Abstract
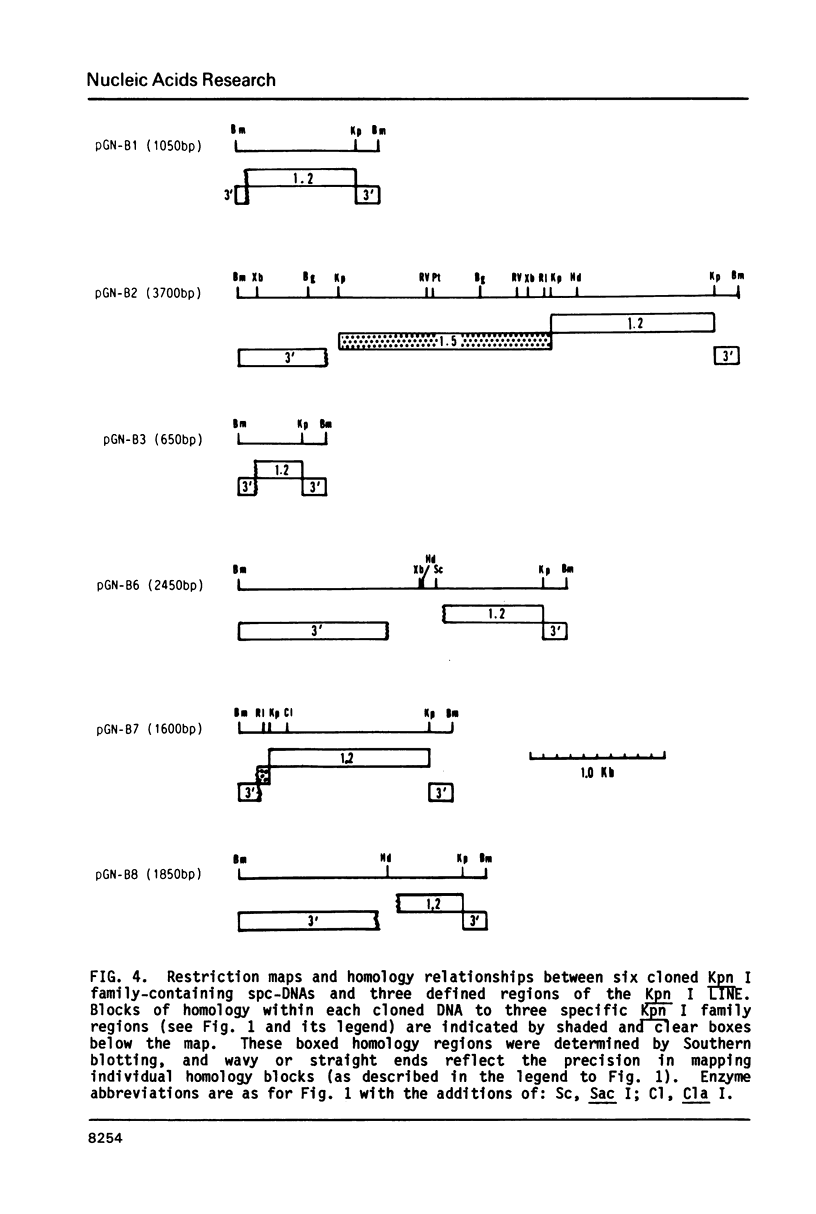
The L1 family of long interspersed nucleotide sequences (LINES) has recently been identified and characterized in the small polydisperse circular DNA (spc-DNA) populations of monkey (1), human (2) and mouse (3) cells. In monkey spc-DNA, the L1 (also known as Kpn I) family is present in discrete size classes (ranging from 300 to 6000 base pairs (bp)) which appear to be generated by non homologous recombination events within chromosomal elements. In this communication it is shown that different regions of the consensus L1 family are present at different frequencies in monkey spc-DNA (as they are in chromosomal DNA), that all regions of the family are present in extrachromosomal DNA, and that each region appears to be represented in an identical discrete spc-DNA size distribution. This size distribution reflects a non-sequence specific mechanism that generates spc-DNA size classes by chromosomal DNA recombination events that are in some way constrained to occur between sites separated by relatively defined lengths.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ballario P., Filetici P., Junakovic N., Pedone F. Ty1 extrachromosomal circular copies in Saccharomyces cerevisiae. FEBS Lett. 1983 May 8;155(2):225–229. doi: 10.1016/0014-5793(82)80608-x. [DOI] [PubMed] [Google Scholar]
- Bertelsen A. H., Humayun M. Z., Karfopoulos S. G., Rush M. G. Molecular characterization of small polydisperse circular deoxyribonucleic acid from an African green monkey cell line. Biochemistry. 1982 Apr 27;21(9):2076–2085. doi: 10.1021/bi00538a015. [DOI] [PubMed] [Google Scholar]
- DeLap R. J., Rush M. G., Zouzias D., Khan S. Isolation and preliminary characterization of the small circular DNA present in African green monkey kidney (BSC-1) cells. Plasmid. 1978 Sep;1(4):508–521. doi: 10.1016/0147-619x(78)90008-2. [DOI] [PubMed] [Google Scholar]
- DiGiovanni L., Haynes S. R., Misra R., Jelinek W. R. Kpn I family of long-dispersed repeated DNA sequences of man: evidence for entry into genomic DNA of DNA copies of poly(A)-terminated Kpn I RNAs. Proc Natl Acad Sci U S A. 1983 Nov;80(21):6533–6537. doi: 10.1073/pnas.80.21.6533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell A. J., Ish-Horowicz D. The origin of extrachromosomal circular copia elements. Cell. 1983 Sep;34(2):415–419. doi: 10.1016/0092-8674(83)90375-6. [DOI] [PubMed] [Google Scholar]
- Flavell A. J. Role of reverse transcription in the generation of extrachromosomal copia mobile genetic elements. Nature. 1984 Aug 9;310(5977):514–516. doi: 10.1038/310514a0. [DOI] [PubMed] [Google Scholar]
- Fujimoto S., Tsuda T., Toda M., Yamagishi H. Transposon-like sequences in extrachromosomal circular DNA from mouse thymocytes. Proc Natl Acad Sci U S A. 1985 Apr;82(7):2072–2076. doi: 10.1073/pnas.82.7.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiev G. P. Mobile genetic elements in animal cells and their biological significance. Eur J Biochem. 1984 Dec 3;145(2):203–220. doi: 10.1111/j.1432-1033.1984.tb08541.x. [DOI] [PubMed] [Google Scholar]
- Grimaldi G., Singer M. F. Members of the KpnI family of long interspersed repeated sequences join and interrupt alpha-satellite in the monkey genome. Nucleic Acids Res. 1983 Jan 25;11(2):321–338. doi: 10.1093/nar/11.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaldi G., Skowronski J., Singer M. F. Defining the beginning and end of KpnI family segments. EMBO J. 1984 Aug;3(8):1753–1759. doi: 10.1002/j.1460-2075.1984.tb02042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kole L. B., Haynes S. R., Jelinek W. R. Discrete and heterogeneous high molecular weight RNAs complementary to a long dispersed repeat family (a possible transposon) of human DNA. J Mol Biol. 1983 Apr 5;165(2):257–286. doi: 10.1016/s0022-2836(83)80257-5. [DOI] [PubMed] [Google Scholar]
- Krolewski J. J., Bertelsen A. H., Humayun M. Z., Rush M. G. Members of the Alu family of interspersed, repetitive DNA sequences are in the small circular DNA population of monkey cells grown in culture. J Mol Biol. 1982 Jan 25;154(3):399–415. doi: 10.1016/s0022-2836(82)80003-x. [DOI] [PubMed] [Google Scholar]
- Krolewski J. J., Rush M. G. Some extrachromosomal circular DNAs containing the Alu family of dispersed repetitive sequences may be reverse transcripts. J Mol Biol. 1984 Mar 25;174(1):31–40. doi: 10.1016/0022-2836(84)90363-2. [DOI] [PubMed] [Google Scholar]
- Kunisada T., Yamagishi H., Sekiguchi T. Intracellular location of small circular DNA complexes in mammalian cell lines. Plasmid. 1983 Nov;10(3):242–250. doi: 10.1016/0147-619x(83)90038-0. [DOI] [PubMed] [Google Scholar]
- Kunisada T., Yamagishi H. Sequence repetition and genomic distribution of small polydisperse circular DNA purified from HeLa cells. Gene. 1984 Nov;31(1-3):213–223. doi: 10.1016/0378-1119(84)90212-9. [DOI] [PubMed] [Google Scholar]
- Lerman M. I., Thayer R. E., Singer M. F. Kpn I family of long interspersed repeated DNA sequences in primates: polymorphism of family members and evidence for transcription. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3966–3970. doi: 10.1073/pnas.80.13.3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutchan T., Hsu H., Thayer R. E., Singer M. F. Organization of African green monkey DNA at junctions between alpha-satellite and other DNA sequences. J Mol Biol. 1982 May 15;157(2):195–211. doi: 10.1016/0022-2836(82)90230-3. [DOI] [PubMed] [Google Scholar]
- Mossie K. G., Young M. W., Varmus H. E. Extrachromosomal DNA forms of copia-like transposable elements, F elements and middle repetitive DNA sequences in Drosophila melanogaster. Variation in cultured cells and embryos. J Mol Biol. 1985 Mar 5;182(1):31–43. doi: 10.1016/0022-2836(85)90025-7. [DOI] [PubMed] [Google Scholar]
- Paulson K. E., Deka N., Schmid C. W., Misra R., Schindler C. W., Rush M. G., Kadyk L., Leinwand L. A transposon-like element in human DNA. Nature. 1985 Jul 25;316(6026):359–361. doi: 10.1038/316359a0. [DOI] [PubMed] [Google Scholar]
- Potter S. S. Rearranged sequences of a human Kpn I element. Proc Natl Acad Sci U S A. 1984 Feb;81(4):1012–1016. doi: 10.1073/pnas.81.4.1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers J. H. The origin and evolution of retroposons. Int Rev Cytol. 1985;93:187–279. doi: 10.1016/s0074-7696(08)61375-3. [DOI] [PubMed] [Google Scholar]
- Rose A. M., Snutch T. P. Isolation of the closed circular form of the transposable element Tc1 in Caenorhabditis elegans. Nature. 1984 Oct 4;311(5985):485–486. doi: 10.1038/311485a0. [DOI] [PubMed] [Google Scholar]
- Ruan K., Emmons S. W. Extrachromosomal copies of transposon Tc1 in the nematode Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1984 Jul;81(13):4018–4022. doi: 10.1073/pnas.81.13.4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler C. W., Rush M. G. The KpnI family of long interspersed nucleotide sequences is present on discrete sizes of circular DNA in monkey (BSC-1) cells. J Mol Biol. 1985 Jan 20;181(2):161–173. doi: 10.1016/0022-2836(85)90082-8. [DOI] [PubMed] [Google Scholar]
- Schmeckpeper B. J., Scott A. F., Smith K. D. Transcripts homologous to a long repeated DNA element in the human genome. J Biol Chem. 1984 Jan 25;259(2):1218–1225. [PubMed] [Google Scholar]
- Shafit-Zagardo B., Maio J. J., Brown F. L. KpnI families of long, interspersed repetitive DNAs in human and other primate genomes. Nucleic Acids Res. 1982 May 25;10(10):3175–3193. doi: 10.1093/nar/10.10.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd B. M., Finnegan D. J. Structure of circular copies of the 412 transposable element present in Drosophila melanogaster tissue culture cells, and isolation of a free 412 long terminal repeat. J Mol Biol. 1984 Nov 25;180(1):21–40. doi: 10.1016/0022-2836(84)90428-5. [DOI] [PubMed] [Google Scholar]
- Stanfield S. W., Helinski D. R. Cloning and characterization of small circular DNA from Chinese hamster ovary cells. Mol Cell Biol. 1984 Jan;4(1):173–180. doi: 10.1128/mcb.4.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Paulson K. E., Schmid C. W., Kadyk L., Leinwand L. Non-Alu family interspersed repeats in human DNA and their transcriptional activity. Nucleic Acids Res. 1984 Mar 26;12(6):2669–2690. doi: 10.1093/nar/12.6.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thayer R. E., Singer M. F. Interruption of an alpha-satellite array by a short member of the KpnI family of interspersed, highly repeated monkey DNA sequences. Mol Cell Biol. 1983 Jun;3(6):967–973. doi: 10.1128/mcb.3.6.967. [DOI] [PMC free article] [PubMed] [Google Scholar]