Abstract
Targeted gene replacement in plastids was used to explore whether the rbcL gene that codes for the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase, the key enzyme of photosynthetic CO2 fixation, might be replaced with altered forms of the gene. Tobacco (Nicotiana tabacum) plants were transformed with plastid DNA that contained the rbcL gene from either sunflower (Helianthus annuus) or the cyanobacterium Synechococcus PCC6301, along with a selectable marker. Three stable lines of transformants were regenerated that had altered rbcL genes. Those containing the rbcL gene for cyanobacterial ribulose-1,5-bisphosphate carboxylase/oxygenase produced mRNA but no large subunit protein or enzyme activity. Those tobacco plants expressing the sunflower large subunit synthesized a catalytically active hybrid form of the enzyme composed of sunflower large subunits and tobacco small subunits. A third line expressed a chimeric sunflower/tobacco large subunit arising from homologous recombination within the rbcL gene that had properties similar to the hybrid enzyme. This study demonstrated the feasibility of using a binary system in which different forms of the rbcL gene are constructed in a bacterial host and then introduced into a vector for homologous recombination in transformed chloroplasts to produce an active, chimeric enzyme in vivo.
Rubisco is the key enzyme of photosynthetic CO2 fixation, catalyzing two competing reactions that involve the carboxylation and oxygenation of ribulose-P2, and initiating the primary steps of photosynthetic C reduction and photorespiration. The relative specificities of these two reactions is not a fixed constraint and varies by at least a factor of 10 in Rubisco enzymes from divergent species (Jordan and Ogren, 1981, 1983; Parry et al., 1989; Read and Tabita, 1992). Even enzymes from similar organisms exhibit significant differences in relative specificity. Because the ratio of these two reactions varies naturally or can be manipulated using in vivo and in vitro mutational techniques (for review, see Hartman and Harpel, 1994; Gutteridge and Gatenby, 1995), it has long been considered an attractive goal to change the properties of the enzyme to favor carboxylation.
In higher plants Rubisco is composed of two distinct subunits. The large subunit is encoded on the plastid genome and synthesized on plastid ribosomes in the stroma, where it aggregates as octameric cores with the aid of chaperonins (Gatenby and Ellis, 1990). Each core associates with eight small subunits that are encoded on the nuclear genome, synthesized on cytoplasmic ribosomes, and subsequently imported into the chloroplast (for review, see Gatenby and Ellis, 1990; Gutteridge and Gatenby, 1995). The elements required to catalyze the reactions of the enzyme, substrate binding and product formation, are located on the large subunit (Andrews, 1988; Cleland et al., 1998). Investigations of the isolated octameric core of cyanobacterial Rubisco showed that partitioning of the bisphosphate substrate between carboxylation and oxygenation is similar to the holoenzyme, indicating that this discrimination is also inherently determined by structural elements of the large subunit (Andrews and Lorimer, 1985; Gutteridge, 1990, 1991). These studies indicated that altering the relative specificity of Rubisco might best be achieved by modifying the large subunit rather than the small subunit (Read and Tabita, 1992). Furthermore, unlike the small subunit, which is coded for by a multigene family in plants, the large subunits are a homogeneous population encoded by the plastid genome.
There are now a number of examples of mutants of Rubisco generated in vivo and in vitro that have altered activities both favorably and less so. Nearly all examples have involved mutagenesis of the enzyme expressed in a heterologous bacterial host or in the green alga Chlamydomonas reinhardtii (for review, see Spreitzer, 1993; Hartman and Harpel, 1994; Gutteridge and Gatenby, 1995). A cyanobacterial operon from Synechococcus PCC6301 coding for both large and small subunits has been used the most because the enzyme readily assembles in Escherichia coli. Unlike the cyanobacterial enzyme, higher-plant Rubisco is not amenable to heterologous expression (Cloney et al., 1993; Gutteridge and Gatenby, 1995), and attempts at modifying the properties of higher-plant enzymes have been thwarted. However, chloroplast-transformation technology has now developed to the point where it is possible to manipulate the plastid genome in both C. reinhardtii (Boynton et al., 1988) and tobacco (Nicotiana tabacum) (Svab et al., 1990; Maliga, 1993; Svab and Maliga, 1993), including the rbcL (Rubisco large subunit) gene (Kanevski and Maliga, 1994; Zhu and Spreitzer, 1994, 1996; for review, see Rochaix, 1997). To determine if the Rubisco large subunit can be replaced with the subunit of other species, yet still fold into an active holoenzyme by assembling with the indigenous small subunits, tobacco plastids were transformed with vectors harboring the rbcL genes from other photosynthetic organisms. Two were chosen that represent enzymes having distinct activities compared with the tobacco enzyme. Our aim was to determine whether these characteristics are also transferred to a hybrid enzyme. The rbcL gene from the cyanobacterium Synechococcus PCC6301 (rbcL-C) generates Rubisco with lower relative specificity than the tobacco enzyme (Andrews and Lorimer, 1985), whereas the gene from sunflower (Helianthus annuus) (rbcL-S) was used to try to enhance the activity of the enzyme (Parry et al., 1989).
MATERIALS AND METHODS
Plasmid Construction
To obtain plasmid pIK28, a 5.3-kb PvuII/XhoI DNA fragment of the tobacco plastid DNA (sites at nucleotides 55147 and 60484; Shinozaki et al., 1986) was cloned in Ecl136II/XhoI-digested pBluescript KS(+) phagemid (Stratagene). Linker-ligation was used to convert the XbaI site at nucleotide 59,234 (Shinozaki et al., 1986) into a HindIII site (5′-CAAGCTTG-3′), and an AccI site at nucleotide position 59,026 (Shinozaki et al., 1986) into an XbaI site (5′-GCTCTAGAGC-3′) to obtain plasmid pIK76. The rbcL sequences of sunflower (Helianthus annuus) (B. Ranty and S. Gutteridge, unpublished data, accession no. AF097517), cyanobacteria (Synechococcus PCC6301) (Shinozaki and Sugiura, 1985), and tobacco (Nicotiana tabacum) (Shinozaki et al., 1986) are relatively compatible in the region encoding residue 10, which allows introduction of a unique NheI site without causing amino acid changes.
In the 3′-flanking region of the gene, a XbaI restriction site was introduced to allow much of the rbcL genes to be ligated in-frame with the 5′ end of the tobacco gene (rbcL-T), yielding plasmids pSGPM1 and pSGPM2, which harbor the rbcL-S and rbcL-C genes, respectively. The 1.6-kb NcoI/XbaI fragment in plasmid pIK76 was replaced with a 1.6-kb NcoI/XbaI fragment from plasmids pSGPM1 and pSGPM2 to obtain plasmids pIK80 and pIK81, respectively. Plastid-transformation vectors pIK83 and pIK84 were obtained by cloning the spectinomycin-resistance gene (aadA) on a HindIII fragment into the HindIII site of plasmids pIK80 and pIK81, respectively. The chimeric aadA gene derives from plasmid pIK82 and is identical to aadA in plasmid pZS197 (Svab and Maliga, 1993) except that (a) the first five amino acids of the tobacco large subunit are translationally fused with the aadA-coding region; (b) the XbaI site between the aadA-coding region and the psbA 3′-untranslated region was filled in by the Klenow fragment of DNA polymerase I; and (c) the Ecl136II site upstream of the promoter was converted into a HindIII site by linker-ligation (5′-CAAGCTTG-3′).
Transformation and Regeneration of Transgenic Plants
Tobacco plants were grown aseptically on agar-solidified medium containing Murashige-Skoog salts (Murashige and Skoog, 1962) and 30 g L−1 Suc. DNA for plastid transformation was introduced into tobacco leaves on the surface of microscopic tungsten particles using a biolistic gun (PDS1000He, DuPont) (Svab and Maliga, 1993; Kanevski and Maliga, 1994). Spectinomycin-resistant calli and shoots were selected on regeneration medium containing 500 mg L−1 spectinomycin dihydrochloride (Svab and Maliga, 1993). Homoplastomic, spectinomycin-resistant plants were obtained by a repeated cycle of shoot regeneration from leaves on the same selective medium and then rooting the shoots on antibiotic-free Murashige-Skoog agar.
DNA and RNA Gel-Blot Analysis
Total cellular DNA was isolated (Mettler, 1987) and digested with the appropriate restriction enzymes, electrophoresed on 0.7% agarose gels, and transferred to nylon membrane (Amersham) using the PosiBlot Transfer apparatus (Stratagene). Blots were probed using Rapid Hybridization Buffer (Amersham) with 32P-labeled probes generated by random priming (Boehringer Mannheim). RNA was extracted using TRIzol reagent (GIBCO-BRL) according to the manufacturer's protocol. RNA gel blots were derived as described previously (Staub and Maliga, 1994).
Immunoblotting
Leaf protein extracts were isolated and resolved in 10% polyacrylamide/SDS gels and immunoblotted with rabbit polyclonal antibody raised against spinach Rubisco large and small subunits (dilution 1:2000 and 1:1000, respectively) using the ECL detection system (Amersham), as described previously (Kanevski and Maliga, 1994).
DNA Sequencing
To sequence the recombinant gene (rbcL-R), total cellular DNA was isolated from the Nt-pIK83-4 line and a 505-bp DNA fragment was amplified according to the standard protocol (1 min at 95°C, 2 min at 56°C, and 1.5 min at 72°C, for 30 cycles) using tobacco-specific (5′-CCTGAGTACCAAACCAAG-3′) and sunflower-specific (5′-CCACGAAGACATTGATAACAA-3′) primers. The amplification product was separated in 1.5% agarose gel, purified with a Geneclean II kit (BIO101, Vista, CA), and directly sequenced (Bachmann et al., 1990) using the Sequenase kit (United States Biochemical). Sequencing primers were the same as for PCR amplification.
Rubisco Isolation and Activity
Rubisco was isolated from transgenic plants grown in sterile culture on Murashige-Skoog medium (Murashige and Skoog, 1962) containing 3% Suc. The enzyme was purified from 5 g of leaves ground to a fine powder in liquid nitrogen. The powder was resuspended in 50 mm Tris-HCl, pH 8.0, containing 1 mm DTT, 0.1 mm EDTA, and 0.1 mm benzamidine (buffer A). The solution was clarified by filtration through a fine-mesh cheesecloth and centrifugation. Separation of Rubisco from other proteins was achieved initially using an EconoQ cartridge (Bio-Rad) equilibrated with buffer A and developed with a step gradient of the same buffer containing 0.5 or 1.0 m KCl. All Rubisco activity was collected in the 0.5 m fraction, concentrated, and desalted by pressure dialysis before further ion-exchange chromatographic purification using a Resource-Q column (Pharmacia-Biotech). The column was equilibrated with buffer A and developed with a 0 to 0.5 m KCl linear gradient. Those fractions containing Rubisco activity were pooled, concentrated, and desalted into 0.1 m triethanolamine, pH 8.0, and stored as 12% glycerol solutions at −80°C. Further purification resulted in a significant loss in activities. Based on 2′-carboxyarabinitol-bisphosphate binding, Rubisco was determined to be about 90% of the total protein in the stored samples.
The specificities of carboxylation relative to oxygenation were determined using [1-14C]ribulose-P2 (5 Ci mol−1) consumed by the enzyme in solutions containing set ratios of CO2:O2. The solutions were buffered with CO2-free 0.1 m triethanolamine HCl, pH 8.2, containing MgCl2. The buffer was then equilibrated with the appropriate ratio of CO2:O2 using a precision Wosthoff gas-mixing pump. The enzyme solution to be used in the assays was also equilibrated with the same gas prior to adding up to 32 μg of a 1.6 mg mL−1 solution to the reaction tube. The reaction was started by the addition of the labeled ribulose-P2 (10 nmol), and the vials were immediately sealed with caps that contained a small amount of silicon grease to remove any dead space from above the solution.
To achieve good precision of the specificity results, enough of the buffer solution was made so that spinach Rubisco, which has well-determined specificity, could be assayed in parallel with the tobacco variants. The assay solutions (300 μL) also contained 0.2 mm vanadate to inhibit unwanted phosphatase activity that tended to deplete the ribulose-P2 during the reactions and 5 μg of carbonic anhydrase to maintain the HCO3-CO2 equilibrium. The reactions were stopped by addition of acetic acid (10%) after at least 90% of the ribulose-P2 had been consumed. The labeled products of carboxylation and oxygenation were collected as filtrates from small Dowex 50 H+ spin columns that also removed the proteins and then separated chromatographically, as described previously (Gutteridge et al., 1989b, 1993). The amount of 2-phosphoglycolate to 3-phosphoglycerate produced from the labeled ribulose-P2 was determined by liquid-scintillation counting.
The standard Michaelis-Menten kinetic parameters of the enzyme were based on measurements of the carboxylase activity determined from the incorporation of 14CO2 into an acid-stable product (Lorimer et al., 1976) at various ribulose-P2 and CO2 concentrations.
RESULTS
Chimeric rbcL Genes
Foreign rbcL genes were introduced into a tobacco plastid DNA fragment that contained the rbcL-T gene and the flanking atpB and accD genes cloned in a Bluescript KS+ phagemid (Fig. 1). Replacement of the majority of the coding region of the rbcL-T gene with those of the sunflower and cyanobacterial genes yielded rbcL-S and rbcL-C chimeras. With the constructs organized as shown in Figure 1A, the transcription of the rbcL-S and rbcL-C genes is from the natural rbcL-T promoter and the mRNA includes the 3′-untranslated region downstream of the rbcL-T gene. The coding region of the chimeric genes was obtained by replacing all but the first 25 nucleotides of rbcL-T with the same region from the heterologous gene via a convenient NheI restriction site that was engineered into all of the genes. Because the first 10 amino acids of the tobacco and sunflower large subunits are identical, the rbcL-S construct encodes the full-length sunflower polypeptide (Fig. 2A). In the rbcL-C gene, however, the translational fusion with the cyanobacterial gene results in the replacement of the large subunit N terminus (8 residues) with that of the first 11 amino acids of the tobacco large subunit (not shown).
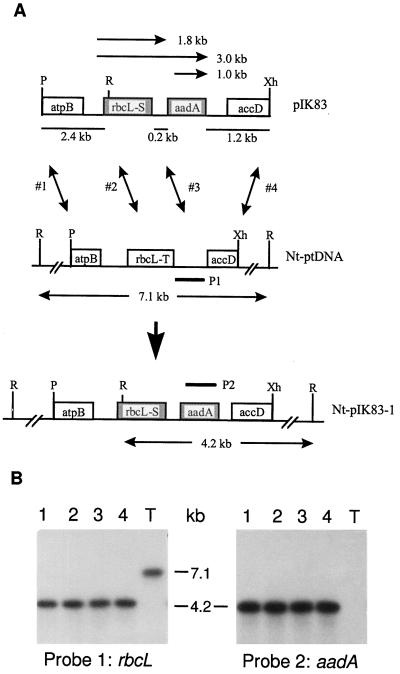
Figure 1.
Targeted replacement of rbcL-T in the tobacco plastid genome with the rbcL-S gene. A, Plastid-targeting region in plasmid pIK83 (plastid DNA is underlined) and the cognate region of the plastid genome (Nt-ptDNA) and of the Nt-pIK83-1 line. atpB (Shinozaki et al., 1986) and accD (Sasaki et al., 1993) are plastid genes. aadA is the plastid-selectable spectinomycin-resistance gene. Recombination endpoints (1–4) discussed in the text are marked by vertical arrows. Horizontal arrows represent mRNAs detected by the rbcL (P1) and aadA (P2) probes. Restriction enzyme recognition sites: P, PvuII; R, EcoRV; Xh, XhoI. B, Wild-type plastid genome copies are absent in four plants regenerated independently (lanes 1–4) from the Nt-pIK83-1 line. Data for wild-type tobacco (T) are also shown. DNA blots of EcoRV-digested total cellular DNA (1 μg per lane) were hybridized with the rbcL (P1) and aadA (P2) probes. The rbcL probe hybridized to a 7.1-kb DNA fragment of the wild-type Nt-ptDNA and a 4.2-kb fragment of the transplastome (Fig. 1A). The 4.2-kb transgenic fragment also hybridized with the aadA probe. Note the lack of wild-type 7.1-kb DNA fragments in the plastid transformants.
Figure 2.
Amino acid sequences of the large (A) and small (B) subunits of tobacco (Mazur and Chui, 1985; Shinozaki et al., 1986) and sunflower (B. Ranty and S. Gutteridge, unpublished data, accession no. AF097517; Waksman and Freyssinet, 1987) Rubisco showing the positions where there are amino acid differences. A significant feature of the sunflower large subunit is the C terminus extended by eight residues. (Accession numbers for the tobacco large and small subunits are P00876 and P26666, respectively. Those for the sunflower large and small subunits are P45738 and P08705, respectively.)
A selectable spectinomycin-resistance gene (aadA) was cloned between the chimeric rbcL gene and the accD gene in the plastid DNA fragment to yield plasmids pIK83 and pIK84, carrying the rbcL-S and rbcL-C genes, respectively. The map of the plastid DNA fragment in the pIK83 plasmid with the chimeric gene is shown in Figure 1A.
Plastid Transformation
In plasmid pIK83 the linked rbcL-S and aadA genes are flanked by a 2.4-kb segment of plastid DNA at the 5′ end and a 1.2-kb segment at the 3′ end that encode atpB and accD, respectively. These sequences target the insertion of the chimeric genes (Fig. 1A) into the plastid genome. Introduction of the plasmid DNA into the chloroplasts of tobacco leaves was accomplished by coating tungsten particles (approximately 1 mm) with DNA and delivering them biolistically into the cells of freshly excised leaves. Selection for spectinomycin resistance during the regeneration of shoots (Svab and Maliga, 1993; Kanevski and Maliga, 1994) yielded transplastomic lines in which the aadA selection cassette had become integrated into the plastid genome by two homologous recombination events via the flanking plastid DNA sequences of the vectors. For example, recombination via atpB and accD (Fig. 1A, sites 1 and 4) led to the integration of both rbcL-S and aadA in line Nt-pIK83-1 (Fig. 1A). Recombination via rbcL-S and accD (Fig. 1A, sites 2 and 4) in line Nt-pIK83-4 yielded a new recombinant rbcL gene (rbcL-R) linked to the aadA gene, whereas recombination via the rbcL 3′-untranslated region and accD (Fig. 1A, sites 3 and 4) led to the integration of aadA only (e.g. line Nt-pIK83-10).
Out of six transplastomic lines, two carried the rbcL-S and aadA genes and three carried the aadA gene alone. Because the regenerated plants within each group were indistinguishable, only one line from each group was studied further. A novel rbcL was also identified in only one line, Nt-pIK83-4, which had formed by another homologous recombination event. In this variant the rbcL-R gene has 233 nucleotides of the tobacco large subunit coding region fused with the sunflower gene. Given the significant (90.4%) DNA sequence conservation between the tobacco and sunflower genes, recombination at this particular site was a chance event, and indicates the feasibility of obtaining different chimeric rbcL genes by in vivo recombination during transformation. The product of rbcL-R in the Nt-pIK83-4 line is identical to the sunflower large subunit except for a conservative change of Glu to Asp at position 19 and a change of Glu to Gln at residue 30 (Fig. 2A).
Transformation with plasmid pIK84 yielded seven transplastomic lines. The plastid genome of two lines (Nt-pIK84-7 and Nt-pIK84-15) carried integrated copies of both rbcL-C and aadA, whereas in five lines only the aadA gene integrated. Since plants representing the independently transformed Nt-pIK84-7 and Nt-pIK84-15 lines were indistinguishable, data are shown for the Nt-pIK84-15 line only.
The rbcL-T/rbcL-S and rbcL-T/rbcL-C gene pairs can be distinguished by 9 and 12 restriction fragment-length polymorphic markers, respectively (not shown). The absence of wild-type plastid DNA copies in the regenerated plants was verified by DNA gel-blot analysis utilizing six restriction fragment-length polymorphic markers for each of the chimeric genes. An example for shoots regenerated from the Nt-pIK83-1 line is shown in Figure 3.
Figure 3.
Tobacco plants with heterologous rbcL-S and rbcL-C genes. A, Leaves of wild-type (top) and of Nt-pIK83-1 (rbcL-S, left) plants are green. The leaves of Nt-pIK84-15 plants (rbcL-C, right) are yellow in sterile culture on 3% Suc RM medium. Illumination was for 16 h at 180 μmol m−2 s−1. B, Nt-pIK83-1 (rbcL-S) shoot grafted on a wild-type plant. Note the pale-green color of transgenic leaves in contrast to the green of the rootstock in the greenhouse.
The leaves of tobacco plants carrying rbcL-S (line Nt-pIK83-1) and the novel chimeric rbcL-R gene (line Nt-pIK83-4) were indistinguishable from wild-type leaves on a medium containing 3% Suc in sterile culture (Fig. 3A). In contrast, the leaves of the same plants were pale green when grown in the greenhouse as grafts (Fig. 3B). Seeds were obtained from shoots grafted onto a wild-type plant. The leaves of homoplasmic Nt-pIK84-15 tobacco plants with rbcL-C were yellow even in sterile culture on Suc (Fig. 3A), and could not be grown into fertile plants in the greenhouse even if grafted onto wild-type root stock.
Expression of the rbcL-C Gene
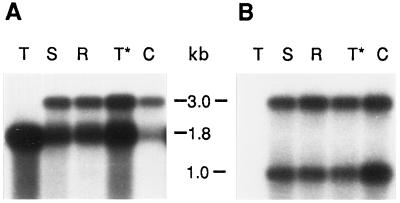
Accumulation of mRNA from the rbcL-C genes was tested in the leaves of Nt-pIK84-15 plants (Fig. 4A). A 0.2-kb DNA probe (Fig. 1A) encoding the tobacco rbcL 3′-untranslated region detected two RNA species transcribed from the rbcL promoter, a 1.8-kb monocistronic rbcL-C mRNA and a 3.0-kb dicistronic rbcL-C/aadA mRNA. The two mRNA species formed due to inefficient transcription termination and/or processing downstream of the rbcL-C coding region (Fig. 1A). The identity of the dicistronic mRNA was confirmed by hybridization with the aadA probe (Fig. 4B). The steady-state level of rbcL-C mRNA in the monocistronic and dicistronic mRNAs was approximately 10% of the wild-type level when quantified using a phosphor imager (Molecular Dynamics, Sunnyvale, CA). Since the rbcL-C gene is transcribed from the wild-type tobacco rbcL promoter, lower levels of mRNA might result from faster mRNA turnover. The increased mRNA turnover was certainly not due to the formation of a dicistronic mRNA, because the plants that carried the native rbcL gene along with aadA (line Nt-pIK83-10) accumulated wild-type levels of mRNA (Fig. 4A).
Figure 4.
Steady-state level of rbcL mRNA in transgenic leaves. Total cellular RNA (1 μg per lane) was from wild-type tobacco (lane T, rbcL-T), Nt-pIK83-1 (lane S, rbcL-S), Nt-pIK83-4 (lane R, rbcL-R), Nt-pIK83-10 (lane T*, rbcL-T), and Nt-pIK84-15 (lane C, rbcL-C) plants. RNA gel blots were hybridized (Staub and Maliga, 1994) with the rbcL probe (Fig. 1A, P1) (A) and with the aadA coding region probe (Fig. 1A, P2) (B). The position of monocistronic rbcL (1.8 kb) and aadA (1.0 kb) transcripts and of the dicistronic rbcL-aadA transcript (3.0 kb) are shown in Figure 1A.
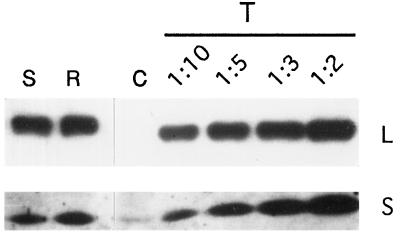
Western analysis was carried out to follow the accumulation of the Rubisco large and small subunits. No large subunit polypeptide could be detected with antibodies raised to either the spinach (Fig. 5) or cyanobacterial large subunit (not shown). Furthermore, no ribulose-P2 dependent CO2 fixation was detected with standard assays for Rubisco activity. Therefore, if the large subunit is synthesized, it must be rapidly degraded. Minor amounts of the tobacco small subunit were detectable in these leaves at about the same level found in tobacco leaves, which lack the rbcL gene (Kanevski and Maliga, 1994).
Figure 5.
Immunoblot analysis of Rubisco accumulation in transgenic leaves. Total cellular protein (1 μg per lane) was from Nt-pIK83-1 (lane S, rbcL-S), Nt-pIK83-4 (lane R, rbcL-R), and Nt-pIK84-15 (lane C, rbcL-C) plants. For comparison, a dilution series (1:10, 1:5, 1:3, and 1:2) of a wild-type tobacco extract (T) is also shown. The blots were probed with antibodies to the spinach Rubisco large (L) and small (S) subunits (Kanevski and Maliga, 1994).
Expression of the rbcL-S Gene
Accumulation of mRNA from the rbcL-S and rbcL-R genes was studied in the leaves of Nt-pIK83-1 and Nt-pIK83-4 plants, respectively. The rbcL 3′-region probe detected monocistronic and dicistronic mRNAs, as discussed above. The level of rbcL-S and rbcL-R mRNA was about 30% of the wild type (Fig. 4A). Apparently, the more closely related sunflower RNA is less susceptible to degradation than the rbcL-C mRNA in tobacco plastids. The rbcL-S mRNA was apparently translatable at wild-type levels, since the Rubisco large and small subunits accumulated to about 30% of the wild-type level (Fig. 5). However, the Rubisco activity in initial extracts from the same plants was only about 12% of the wild-type level. This discrepancy between the amount of enzyme and Rubisco activity in the Nt-pIK83-1 and Nt-pIK83-4 lines could be explained by incompatibility between the sunflower large subunit and the tobacco small subunit in the hybrid holoenzyme.
Rubisco Properties
Rubisco was purified from the leaves of transgenic Nt-pIK83-1 and, for comparison, from the “parental” sunflower and tobacco leaves. The kinetic parameters of the enzymes are shown in Table I. The hybrid Rubisco, containing sunflower large subunits and tobacco small subunits, showed compromised affinities for both ribulose-P2 and CO2. The overall turnover of the enzyme was also diminished by about a factor of four. The relative specificity of the hybrid enzyme was not significantly different from the value for the tobacco enzyme and certainly not lower than that expected for a higher plant variant. Rubisco must be activated by carbamylation of an active-site Lys residue. If this process were also compromised, this would be the basis for the lower overall turnover rate. However, when the extent of activation of the hybrid was determined by trapping 14CO2 at the active site with the tight-binding inhibitor 2′-carboxyarabinitol bisphosphate, 80% of potential active sites contained the activating cofactor (Table I).
Table I.
Comparison of the properties of the various forms of Rubiscoa
| Source | CO2 |
Km
|
Vmax Carboxylation | Relative Specificity | |
|---|---|---|---|---|---|
| CO2 | Ribulose-P2 | ||||
| mol CO2 large subunit−1 | μm | s−1 | |||
| Sunflower | 0.97 ± 0.07 | 8.2 ± 0.7 | 16.3 ± 2.1 | 2.2 ± 0.2 | 98 ± 2.1 |
| Hybrid | 0.80 ± 0.08 | 60.4 ± 8.2 | 100.1 ± 8.4 | 0.5 ± 0.1 | 89 ± 2.2 |
| Tobacco | 1.10 ± 0.08 | 12.2 ± 1.0 | 20.3 ± 3.0 | 1.9 ± 0.2 | 85 ± 2.5 |
Values are ±se; n = 3.
DISCUSSION
We report here replacement of the 1.8-kb rbcL-coding region with cognate sequences from heterologous sources. In higher plants manipulation of the plastid genome thus far has involved insertion of chimeric genes in intergenic regions (Staub and Maliga, 1993; Svab and Maliga, 1993; Carrer and Maliga, 1995), introduction of point mutations (Bock et al., 1994; Bock and Maliga, 1995), and targeted gene deletions (Kanevski and Maliga, 1994; Sugita et al., 1997; Burrows et al., 1998). As to the practicality of coding region replacement, valuable information was obtained concerning separation of the selectable marker from the engineered coding region via short homologous sequences between the two (approximately 0.2 kb of the rbcL 3′-untranslated region).
Incorporation of the selectable marker aadA alone was observed in three of six pIK83-transformed lines and two of seven pIK84-transformed lines. Thus, in 50% to 70% of the lines aadA integration occurred without incorporation of any of the heterologous rbcL sequences. Integration of aadA alone indicated that homologous recombination events via the approximately 0.2-kb rbcL 3′-untranslated region adjacent to the selectable marker must have occurred. However, when both aadA and the engineered rbcL integrated, recombination in all but one clone occurred upstream of the coding region. Therefore, recombination events in the heterologous coding region were relatively rare (one in five clones). Minor sequence variations in the coding region of heterologous rbcL genes may suppress recombination via short stretches of homology.
Plastid transformation and targeting the rbcL gene for replacement is therefore feasible at an acceptable frequency. Even the gene from a photosynthetic bacterium with only 50% homology can be stably incorporated into the plastid genome. This allows a binary procedure to be used to construct plant large subunit chimeras and mutant variants. The first step involves a bacterial host such as E. coli as a means to construct variants of the rbcL gene from any source. This is followed by transformation of the plant system to express the chimeric genes and produce protein. The full gene from Synechococcus PCC6301 was transcribed into mRNA, but the mRNA was not translated into protein. At this stage, it is unknown whether there is incompatibility at the level of translation, or if the protein, once produced, is unable to assemble correctly using the indigenous folding machinery. Western analysis indicated that if protein is produced it is transient and does not accumulate enough to be detected by antibodies raised against the cyanobacterial large subunit. However, in those cases in which higher plant/cyanobacterial rbcL chimeras are of interest (Gutteridge et al., 1989a) and have proved intractable using E. coli expression and refolding, this system might provide an alternative approach, one that can supply the amounts of enzyme required for detailed structural analysis using crystallography.
Incorporation of a rbcL-S gene in the plastid genome of tobacco did result in protein that was able to assemble with tobacco small subunits to form an active hybrid Rubisco. The transformants produced about 30% of the wild-type level of Rubisco, and the hybrid enzyme had about 20% of the carboxylase activity at saturating substrate concentrations. This was not enough activity for the plants to withstand greenhouse growing conditions to reach maturity and supply seed unless grafted onto wild-type stock. Nevertheless, enough leaf material was obtained from the plants grown on Suc for an analysis of the properties of the chimeric protein. Naturally, the holoenzyme must be a hybrid of sunflower large subunits and one or more small subunits encoded by the rbcS-T gene family (see Fig. 5). Assuming that the hybrid retains a significant number of small subunits during purification, the lower affinities for the substrates may simply reflect some binding incompatibility at the interface between the large and small subunits.
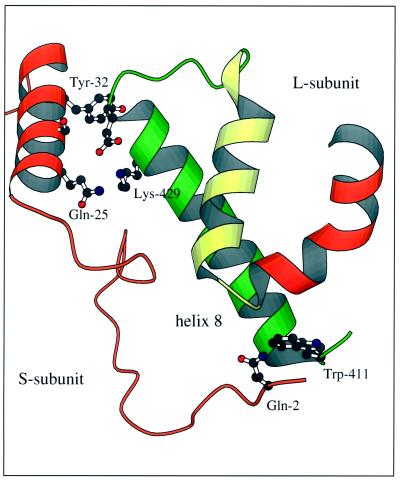
Although large subunits contain the active site and are catalytically active, even some distance (15 Å) from the active site, small subunits influence both turnover and substrate affinities (Andrews, 1988; Gutteridge, 1991; Read and Tabita, 1992). Figure 6 shows the structural elements that form one of the interfaces between the large and small subunits of the tobacco hexadecamer. Of the 32 differences in primary structure between the tobacco and sunflower large subunits, several residues are within or directly adjacent to residues at the large/small subunit interfaces (Knight et al., 1990; Curmi et al., 1992). For example (Fig. 6), Lys-429, which resides in helix 8 of the C-terminal α/β-barrel domain of the tobacco large subunit, is a Gln in the sunflower enzyme. The contacts that would normally exist between this side chain and Glu-25 of the A helix of the small subunit may be less than ideal in the hybrid enzyme. This helix has multiple differences in primary structure (Waksman and Freysinnet, 1987), including a Ser at position 28 in tobacco that is occupied by Lys in the sunflower small subunit (Fig. 2B). Presumably, the loss of a basic residue that would be expected to readily form ionic interactions with Gln-429 and Glu-433 in the native sunflower large subunit may disrupt the association with the small subunits in the hybrid.
Figure 6.
Amino acids at one of the interfaces of the large and small subunits. The region shown involves residues of helix 8 of the C-terminal domain of the large subunit (Knight et al., 1990) and the N-terminal segment of the small subunit. The side chains of amino acids that differ between the two species, or are adjacent to sequence changes that make contacts at the subunit interface (Fig. 2), are shown in this Molscript (Kraulis, 1991) rendition of tobacco Rubisco (Curmi et al., 1992). The position of the N-terminal end of helix 8 is critical for ribulose-P2 binding at the active site. L-subunit, Large subunit; S-subunit, small subunit.
Sequence differences between tobacco and sunflower small subunits total 28 (Fig. 2B), about 10 of which are in positions to influence large/small subunit interactions. These differences, compounded by the heterogeneous population of the native small subunits and the uncertainty about small subunit content of the purified hybrid, may all contribute to the reason for lower substrate affinities. For example, one role of the small subunit located between adjacent large subunit dimers is to stabilize the large subunit core, the integrity of which may be compromised in the hybrid, contributing to weaker substrate affinities (Andrews, 1988). Another factor that must be considered is whether a hybrid Rubisco variant is able to bind activating CO2. Carbamylation was compromised in a hybrid version of the enzyme, in which Arabidopsis Rubisco large subunits were partnered with pea small subunits in Arabidopsis nuclear transformants (Getzoff et al., 1998). The sunflower-tobacco hybrid also showed some inability to bind activating CO2 at all sites (Table I), but this clearly cannot explain the much larger drop in overall turnover.
Apart from the differences in primary structure, a striking feature of the sunflower large subunit is the C-terminal extension of eight charged residues compared with the tobacco subunit, a region of the protein that affects relative specificity (Gutteridge et al., 1993). The hybrid nature of the enzyme might be such that this extension is unable to interact correctly with other structural elements of the protein. Nevertheless, given the feasibility of plastid transformation, we now have a means of investigating these amino acid differences by introducing specific mutations into the large subunit and/or generating structural chimeras rationally or by chance recombination. The identification of photosynthetic organisms with higher relative specificities than either tobacco or sunflower (Read and Tabita, 1992) make this an exciting prospect. Because small subunits might, at the very least, influence the integrity of the complex quaternary structure of the holoenzyme, and therefore influence turnover, coexpression of the two subunits in the plastid might be attempted to circumvent any subunit heterogeneity. In terms of maintaining the vigor of regenerated transformants, an additional factor that influences Rubisco activity is activase (Portis, 1995; Andrews, 1996), which ensures that Rubisco does not become inactivated during turnover. The interaction between activase and a hybrid enzyme composed of sunflower large subunits may not be ideal (Wang et al., 1992). A combination of plastid transformation to alter tandemly incorporated large and small subunits and nuclear transformation to introduce modified activase would be an interesting system with which to study this elusive but essential interaction.
ACKNOWLEDGMENTS
We thank Zora Svab for the aadA gene, Benoit Ranty for the rbcL-S gene, and Charles Herrmann for technical assistance.
Abbreviation:
- ribulose-P2
ribulose 1,5-bisphosphate
Footnotes
This research was supported by the National Science Foundation (grant no. MCB 93-05037) and by DuPont Science and Engineering (grant to P.M.).
LITERATURE CITED
- Andrews TJ. Catalysis by cyanobacterial ribulose-bisphosphate carboxylase large subunit in the complete absence of small subunits. J Biol Chem. 1988;263:12213–12219. [PubMed] [Google Scholar]
- Andrews TJ. The bait in the rubisco mousetrap. Nature Struct Biol. 1996;3:3–7. doi: 10.1038/nsb0196-3. [DOI] [PubMed] [Google Scholar]
- Andrews TJ, Lorimer GH. Catalytic properties of a hybrid cyanobacterial large subunits and higher plant small subunits of ribulose bisphosphate carboxylase-oxygenase. J Biol Chem. 1985;260:4632–4636. [PubMed] [Google Scholar]
- Bachmann B, Lueke W, Hunsmann G. Improvement of PCR amplified DNA sequencing with the aid of detergents. Nucleic Acids Res. 1990;18:1309. doi: 10.1093/nar/18.5.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock R, Kossel H, Maliga P. Introduction of a heterologous editing site into the tobacco plastid genome: lack of RNA editing leads to a mutant phenotype. EMBO J. 1994;13:4623–4628. doi: 10.1002/j.1460-2075.1994.tb06784.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock R, Maliga P. In vivo testing of a tobacco plastid DNA segment for guide RNA function in psbL editing. Mol Gen Genet. 1995;247:439–443. doi: 10.1007/BF00293145. [DOI] [PubMed] [Google Scholar]
- Boynton JE. Chloroplast transformation in Chlamydomonas with high velocity microprojectiles. Science. 1988;240:1534–1538. doi: 10.1126/science.2897716. [DOI] [PubMed] [Google Scholar]
- Sazanov PL, Burrows A, Svab Z, Maliga P, Nixon PJ. Identification of a functional respiratory complex in chloroplasts through analysis of tobacco mutants containing disrupted plastid ndh genes. EMBO J. 1998;17:868–876. doi: 10.1093/emboj/17.4.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrer H, Maliga P. Targeted insertion of foreign genes into the tobacco plastid genome without physical linkage to the selectable marker gene. Biotechnology. 1995;13:791–794. [Google Scholar]
- Cleland WW, Andrews TJ, Hartman FC, Gutteridge S, Lorimer GH. Mechanism of rubisco: the carbamate as general base. Chem Rev. 1998;98:549–561. doi: 10.1021/cr970010r. [DOI] [PubMed] [Google Scholar]
- Cloney LP, Bekkaoui DR, Hemmingsen SM. Co-expression of plastid chaperonin genes and a synthetic Rubisco operon in Escherichia coli. Plant Mol Biol. 1993;23:1285–1290. doi: 10.1007/BF00042362. [DOI] [PubMed] [Google Scholar]
- Curmi PMG, Cascio D, Sweet RM, Eisenberg D, Schreuder H. Crystal structure of the unactivated form of ribulose-1,5-bisphosphate carboxylase/oxygenase from tobacco refined at 2.0-Å resolution. J Biol Chem. 1992;267:16980–16989. [PubMed] [Google Scholar]
- Gatenby AA, Ellis RJ. Chaperone function: the assembly of ribulose bisphosphate carboxylase-oxygenase. Annu Rev Cell Biol. 1990;6:125–149. doi: 10.1146/annurev.cb.06.110190.001013. [DOI] [PubMed] [Google Scholar]
- Getzoff TP, Zhu G, Bohnert HJ, Jensen RG. Chimeric Arabidopsis thaliana Rubisco containing pea small subunit protein is compromised in carbamylation. Plant Physiol. 1998;116:695–702. doi: 10.1104/pp.116.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutteridge S. Limitations of the primary events of CO2 fixation in photosynthetic organisms: the structure and mechanism of rubisco. Biochem Biophys Acta. 1990;1015:1–14. [Google Scholar]
- Gutteridge S. The relative catalytic specificities of the large subunit core of Synechococcus ribulose bisphosphate carboxylase/oxygenase. J Biol Chem. 1991;266:7359–7362. [PubMed] [Google Scholar]
- Gutteridge S, Gatenby AA. Rubisco synthesis, assembly, mechanism and regulation. Plant Cell. 1995;7:809–819. doi: 10.1105/tpc.7.7.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutteridge S, Pierce J, Lorimer GH. Details of the reactions catalysed by mutant forms of rubisco. Plant Physiol Biochem. 1989a;26:675–682. [Google Scholar]
- Gutteridge S, Reddy GS, Lorimer G. The synthesis and purification of 2′-carboxyarabinitol 1-phosphate, a natural inhibitor of ribulose 1,5-bisphosphate carboxylase, investigated by 31P NMR. Biochem J. 1989b;260:711–716. doi: 10.1042/bj2600711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutteridge S, Rhoades DF, Herrmann C. Site-specific mutations in a loop region of the C-terminal domain of the large subunit of ribulose bisphosphate carboxylase/oxygenase that influence substrate partitioning. J Biol Chem. 1993;268:7818–7824. [PubMed] [Google Scholar]
- Hartman FC, Harpel MR. Structure, function, regulation and assembly of ribulose-1,5-bisphosphate carboxylase/oxygenase. Annu Rev Biochem. 1994;63:157–234. doi: 10.1146/annurev.bi.63.070194.001213. [DOI] [PubMed] [Google Scholar]
- Jordan DB, Ogren WL. Species variation in the specificity of ribulose bisphosphate carboxylase/oxygenase. Nature. 1981;291:513–515. [Google Scholar]
- Jordan DB, Ogren WL. Species variation in kinetic properties of ribulose 1,5-bisphosphate carboxylase/oxygenase. Arch Biochem Biophys. 1983;227:425–433. doi: 10.1016/0003-9861(83)90472-1. [DOI] [PubMed] [Google Scholar]
- Kanevski I, Maliga P. Relocation of the plastid rbcL gene to the nucleus yields functional ribulose-1,5-bisphosphate carboxylase in tobacco chloroplasts. Proc Natl Acad Sci USA. 1994;91:1969–1973. doi: 10.1073/pnas.91.5.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight S, Andersson I, Branden C-I. Crystallographic analysis of ribulose 1,5-bisphosphate carboxylase from spinach at 2.4 Å resolution. J Mol Biol. 1990;215:113–160. doi: 10.1016/S0022-2836(05)80100-7. [DOI] [PubMed] [Google Scholar]
- Kraulis P. Molscript: a program to produce both detailed and schematic plots of protein structures. J Appl Crystallog. 1991;24:946–950. [Google Scholar]
- Lorimer GH, Badger MR, Andrews TJ. The activation of ribulose-1,5-bisphosphate carboxylase by carbon dioxide and magnesium ions: equilibria, kinetics, a suggested mechanism and physiological implications. Biochemistry. 1976;15:529–538. doi: 10.1021/bi00648a012. [DOI] [PubMed] [Google Scholar]
- Maliga P. Towards plastid transformation in flowering plants. Trends Biotechnol. 1993;11:101–106. [Google Scholar]
- Mazur B, Chui C-F. Sequence of a genomic DNA clone for the small subunit of ribulose bisphosphate carboxylase-oxygenase from tobacco. Nucleic Acids Res. 1985;13:2373–2386. doi: 10.1093/nar/13.7.2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettler IJ. A simple and rapid method for minipreparation of DNA from tissue cultured cells. Plant Mol Biol Rep. 1987;5:346–349. [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant. 1962;15:493–497. [Google Scholar]
- Parry MAJ, Keys AJ, Gutteridge S. Variation in the specificity factor of C3 higher plant Rubiscos determined by the total consumption of ribulose-P2. J Exp Bot. 1989;40:317–320. [Google Scholar]
- Portis A. Regulation of rubisco by rubisco activase. J Exp Bot. 1995;46:1285–1291. doi: 10.1093/jxb/erm240. [DOI] [PubMed] [Google Scholar]
- Read BA, Tabita FR. A hybrid ribulose bisphosphate carboxylase/oxygenase enzyme exhibiting a substantial increase in substrate specificity factor. Biochemistry. 1992;31:5553–5560. doi: 10.1021/bi00139a018. [DOI] [PubMed] [Google Scholar]
- Rochaix J-D. Chloroplast reverse genetics: new insights into the function of plastid genes. Trends Plant Sci. 1997;2:419–425. [Google Scholar]
- Sasaki Y, Hakamada K, Suama Y, Nagano Y, Furusawa I, Matsuno R. Chloroplast-encoded protein as a subunit of acetyl-CoA carboxylase in pea plant. J Biol Chem. 1993;268:25118–25123. [PubMed] [Google Scholar]
- Shinozaki K, Ohme M, Tanaka M, Wakasugi T, Hayashida N, Matsubayashi T, Zaita N, Chunwongse J, Obokata J, Yamaguchi-Schinozaki K and others. The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J. 1986;5:2043–2049. doi: 10.1002/j.1460-2075.1986.tb04464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, Sugiura M. Genes for the large and small subunits of ribulose-1,5-bisphosphate carboxylase/oxygenase constitute a single operon in a cyanobacterium Anacystis nidulans 6301. Mol Gen Genet. 1985;200:27–32. [Google Scholar]
- Spreitzer RJ. Genetic dissection of Rubisco structure and function. Annu Rev Plant Physiol Plant Mol Biol. 1993;44:411–434. [Google Scholar]
- Staub J, Maliga P. Accumulation of D1 polypeptide in tobacco plastids is regulated via the untranslated region of the psbA mRNA. EMBO J. 1993;12:601–606. doi: 10.1002/j.1460-2075.1993.tb05692.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staub JM, Maliga P. Translation of psbA mRNA is regulated by light via the 5′-untranslated region in tobacco plastids. Plant J. 1994;6:547–553. doi: 10.1046/j.1365-313x.1994.6040547.x. [DOI] [PubMed] [Google Scholar]
- Sugita M, Svab Z, Maliga P, Sugiura M. Targeted deletion of sprA from the tobacco plastid genome indicates that the encoded small RNA is not essential for pre-16S rRNA maturation in plastids. Mol Gen Genet. 1997;257:23–27. doi: 10.1007/s004380050619. [DOI] [PubMed] [Google Scholar]
- Svab Z, Hajdukiewicz P, Maliga P. Stable transformation of plastids in higher plants. Proc Natl Acad Sci USA. 1990;87:8526–8530. doi: 10.1073/pnas.87.21.8526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svab Z, Maliga P. High-frequency plastid transformation in tobacco by selection for a chimeric aadA gene. Proc Natl Acad Sci USA. 1993;90:913–917. doi: 10.1073/pnas.90.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waksman G, Freyssinet G. Nucleotide sequence of a cDNA encoding the ribulose-1,5-bisphosphate carboxylase/oxygenase from sunflower (Helianthus annuus) Nucleic Acids Res. 1987;15:1328. doi: 10.1093/nar/15.3.1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Snyder GW, Esau BD, Portis AR, Ogren WL. Species-dependent variation in the interaction of substrate-bound ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) and Rubisco activase. Plant Physiol. 1992;100:1858–1862. doi: 10.1104/pp.100.4.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G, Spreitzer RJ. Directed mutagenesis of chloroplast ribulose-bisphosphate carboxylase/oxygenase: substitution at large-subunit asparagine 123 and serine 379 decrease CO2/O2 specificity. J Biol Chem. 1994;269:3952–3956. [PubMed] [Google Scholar]
- Zhu G, Spreitzer RJ. Directed mutagenesis of chloroplast ribulose-bisphosphate carboxylase/oxygenase: loop-6 substitutions complement for structural stability but decrease catalytic efficiency. J Biol Chem. 1996;271:18494–18498. doi: 10.1074/jbc.271.31.18494. [DOI] [PubMed] [Google Scholar]