Abstract
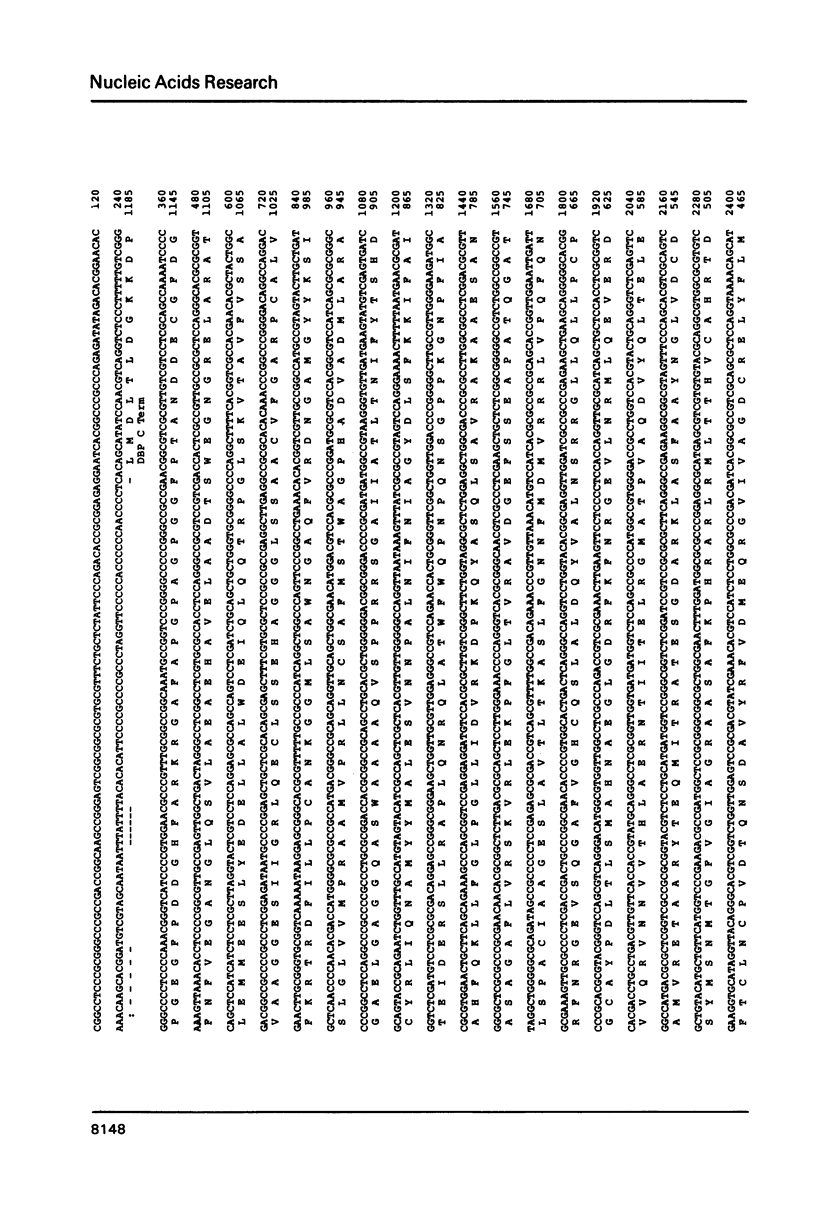
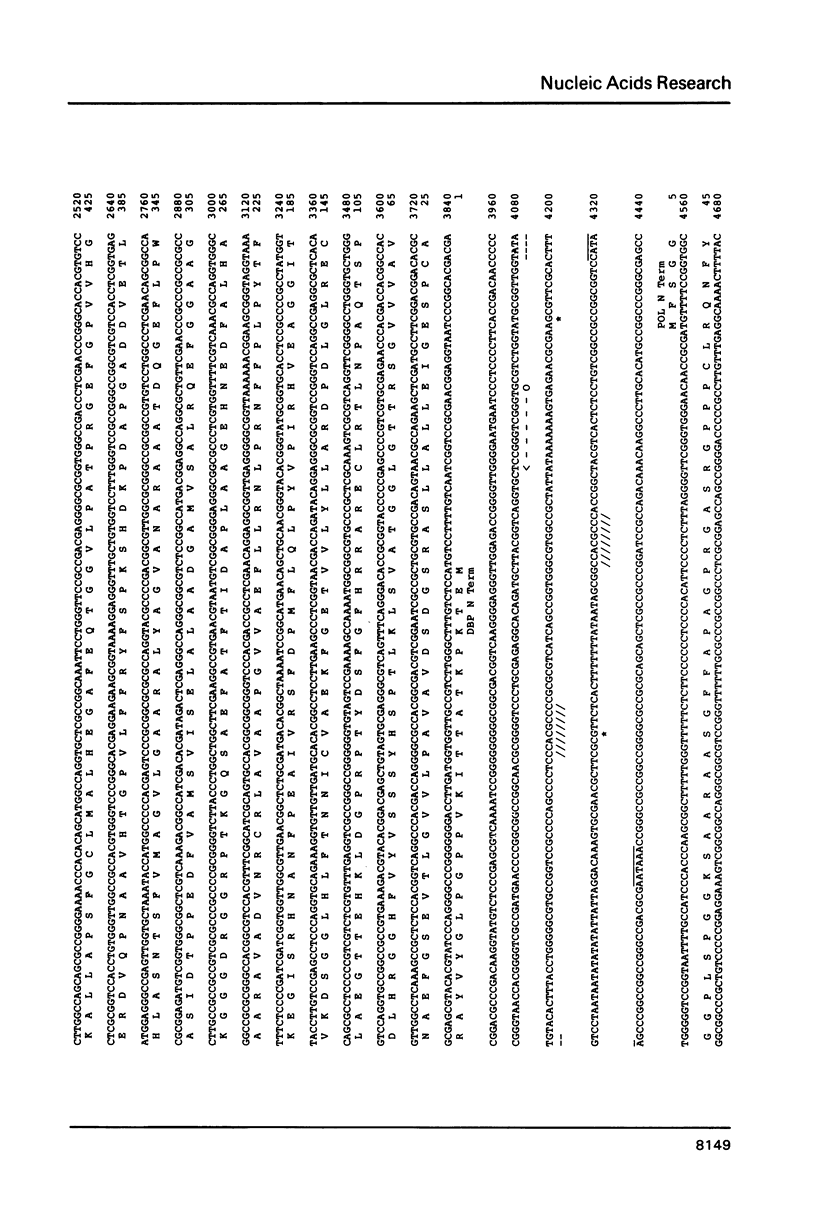
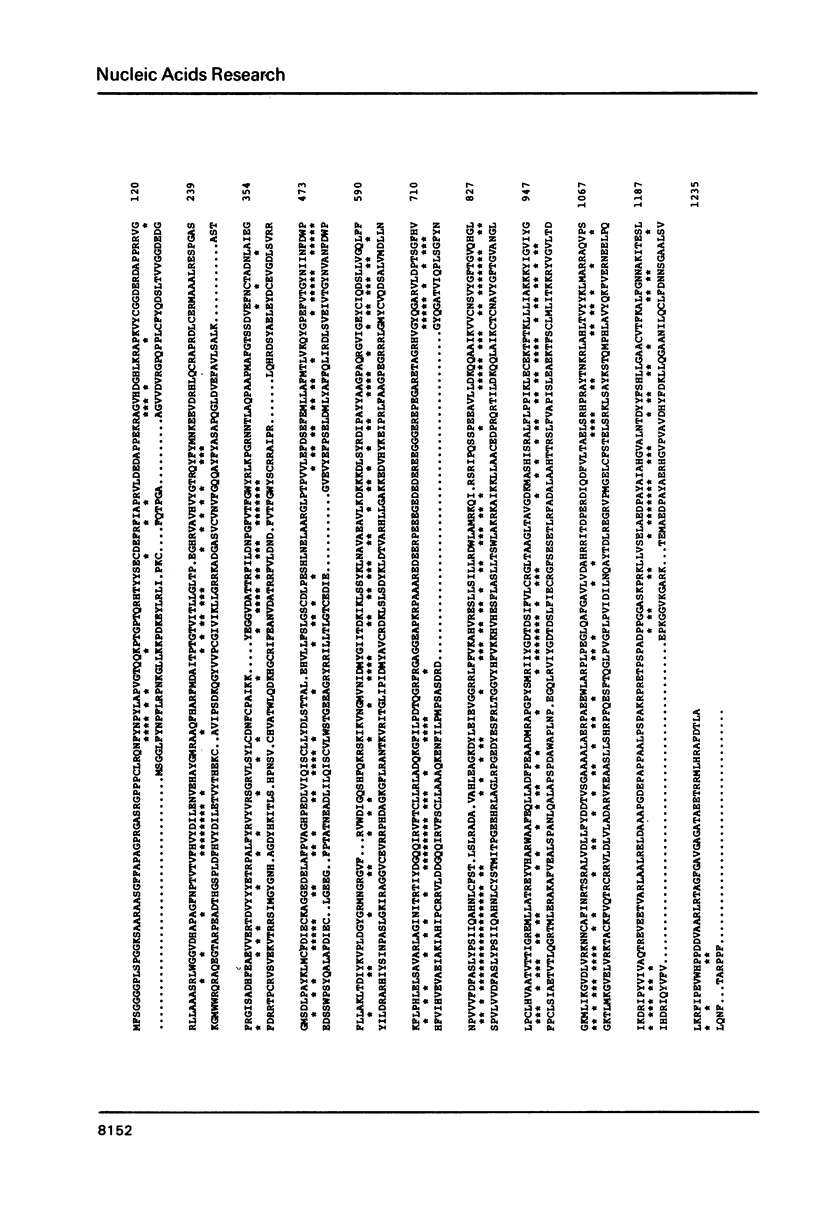
In the long unique region of the genome of herpes simplex virus type 1 (HSV-1), the genes for DNA polymerase and the major DNA binding protein are arranged in a head to head manner, with an origin of DNA replication (termed OriL) located between them. This paper reports an 8400 base pair DNA sequence containing both genes and the origin, obtained mostly by M13/dideoxy analysis of plasmid cloned fragments. Amino acid sequences of the two proteins were deduced. Homologues of both genes were detected in the genome sequence of the distantly related Epstein-Barr virus (EBV). Arrangement of these HSV-1 and EBV genes differs in genome location and in relative orientation. A part of HSV-1 DNA polymerase was found to be similar to a sequence in adenovirus 2 DNA polymerase, but the significance of this is unclear. Since a DNA sequence in the locality of OriL deletes on plasmid cloning, this region was analysed using virus DNA. A palindrome with 72-residue arms was found, which shows great similarity to the better characterized origin, OriS.
Full text
PDF

















Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baer R., Bankier A. T., Biggin M. D., Deininger P. L., Farrell P. J., Gibson T. J., Hatfull G., Hudson G. S., Satchwell S. C., Séguin C. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984 Jul 19;310(5974):207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- Bayliss G. J., Marsden H. S., Hay J. Herpes simplex virus proteins: DNA-binding proteins in infected cells and in the virus structure. Virology. 1975 Nov;68(1):124–134. doi: 10.1016/0042-6822(75)90154-3. [DOI] [PubMed] [Google Scholar]
- Bergsma D. J., Olive D. M., Hartzell S. W., Subramanian K. N. Territorial limits and functional anatomy of the simian virus 40 replication origin. Proc Natl Acad Sci U S A. 1982 Jan;79(2):381–385. doi: 10.1073/pnas.79.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bzik D. J., Fox B. A., DeLuca N. A., Person S. Nucleotide sequence specifying the glycoprotein gene, gB, of herpes simplex virus type 1. Virology. 1984 Mar;133(2):301–314. doi: 10.1016/0042-6822(84)90397-0. [DOI] [PubMed] [Google Scholar]
- Chartrand P., Stow N. D., Timbury M. C., Wilkie N. M. Physical mapping of paar mutations of herpes simplex virus type 1 and type 2 by intertypic marker rescue. J Virol. 1979 Aug;31(2):265–276. doi: 10.1128/jvi.31.2.265-276.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen D. M., Aschman D. P., Gelep P. T., Retondo M. J., Weller S. K., Schaffer P. A. Fine mapping and molecular cloning of mutations in the herpes simplex virus DNA polymerase locus. J Virol. 1984 Jan;49(1):236–247. doi: 10.1128/jvi.49.1.236-247.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen D. M., Furman P. A., Gelep P. T., Schaffer P. A. Mutations in the herpes simplex virus DNA polymerase gene can confer resistance to 9-beta-D-arabinofuranosyladenine. J Virol. 1982 Mar;41(3):909–918. doi: 10.1128/jvi.41.3.909-918.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conley A. J., Knipe D. M., Jones P. C., Roizman B. Molecular genetics of herpes simplex virus. VII. Characterization of a temperature-sensitive mutant produced by in vitro mutagenesis and defective in DNA synthesis and accumulation of gamma polypeptides. J Virol. 1981 Jan;37(1):191–206. doi: 10.1128/jvi.37.1.191-206.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crumpacker C. S., Chartrand P., Subak-Sharpe J. H., Wilkie N. M. Resistance of herpes simplex virus to acycloguanosine--genetic and physical analysis. Virology. 1980 Aug;105(1):171–184. doi: 10.1016/0042-6822(80)90165-8. [DOI] [PubMed] [Google Scholar]
- Deininger P. L. Random subcloning of sonicated DNA: application to shotgun DNA sequence analysis. Anal Biochem. 1983 Feb 15;129(1):216–223. doi: 10.1016/0003-2697(83)90072-6. [DOI] [PubMed] [Google Scholar]
- Dunn J. J., Studier F. W. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol. 1983 Jun 5;166(4):477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- Gait M. J., Matthes H. W., Singh M., Sproat B. S., Titmas R. C. Rapid synthesis of oligodeoxyribonucleotides. VII. Solid phase synthesis of oligodeoxyribonucleotides by a continuous flow phosphotriester method on a kieselguhr-polyamide support. Nucleic Acids Res. 1982 Oct 25;10(20):6243–6254. doi: 10.1093/nar/10.20.6243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingeras T. R., Sciaky D., Gelinas R. E., Bing-Dong J., Yen C. E., Kelly M. M., Bullock P. A., Parsons B. L., O'Neill K. E., Roberts R. J. Nucleotide sequences from the adenovirus-2 genome. J Biol Chem. 1982 Nov 25;257(22):13475–13491. [PubMed] [Google Scholar]
- Gray C. P., Kaerner H. C. Sequence of the putative origin of replication in the UL region of herpes simplex virus type 1 ANG DNA. J Gen Virol. 1984 Dec;65(Pt 12):2109–2119. doi: 10.1099/0022-1317-65-12-2109. [DOI] [PubMed] [Google Scholar]
- Hay J., Subak-Sharpe J. H. Mutants of herpes simplex virus types 1 and 2 that are resistant to phosphonoacetic acid induce altered DNA polymerase activities in infected cells. J Gen Virol. 1976 Apr;31(1):145–148. doi: 10.1099/0022-1317-31-1-145. [DOI] [PubMed] [Google Scholar]
- Hoffmann P. J., Cheng Y. C. The deoxyribonuclease induced after infection of KB cells by herpes simplex virus type 1 or type 2. I. Purification and characterization of the enzyme. J Biol Chem. 1978 May 25;253(10):3557–3562. [PubMed] [Google Scholar]
- Holland L. E., Sandri-Goldin R. M., Goldin A. L., Glorioso J. C., Levine M. Transcriptional and genetic analyses of the herpes simplex virus type 1 genome: coordinates 0.29 to 0.45. J Virol. 1984 Mar;49(3):947–959. doi: 10.1128/jvi.49.3.947-959.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honess R. W., Purifoy D. J., Young D., Gopal R., Cammack N., O'Hare P. Single mutations at many sites within the DNA polymerase locus of herpes simplex viruses can confer hypersensitivity to aphidicolin and resistance to phosphonoacetic acid. J Gen Virol. 1984 Jan;65(Pt 1):1–17. doi: 10.1099/0022-1317-65-1-1. [DOI] [PubMed] [Google Scholar]
- Joyce C. M., Kelley W. S., Grindley N. D. Nucleotide sequence of the Escherichia coli polA gene and primary structure of DNA polymerase I. J Biol Chem. 1982 Feb 25;257(4):1958–1964. [PubMed] [Google Scholar]
- Knopf K. W. Properties of herpes simplex virus DNA polymerase and characterization of its associated exonuclease activity. Eur J Biochem. 1979 Jul;98(1):231–244. doi: 10.1111/j.1432-1033.1979.tb13181.x. [DOI] [PubMed] [Google Scholar]
- Littler E., Purifoy D., Minson A., Powell K. L. Herpes simplex virus non-structural proteins. III. Function of the major DNA-binding protein. J Gen Virol. 1983 May;64(Pt 5):983–995. doi: 10.1099/0022-1317-64-5-983. [DOI] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Murchie M. J., McGeoch D. J. DNA sequence analysis of an immediate-early gene region of the herpes simplex virus type 1 genome (map coordinates 0.950 to 0.978). J Gen Virol. 1982 Sep;62(Pt 1):1–15. doi: 10.1099/0022-1317-62-1-1. [DOI] [PubMed] [Google Scholar]
- Ohlendorf D. H., Matthews B. W. Structural studies of protein-nucleic acid interactions. Annu Rev Biophys Bioeng. 1983;12:259–284. doi: 10.1146/annurev.bb.12.060183.001355. [DOI] [PubMed] [Google Scholar]
- Powell K. L., Littler E., Purifoy D. J. Nonstructural proteins of herpes simplex virus. II. Major virus-specific DNa-binding protein. J Virol. 1981 Sep;39(3):894–902. doi: 10.1128/jvi.39.3.894-902.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell K. L., Purifoy D. J. DNA-binding proteins of cells infected by herpes simplex virus type 1 and type 2. Intervirology. 1976;7(4-5):225–239. doi: 10.1159/000149955. [DOI] [PubMed] [Google Scholar]
- Purifoy D. J., Lewis R. B., Powell K. L. Identification of the herpes simplex virus DNA polymerase gene. Nature. 1977 Oct 13;269(5629):621–623. doi: 10.1038/269621a0. [DOI] [PubMed] [Google Scholar]
- Purifoy D. J., Powell K. L. Temperature-sensitive mutants in two distinct complementation groups of herpes simplex virus type 1 specify thermolabile DNA polymerase. J Gen Virol. 1981 May;54(Pt 1):219–222. doi: 10.1099/0022-1317-54-1-219. [DOI] [PubMed] [Google Scholar]
- Pustell J., Kafatos F. C. A high speed, high capacity homology matrix: zooming through SV40 and polyoma. Nucleic Acids Res. 1982 Aug 11;10(15):4765–4782. doi: 10.1093/nar/10.15.4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Spaete R. R., Frenkel N. The herpes simplex virus amplicon: a new eucaryotic defective-virus cloning-amplifying vector. Cell. 1982 Aug;30(1):295–304. doi: 10.1016/0092-8674(82)90035-6. [DOI] [PubMed] [Google Scholar]
- Staden R., McLachlan A. D. Codon preference and its use in identifying protein coding regions in long DNA sequences. Nucleic Acids Res. 1982 Jan 11;10(1):141–156. doi: 10.1093/nar/10.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stow N. D., McMonagle E. C. Characterization of the TRS/IRS origin of DNA replication of herpes simplex virus type 1. Virology. 1983 Oct 30;130(2):427–438. doi: 10.1016/0042-6822(83)90097-1. [DOI] [PubMed] [Google Scholar]
- Taylor P. A fast homology program for aligning biological sequences. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):447–455. doi: 10.1093/nar/12.1part2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller S. K., Lee K. J., Sabourin D. J., Schaffer P. A. Genetic analysis of temperature-sensitive mutants which define the gene for the major herpes simplex virus type 1 DNA-binding protein. J Virol. 1983 Jan;45(1):354–366. doi: 10.1128/jvi.45.1.354-366.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller S. K., Spadaro A., Schaffer J. E., Murray A. W., Maxam A. M., Schaffer P. A. Cloning, sequencing, and functional analysis of oriL, a herpes simplex virus type 1 origin of DNA synthesis. Mol Cell Biol. 1985 May;5(5):930–942. doi: 10.1128/mcb.5.5.930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitton J. L., Clements J. B. Replication origins and a sequence involved in coordinate induction of the immediate-early gene family are conserved in an intergenic region of herpes simplex virus. Nucleic Acids Res. 1984 Feb 24;12(4):2061–2079. doi: 10.1093/nar/12.4.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]



