M, Apweiler R, Martens L, et al. Automated reprocessing pipeline for searching heterogeneous mass spectrometric data of the HUPO Brain Proteome Project pilot phase. Proteomics. 2006;6:5015–5029. doi: 10.1002/pmic.200600294. [DOI] [PubMed] [Google Scholar]
- [28].Gupta N, Tanner S, Jaitly N, Adkins J, Lipton M, Edwards R, Romine M, Osterman A, Bafna V, Smith R, Pevzner P. Whole proteome analysis of post-translational modifications: applications of mass-spectrometry for proteogenomic annotation. Genome Res. 2007;17:1362–1377. doi: 10.1101/gr.6427907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Desiere F, Deutsch E, Nesvizhskii A, Mallick P, King N, Eng J, Aderem A, Boyle R, Brunner E, Donohoe S, et al. Integration with the human genome of peptide sequences obtained by high-throughput mass spectrometry. Genome Biology. 2004;6:R9. doi: 10.1186/gb-2004-6-1-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Houmard J, Drapeau GR. Staphylococcal Protease: A Proteolytic Enzyme Specific for Glutamoyl Bonds. Proceedings of the National Academy of Sciences of the United States of America. 1972;69:3506–3509. doi: 10.1073/pnas.69.12.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Sorensen SB, Sorensen TL, Breddam K. Fragmentation of proteins by S. aureus strain V 8 protease: Ammonium bicarbonate strongly inhibits the enzyme but does not improve the selectivity for glutamic acid. FEBS Letters. 1991;294:195–197. doi: 10.1016/0014-5793(91)80667-r. [DOI] [PubMed] [Google Scholar]
- [32].Gross E, Witkop B. Selective cleavage of the methionyl peptide bonds in ribonuclease with cyanogen bromide1. Journal of the American Chemical Society. 1961;83:1510–1511. [Google Scholar]
- [33].Schellenberger V, Braune K, Hofmann HJ, Jakubke HD. The specificity of chymotrypsin. Eur. J. Biochem. 1991;199:623–636. doi: 10.1111/j.1432-1033.1991.tb16163.x. [DOI] [PubMed] [Google Scholar]
- [34].Crooks GE, Hon G, Chandonia J, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Austen BM, Smith EL. Action of staphylococcal proteinase on peptides of varying chain length and composition. Biochem Biophys Res Commun. 1976;72:411–417. doi: 10.1016/s0006-291x(76)80058-7. [DOI] [PubMed] [Google Scholar]
- [36].Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- [37].Georgiou T, Yu YTN, Ekunwe S, Buttner MJ, Zuurmond AM, Kraal B, Kleanthous C, Snyder L. Specific peptide-activated proteolytic cleavage of Escherichia coli elongation factor Tu. Proceedings of the National Academy of Sciences. 1998;95:2891–2895. doi: 10.1073/pnas.95.6.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Igarashi Y, Eroshkin A, Gramatikova S, Gramatiko K, Zhang Y, Smith JW, Osterman AL, Godzik A. CutDB: a proteolytic event database. Nucleic Acids Research. 2007;35:D546. doi: 10.1093/nar/gkl813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Bader O, Krauke Y, Hube B. Processing of predicted substrates of fungal Kex2 proteinases from Candida albicans, C. glabrata, Saccharomyces cerevisiae and Pichia pastoris. BMC microbiology. 2008;8:116. doi: 10.1186/1471-2180-8-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Mallick P, Schirle M, Chen SS, Flory MR, Lee H, Martin D, Ranish J, Raught B, Schmitt R, Werner T, et al. Computational prediction of proteotypic peptides for quantitative proteomics. Nat Biotechnol. 2007;25:125–131. doi: 10.1038/nbt1275. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.