Abstract
Ubiquitination is an important protein post-translational modification, which is involved in various cellular processes in higher plants, and U-box E3 ligases play important roles in diverse functions in eukaryotes. Here, we describe the functions of Arabidopsis thaliana PUB19 (AtPUB19), which we demonstrated in an in vitro assay to encode a U-box type E3 ubiquitin ligase. AtPUB19 was up-regulated by drought, salt, cold, and abscisic acid (ABA). Down-regulation of AtPUB19 led to hypersensitivity to ABA, enhanced ABA-induced stomatal closing, and enhanced drought tolerance, while AtPUB19 overexpression resulted in the reverse phenotypes. Molecular analysis showed that the expression levels of a number of ABA and stress marker genes were altered in both AtPUB19 overexpressing and atpub19-1 mutant plants. In summary, our data show that AtPUB19 negatively regulates ABA and drought responses in A. thaliana.
Keywords: U-box, ABA, drought stress, Arabidopsis
INTRODUCTION
Plants are frequently exposed to various environmental stresses, which greatly affect their growth and development. Drought and high salinity can result in water stress, constituting the main reason for dramatic reduction in crop yield worldwide (Boyer, 1982). To survive such detrimental conditions, plants have developed many defense strategies, often involving rapid accumulation of abscisic acid (ABA), an important phytohormone that directs seed maturation and controls seed dormancy to ensure that seeds germinate under favorable growth conditions. During the growth of seedlings and plant maturation, the accumulation of ABA can protect plants from damage induced by drought, salinity, and pathogenic attack (Lopez-Molina et al., 2001; Finkelstein et al., 2002).
Several ABA receptors have been reported (Razem et al., 2006; Shen et al., 2006; Ma et al., 2009; Pandey et al., 2009; Park et al., 2009). The identification of PYR/PYL/RCAR receptors and the mechanism for ABA action through the PP2Cs and SnRk2s to transmit signals was an important step for our understanding of ABA signal transduction. PYR/PYL/RCAR proteins can bind to ABA and inhibit the negative effects of group A PP2C proteins on SnRK2s. Consequently, the accumulation of active SnRK2s is involved in the direct phosphorylation of bZIP transcription factors, which promote ABA-induced gene expression (Wasilewska et al., 2008; Park et al., 2009). This signaling mechanism has been confirmed in vitro using recombinant PYR1, ABI1, OST1, and ABF2 (Fujii et al., 2009), and a large number of genes regulated by ABA or water stress have been identified. Furthermore, some of them were applied in genetic engineering of drought-tolerant crops (Wang et al., 2009; Yang et al., 2010). Although the biological functions of a few genes in stress sensitivity or tolerance have been characterized, the functions of larger genes are still unknown. Therefore, it is necessary to study the functions of stress-related genes not only to uncover the molecular mechanisms underlying the responses to harsh environmental conditions, but also to understand the development of transgenic plants tolerant to water deficit.
The ubiquitination pathway mediates post-translational modification of proteins or degradation of the target protein. It is an important process during the eukaryotic response to developmental cues or adaption to various environmental stresses. Ubiquitination is carried in the following three steps: (1) ubiquitin (Ub) molecules are activated by E1 (Ub-activating enzyme); (2) activated Ub is transferred to E2 (a Ub-conjugating enzyme), forming an E2–Ub intermediate; (3) Ub is transferred from E2–Ub intermediate to the target protein by E3 (Ub-ligase) or E3 forms a third thioester bond with Ub prior to transfer to the substrate (Moon et al., 2004; Smalle and Vierstra, 2004; Dreher and Callis, 2007). As the E3 ligase confers the substrate specificity to the ubiquitination process, researchers are highly interested in studying the roles of E3 ligases during growth and development of organisms or their responses to different stresses.
Plants have larger numbers of different E3 ligases than other eukaryotes. For example, the Arabidopsis thaliana genome contains >1 300 genes predicted to encode E3 ligases. Based on the mechanism of action and the presence of domains, E3 ligases can be divided into different families: HECT, RING, and U-box ligases (Smalle and Vierstra, 2004; Stone and Callis, 2007). The U-box is a modified RING-finger domain composed of ∼70 conserved amino acids. In comparison to the 2 U-box genes in yeast and 21 U-box genes in the human genome, there are 64 U-box genes predicted in the Arabidopsis genome, and 77 have been annotated in the rice genome, indicating that they play diverse roles in plants (Wiborg et al., 2008; Zeng et al., 2008). U-box proteins participate in many cellular processes, such as self-incompatibility and pseudo-self-incompatibility, plant hormone responses, and abiotic and biotic stresses (Yee and Goring, 2009).
In this study, we identified an Arabidopsis U-box gene, AtPUB19, which was induced by ABA and abiotic stresses. We showed that pub19 mutants were hypersensitive to ABA and drought-tolerant compared with wild-type (WT) plants, while the AtPUB19 overexpressing lines showed the reverse phenotypes. The results show that AtPUB19 is a negative regulator in the A. thaliana response to ABA and drought stress.
RESULTS
Identification of AtPUB19
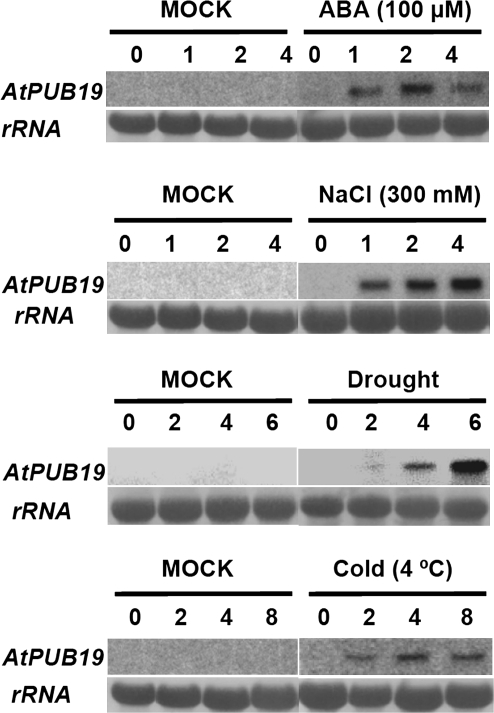
In a previous functional genomic project to identify stress-related Arabidopsis single subunit E3 ligase genes (Zhang et al., 2007), in silico gene expressions of Arabidopsis RING finger and U-box genes were analyzed using several publically available stress-related microarray datasets. We found that the induced expression of the U-box gene AtPUB19 (at1g60190) was the highest among the PUB genes in the Arabidopsis plants treated with ABA and various abiotic stresses. To confirm this microarray analysis, a more extensive expression profile of AtPUB19 was monitored by Northern blot in this study. As shown in Figure 1, AtPUB19 was markedly up-regulated in 2-week-old seedlings by ABA, high salinity, drought, and cold treatments. We observed that the induction profiles of AtPUB19 by ABA and cold were similar, peaking at 2 and 4 h of treatment, respectively. The induction profiles of AtPUB19 by high salinity and drought were also similar, markedly increasing as the treatment time progressed. These findings indicate that AtPUB19 is inducible by ABA and various abiotic stresses.
Figure 1.
Northern Blot Analysis of AtPUB19 Transcript.
Expression of AtPUB19 in response to ABA (0–4 h with 100 μM), high salinity (0–4 h with 300 mM NaCl), drought (0–6 h), and cold (0–8 h at 4°C) treatments. rRNA: Methylene blue-stained ribosome RNA were used as loading controls.
AtPUB19 Is a Functional U-Box E3 Ubiquitin Ligase
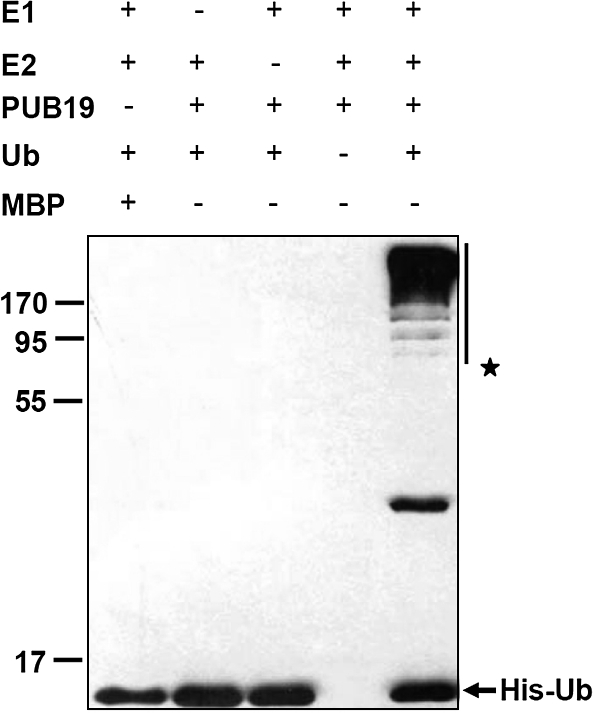
Many studies have identified U-box-containing proteins as functional E3 Ub ligases (Stone et al., 2003; Yan et al., 2003; Zeng et al., 2004; Yang et al., 2006; Cho et al., 2008; Wiborg et al., 2008; Raab et al., 2009). Here, domain analysis showed that AtPUB19 contains a conserved U-box domain (281–344 amino acids), suggesting that it may also have E3 Ub ligase activity. To test this hypothesis, we produced a full-length Arabidopsis PUB19 fused protein with maltose binding protein (MBP) in Escherichia coli and subsequently affinity-purified it (MBP–AtPUB19) from the soluble fraction. In vitro self-ubiquitination assays were performed in the presence of wheat E1, human E2 (UBCh5b), and 6xHis tag Ub, and polyubiquitination was detected only in the presence of E1, E2 and MBP–AtPUB19 (Figure 2, line 5). A negative result was observed if either E1 or E2 was omitted in the reaction. These results indicate that AtPUB19 has E3 Ub ligase activity.
Figure 2.
E3 Ub Ligase Activity of AtPUB19.
MBP–PUB19 fusion proteins were assayed for E3 activity in the presence of wheat E1, human E2 (UBCh5b), and 6xHis tag Ub. The left numbers denote the molecular masses of marker proteins in kilodaltons. MBP was used as a negative control. Samples were resolved by 12% SDS–PAGE. Anti-His antibody was used to detect the His tag Ub.
Genetic Analysis of PUB19 Gene
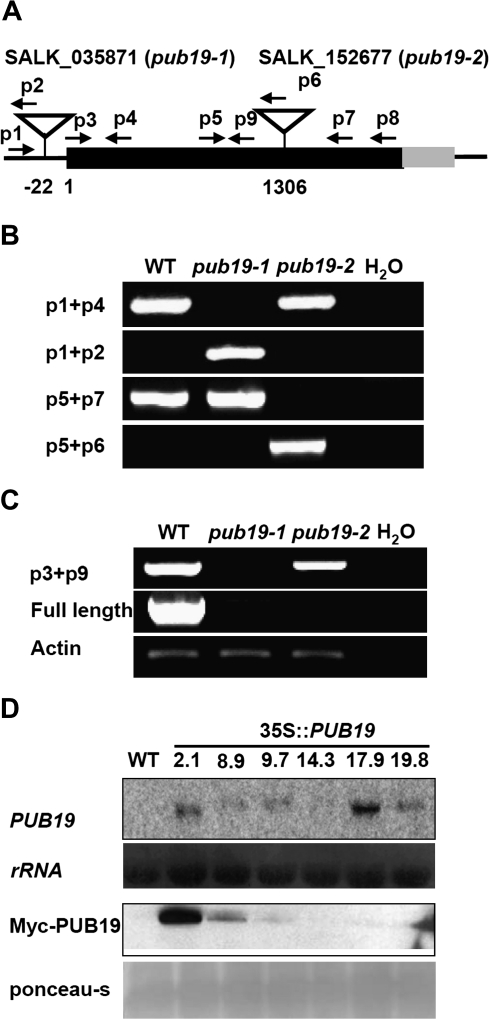
To define the functions of AtPUB19 in vivo, we applied a reverse genetic approach. Two T-DNA insertion lines with different target sites were obtained from the Salk mutant collection: SALK_035871 (pub19-1) and SALK_152677 (pub19-2). The T-DNA insertion positions are indicated in Figure 3A, and homozygous mutants were verified by genomic PCR using four different sets of primers (Figure 3A and 3B). The RT–PCR results also showed that there are truncated transcripts in pub19-2, but the full-length mRNA of AtPUB19 was not detected in the pub19-1 and pub19-2 mutant lines (Figure 3C). The pub19-1 and pub19-2 mutant lines did not differ in appearance from the WT seedlings. In addition, we generated transgenic Arabidopsis plants overexpressing the AtPUB19 gene under the control of the cauliflower mosaic virus 35S promoter. RNA gel blot analysis revealed that most of the transgenic lines expressed AtPUB19 at higher levels compared with WT plants and, in Western blot assays, these lines exhibited high expression levels of the Myc-PUB19 protein (Figure 3D). Thus, the overexpressing lines 2.1 and 8.9, designated as OX-1 and OX-2, respectively, were selected for further analysis. By comparing morphologies in the early stage of seedling growth, we observed that the roots of the AtPUB19 overexpressors were shorter than those of control plants overexpressing empty vector under standard growth conditions.
Figure 3.
Molecular Characterization of the pub19 Mutant Lines and AtPUB19-Overexpressing Transgenic Plants.
(A) Schematic of the AtPUB19 depicting the positions of T-DNA insertions shown as inverted triangles. Arrows indicate the positions of primers used in separate genomic PCR or RT–PCR.
(B) Genomic PCR analysis of WT, pub19-1, and pub19-2 plants. DNA was isolated from 10-day-old seedlings. In this experiment, four different primer sets were used as indicated on the right.
(C) RT–PCR analysis of WT, pub19-1, and pub19-2 plants. RNA was isolated from 2-week-old seedlings treated with 300 mM NaCl.
(D) AtPUB19 transcript and protein levels in transgenic lines. Western blot was performed using an anti-Myc monoclonal antibody.
Mutants of AtPUB19 Are More Sensitive to ABA
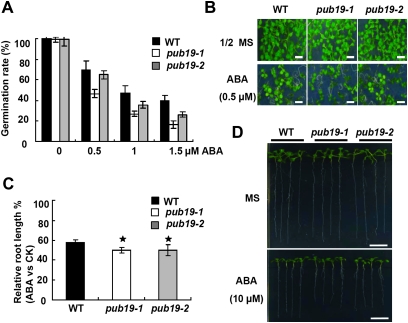
ABA plays a key role in regulating plant responses to different stresses (Finkelstein et al., 2002). Salt and drought often result in increased levels of ABA in plants, which can activate a series of ABA-dependent responses (Zhu, 2002). Inhibitory experiments of seed germination are useful for characterizing components of the signaling pathway(s) involved in the response to ABA (Giraudat, 1995). Here, the dramatic induction of AtPUB19 transcript levels by ABA in the Northern blot assay (Figure 1) suggested that this gene may have potential roles in the ABA response. To elucidate the role of AtPUB19 in ABA signaling, the seeds of pub19-1, pub19-2, and WT plants were germinated on ½MS medium with different concentrations of ABA (0, 0.5, 1, and 1.5 μM). No difference in germination rates was observed among the untreated plants (Figure 4A). However, in the presence of 0.5 μM ABA, the germination rates decreased to 69.82% for WT plants, 46.49% for pub19-1, and 64.92% for pub19-2 (Figure 4A and 4B). The ABA-hypersensitive response of pub19 mutant lines occurred at all concentrations of ABA added to the medium, and the effect was dosage-dependent. To further characterize the role of AtPUB19 in plant sensitivity to ABA during the post-germination stage, root elongation inhibition was analyzed in these plants. Four-day-old seedlings were transferred to MS medium with 10 μM ABA. After 5 d, root growth inhibition of the pub19 mutant lines by ABA was more severe than that of the WT plants (Figure 4C and 4D). Furthermore, the pub19-1 plants were more sensitive to ABA than pub19-2 plants. These results indicated that the truncation mutant of pub19-2 gene may function partially in the plants. Therefore, it can be concluded that the pub19 mutant plants are sensitive to ABA, and AtPUB19 is involved in the ABA response.
Figure 4.
ABA Sensitivity of pub19 Plants.
(A) ABA dose-response analysis of germination in different genotypes of plants. Seeds were germinated for 72 h on plates containing a range of ABA concentrations (0, 0.5, 1, and 1.5 μM). Germination rates represented by the chart are based on three repeated experiments (n = 90).
(B) Growth of different genotypes of plants on ½MS medium containing 0.5 μM ABA. Seeds were germinated and grown for 11 d. Bar = 0.5 cm.
(C) Quantitative analysis of root growth. The experiment was performed in triplicate (n = 12) and repeated three times. Bars indicate standard errors.
(D) Phenotypic comparison of root length in the MS medium with or without ABA (Bar = 1 cm).
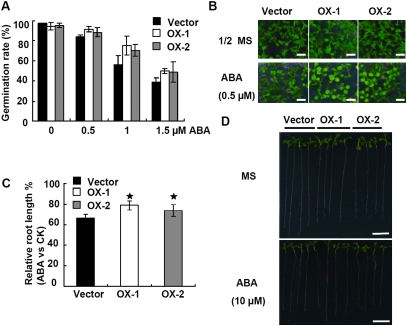
Overexpression of AtPUB19 Decreases Plant Sensitivity to ABA
To further characterize the in vivo functions of AtPUB19, we analyzed the capacity of the WT and the transgenic plants overexpressing this gene, PUB19-OX1 and PUB19-OX2, to respond to ABA. As shown in Figure 5A and 5B, the germination rates of PUB19-OX1 and PUB19-OX2 were similar to those of vector control plants under standard conditions. However, in the presence of different concentrations of ABA, the germination rates of PUB19-OX1 and PUB19-OX2 were higher than those of vector control plants (Figure 5A and 5B). For example, in the presence of 1 μM ABA, the germination rates decreased to 56.3% for vector control plants, 75.35% for PUB19-OX1, and 70.57% for PUB19-OX2 plants. Furthermore, root growth inhibition by ABA was less severe for the PUB19-OX1 and PUB19-OX2 plants compared to the vector control plants (Figure 5C and 5D). These results indicated that enhanced expression of AtPUB19 decreased the plant sensitivity to ABA. We conclude from these results, together with the observed ABA phenotype of the pub19 mutants, that AtPUB19 is a negative regulator of ABA signaling.
Figure 5.
ABA Sensitivity of PUB19 Overexpressor Plants.
(A) ABA dose-response analysis of germination in plants of different genotypes. Seeds were germinated for 84 h on plates containing a range of concentrations of ABA (0, 0.5, 1, and 1.5 μM). Germination rates represented by the chart are based on three repeated experiments (n = 90).
(B) Growth of different genotypes of plants on ½MS medium containing 0.5 μM ABA. Seeds were germinated and grown for 11 d. Bar = 0.5 cm.
(C) Quantitative analysis of root growth. The experiment was performed in triplicate (n = 12) and repeated three times. Bars indicate standard errors.
(D) Phenotypic comparison of root length in the MS medium with or without ABA. Bar = 1 cm.
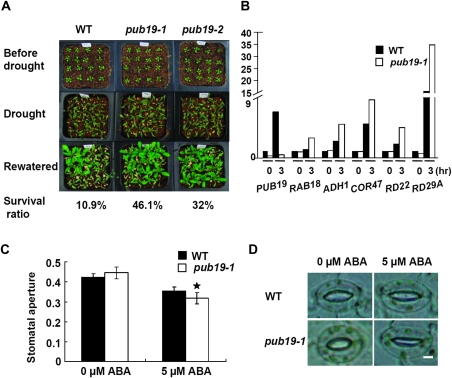
pub19 Plants Are Relatively More Resistant to Drought Stress than Wild-Type Plants
Because AtPUB19 is a drought-inducible gene, it is likely involved in the plant response to drought. To test this hypothesis, a whole-plant drought assay was performed in soil. Two-week-old seedlings were transferred from plates to water-saturated soil, and then the water was withheld to create a water deficit in the soil. After 15 d, most of the WT plants and mutant plants were wilted due to water deprivation. After re-watering, the pub19-1 and pub19-2 plants exhibited a high survival rate (46.1% for pub19-1 and 32% for pub19-2 plants), whereas the corresponding survival rate was 10.9% for WT plants (Figure 6A). To reveal how AtPUB19 affects drought-responsive genes, we examined the expression of stress-related marker genes in the drought-stressed plants. As shown in Figure 6B and Supplemental Figure 1A, the transcript levels of RAB18, ADH1, COR47, RD22, and RD29A were significantly up-regulated in the pub19-1 mutant relative to the WT plants. These results showed that pub19 mutant plants exhibited enhanced tolerance to water deficit and that AtPUB19 is probably a negative regulator of drought tolerance.
Figure 6.
Drought-Tolerance Assays of Control and pub19 mutant Plants and Effects of ABA on Stomatal Aperture of WT and pub19-1 Mutant Plants.
(A) Two-week-old plants were used for the drought-tolerance assays. Plants were grown in soil in the same container, withheld from water for 15 d and then re-watered. The photographs were taken 36 h after re-watering.
(B) Expression of ABA and stress-responsive genes in WT and pub19-1 mutant seedlings treated by drought stress for 3 h. Total RNA was extracted from seedlings after different treatment times. Total RNA (10 μg) was loaded in each lane and analyzed by RNA gel blots hybridized with gene-specific probes.
(C) Ten fresh leaves from WT or pub19-1 mutant plants were incubated under light in buffer for 3 h and then treated with 0 and 5 μM ABA for 5 h before aperture measurements. Data are mean ratios of width to length ± SE of three independent experiments (n = 30–40).
(D) Stomatal apertures of WT and pub19-1 mutant plants. Stomatal guard cells were observed in the epidermal peels treated with 0 or 5 μM ABA. Bar = 5 μm.
Stomatal closure is also an ABA-controlled process that determines the rate of transpiration under water-deficit conditions (Leung and Giraudat, 1998). To investigate whether AtPUB19 plays a role in ABA-mediated stomatal closure, we treated leaves of pub19-1 plants and WT plants with ABA to analyze the stomatal apertures. As shown in Figure 6C and 6D, treating the leaf epidermis of pub19-1 plants with ABA caused more pronounced closure of stomata than that in WT plants. This result further supports AtPUB19 as a negative regulator of drought tolerance through regulation of ABA-controlled stomata movement.
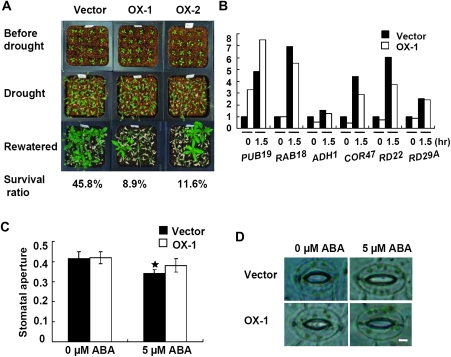
Overexpression of AtPUB19 Increases Sensitivity to Drought Stress
In contrast to the drought-tolerant pub19-1 and pub19-2 plants, the PUB19-OX1 and PUB19-OX2 plants were hypersensitive to dehydration. Two-week-old seedlings were subjected to water-deficit treatment. After 15 d, all plants, regardless of genotype, were wilted. Five days after re-watering, 45% (44 of 96) of vector control plants were able to continue to grow. By contrast, the survival ratios of the PUB19-OX1 and PUB19-OX2 plants were 8% (10 of 112) and 11% (13 of 112), respectively (Figure 7A). When the bolting plants were used for drought-tolerance assays, similar results were obtained. The mRNA levels of RAB18, ADH1, COR47, and RD22 were slightly down-regulated in transgenic plants relative to the vector control plants in drought conditions, and the transcript levels of RD29A showed no difference between the plants of either genotype (Figure 7B and Supplemental Figure 1B). Because overexpression of AtPUB19 increased sensitivity to dehydration, and its mutation enhanced tolerance to the stress, these results provide evidence that the AtPUB19 expression level is inversely correlated with the degree of tolerance to water stress and that AtPUB19 is a negative regulator of the plant response to drought. This result is also correlated with the ABA insensitivity of AtPUB19 overexpressing plants.
Figure 7.
Drought-Tolerance Assays of Control and Overexpressing Plants and Effects of ABA on Stomatal Aperture of Vector Control and PUB19-OX1 Plants.
(A) Two-week-old plants were used for drought-tolerance assays. Plants were grown in soil in the same container, withheld from water for 15 d, and then re-watered. The photographs were taken 5 d after re-watering. Vector: control plants, OX-1 and OX-2: transgenic plants.
(B) Expression of ABA and stress-responsive genes in vector control and PUB19-OX1 seedlings treated by drought stress at different times. Total RNA was extracted from seedlings for 1.5 h. Total RNA (10 μg) was loaded in each lane and analyzed by RNA gel blots hybridized to gene-specific probes.
(C) Ten fresh leaves from vector control or PUB19-OX1 plants were incubated under light in buffer for 3 h and then treated with 0 and 5 μM ABA for 5 h before aperture measurements. Data are mean ratios of width to length ± SE of three independent experiments (n = 30–40).
(D) Stomatal apertures of vector control and PUB19-OX1 plants. Stomatal guard cells were observed in the epidermal peels treated with 0 or 5 μM ABA. Bar = 5 μm.
When the leaf epidermis of PUB19-OX1 and vector control plants were treated with ABA, the stomatal apertures of vector were smaller than those of PUB19-OX1 (Figure 7C and 7D). This result showed that the PUB19-OX1 guard cells have an impaired response to ABA. Thus, AtPUB19 may play a key role in ABA-related stomatal closure under drought stress.
DISCUSSION
A number of studies have shown that not only the expression and regulation of genes, but also the protein turnover are involved in stress signaling. Ubiquitination plays an important role in the perception and signal transduction of hormone and various stress responses (Hellmann and Estelle, 2002). Many U-box-containing proteins, demonstrated to function as E3 ligases, have been shown to be involved in various physiological processes. For example, Arabidopsis pub9 knockout lines are hypersensitive to ABA during seed germination, and AtPUB9 can re-localize from the nucleus to the plasma membrane with ABA treatment (Samuel et al., 2008). Recent studies on AtCHIP, HOS1, AFP, AIP2, SDIR1, PUB22, PUB23, and OsPUB15 support that ubiquitination plays an important role in the plant response to abiotic stresses (Lee et al., 2001; Yan et al., 2003; Zhang et al., 2005; Stone et al., 2006; Zhang et al., 2007; Cho et al., 2008; Park et al., 2011). AtCHIP, PUB22, PUB23, or OsPUB15, encoding a protein with a U-box domain, has E3 ubiquitin activity in vitro. Overexpression of AtCHIP in Arabidopsis rendered plants more sensitive to both low- and high-temperature treatment (Yan et al., 2003). It was shown that PUB22- and PUB23-overexpressing transgenic plants are hypersensitive to drought, while pub22 and pub23 mutant plants are more drought-tolerant (Cho et al., 2008). As OsPUB15 is a negative regulator of cell death and reactive oxygen species (ROS) stress, plants overexpressing OsPUB15 were observed to grow better than the WT under salt and paraquat stresses (Park et al., 2011). Thus, the existence of a large number of U-box proteins reflects the diversity of function of these proteins.
In our previous work, we found AtPUB19 was rapidly induced by ABA and drought. Our detailed functional analysis showed that AtPUB19 is involved in the plants’ response to ABA and drought stress. PUB22, PUB23, and AtPUB19 have the similar functions during plant response to drought stress. Different from PUB22 and PUB23, which are not induced by ABA, the expression of AtPUB19 increased when plants are treated with ABA. Furthermore, both PUB22- and PUB23-overexpressing transgenic plants and pub22 and pub23 mutant plants showed similar phenotypes compared with WT plants in germination rate and stomatal closure in the presence of exogenously applied ABA (Cho et al., 2008). In our study, AtPUB19 overexpressing transgenic plants were hypersensitive to ABA during germination, post-germination growth, and stomatal closure, while the pub19-1 mutant plants showed the contrary phenotype when treated with ABA. As the pub19-2 mutant plants showed similar germination rates to WT plants in the presence of ABA, it is possible that the truncated transcripts in pub19-2 can still function in the ABA signaling pathway.
We were able to confirm that AtPUB19 has E3 ligase activity by overexpressing and purified MBP-tagged PUB19 from E. coli and performing ubiquitination assays. Furthermore, the pub19-1 and pub19-2 mutant plants showed similar phenotypes to the WT seedlings, indicating that functional redundancy exists in Arabidopsis. In spite of the functional similarities between PUB22, PUB23, and AtPUB19, the phenotype of AtPUB19 overexpressors was not identical to that of PUB22 or PUB23 overexpressors, which have significantly longer roots than WT plants (Cho et al., 2008). We observed that the roots of the AtPUB19 overexpressors were shorter than those of control plants. It is plausible that PUB22, PUB23, and AtPUB19 take part in root development with different substrates. RPN12a is a known target protein for ubiquitination by PUB22 and PUB23 (Cho et al., 2008), but more detailed studies on the target of AtPUB19 are necessary to explain the differences between these proteins.
In conclusion, our data show that AtPUB19 is a functional E3 ligase and a negative regulator of the Arabidopsis response to ABA and drought stress. Its mutant displayed altered expression of stress-related genes, enhanced drought tolerance, and ABA sensitivity, while overexpressors of AtPUB19 displayed reduced plant sensitivity to ABA and hypersensitivity to dehydration. This study contributes to our understanding of the molecular factors involved in the responses of plants to abiotic stresses.
METHODS
Plant Materials and Growth Conditions
A. thaliana ecotype Columbia (Col-0) was used in this study. SALK_035871 and SALK_152677 (PUB19 T-DNA insertion mutants) were obtained from the Arabidopsis Biological Resource Center (ABRC, Ohio State University, Columbus, OH). Seeds were sterilized with 10% bleach and washed three times with sterile water. Sterile seeds were suspended in 0.2% agar and plated on ½MS medium plus 1.5% sucrose. Seeds were stratified in the dark for 2–4 d at 4°C and then transferred to rooms at 22°C. Plant growth conditions were described previously (Xie et al., 2000).
E3 Ub Activity of AtPUB19
The entire AtPUB19 open reading frame (2061 bp) was cloned into the pMAL-c2 vector (New England Biolabs) with the primers 5'-ACCTCGAGATGATCCATACACCAACCG-3' and 5'-ACAAGCTTTCACCAGGCGTGGACAAACC-3' and expressed in E. coli strain BL21. The fusion proteins were prepared using MBP beads according to the manufacturer's instructions. For the E3 ubiquitin ligase activity assay of the fusion proteins, crude extract containing recombinant wheat (Triticum aestivum) E1 (GI: 136632), human E2 (UBCh5b), Arabidopsis Ub (UBQ14, At4g02890) fused with the 6×His tag and purified E3 fused with the MBP tag were used for the assay. The in vitro E3 ligase assays were performed as described (Xie et al., 2002). After the reaction, proteins were separated by SDS–PAGE followed by Western blot analysis with an anti-His antibody and visualized using chemiluminescence as instructed by the manufacturer (Millipore, Immobilon Western Chemiluminescent HRP Substrate).
Gene Expressing Analysis
Two-week-old seedlings grown on agar plates were treated with NaCl (300 mM), ABA (100 μM), cold (4°C), and drought. For drought stress, 2-week-old seedlings from the agar plate were transferred onto a filter paper in a covered Petri dish and treated with fresh liquid ½MS medium for about 12 h, and then the seedlings were subjected to drought treatment for different times. Total RNA was isolated using the hot phenol method (Xie et al., 1999), and 10 μg was applied in each lane for RNA gel analysis. Hybridizations were performed with the α-32p-labeled PUB19, RD22, ADH1, COR47, RD29A, and RAB18. The relative expression level of each sample was quantified by ImageJ software. Values in the figures represent the ratio of target gene to rRNA.
Genomic PCR and RT–PCR Amplification
The pub19-1 (SALK_035871) and pub19-2 (SALK_152677) seeds were obtained from the ABRC. Homozygous mutants were identified by PCR from genomic DNA using AtPUB19 gene-specific primers (P1, 5'-TCTTCGAAGCAGTGTCTA-AAACC-3'; P4, 5'-TTCTGGAAGATGACGTGAAGC-3'; P5, 5'-TGATCTTCGTTGTCCGATTTC-3'; P7, 5'-CTTTCTAGAACCGGTTCACCC-3'), and T-DNA left border primers (P2 and P6 5'-GCGTGGACCGCTTGCTGCAACT-3'). To examine the expression of AtPUB19 by RT–PCR, DNase I-treated total RNA (2 μg) was denatured and reverse transcribed using the M-MLV Reverse Transcriptase (Promega) at 42°C for 60 min. Full-length CDS amplification was performed using AtPUB19-specific primers P3 (5'-ATGATCCATACACCAACCG-3') and P8 (5'-TCACCAGGCGTGGACAAACC-3'). 5' end of CDS was amplification using primers P3 and P9 (5'-CGAAGAATCTTCATCAACGATTCA-3'). Expression levels of Actin1 were monitored with forward (5'-CATCAGGAAGGACTTGTACGG-3') and reverse (5'-GATGGACCTGACTCGTCATAC-3') primers to serve as an internal control.
Construction of Transgenic Plants
To create the transgenic construct for overexpressing AtPUB19 in Arabidopsis, the cDNA was amplified by RT–PCR with specific primers (5'-AAGGTACCCATGATCCATACACCAACCG-3' and 5'-CCACTAGTTCACCAGGCGTGGACAA-3') and cloned into the KpnI and SpeI site in the pCanGMyc vector. In this construct, AtPUB19 was fused in frame to the 6X myc tag at the C-terminus, and the expression of the fused protein was driven by the CaMV 35S promoter. Transformation of Arabidopsis was performed by the vacuum infiltration method using the Agrobacterium tumefaciens strain EHA105 (Bechtold and Pelletier, 1998). For the phenotypic analysis, T3 or T4 homozygous lines were used.
Abiotic Stress Assays
Seeds harvested at the same time from plants grown simultaneously were sterilized, planted in ½MS medium supplemented with 1.5% sucrose, stratified in the dark at 4°C for 3 d, and then transferred to growth chambers with the same environmental conditions described above. In the germination rate assay, seeds were plated on ½MS medium without or with ABA (0, 0.5, 1, and 1.5 μM). In the root growth assay, seeds sown on MS plates were stratified for 3 d at 4°C and were vertically grown for 4 d under normal conditions. The seedlings were then transferred on vertical square MS plates with or without 10 μM ABA. The treatment was conducted in an environment with 70% humidity. For the soil-grown plant drought-tolerance test, 1-week-old seedlings were transplanted to the soil for 1 week under standard growth conditions, and then the plants were subjected to progressive drought by withholding water for specified times. To minimize experimental variations, the same numbers of plants were grown on the same tray.
Stomatal Aperture Analysis
Ten fresh leaves from 4-week-old soil-grown plants at similar developmental stages under standard growth conditions were incubated in a buffer containing 5 mM KCl, 50 mM CaCl2, and 10 mM MES-Tris, pH 6.1, at 20°C for at least 3 h. To ensure that all stomatals open, the leaves were placed under strong light. The leaves were then transferred to fresh buffer with or without 5 μM ABA for 5 h. Stomatals were photographed using a microscope with a camera, and stomatal apertures were measured using Axiovs40 4.6.3.0 software.
SUPPLEMENTARY DATA
Supplementary Data are available at Molecular Plant Online.
FUNDING
This research was supported by the Chinese Ministry of Science and Technology 973-2011CB915402 grant and the National Natural Science Foundation of China (CNSF30670195/31030047/90717006).
Supplementary Material
Acknowledgments
We would like to thank the Arabidopsis Biological Resource Center (ABRC) at Ohio State University for providing the T-DNA insertion lines. No conflict of interest declared.
References
- Bechtold N, Pelletier G. In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 1998;82:259–266. doi: 10.1385/0-89603-391-0:259. [DOI] [PubMed] [Google Scholar]
- Boyer JS. Plant productivity and environment. Science. 1982;218:443–448. doi: 10.1126/science.218.4571.443. [DOI] [PubMed] [Google Scholar]
- Cho SK, Ryu MY, Song C, Kwak JM, Kim WT. Arabidopsis PUB22 and PUB23 are homologous U-Box E3 ubiquitin ligases that play combinatory roles in response to drought stress. Plant Cell. 2008;20:1899–1914. doi: 10.1105/tpc.108.060699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher K, Callis J. Ubiquitin, hormones and biotic stress in plants. Ann. Bot. 2007;99:787–822. doi: 10.1093/aob/mcl255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Gampala SS, Rock CD. Abscisic acid signaling in seeds and seedlings. Plant Cell. 2002;14 Supp:S15–S45. doi: 10.1105/tpc.010441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, et al. In vitro reconstitution of an abscisic acid signalling pathway. Nature. 2009;462:660–664. doi: 10.1038/nature08599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudat J. Abscisic acid signaling. Curr. Opin. Cell Biol. 1995;7:232–238. doi: 10.1016/0955-0674(95)80033-6. [DOI] [PubMed] [Google Scholar]
- Hellmann H, Estelle M. Plant development: regulation by protein degradation. Science. 2002;297:793–797. doi: 10.1126/science.1072831. [DOI] [PubMed] [Google Scholar]
- Lee H, Xiong L, Gong Z, Ishitani M, Stevenson B, Zhu JK. The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a RING finger protein that displays cold-regulated nucleo–cytoplasmic partitioning. Genes Dev. 2001;15:912–924. doi: 10.1101/gad.866801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J, Giraudat J. Abscisic acid signal transduction. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998;49:199–222. doi: 10.1146/annurev.arplant.49.1.199. [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, Chua NH. A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl Acad. Sci. U S A. 2001;98:4782–4787. doi: 10.1073/pnas.081594298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, et al. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324:1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- Moon J, Parry G, Estelle M. The ubiquitin–proteasome pathway and plant development. Plant Cell. 2004;16:3181–3195. doi: 10.1105/tpc.104.161220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey S, Nelson DC, Assmann SM. Two novel GPCR-type G proteins are abscisic acid receptors in Arabidopsis. Cell. 2009;136:136–148. doi: 10.1016/j.cell.2008.12.026. [DOI] [PubMed] [Google Scholar]
- Park JJ, et al. OsPUB15, an E3 ubiquitin ligase, functions to reduce cellular oxidative stress during seedling establishment. Plant J. 2011;65:194–205. doi: 10.1111/j.1365-313X.2010.04416.x. [DOI] [PubMed] [Google Scholar]
- Park SY, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab S, et al. Identification of a novel E3 ubiquitin ligase that is required for suppression of premature senescence in Arabidopsis. Plant J. 2009;59:39–51. doi: 10.1111/j.1365-313X.2009.03846.x. [DOI] [PubMed] [Google Scholar]
- Razem FA, El-Kereamy A, Abrams SR, Hill RD. The RNA-binding protein FCA is an abscisic acid receptor. Nature. 2006;439:290–294. doi: 10.1038/nature04373. [DOI] [PubMed] [Google Scholar]
- Samuel MA, et al. Interactions between the S-domain receptor kinases and AtPUB–ARM E3 ubiquitin ligases suggest a conserved signaling pathway in Arabidopsis. Plant Physiol. 2008;147:2084–2095. doi: 10.1104/pp.108.123380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen YY, et al. The Mg-chelatase H subunit is an abscisic acid receptor. Nature. 2006;443:823–826. doi: 10.1038/nature05176. [DOI] [PubMed] [Google Scholar]
- Smalle J, Vierstra RD. The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 2004;55:555–590. doi: 10.1146/annurev.arplant.55.031903.141801. [DOI] [PubMed] [Google Scholar]
- Stone SL, Callis J. Ubiquitin ligases mediate growth and development by promoting protein death. Curr. Opin. Plant Biol. 2007;10:624–632. doi: 10.1016/j.pbi.2007.07.010. [DOI] [PubMed] [Google Scholar]
- Stone SL, Anderson EM, Mullen RT, Goring DR. ARC1 is an E3 ubiquitin ligase and promotes the ubiquitination of proteins during the rejection of self-incompatible Brassica pollen. Plant Cell. 2003;15:885–898. doi: 10.1105/tpc.009845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Williams LA, Farmer LM, Vierstra RD, Callis J. KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell. 2006;18:3415–3428. doi: 10.1105/tpc.106.046532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, et al. Shoot-specific down-regulation of protein farnesyltransferase (alpha-subunit) for yield protection against drought in canola. Mol. Plant. 2009;2:191–200. doi: 10.1093/mp/ssn088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasilewska A, et al. An update on abscisic acid signaling in plants and more. Mol. Plant. 2008;1:198–217. doi: 10.1093/mp/ssm022. [DOI] [PubMed] [Google Scholar]
- Wiborg J, O'Shea C, Skriver K. Biochemical function of typical and variant Arabidopsis thaliana U-box E3 ubiquitin–protein ligases. Biochem. J. 2008;413:447–457. doi: 10.1042/BJ20071568. [DOI] [PubMed] [Google Scholar]
- Xie Q, Frugis G, Colgan D, Chua NH. Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 2000;14:3024–3036. doi: 10.1101/gad.852200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q, Guo HS, Dallman G, Fang S, Weissman AM, Chua NH. SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature. 2002;419:167–170. doi: 10.1038/nature00998. [DOI] [PubMed] [Google Scholar]
- Xie Q, Sanz-Burgos AP, Guo H, Garcia JA, Gutierrez C. GRAB proteins, novel members of the NAC domain family, isolated by their interaction with a geminivirus protein. Plant Mol. Biol. 1999;39:647–656. doi: 10.1023/a:1006138221874. [DOI] [PubMed] [Google Scholar]
- Yan J, Wang J, Li Q, Hwang JR, Patterson C, Zhang H. AtCHIP, a U-box-containing E3 ubiquitin ligase, plays a critical role in temperature stress tolerance in Arabidopsis. Plant Physiol. 2003;132:861–869. doi: 10.1104/pp.103.020800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CW, et al. The E3 ubiquitin ligase activity of Arabidopsis PLANT U-BOX17 and its functional tobacco homolog ACRE276 are required for cell death and defense. Plant Cell. 2006;18:1084–1098. doi: 10.1105/tpc.105.039198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Vanderbeld B, Wan J, Huang Y. Narrowing down the targets: towards successful genetic engineering of drought-tolerant crops. Mol. Plant. 2010;3:469–490. doi: 10.1093/mp/ssq016. [DOI] [PubMed] [Google Scholar]
- Yee D, Goring DR. The diversity of plant U-box E3 ubiquitin ligases: from upstream activators to downstream target substrates. J. Exp. Bot. 2009;60:1109–1121. doi: 10.1093/jxb/ern369. [DOI] [PubMed] [Google Scholar]
- Zeng LR, et al. Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell. 2004;16:2795–2808. doi: 10.1105/tpc.104.025171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng LR, Park CH, Venu RC, Gough J, Wang GL. Classification, expression pattern, and E3 ligase activity assay of rice U-box-containing proteins. Mol. Plant. 2008;1:800–815. doi: 10.1093/mp/ssn044. [DOI] [PubMed] [Google Scholar]
- Zhang X, Garreton V, Chua NH. The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation. Genes Dev. 2005;19:1532–1543. doi: 10.1101/gad.1318705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, et al. SDIR1 is a RING finger E3 ligase that positively regulates stress-responsive abscisic acid signaling in Arabidopsis. Plant Cell. 2007;19:1912–1929. doi: 10.1105/tpc.106.048488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.