Abstract
Background/Aim:
Resistance to clarithromycin in H. pylori isolates is accepted as a main cause of treatment failure in developing countries. We aimed to determine the prevalence of clarithromycin-resistant strains isolated from dyspeptic patients in northern Iran, furthermore we aimed to assess the relationship between clinical outcomes of infection with point mutations.
Materials and Methods:
A total of 147 consecutive patients infected with H. pylori were included for determining the status of resistant H. pylori strains. With upper gastroscopy, three antral biopsies were taken from each patient, first section for rapid urea test, second for pathology and third section was used for bacterial culture in microbiologic lab. The antimicrobial susceptibility tests in this examination were agar dilution, in accordance with clinical and laboratory standards institue guidelines. Restriction fragment length polymorphism-PCR (RFLP-PCR) method was applied to determine the frequency of point mutations in 23s rRNA gene. Statistical analysis was performed using SPSS software (15.0) (SPSS, Inc., Chicago, Ill). Chi-square and Fisher's exact tests were applied to our analysis. A P value less than 5% was considered as statistically significant.
Results:
Our results showed that there was no point mutation in clarithromycin-susceptible strains of H. pylori.
Conclusion:
The important findings in our study indicate that A2143G is the most prevalent point mutation (30/32: 93.7%) attributed in clarithromycin resistance among the H. pylori strains. The current study concluded that clarithromycin could still be involved in the empirical treatment of H. pylori infection, although a high frequency of A2143G mutation may increase the concerns regarding treatment failure.
Keywords: Clarithromycin, Helicobacter pylori, Iran
Helicobacter pylori is a Gram-negative, urease-positive and microaerophilic bacteria which colonizes the gastric mucosa of 50% of the world's population.[1] The rate of H. pylori infection in developing countries such as Iran is up to 80-90%.[2,3] Infection with H. pylori is considered a major cause of chronic gastritis and peptic ulcer disease.[1] Resistance to clarithromycin among H. pylori isolates is accepted as a main explanation of treatment failure.[4,5] Although there have been supportive reports regarding the importance of effective therapeutic regimens against H. pylori, other data have shown that current guidelines are not optimized enough.[5,6] Emergence of drug resistance to H. pylori isolates reduces the eradication rate of treatment.[7,8] Amoxicillin, metronidazole, clarithromycin and tetracycline are the most recommended antibiotics used in H. pylori eradication regimens. Clarithromycin resistance in H. pylori is associated with treatment failure, although geographical variations were observed.[5,6,9] Eradication of H. pylori is recommended for the treatment of disorders including MALT lymphoma, gastric ulcer, gastritis, duodenal ulcer and gastric cancer.[4,6] The prevalence of H. pylori resistant to clarithromycin is strongly dependent on the location of the study.[10,11] The prevalence of A2143G and A2144G in clinical H. pylori isolates has not been investigated in Iran and other Mid-Eastern countries. To date, no published data is available which investigates the frequency of point mutations attributed with clarithromycin resistance in Iran. The current study was undertaken to 1) determine the prevalence of clarithromycin-resistant strains isolated from dyspeptic patients in northern Iran, 2) assess the relationship between clinical outcomes of infection and point mutations.
MATERIALS AND METHODS
Patients and H. pylori strains
Clinical isolates of H. pylori were taken from 147 consecutive patients with digestive disorders and were investigated for the presence of point mutations in the 23s rRNA gene. Our sample collection began in December 2008, and lasted till November 2010 in Tooba Medical Center, Sari, Iran. With upper gastroscopy, three antral biopsies were taken, the first section for rapid urea test (RUT), second for pathology and the third section was used for bacterial culture in a microbiologic laboratory. All participants signed the informed consent forms and our study was approved by the ethical committee of Tarbiat Modares University, Tehran, Iran. Our exclusion criteria were: treatment with omeperazole and H2-blocker for at least three months prior to endoscopy, gastrointestinal surgery, history of allergic reaction, and consumption of anti-H. pylori antibiotics during the last four months. The severity of gastroduodenal diseases was diagnosed by pathology and endoscopic findings.
Susceptibility tests
For bacterial culture, antral biopsies were ground softly and then 100 μl of homogenate was spread on Colombia agar (Merck, Germany) plates. Culture medium consisted of Columbia agar (Merck, Germany), supplemented with 10% FBS (Fetal Bovian Serum) (Gibco, USA), 7% defibrinated sheep blood (Jihad Daneshgahi, Iran) and selectab disks (MAST, UK).[5] Plates were incubated at 37°C, 8% CO2 and 90% humidity conditions.[5] All plates were checked after five days while H. pylori identity was confirmed by biochemical tests such as oxidase, catalase, urease and Gram staining.[5] The antimicrobial susceptibility tests in this examination were agar dilution, in accordance with CLSI guidelines.[12] Plates were incubated for four days, and the Minimum inhibitory concentration (MIC) was recorded as the lowest concentration of the antibiotic inhibiting visible growth. The resistance breakpoints for clarithromycin were ≥1 mg/L, although other breakpoints are reported.[5] Chromosomal DNA was extracted from a single colony H. pylori.[5]
DNA Extraction
Genomic content of bacteria was extracted from a fresh culture using the QIAamp DNA mini kit (Qiagen GmbH, Hilden, Germany), according to manufacturer's instructions. Thereafter, we stored DNA at -20°C for further analysis.
Determination of point mutations in 23s rRNA
We confirmed our H. pylori identity with PCR assay for glmM gene amplification.[13] Presence of point mutations (A2143G and A2144G) in 23s rRNA were analyzed by PCR-RFLP method as described before.[14–16] In this study, following 23s rRNA gene amplification,[14] we performed RFLP for detection of 23s rRNA mutations in the amplified sequences.[14–17] In brief, we used the BpiI enzyme (Fermantas, Germany) and BsaI (New England Biolabs, USA) to detect the A2143G and A2144G mutations, respectively. Finally, the products were visualized under UV-illuminator after electrophoresis with 2% agarose gel stained with ethidium bromide.
Analysis
Statistical analysis was performed using SPSS software (15.0) (SPSS, Inc., Chicago, Ill). Chi-square and Fisher's exact tests were applied to our analysis. A P value less than 0.05 was considered as statistically significant.
RESULTS
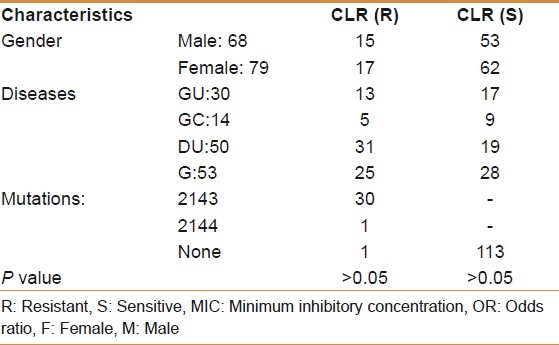
Of 147 consecutive patients (with both RUT and culture being positive) in this study, 53 were diagnosed with gastritis, 50 with duodenal ulcer, 30 with gastric ulcer and 14 with gastric cancer. Sixty-eight out of 147 patients were males (46.2%); age range (21-72) with mean age 34.5 years. Our findings showed that 32 patients (21.7%) were harboring a single colony of resistant H. pylori strains to clarithromycin [Table 1]. The frequency of clarithromycin resistance was similar between gastric ulcer (7/30, 23.3%), gastric cancer (3/14, 21.4%), duodenal ulcer (11/50, 22%) and gastritis patients (11/53, 20.7%). We found no significant association between presence of resistant strains and clinical outcomes of infection (P>0.05). In our examinations, we did not detect any point mutation in the 23s rRNA in all the clarithromycin-susceptible H. pylori strains. In total, 31 of 147 strains were harboring the mutations in 23s rRNA gene and just one of them was in the wild-type. The predominant mutations among the 32 H. pylori clarithromycin-resistant strains were at A2143G in 30 cases (93.7%) and A2144G in one case (3.1%). However, no dual point mutations were observed in the same H. pylori isolate. In other words, among all the resistant isolates, only one point mutation was detected, either A2143G or A2144G. No associations were observed between items like age, gender and dyspeptic disorders and resistance status of H. pylori isolates (P>0.05). According to the agar dilution method in this study, a total of 32 strains were resistant, while in the RFLP-PCR assay, we detected 31 of them as resistant strains. A significant correlation was observed between the two mentioned methods (P=0.002).
Table 1.
Distribution of 147 consecutive patients infected with H. pylori based on age and resistant strains
DISCUSSION
It has been stated that, if properly followed, efficient therapeutic practice can result in acceptable eradication rate of H. pylori infection in the mucosal surface of the gastric epithelium.[5,6]
Clarithromycin is the most important antibiotic included in the standard triple therapies for H. pylori eradication established worldwide.[6] Primary clarithromycin resistance in H. pylori is a considerable cause of treatment failure in all eradication therapies, while its prevalence varies geographically, from Western to Eastern countries.[2,6,10] Determination of the exact prevalence of clarithromycin-resistant H. pylori in each area in patients with gastroduodenal disorders can elucidate the efficacy of this critical antibiotic. Since clarithromycin shows strong antibacterial effects against H. pylori infection, it can be considered as a key antibiotic in the treatment of this gastric pathogen. Remarkably, the global resistance rate of clarithromycin is increasing; a fact which clarifies the need to have up-to-date information regarding its resistance status in our region. H. pylori resistance to clarithromycin is attributed to A2143G and A2144G transition mutations in the 23s rRNA gene, a mutation which results in a decrease in the affinity of clarithromycin binding to the ribosome. In Europe, the prevalence of clarithromycin-resistant H. pylori seems to range between 10-45%,[11,18–20] a rate which is lower in other countries of the world.[9,21,22] Our results showed that there was no point mutation in clarithromycin-susceptible strains of H. pylori.
The increasing usage of clarithromycin in developing countries, mostly for respiratory tract infections, has caused a large number of clarithromycin-resistant H. pylori strains. A few studies have worked on identifying the mutations involved in clarithromycin resistance in Iran,[22] and our study was the first examination for determining the point mutations in the 23s rRNA gene of H. pylori strains. In contrast with other studies,[23] our results showed that clarithromycin resistance was higher among gastric ulcer patients (23.3%) than other groups, although we did not observe a significant difference (P>0.05). There is no previous study in Iran to have an accurate comparison but we can declare that this study can be a good basis for further investigations in this country. The variation of the eradication rates obtained by the different studies may be due to the clarithromycin resistance in H. pylori isolates. According to studies in Iran, our resistance rate (21.7%) was high;[24–26] although another study from Turkey[27] was supportive of our result. It appears that clarithromycin resistance among H. pylori strains is strongly population-dependent.[28] According to current findings, no point mutations were detected in clarithromycin-susceptible strains of H. pylori, a finding which has not been reported in Iran yet. The important findings in our study indicate that A2143G is the most prevalent point mutation attributed with clarithromycin resistance among the H. pylori strains. However, more investigations with a bigger sample size are required to obtain a more encompassing image of the prevalence of mutations related to clarithromycin resistance in Iran and other Asian countries. Meantime, the main reason of the high level resistance in H. pylori is unclear, there is a possibility that another mechanism may be active to confering the high clarithromycin resistance in H. pylori. However, it has been reported that the presence of A2143G is the main cause of emergence of resistance in H. pylori strains.[29,30] Remarkably, De Francesco et al., showed that A2143G mutation could be associated with treatment failure in their patients.[30]
In conclusion, the A2143G remains the most prevalent mutation contributing to the resistance phenomena in H. pylori, suggesting that novel therapeutic approaches should be designed.
Nonetheless, several point mutations have been already investigated, the large majority of the primary clarithromycin resistance items depends on the point mutations (A2143G, A2144G), which have been related to different MIC values in vitro.[4,19,29]
The guideline for the eradication of H. pylori in Iran consists of an H2 receptor blocker (such as ranitidine), two antibacterial agents, such as metronidazole, amoxicillin/clarithromycin plus a bismuth salt; however, based on our results it would be clear that we need to urgently modify the current regimen to a more effective therapeutic formula for the treatment of this gastric pathogen.
CONCLUSION
Current findings indicate that clarithromycin can be included in the empirical treatment of H. pylori in Iran. Clarithromycin resistance appears to be less prevalent in Iranian dyspeptic patients. Continued surveillance of antibiotic susceptibility tests specially for determining the exact prevalence of A2143G mutation will be essential for the design of the best effective eradication therapy.
ACKNOWLEDGMENT
This work was supported financially by an MSc thesis in Tarbiat Modares University (Tehran, Iran).
Footnotes
Source of Support: This work was supported financially by an MSc thesis in Tarbiat Modares University (Tehran, Iran)
Conflict of Interest: None declared.
REFERENCES
- 1.Kusters JG, van Vliet AH, Kuipers EJ. Pathogenesis of Helicobacter pylori Infection. Clin Microbiol Rev. 2006;19:449–90. doi: 10.1128/CMR.00054-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Massarrat S, Saberi-Firoozi M, Soleimani A, Himmelmann GW, Hitzges M, Keshavarz H. Peptic ulcer disease, irritable bowel syndrome and constipation in two populations in Iran. Eur J Gastroenterol Hepatol. 1995;7:427–33. [PubMed] [Google Scholar]
- 3.Mansour-Ghanaei F, Yousefi Mashhour M, Joukar F, Sedigh M, Bagher-Zadeh AH, Jafarshad R. Prevalence of Helicobacter pylori infection among children in Rasht, Northern Iran. Middle East J Dig Dis. 2009;1:84–8. [Google Scholar]
- 4.Megraud F. H. pylori antibiotic resistance: Prevalence, importance, and advances in testing. Gut. 2004;53:1374–84. doi: 10.1136/gut.2003.022111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Megraud F, Lehours P. Helicobacter pylori detection and antimicrobial susceptibility testing. Clin Microbiol Rev. 2007;20:280–322. doi: 10.1128/CMR.00033-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Connor A, Gisbert JP, McNamara D, O’Morain C. Treatment of Helicobacter pylori Infection 2010. Helicobacter. 2010;15(Suppl 1):46–52. doi: 10.1111/j.1523-5378.2010.00774.x. [DOI] [PubMed] [Google Scholar]
- 7.Alarcon T, Domingo D, Lopez-Brea M. Antibiotic resistance problems with Helicobacter pylori. Int J Antimicrob Agents. 1999;12:19–26. doi: 10.1016/s0924-8579(99)00051-5. [DOI] [PubMed] [Google Scholar]
- 8.Stone GG, Shortridge D, Flamm RK, Versalovic J, Beyer J, Idler K, et al. Identification of a 23S rRNA gene mutation in clarithromycin-resistant Helicobacter pylori. Helicobacter. 1996;1:227–8. doi: 10.1111/j.1523-5378.1996.tb00043.x. [DOI] [PubMed] [Google Scholar]
- 9.Haghi Tomatari F, Mohabbati Mobarez A, Amini M, Hosseini D, Talebi Bezmin Abadi A. Helicobacter pylori resistance to metronidazole and clarithromycin in dyspeptic patients in Iran. IRCMJ. 2010;12:409–12. [Google Scholar]
- 10.Boyanova L, Nikolov R, Lazarova E, Gergova G, Katsarov N, Kamburov V, et al. Antibacterial resistance in Helicobacter pylori strains isolated from Bulgarian children and adult patients over 9 years. J Med Microbiol. 2006;55:65–8. doi: 10.1099/jmm.0.46208-0. [DOI] [PubMed] [Google Scholar]
- 11.Agudo S, Peerez-Perez G, Alarcon T, Lopez-Brea M. High prevalence of clarithromycin-resistant Helicobacter pylori strains and risk factors associated with resistance in Madrid, Spain. J Clin Microbiol. 2010;48:3703–7. doi: 10.1128/JCM.00144-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Performance standards for antimicrobial susceptibility testing; fifteenth information supplement. Wayne, PA, USA: 2007. Clinical and Laboratory Standards Institute (CLSI) [Google Scholar]
- 13.Ge Z, Taylor DE. Rapid PCR screening of Helicobacter pylori chromosomal point mutations. Helicobacter. 1997;2:127–31. doi: 10.1111/j.1523-5378.1997.tb00073.x. [DOI] [PubMed] [Google Scholar]
- 14.Taylor DE, Ge Z, Purych D, Lo T, Hiratsuka K. Cloning and sequence analysis of two copies of a 23S rRNA gene from Helicobacter pylori and association of clarithromycin resistance with 23S rRNA mutations. Antimicrob Agents Chemother. 1997;41:2621–8. doi: 10.1128/aac.41.12.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Versalovic J, Shortridge D, Kibler K, Griffy MV, Beyer J, Flamm RK, et al. Mutations in 23S rRNA are associated with clarithromycin resistance in Helicobacter pylori. Antimicrob Agents Chemother. 1996;40:477–80. doi: 10.1128/aac.40.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sevin E, Lamarque D, Delchier JC, Soussy CJ, Tankovic J. Co-detection of Helicobacter pylori and of its resistance to clarithromycin by PCR. FEMS Microbiol Lett. 1998;165:369–72. doi: 10.1111/j.1574-6968.1998.tb13172.x. [DOI] [PubMed] [Google Scholar]
- 17.Miehlke S, Yu J, Schuppler M, Frings C, Kirsch C, Negraszus N, et al. Helicobacter pylori vacA, iceA and cagA status and pattern of gastritis in patients with malignant and benign gastroduodenal disease. Am J Gastroentrol. 2001;96:1008–13. doi: 10.1111/j.1572-0241.2001.03685.x. [DOI] [PubMed] [Google Scholar]
- 18.Glupczynski Y, Megraud F, Lopez-Brea M, Andersen LP. European multicenter survey of in vitro antimicrobial resistance in Helicobacter pylori. Eur J Clin Microbiol Infect Dis. 2001;20:820–3. doi: 10.1007/s100960100611. [DOI] [PubMed] [Google Scholar]
- 19.Van Doorn LJ, Figueiredo C, Sanna R, Plaisier A, Schneeberger P, de Boer W, et al. Clinical relevance of the cagA, vacA, and iceA status of Helicobacter pylori. Gastroenterology. 1998;115:58–66. doi: 10.1016/s0016-5085(98)70365-8. [DOI] [PubMed] [Google Scholar]
- 20.Simsek H, Balaban YH, Gunes DD, Hascelik G, Ozarslan E, Tatar G. Alarming clarithromycin resistance of Helicobacter pylori in Turkish population. Helicobacter. 2005;10:360–1. doi: 10.1111/j.1523-5378.2005.00325.x. [DOI] [PubMed] [Google Scholar]
- 21.Khan R, Nahar S, Sultana J, Ahmad MM, Rahman M. T2182C Mutation in 23S rRNA is associated with clarithromycin resistance in Helicobacter pylori isolates obtained in Bangladesh. Antimicrob Agents Chemother. 2004;48:3567–9. doi: 10.1128/AAC.48.9.3567-3569.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mohammadi M, Doroud D, Mohajerani N, Massarrat S. Helicobacter pylori antibiotic resistance in Iran. World J Gastroenterol. 2005;11:6009–13. doi: 10.3748/wjg.v11.i38.6009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Broutet N, Tchamgoue S, Pereira E, Lamouliatte H, Salamon R, Megraud F. Risk factors for failure of Helicobacter pylori therapy-results of an individual data analysis of 2751 patients. Aliment Pharmacol Ther. 2003;17:99–109. doi: 10.1046/j.1365-2036.2003.01396.x. [DOI] [PubMed] [Google Scholar]
- 24.Khashei R, Shojaei H, Adibi P, Shavakhi A, Aslani MM, Naser AD. Genetic diversity and drug resistance of Helicobacter pylori strains in Isfahan, Iran. Iran J Basic Med Sci. 2008;11:174–82. [Google Scholar]
- 25.Siavoshi F, Pourkhajeh AH, Merat S, Asl-Soleimani H, Heidarian E, Khatibian M, et al. Susceptibility of various strains of Helicobacter pylori to selected agents. Arch Iran Med. 2000;3:60–3. [Google Scholar]
- 26.Falsafi T, Mobasheri F, Nariman F, Najafi M. Susceptibilities to different antibiotics of Helicobacter pylori strains isolated from patients at the pediatric medical center of Tehran, Iran. J Clin Microbiol. 2004;42:387–9. doi: 10.1128/JCM.42.1.387-389.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bakir Ozbey S, Ozakin C, Keskin M. Antibiotic resistance rates of Helicobacter pylori isolates and the comparison of E-test and fluorescent in situ hybridization methods for the detection of clarithromycin resistant strains. Mikrobiyol Bul. 2009;43:227–34. [PubMed] [Google Scholar]
- 28.Kalach N, Serhal L, Asmar E, Campeotto F, Bergeret M, Dehecq E, et al. Helicobacter pylori primary resistant strains over 11 years in French children. Diagn Microbiol Infect Dis. 2007;59:217–22. doi: 10.1016/j.diagmicrobio.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 29.De Francesco V, Margiotta M, Zullo A, Hassan C, Giorgio F, Burattini O, et al. Prevalence of primary clarithromycin resistance in Helicobacter pylori strains over a 15 year period in Italy. J Antimicrob Chemother. 2007;59:783–5. doi: 10.1093/jac/dkm005. [DOI] [PubMed] [Google Scholar]
- 30.De Francesco V, Margiotta M, Zullo A, Hassan C, Troiani L, Burattini O, et al. Clarithromycin-resistant genotypes and eradication of Helicobacter pylori. Ann Intern Med. 2006;144:94–100. doi: 10.7326/0003-4819-144-2-200601170-00006. [DOI] [PubMed] [Google Scholar]


