Abstract
Biologists have long been interested in both the regularities and the deviations in the relationship between brain, development, ecology, and behavior between taxa. We first examine some basic information about the observed ranges of fundamental changes in developmental parameters (i.e. neurogenesis timing, cell cycle rates, and gene expression patterns) between taxa. Next, we review what is known about the relative importance of different kinds of developmental mechanisms in producing brain change, focusing on mechanisms of segmentation, local and general features of neurogenesis, and cell cycle kinetics. We suggest that a limited set of developmental alterations of the vertebrate nervous system typically occur and that each kind of developmental change may entail unique anatomical, functional, and behavioral consequences for the organism. Thus, neuroecologists who posit a direct mapping of brain size to behavior should consider that not any change in brain anatomy is possible.
Key Words: Neurogenesis, Evolution, Development, Mammals, Birds
Introduction
Historically, studies of brain allometry have focused on variations in adult brains and how they might be related to sensory and motor specializations, as well as ecological and behavioral niches [Jerison, 1973; Stephan et al., 1981; Nudo et al., 1995; Lefebvre et al., 2002; Sol et al., 2005; Lefebvre and Sol, 2008]. Following Bolhuis and Macphail [2001], we term this approach ‘neuroecology’. More recently, the evo-devo approach has identified broadly conserved developmental mechanisms and patterns across taxa [Rubenstein et al., 1994; Northcutt, 2001; Puelles and Rubenstein, 2003; Striedter, 2005]. The latter endeavor has revealed a previously unimagined degree of conservation in the developmental mechanisms that set up brain and body plans across vertebrates and invertebrates [Carroll, 2000]. Thus, the evo-devo approach has been associated with the description of important homologies across taxa [Puelles et al., 2000].
The apparent contradiction posed by the conservation identified by the evo-devo approach and the variation described by the neuroecological approach is illusory, however. Evo-devo researchers argue that developmental mechanisms are conserved precisely because they are robust and flexible, capable of generating the breathtaking diversity of vertebrate morphology and behavior. Evo-devo emphasizes that developmental mechanisms have themselves been targets of intense selection. By contrast, neuroecologists tend to posit a fairly direct mapping of the importance of sensory, motor, and behavioral specializations onto the size of the neural systems thought to subserve them. They have generally remained agnostic about developmental mechanisms of brain change, assuming at first pass that any change in brain anatomy is possible and behaviorally advantageous.
Even at a gross level of observation, however, it is clear that only a limited set of potential alterations of the vertebrate nervous system typically occurs [Kirschner and Gerhart, 2005]. Variations in the number of central nervous system segments [Bergquist and Källén, 1954; Gilland and Baker, 1993; Northcutt, 2001, 2002; Puelles and Rubenstein, 2003], basic segmental structure [Northcutt, 2002; Urbach and Technau, 2008], and circuit organization are relatively rare [Medina and Reiner, 1995; Katz and Harris-Warrick, 1999; Reichert, 2009], as are changes in the complement of the small molecules which serve as neurotransmitters and neuromodulators [Venter et al., 1988; Medina and Reiner, 1995; Wong et al., 2011]. On the other hand, changes in the relative brain size or its parts [Stephan et al., 1981; Northcutt, 2002; Kaskan et al., 2005; Reep et al., 2007; Yopak et al., 2010], large morphological diversification of single neuronal types [Arendt, 2008], as well as changes in neurotransmitter receptor structure and distribution appear to occur commonly [Insel et al., 1991; Katz and Harris-Warrick, 1999; Goodson and Bass, 2001; Callier et al., 2003; Lim et al., 2004; Goodson, 2008].
It is now time to integrate evo-devo with patterns of taxonomic variation in the vertebrate brain in greater detail. The first aim of this article is to describe the ‘toolbox’ of developmental mechanisms of variability inherent in conserved brain and body plans. The second part aims to describe how brains do change, as our understanding of the complexities of neurons and circuits has grown immensely since the conjecture that there should be a direct relationship between behavioral specialization and the size of the relevant brain part (i.e. the principle of ‘proper mass’ [Jerison, 1973]). Here we will discuss how evolutionary changes in neurogenesis timing, cell cycle rates, and the ‘founder’ pool population during early brain segmentation give rise to variations in adult phenotypes. Finally, we will point out some future routes for mapping useful variability in neurodevelopmental processes.
Basic Plan of Development
Vertebrate brains have a fundamental segmental architecture [Bergquist and Källén, 1954; Pombal et al., 2009]. For instance, in the spinal cord, sensory, motor, and interneurons reside in homologous divisions of each segment. These segments, which may give rise to various cell types, are repeated and elaborated in the rhombomeres of the medulla and metencephalon (i.e. pons and cerebellum) as well as in the prosomeres of the forebrain [Lumsden and Keynes, 1989; Clarke and Lumsden, 1993; Rubenstein et al., 1994; Puelles and Rubenstein, 2003; Lumsden, 2004]. In spinal cord segments and rhombomeres, little cell migration occurs between segments, though there appears to be considerable intrasegmental migration in the forebrain [Anderson et al., 2001; Cobos et al., 2001; Corbin et al., 2001].
Position within and between segments, in both the basal-to-alar and the rostral-to-caudal direction, is associated with the duration of stem cell and neuronal production in mammals and perhaps all vertebrates [Finlay and Darlington, 1995; Finlay et al., 1998, 2001]. In understanding local variations in the generation of neurons in the brain, it is useful to distinguish ‘cytogenesis’, cell division in the ventricular zone establishing the founder population of progenitor cells in each region or segment, and ‘neurogenesis’, a terminal asymmetric or symmetric division in which neuroblasts incapable of further division are produced. Progenitor cells in nervous system segments generally begin their divisions at similar times in development. For instance, cytogenesis has its onset at approximately the same time in the telencephalon as in the spinal cord, at least in some mammals [Finlay and Darlington, 1995; Clancy et al., 2001]. Termination of cytogenesis and the onset of neurogenesis, however, varies by segmental position. The pattern of roughly simultaneous onset and staggered offset can be seen both between regions and within a brain region. For instance, neurogenesis onset occurs earlier in the spinal cord compared with the isocortex. In addition, there are maturational gradients of neurogenesis in the mammalian retina [Dyer et al., 2009] and the isocortex [Sanderson and Weller, 1990].
The staggered onset of neurogenesis reflects the staggered termination of cytogenesis, with marked global patterns. For example, neurons in alar regions are ‘born’ after neurons in basal regions over the entire rostral-caudal extent of the nervous system [Finlay et al., 1998]. In caudal regions of the neuraxis, the spinal cord and medulla terminate cytogenesis and begin producing neurons before the telencephalon: the ‘average’ neuron in the medulla becomes postmitotic before the ‘average’ neuron in the telencephalon [Altman and Bayer, 1980; Finlay and Darlington, 1995; Clancy et al., 2001]. However, the basic medial-to-lateral and caudal-to-rostral gradient does not dominate every aspect of neurogenesis. For instance, the ‘average’ neuron in the cerebellum is born after the ‘average’ neuron in the mesencephalon [Das and Nornes, 1972; Altman and Bayer, 1980].
The principal way (but not the only way, as we will discuss) to expand a brain is to prolong the duration of neuronal and glial cell production. If development is extended to produce a larger brain (everything else being equal), the gradients and local differences in termination in cytogenesis appear to generate predictable, nonlinear changes in the relative sizes of brain parts. The basic kinetics of cytogenesis dictate that precursor pools with extended cytogenesis (with correspondingly late neurogenesis or ‘neuronal birthdays’) will show the exponential increase due to the multiplicative expansion of their stem cell pools and become disproportionately large (i.e. late equals large) [Finlay and Darlington, 1995; Striedter, 2005; Finlay et al., 2010].
In accord with this prediction, Finlay and Darlington [1995] showed that brain regions with different mean birthdays (e.g. neocortex vs. medulla) enlarge with different allometric slopes with respect to each other. That is, brain regions with later mean birthdays exhibit steeper allometric slopes relative to brain regions with early mean birthdates [Finlay et al., 1998, 2001; Reep et al., 2007]. Interestingly, as developmental schedules lengthen or shorten in mammals, the ‘birth order’ of various brain regions is generally conserved [Clancy et al., 2001, 2007]. Alternate scenarios for evolutionary brain change are generally not observed. For example, cells do not simply undergo one more round of cell division throughout the neuraxis resulting in isometric enlargement, nor is all additional cytogenesis restricted to a terminal segment, nor are cells added in random locations by species (though local, ‘mosaic’ differentiation can be superimposed in this general pattern). Instead, later born regions undergo proportionally more rounds of cell division than early born regions, producing a predictably greater increase in the size of the telencephalon and cerebellum compared to the brainstem in every vertebrate taxa yet examined [Yopak et al., 2010]. The mechanism through which such a coordinated stretching or compressing of neurogenetic schedules might be accomplished is still unknown.
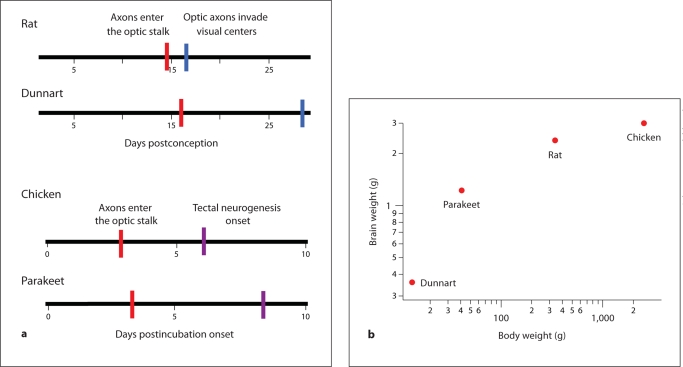
Passingham [1985] showed that, if brain size is plotted against gestation length in a wide variety of mammals, a surprising linear relationship between brain size and gestation length emerges. This observation suggests that brains expand at a constant rate in development. Further work relating the duration of neurogenesis, rather than gestational length in eutherian (i.e. placental) mammals, improves the prediction of adult brain mass. However, it also reveals taxonomic differences [Finlay and Darlington, 1995; Finlay et al., 2001]. The production of neurons per unit of time is generally slower in marsupials than in placental mammals (fig. 1) [Finlay and Darlington, 1995; Darlington et al., 1999]. For instance, the brush-tailed possum (Trichosurus vulpecula) and the rabbit (Oryctolagus cuniculus) have similar mature brain sizes, i.e. about 11–12 g, but it takes the rabbit only about 21 days to generate its full complement of neurons, while the possum takes over 60 days [Darlington et al., 1999]. Growth rates might also vary within and between other amniote classes. Among birds, parakeets (Melopsittacus undulatus) have a long developmental schedule relative to chickens (Gallus gallus domesticus) [Portmann, 1947b; Boire and Baron, 1994; Striedter and Charvet, 2008; Charvet and Striedter, 2009b], but chickens have a bigger brain than parakeets (fig. 1). Moreover, chickens produce brain mass faster than rats do. Thus, it is likely that species differences in cell cycle rates account for variations in the size of adult brains within and between classes. More generally, these observations suggest that the rate of neuron production and the duration of neuron production can evolve independently from one another.
Fig. 1.
The absolute duration of brain development does not always predict the overall brain size. a Among mammals, the developmental schedule of the dunnart (Sminthopsis crassicaudata) is longer than that of the rat (Rattus norvegicus). As developmental schedules lengthen, later events (e.g. optic axons invading visual centers) occur disproportionately later than earlier events (e.g. optic axons entering the optic stalk). For instance, optic axons reach the optic stalk on postconception day 16 in dunnarts whereas this event occurs at postconception day 14.5 in rats. Furthermore, optic axons invade visual centers at postconception day 28.5 in dunnarts but do so already at postconception day 16.5 in rats. b However, the brains of rats are larger than those of dunnarts. Given that the developmental schedules are longer in dunnarts than in rats but rat brains are smaller, we hypothesize that growth rates vary between marsupial and placental mammals. a Among birds, the schedule of brain development is longer in parakeets than in chickens. For instance, optic axons reach the optic stalk at approximately 4 days after incubation onset in parakeets, but this event occurs 3 days after incubation onset in chickens. b Although the duration of brain development is relatively short in chickens, the brain is larger in chickens than in parakeets, dunnarts, or rats. Thus, growth rates can vary independently of neurogenetic schedules within and between amniote classes.
Among vertebrates, a systematic relationship exists between early and late neurogenesis and cell cycle rates. Early cell cycles are relatively quick, and later cycles lengthen [Caviness et al., 2003; Charvet and Striedter, 2008]. For instance, at the onset of neurogenesis, the cell cycle rate in the isocortex of a mouse lasts approximately 8 h. By the time isocortical neurogenesis is close to complete, the cell cycle length lasts approximately 18 h [Caviness et al., 2003]. In rhesus monkeys (Macaca mulatta), the cell cycle lasts approximately 22.7 h at the onset of isocortical neurogenesis [Kornack and Rakic, 1998]. By the time neurogenesis in layer IV of the isocortex is complete, the cell cycle in the isocortical ventricular zone lasts approximately 27.4 h. These data suggest that the cell cycle lengthens as cells exit the cell cycle. Although general trends across taxa can be described, the data are not complete enough to specify any particular function relating the order of neurogenesis to cell cycle duration. However, substantial differences in cell cycle rates across species and brain regions can reasonably be expected.
Evolutionary Alterations in Neurogenesis Timing
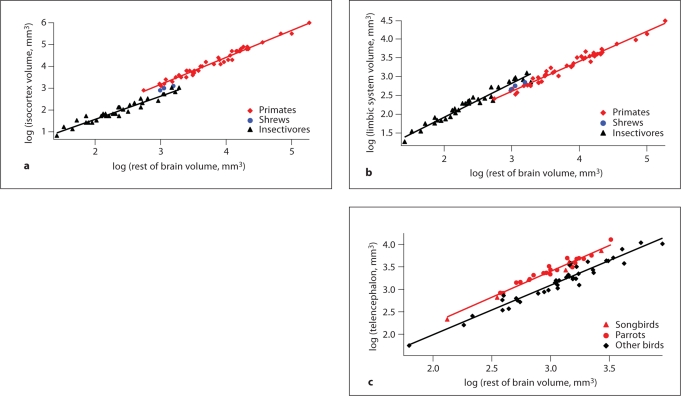
Evolutionary alterations in absolute brain size in mammals are generally associated with changes in brain region proportions as well as changes in a region's birth date, as we have discussed [Finlay and Darlington, 1995; Finlay et al., 2001; Reep et al., 2007]. Although brain regions tend to scale with the rest of the brain, partially independent scaling of some regions, notably the olfactory bulb and limbic system (e.g. hippocampus, amygdala), has been documented as well [Reep et al., 2007; Yopak et al., 2010; Finlay et al., 2011]. In studies of brain allometry, a persistent observation is that of a ‘grade shift’, which is a change in the relative size of a brain structure between taxonomic groups [Barton and Harvey, 2000; Striedter, 2005]. Among mammals, for instance, the isocortex of primates (i.e. lemurs, lorises, galagos, tarsiers, and simians) is disproportionately enlarged relative to most other mammals, whereas the limbic system (e.g. amygdala) is smaller in primates relative to many other mammals (fig. 2) [Stephan et al., 1981; Reep et al., 2007]. Among birds, the telencephalon of parrots and songbirds is disproportionately enlarged relative to that of many other birds (fig. 2) [Portmann, 1947a; Boire and Baron, 1994; Iwaniuk and Hurd, 2005].
Fig. 2.
Although brain regions generally scale with the rest of the brain, some brain regions exhibit grade shifts in birds and in mammals. a A plot of isocortex volume versus rest of brain size (i.e. brain minus telencephalon) shows that primates (e.g. gray mouse lemurs, rhesus monkeys, and humans) exhibit a disproportionately enlarged isocortex relative to many other mammals. b In contrast, a plot of limbic system volume (i.e. amygdala, hippocampus, schizocortex, and septum) regressed against rest of brain size shows that the limbic system is disproportionately smaller in primates than in many other mammals. c A plot of telencephalon size versus rest of brain size shows that parrots, songbirds, as well as other birds exhibit a disproportionately enlarged telencephalon relative to many other birds (e.g. pigeons, shorebirds, falcons, and galliform birds). a, b Separate linear regressions were fitted through the data in primates and insectivores. c A linear regression was fitted through the data in parrots and songbirds and another linear regression was fitted through the data of the remaining birds. a, b Data are from Stephan et al. [1981]. c Data are from Iwaniuk and Hurd [2005].
Studies of birds and mammals indicate that the size of a brain region's initial founder pool, progenitor cell cycle rates, and the duration of neurogenesis may all vary across species, giving rise to species differences in brain region size. In the following paragraphs, we examine how evolutionary alterations in developmental mechanisms give rise to selective changes in brain region size and how these evolutionary changes in developmental parameters influence brain anatomy in birds and in mammals.
Among birds, songbirds (i.e. zebra finches; Taeniopygia guttata) and parrots (i.e. parakeets; Melopsittacus undulatus) delay telencephalic neurogenesis relative to galliform birds (i.e. bobwhite quail; fig. 2; Colinus virginianus) [Charvet and Striedter, 2008, 2009a; Striedter and Charvet, 2008]. More specifically, parakeets and zebra finches delay telencephalic neurogenesis onset and offset relative to galliform birds. The latter is delayed well into the posthatching period. In contrast, neurogenesis timing in the medulla is not delayed in zebra finches and parakeets relative to galliform birds. Therefore, the delay in telencephalic neurogenesis is not the result of a uniform stretching of neurogenetic schedules but is relatively specific to the telencephalon. These findings imply that parakeets and zebra finches enlarge their telencephalon by delaying and prolonging telencephalic neurogenesis relative to some other birds.
One consequence of delaying neurogenesis is that it also delays neuronal maturation. Importantly, delaying maturation into the posthatching period probably makes neurons more susceptible to environmental influences. Parrots and songbirds have evolved specialized telencephalic circuits that allow them to produce learned vocalizations [Nottebohm, 1972; Nottebohm et al., 1976; Striedter, 1994; Pepperberg, 2010]. Perhaps extending the period of posthatchling telencephalic maturation into the posthatchling period is necessary for parrots and songbirds to learn vocalizations during the juvenile period [Kirn, 2010].
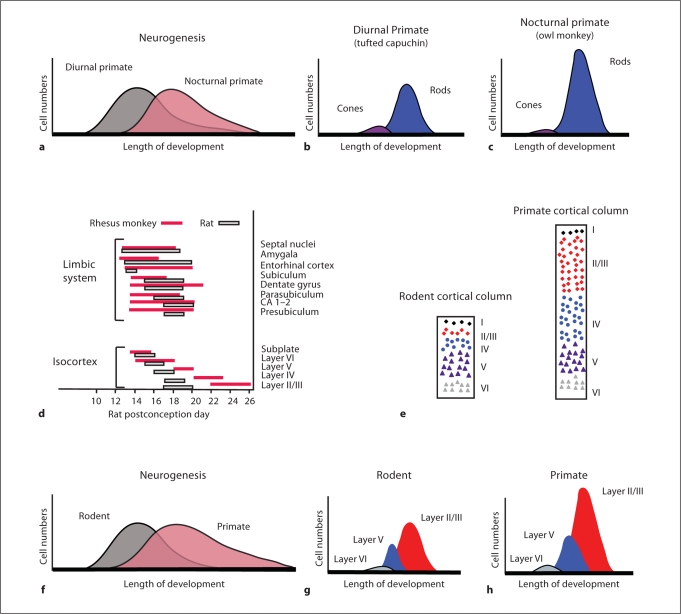
Alteration of neurogenesis timing in the predicted direction correlates with the grade shifts of a larger isocortex and reduced olfactory and limbic systems in primates compared with many other mammals. More specifically, comparative analyses of birth-dating data have shown that cortical neurogenesis is protracted, whereas limbic neurogenesis is shortened, in primates (i.e. rhesus monkey) compared with rodents (i.e. rats, mice, hamsters, spiny mice, and guinea pigs; fig. 3) [Clancy et al., 2000, 2001, 2007]. These observations suggest that appropriate changes in neurogenesis duration account, at least in part, for the expansion of the isocortex and reduced limbic system in primates.
Fig. 3.
Diagrams show the anatomical consequences of delaying neurogenesis in the retina (a–c) and the isocortex (d–h). The owl monkey is nocturnal, whereas the capuchin monkey is diurnal. a During development, peak retinal neurogenesis is delayed in the owl monkey relative to that of the capuchin monkey [Dyer et al., 2009]. b, c Because rods are born after cones [Rapaport et al., 1992], a delay in retinal neurogenesis entails that the adult retina will contain a disproportionately larger number of rods relative to cones. d The timing of neural events in rhesus monkeys has been adjusted to fit the period of neural events in rats [Finlay et al., 1998]. Such a comparison shows that the onset of terminal neurogenesis for limbic structures (e.g. amygdala) is advanced in rhesus monkeys relative to rats. In contrast, isocortical neurogenesis onset and offset (e.g. layer II/III) is delayed in rhesus monkeys relative to rats. e The isocortex of primates (i.e. lemurs, lorises, galagos, tarsiers, and simians) contains a disproportionately expanded layer II–III compared with rodents (e.g. rat, porcupine, and capybara) in adulthood [Hutsler et al., 2005]. f–h Because layer II–III cells are born after layer V–VI cells, delaying isocortical neurogenesis should cause layers II–III to become disproportionately enlarged relative to deeper cortical layers.
Delaying isocortical neurogenesis entails that neurons born late in cortical development become disproportionately more numerous than neurons born early during cortical neurogenesis. Given that upper layer (e.g. layers II-III) cells are born after deeper layers of the isocortex (e.g. layers V-VI; fig. 3) [Rakic, 1977; Noctor et al., 1997], delaying isocortical neurogenesis should expand the pool of cells in layers II-III relative to cells in the deeper isocortical layers (fig. 3). This hypothesis is confirmed by the finding that mammals that delay isocortical neurogenesis (e.g. primates) exhibit a proportionally expanded layer II–III relative to many other mammals [Hutsler et al., 2005; Kriegstein et al., 2006]. These observations suggest that the ‘clock’ for cell type determination can be shifted relative to the clock for neurogenesis timing.
This hypothesis was examined further in a study of two closely related New World monkeys, the nocturnal owl monkey (Aotus azarae) and the diurnal capuchin monkey (Cebus apella). These two species exhibit shifts in the numbers of all classes of retinal neurons and photoreceptors appropriate for a diurnal or nocturnal niche. Recent work by Dyer et al. [2009] has shown that retinal neurogenesis is delayed in the owl monkey relative to the capuchin monkey. Because cones and retinal ganglion cells are born early in retinal development, but rods and rod bipolar cells are born late in development [Rapaport et al., 1992], a delay in retinal neurogenesis will systematically reduce cone numbers and their associated neurons and increase rod numbers and their transmitting neurons (fig. 3) [Finlay, 2008]. Thus, a single change in developmental mechanism (i.e. shifting the envelope of neurogenesis) may result in a coordinated change in all of the cell types contributing to nocturnal versus diurnal vision. More generally, these observations suggest that the order of neurogenesis in the retina has been subject to selection, perhaps due to the multiple transitions from nocturnal to diurnal niches over vertebrate phylogeny, with the result that a nocturnal-type retina is ‘easy to evolve’ even in recently evolved primates.
Evolutionary Alterations in Cell Cycle Rates
Whereas most work examining developmental mechanisms underlying changes in brain size focuses on species differences in the duration and relative timing of neurogenesis, species differences in the founder pool population have also been reported [Charvet, 2010; Charvet et al., 2010]. We here illustrate how species differences in cell cycle rates account for variations in the founder pool population, which accounts for variations in adult brain size among galliform birds (e.g. chickens and bobwhite quail). The chicken (Gallus gallus domesticus) brain is larger than that of the bobwhite quail (Colinus virginianus) in adulthood. The expansion of the chicken brain relative to that of the bobwhite quail can be traced back to species differences in cell cycle rates [Charvet and Striedter, 2010]. That is, brain precursor cells in the presumptive telencephalon and tectum cycle more rapidly at early stages of development in chickens than in bobwhite quail, and this acceleration is limited to a brief period of embryonic development before neurogenesis begins. After neurogenesis onset, the rate of brain growth is similar in both species [Charvet and Striedter, 2010]. Thus, chickens enlarge their brain relative to quail by expanding the precursor pool population before cells exit the cell cycle. At this point, however, it is not clear how common alterations in cell cycle rates are as mechanisms for changing brain or brain region size. Although chickens and quail differ in the duration of the cell cycle rate early in development, developmental schedules (e.g. neurogenesis timing) are uniformly compressed in chickens relative to bobwhite quail [Charvet and Striedter, 2010]. These findings suggest that cell cycle rates and the timing of neurogenetic schedules can evolve independently of one another.
Evolutionary Alterations in Brain Patterning
Major anatomical divisions tend to be established prior to neurogenesis. That is, regionalization is due to dynamic patterns of gene expression within precursor tissues. Most importantly, boundaries are set up by mutually interacting and repressive genes [Bishop et al., 2000; O’Leary and Sahara, 2008]. For example, the sharp midbrain-hindbrain boundary is thought to arise through a mutual repression between Gbx2 and Otx2 [Katahira et al., 2000; Nakamura, 2001]. The boundaries may then become signal sources themselves so that patterning tends to be progressive. Major divisions and boundaries are specified first; as development progresses, smaller divisions are specified.
Evo-devo neurobiologists have only recently begun to examine species differences in developmental gene expression [Bachy et al., 2001; Menuet et al., 2007; Abellan et al., 2010; McGowan et al., 2010; Sylvester et al., 2010]. Among cichlid fishes, Sylvester et al. [2010] showed that telencephalic enlargement in cichlids of the mbuna lineage is associated with a shift in gene expression patterns [Sylvester et al., 2011]. More specifically, the telencephalic expansion of mbunas is associated with a shift in the anterior-posterior boundaries mediated by the differential expression of sonic hedgehog (SHH) at the zona limitans.
An interesting nonneurobiological example in which changes in the timing of gene expression are known to influence adult form and function is vertebrate somite formation [Cooke and Zeeman, 1976; Pourquié, 2003; Dequéant et al., 2006; Gomez et al., 2008]. Somite numbers may vary widely among vertebrates, ranging from 10 in some frogs to over 300 in snakes [Richardson et al., 1998]. The rate of oscillation in gene expression patterns (e.g. lunatic fringe) is thought to account for the diversity in the number of somites across vertebrates [Dequéant et al., 2006; Gomez et al., 2008]. Such changes in the timing of gene expression patterns are not yet known to account for any species differences in the nervous system, but their existence in somites suggests that similar developmental mechanisms might account for some variation among adult brains.
Other Variations in Brain Region Size
We have not discussed the myriad examples of other variations in brain region size, which are typically much smaller in magnitude than the whole brain scaling and taxonomic grade shifts we have discussed here. Some of these changes may be associated with general abilities such as home range size [Sherry et al., 1992] and food-storing behavior [Smulders et al., 2000; Sherry and Hoshooley, 2010] or very specific skills like learned vocalizations. For instance, the finding that the size of song nuclei in some songbirds exhibits seasonal variations [Nottebohm, 1981; De Groof et al., 2009] suggests that such alterations in brain region size underlie changes in seasonal behaviors. Some of these changes may involve neuronal death, induced changes in postnatal or posthatching neurogenesis, or volumetric changes related to trophic effects, and differential use [Nottebohm, 1981; Kirn, 2010]. In time, it will be useful to embed these aspects of neuronal development in the overall framework of variations in development as we have described here.
Conclusion
We have contrasted the variability of the adult vertebrate brain with the conservation of developmental mechanisms that are used to produce it. The adult brain shows very consistent patterns of variability, suggesting that some types of changes may be disadvantageous for reasons we do not yet understand. The central nervous system may vary little in some dimensions (e.g. fundamental divisions), but it may vary considerably in the absolute and relative sizes of its divisions. Three major features predominate in studies of brain allometry. First, we find close coordination in the size of brain regions across taxa. Second, there are characteristic differences in the relative size of individual brain structures as the overall brain size increases. Finally, we find ‘grade shifts’ in that the relative size of some brain regions varies between taxa. For the interesting case of grade shifts, we have discussed possible developmental mechanisms with a few examples. However, we know very little as yet about the relative frequency of these various developmental mechanisms. Grade shifts between taxa might arise because of alterations in neurogenesis timing, changes in cell cycle rates, or boundary shifts. However, these parameters are not set up randomly, and it is essential to look at the overall pattern of developmental mechanisms, rather than a simple checklist, to discover the principles through which brains change.
Acknowledgements
This work was supported by NSF grant No. 0849612 to B.L.F., and NSF grant No. IOS-0744332 to G.F.S. This work was also supported by Eunice Kennedy Shriver National Institute of Child Health and Human Development fellowship No. F32HD067011 to C.J.C. The content is solely the responsibility of the authors and does not necessarily represent the official views of the Eunice Kennedy Shriver National Institute of Child Health and Human Development or the National Institutes of Health.
References
- Abellan A, Menuet A, Dehay C, Medina L, Rétaux S. Differential expression of LIM-homeodomain factors in Cajal-Retzius cells of primates, rodents, and birds. Cereb Cortex. 2010;20:1788–1798. doi: 10.1093/cercor/bhp242. [DOI] [PubMed] [Google Scholar]
- Altman J, Bayer SA. Development of the brain stem in the rat. 1. Thymidine-radiographic study of the time of origin of neurons of the lower medulla. J Comp Neurol. 1980;194:1–35. doi: 10.1002/cne.901940102. [DOI] [PubMed] [Google Scholar]
- Anderson SA, Marín O, Horn C, Jennings K, Rubenstein JL. Distinct cortical migrations from the medial and lateral ganglionic eminences. Development. 2001;128:353–363. doi: 10.1242/dev.128.3.353. [DOI] [PubMed] [Google Scholar]
- Arendt D. The evolution of cell types in animals: emerging principles from molecular studies. Nat Rev Genet. 2008;9:868–882. doi: 10.1038/nrg2416. [DOI] [PubMed] [Google Scholar]
- Bachy I, Vernier P, Retaux S. The LIM-homeodomain gene family in the developing Xenopus brain: conservation and divergences with the mouse related to the evolution of the forebrain. J Neurosci. 2001;21:7620–7629. doi: 10.1523/JNEUROSCI.21-19-07620.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton RA, Harvey PH. Mosaic evolution of brain structure in mammals. Nature. 2000;405:1055–1058. doi: 10.1038/35016580. [DOI] [PubMed] [Google Scholar]
- Bergquist H, Källén B. Notes on the early histogenesis and morphogenesis of the central nervous system in vertebrates. J Comp Neurol. 1954;100:627–659. doi: 10.1002/cne.901000308. [DOI] [PubMed] [Google Scholar]
- Bishop KM, Goudreau G, O'Leary DD. Regulation of area identity in the mammalian neocortex by Emx2 and Pax6. Science. 2000;288:344–349. doi: 10.1126/science.288.5464.344. [DOI] [PubMed] [Google Scholar]
- Boire D, Baron G. Allometric comparison of brain and main brain subdivisions in birds. J Hirnforsch. 1994;35:49–66. [PubMed] [Google Scholar]
- Bolhuis JJ, Macphail EM. A critique of the neuroecology of learning and memory. Trends Cogn Sci. 2001;5:426–433. doi: 10.1016/s1364-6613(00)01753-8. [DOI] [PubMed] [Google Scholar]
- Callier S, Snapyan M, Le Crom S, Prou D, Vincent JD, Vernier P. Evolution and cell biology of dopamine receptors in vertebrates. Biol Cell. 2003;95:489–502. doi: 10.1016/s0248-4900(03)00089-3. [DOI] [PubMed] [Google Scholar]
- Carroll SB. Endless forms: the evolution of gene regulation and morphological diversity. Cell. 2000;101:577–580. doi: 10.1016/s0092-8674(00)80868-5. [DOI] [PubMed] [Google Scholar]
- Caviness VS, Goto T, Tarui T, Takahashi T, Bhide PG, Nowakowski RS. Cell output, cell cycle duration and neuronal specification: a model of integrated mechanisms of the neocortical proliferative process. Cereb Cortex. 2003;13:592–598. doi: 10.1093/cercor/13.6.592. [DOI] [PubMed] [Google Scholar]
- Charvet CJ. A reduced progenitor pool population accounts for the rudimentary appearance of the septum, medial pallium and dorsal pallium in birds. Brain Behav Evol. 2010;76:289–300. doi: 10.1159/000322102. [DOI] [PubMed] [Google Scholar]
- Charvet CJ, Sandoval AL, Striedter GF. Phylogenetic origins of early alterations in brain region proportions. Brain Behav Evol. 2010;75:104–110. doi: 10.1159/000300573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charvet CJ, Striedter GF. Developmental species differences in brain cell cycle rates between bobwhite quail (Colinus virginianus) and parakeets (Melopsittacus undulatus): implications for mosaic brain evolution. Brain Behav Evol. 2008;72:295–306. doi: 10.1159/000184744. [DOI] [PubMed] [Google Scholar]
- Charvet CJ, Striedter GF. Developmental origins of mosaic brain evolution: morphometric analysis of the developing zebra finch brain. J Comp Neurol. 2009a;514:203–213. doi: 10.1002/cne.22005. [DOI] [PubMed] [Google Scholar]
- Charvet CJ, Striedter GF. Developmental basis for telencephalon expansion in waterfowls: enlargement prior to neurogenesis. Proc Biol Sci. 2009b;276:3241–3247. doi: 10.1098/rspb.2009.0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charvet CJ, Striedter GF. Bigger brains cycle faster before neurogenesis begins: a comparison in brain development between chickens and bobwhite quail. Proc Biol Sci. 2010;277:3469–3475. doi: 10.1098/rspb.2010.0811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy B, Darlington RB, Finlay BL. The course of human events: predicting the timing of primate neural development. Dev Sci. 2000;3:57–66. [Google Scholar]
- Clancy B, Darlington RB, Finlay BL. Translating developmental time across mammalian species. Neuroscience. 2001;105:7–17. doi: 10.1016/s0306-4522(01)00171-3. [DOI] [PubMed] [Google Scholar]
- Clancy B, Kersh B, Hyde J, Anand KJS, Darlington RB, Finlay BL. Web-based method for translating neurodevelopment from laboratory species to humans. Neuroinformatics. 2007;5:79–94. doi: 10.1385/ni:5:1:79. [DOI] [PubMed] [Google Scholar]
- Clarke JD, Lumsden A. Segmental repetition of neuronal phenotype sets in the chick embryo hindbrain. Development. 1993;118:151–162. doi: 10.1242/dev.118.1.151. [DOI] [PubMed] [Google Scholar]
- Cobos I, Puelles L, Martínez S. The avian telencephalic subpallium originates inhibitory neurons that invade tangentially the pallium (dorsal ventricular ridge and cortical areas) Dev Biol. 2001;239:30–45. doi: 10.1006/dbio.2001.0422. [DOI] [PubMed] [Google Scholar]
- Cooke J, Zeeman EC. A clock and wavefront model for control of the number of repeated structures during animal morphogenesis. J Theor Biol. 1976;58:455–476. doi: 10.1016/s0022-5193(76)80131-2. [DOI] [PubMed] [Google Scholar]
- Corbin JG, Nery S, Fishell G. Telencephalic cells take a tangent: non-radial migration in the mammalian forebrain. Nat Neurosci. 2001;4(suppl):1177–1182. doi: 10.1038/nn749. [DOI] [PubMed] [Google Scholar]
- Darlington RB, Dunlop SA, Finlay BL. Neural development in metatherian and eutherian mammals: variation and constraint. J Comp Neurol. 1999;411:359–368. [PubMed] [Google Scholar]
- Das GD, Nornes HO. Neurogenesis in the cerebellum of the rat: an autoradiographic study. Z Anat Entwicklungsgesch. 1972;38:155–156. doi: 10.1007/BF00519378. [DOI] [PubMed] [Google Scholar]
- De Groof G, Verhoye M, Poirier C, Leemans A, Eens M, Darras VM, Van der Linden A. Structural changes between seasons in the songbird auditory forebrain. J Neurosci. 2009;29:13557–13565. doi: 10.1523/JNEUROSCI.1788-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dequéant ML, Glynn E, Gaudenz K, Wahl M, Chen J, Mushegian A, Pourquié O. A complex oscillating network of signaling genes underlies the mouse segmentation clock. Science. 2006;314:1595–1598. doi: 10.1126/science.1133141. [DOI] [PubMed] [Google Scholar]
- Dyer MA, Martins R, da Silva Filho M, Muniz JA, Silveira LC, Cepko CL, Finlay BL. Developmental sources of conservation and variation in the evolution of the primate eye. Proc Natl Acad Sci USA. 2009;106:8963–8968. doi: 10.1073/pnas.0901484106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay BL. The developing and evolving retina: using time to organize form. Brain Res. 2008;1192:5–16. doi: 10.1016/j.brainres.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Finlay BL, Clancy B, Darlington RB. Late still equals large. Brain Behav Evol. 2010;75:4–6. [Google Scholar]
- Finlay BL, Darlington RB. Linked regularities in the development and evolution of mammalian brains. Science. 1995;268:1578–1584. doi: 10.1126/science.7777856. [DOI] [PubMed] [Google Scholar]
- Finlay BL, Darlington RB, Nicastro N. Developmental structure in brain evolution. Behav Brain Sci. 2001;24:263–278. [PubMed] [Google Scholar]
- Finlay BL, Hersman MN, Darlington RB. Patterns of vertebrate neurogenesis and the paths of vertebrate evolution. Brain Behav Evol. 1998;52:232–242. doi: 10.1159/000006566. [DOI] [PubMed] [Google Scholar]
- Finlay BL, Hinz F, Darlington RB. Mapping behavioural evolution onto brain evolution: the strategic roles of conserved organization in individuals and species. Phil Trans R Soc Lond B Biol Sci. 2011;366:2111–2123. doi: 10.1098/rstb.2010.0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilland E, Baker R. Conservation of neuroepithelial and mesodermal segments in the embryonic vertebrate head. Acta Anat (Basel) 1993;148:110–123. doi: 10.1159/000147530. [DOI] [PubMed] [Google Scholar]
- Gomez C, Ozbudak EM, Wunderlich J, Baumann D, Lewis J, Pourquié O. Control of segment number in vertebrate embryos. Nature. 2008;454:335–339. doi: 10.1038/nature07020. [DOI] [PubMed] [Google Scholar]
- Goodson JL. Nonapeptides and the evolutionary patterning of sociality. Prog Brain Res. 2008;170:3–15. doi: 10.1016/S0079-6123(08)00401-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodson JL, Bass AH. Social behavior functions and related anatomical characteristics of vasotocin/vasopressin systems in vertebrates. Brain Res Brain Res Rev. 2001;35:246–265. doi: 10.1016/s0165-0173(01)00043-1. [DOI] [PubMed] [Google Scholar]
- Hutsler JJ, Lee DG, Porter KK. Comparative analysis of cortical layering and supragranular layer enlargement in rodent carnivore and primate species. Brain Res. 2005;1052:71–81. doi: 10.1016/j.brainres.2005.06.015. [DOI] [PubMed] [Google Scholar]
- Insel TR, Gelhard R, Shapiro LE. The comparative distribution of forebrain receptors for neurohypophyseal peptides in monogamous and polygamous mice. Neuroscience. 1991;43:623–630. doi: 10.1016/0306-4522(91)90321-e. [DOI] [PubMed] [Google Scholar]
- Iwaniuk AN, Hurd PL. The evolution of cerebrotypes in birds. Brain Behav Evol. 2005;65:215–230. doi: 10.1159/000084313. [DOI] [PubMed] [Google Scholar]
- Jerison HJ. Evolution of the Brain and Intelligence. New York: Academic Press; 1973. [Google Scholar]
- Kaskan PM, Franco EC, Yamada ES, Silveira LC, Darlington RB, Finlay BL. Peripheral variability and central constancy in mammalian visual system evolution. Proc Biol Sci. 2005;272:91–100. doi: 10.1098/rspb.2004.2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katahira T, Sato T, Sugiyama S, Okafuji T, Araki I, Funahashi J, Nakamura H. Interaction between Otx2 and Gbx2 defines the organizing center for the optic tectum. Mech Dev. 2000;91:43–52. doi: 10.1016/s0925-4773(99)00262-2. [DOI] [PubMed] [Google Scholar]
- Katz PS, Harris-Warrick RM. The evolution of neuronal circuits underlying species-specific behavior. Curr Opin Neurobiol. 1999;9:628–633. doi: 10.1016/S0959-4388(99)00012-4. [DOI] [PubMed] [Google Scholar]
- Kirn JR. The relationship of neurogenesis and growth of brain regions to song learning. Brain Lang. 2010;115:29–44. doi: 10.1016/j.bandl.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner M, Gerhart J. The Plausibility of Life. New Haven: Yale University Press; 2005. [Google Scholar]
- Kornack DR, Rakic P. Changes in cell-cycle kinetics during the development and evolution of primate neocortex. Proc Natl Acad Sci USA 95:12421246. 1998;95:1242–1246. doi: 10.1073/pnas.95.3.1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegstein A, Noctor S, Martínez-Cerdeño V. Patterns of neural stem and progenitor cell division may underlie evolutionary cortical expansion. Nat Rev Neurosci. 2006;7:883–890. doi: 10.1038/nrn2008. [DOI] [PubMed] [Google Scholar]
- Lefebvre L, Nicolakakis N, Boire D. Tools and brains in birds. Behaviour. 2002;139:939–973. [Google Scholar]
- Lefebvre L, Sol D. Brains, lifestyles and cognition: are there general trends? Brain Behav Evol. 2008;72:135–144. doi: 10.1159/000151473. [DOI] [PubMed] [Google Scholar]
- Lim MM, Wang Z, Olazábal DE, Ren X, Terwilliger EF, Young LJ. Enhanced partner preference in a promiscuous species by manipulating the expression of a single gene. Nature. 2004;429:754–757. doi: 10.1038/nature02539. [DOI] [PubMed] [Google Scholar]
- Lumsden A. Segmentation and compartition in the early avian hindbrain. Mech Dev. 2004;121:1081–1088. doi: 10.1016/j.mod.2004.04.018. [DOI] [PubMed] [Google Scholar]
- Lumsden A, Keynes R. Segmental patterns of neuronal development in the chick hindbrain. Nature. 1989;337:424–428. doi: 10.1038/337424a0. [DOI] [PubMed] [Google Scholar]
- McGowan L, Kuo E, Martin A, Monuki ES, Striedter G. Species differences in early patterning of the avian brain. Evolution. 2010;65:907–911. doi: 10.1111/j.1558-5646.2010.01126.x. [DOI] [PubMed] [Google Scholar]
- Medina L, Reiner A. Neurotransmitter organization and connectivity of the basal ganglia in vertebrates: implications for the evolution of basal ganglia. Brain Behav Evol. 1995;46:235–258. doi: 10.1159/000113277. [DOI] [PubMed] [Google Scholar]
- Menuet A, Alunni A, Joly JS, Jeffrey WR, Rétaux S. Expanded expression of Sonic Hedgehog in Astyanax cavefish: multiple consequences on forebrain development and evolution. Development. 2007;134:845–855. doi: 10.1242/dev.02780. [DOI] [PubMed] [Google Scholar]
- Nakamura H. Regionalization of the optic tectum: combinations of gene expression that define the tectum. Trends Neurosci. 2001;24:32–39. doi: 10.1016/s0166-2236(00)01676-3. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Scholnicoff NJ, Juliano SL. Histogenesis of ferret somatosensory cortex. J Comp Neurol. 1997;387:179–193. [PubMed] [Google Scholar]
- Northcutt RG. Changing views of brain evolution. Brain Res Bull. 2001;55:663–674. doi: 10.1016/s0361-9230(01)00560-3. [DOI] [PubMed] [Google Scholar]
- Northcutt RG. Understanding vertebrate brain evolution. Integr Comp Biol. 2002;42:743–756. doi: 10.1093/icb/42.4.743. [DOI] [PubMed] [Google Scholar]
- Nottebohm F. The origins of vocal learning. Am Nat. 1972;106:116–140. [Google Scholar]
- Nottebohm F. A brain for all seasons: cyclical anatomical changes in song control nuclei of the canary brain. Science. 1981;214:1368–1370. doi: 10.1126/science.7313697. [DOI] [PubMed] [Google Scholar]
- Nottebohm F, Stokes TM, Leonard CM. Central control of song in the canary, Serinus canarius. J Comp Neurol. 1976;165:457–486. doi: 10.1002/cne.901650405. [DOI] [PubMed] [Google Scholar]
- Nudo RJ, Sutherland DP, Masterton RB. Variation and evolution of mammalian corticospinal somata with special reference to primates. J Comp Neurol. 1995;358:181–205. doi: 10.1002/cne.903580203. [DOI] [PubMed] [Google Scholar]
- O'Leary DD, Sahara S. Genetic regulation of arealization of the neocortex. Curr Opin Neurobiol. 2008;18:90–100. doi: 10.1016/j.conb.2008.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passingham RE. Rates of brain development in mammals including man. Brain Behav Evol. 1985;26:167–175. doi: 10.1159/000118773. [DOI] [PubMed] [Google Scholar]
- Pepperberg IM. Vocal learning in grey parrots: a brief review of perception, production, and cross-species comparisons. Brain Lang. 2010;115:81–91. doi: 10.1016/j.bandl.2009.11.002. [DOI] [PubMed] [Google Scholar]
- Pombal MA, Megías M, Bardet SM, Puelles L. New and old thoughts on the segmental organization of the forebrain in lampreys. Brain Behav Evol. 2009;74:7–19. doi: 10.1159/000229009. [DOI] [PubMed] [Google Scholar]
- Portmann A. Étude sur la cérébralisation chez les oiseaux. 2. Les indices intracérébraux. Alauda. 1947a;15:1–15. [Google Scholar]
- Portmann A. Étude sur la cérébralisation chez les oiseaux. 3. Cérébralisation et mode ontogénétique. Alauda. 1947b;15:161–171. [Google Scholar]
- Pourquié O. The segmentation clock: converting embryonic time into spatial pattern. Science. 2003;301:328–330. doi: 10.1126/science.1085887. [DOI] [PubMed] [Google Scholar]
- Puelles L, Kuwana E, Puelles E, Bulfone A, Shimamura K, Keleher J, Smiga S, Rubenstein JL. Pallial and subpallial derivatives in the embryonic chick and mouse telencephalon, traced by the expression of the genes Dlx-2, Emx-1, Nkx-2.1, Pax-6, and Tbr-1. J Comp Neurol. 2000;424:409–438. doi: 10.1002/1096-9861(20000828)424:3<409::aid-cne3>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Puelles L, Rubenstein JL. Forebrain gene expression domains and the evolving prosomeric model. Trends Neurosci. 2003;26:469–476. doi: 10.1016/S0166-2236(03)00234-0. [DOI] [PubMed] [Google Scholar]
- Rakic P. Prenatal development of the visual system in rhesus monkey. Philos Trans R Soc Lond B Biol Sci. 1977;278:245–260. doi: 10.1098/rstb.1977.0040. [DOI] [PubMed] [Google Scholar]
- Rapaport DH, Fletcher JT, LaVail MM, Rakic P. Genesis of neurons in the retinal ganglion cell layer of the monkey. J Comp Neurol. 1992;322:577–588. doi: 10.1002/cne.903220411. [DOI] [PubMed] [Google Scholar]
- Reep RL, Finlay BL, Darlington RB. The limbic system in mammalian brain evolution. Brain Behav Evol. 2007;70:57–70. doi: 10.1159/000101491. [DOI] [PubMed] [Google Scholar]
- Reichert H. Evolutionary conservation of mechanisms for neural regionalization, proliferation and interconnection in brain development. Biol Lett. 2009;5:112–116. doi: 10.1098/rsbl.2008.0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson MK, Allen SP, Wright GM, Raynaud A, Hanken J. Somite number and vertebrate evolution. Development. 1998;125:151–160. doi: 10.1242/dev.125.2.151. [DOI] [PubMed] [Google Scholar]
- Rubenstein JL, Martinez S, Shimamura K, Puelles L. The embryonic vertebrate forebrain: the prosomeric model. Science. 1994;266:578–580. doi: 10.1126/science.7939711. [DOI] [PubMed] [Google Scholar]
- Sanderson KJ, Weller WL. Gradients of neurogenesis in possum neocortex. Brain Res Dev Brain Res. 1990;55:269–274. doi: 10.1016/0165-3806(90)90208-g. [DOI] [PubMed] [Google Scholar]
- Sherry DF, Hoshooley JS. Seasonal hippocampal plasticity in food-storing birds. Philos Trans R Soc Lond B Biol Sci. 2010;365:933–943. doi: 10.1098/rstb.2009.0220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry DF, Jacobs LF, Gaulin SJ. Spatial memory and adaptive specialization of the hippocampus. Trends Neurosci. 1992;15:298–303. doi: 10.1016/0166-2236(92)90080-r. [DOI] [PubMed] [Google Scholar]
- Smulders TV, Shiflett MW, Sperling AJ, DeVoogd TJ. Seasonal changes in neuron numbers in the hippocampal formation of a food-hoarding bird: the black-capped chickadee. J Neurobiol. 2000;44:414–422. doi: 10.1002/1097-4695(20000915)44:4<414::aid-neu4>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- Sol D, Duncan RP, Blackburn TM, Cassey P, Lefebvre L. Big brains, enhanced cognition, and response of birds to novel environments. Proc Natl Acad Sci USA. 2005;102:5460–5465. doi: 10.1073/pnas.0408145102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan H, Frahm H, Baron G. New and revised data on volumes of brain structures on insectivores and primates. Folia Primatol (Basel) 1981;35:1–29. doi: 10.1159/000155963. [DOI] [PubMed] [Google Scholar]
- Striedter GF. The vocal control pathways in budgerigars differ from those in songbirds. J Comp Neurol. 1994;343:35–56. doi: 10.1002/cne.903430104. [DOI] [PubMed] [Google Scholar]
- Striedter GF. Principles of Brain Evolution. Sunderland: Sinauer Associates; 2005. [Google Scholar]
- Striedter GF, Charvet CJ. Developmental origins of species differences in telencephalon and tectum size: morphometric comparisons between a parakeet (Melopsittacus undulatus) and a quail (Colinus virgianus) J Comp Neurol. 2008;507:1663–1675. doi: 10.1002/cne.21640. [DOI] [PubMed] [Google Scholar]
- Sylvester JB, Pottin K, Streelman JT (2011): Integrated brain diversification along the early neuraxes. Brain Behav Evol, DOI 329840. [DOI] [PubMed]
- Sylvester JB, Rich CA, Loh YH, van Staaden MJ, Fraser GJ, Streelman JT. Brain diversity evolves via differences in patterning. Proc Natl Acad Sci USA. 2010;107:9718–9723. doi: 10.1073/pnas.1000395107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbach R, Technau GM. Dorsoventral patterning of the brain: a comparative approach. Adv Exp Med Biol. 2008;628:42–56. doi: 10.1007/978-0-387-78261-4_3. [DOI] [PubMed] [Google Scholar]
- Venter JC, di Porzio U, Robinson DA, Shreeve SM, Lai J, Kerlavage AR, Fracek SP, Jr, Lentes KU, Fraser CM. Evolution of neurotransmitter receptor systems. Prog Neurobiol. 1988;30:105–169. doi: 10.1016/0301-0082(88)90004-4. [DOI] [PubMed] [Google Scholar]
- Wong KK, Ng SY, Lee LT, Ng HK, Chow BK. Orexins and their receptors from fish to mammals: a comparative approach. Gen Comp Endocrinol. 2011;171:124–130. doi: 10.1016/j.ygcen.2011.01.001. [DOI] [PubMed] [Google Scholar]
- Yopak KE, Lisney TJ, Darlington RB, Collin SP, Montgomery JC, Finlay BL. A conserved pattern of brain scaling from sharks to primates. Proc Natl Acad Sci USA. 2010;107:12946–12951. doi: 10.1073/pnas.1002195107. [DOI] [PMC free article] [PubMed] [Google Scholar]