Abstract
The process of dark-induced senescence in plants is not fully understood, however, the functional involvement of an electron-transfer flavoprotein/electron-transfer flavoprotein:ubiquinone oxidoreductase (ETF/ETFQO), has been demonstrated. Recent studies have revealed that the enzymes isovaleryl-coenzyme A (CoA) dehydrogenase and 2-hydroxyglutarate dehydrogenase act as important electron donors to this complex. In addition both enzymes play a role in the breakdown of cellular carbon storage reserves with isovaleryl-CoA dehydrogenase being involved in degradation of the branched-chain amino acids, phytol, and lysine while 2-hydroxyglutarate dehydrogenase is exclusively involved in lysine degradation. Given that the chlorophyll breakdown intermediate phytanoyl-CoA accumulates dramatically both in knockout mutants of the ETF/ETFQO complex and of isovaleryl-CoA dehydrogenase following growth in extended dark periods we have investigated the direct importance of chlorophyll breakdown for the supply of carbon and electrons during this process. For this purpose we isolated three independent Arabidopsis (Arabidopsis thaliana) knockout mutants of phytanoyl-CoA 2-hydroxylase and grew them under the same extended darkness regime as previously used. Despite the fact that these mutants accumulated phytanoyl-CoA and also 2-hydroxyglutarate they exhibited no morphological changes in comparison to the other mutants previously characterized. These results are consistent with a single entry point of phytol breakdown into the ETF/ETFQO system and furthermore suggest that phytol is not primarily metabolized by this pathway. Furthermore analysis of isovaleryl-CoA dehydrogenase/2-hydroxyglutarate dehydrogenase double mutants generated here suggest that these two enzymes essentially account for the entire electron input via the ETF complex.
In Arabidopsis (Arabidopsis thaliana), the electron-transfer flavoprotein/electron-transfer flavoprotein:ubiquinone oxidoreductase (ETF/ETFQO) complex was first identified in mitochondria by the use of gel-based or liquid chromatography tandem mass spectrometry (MS) mitochondrial proteomic analysis (Heazlewood et al., 2004) and shown to be induced at the level of transcription during dark-induced senescence (Buchanan-Wollaston et al., 2005) and oxidative stress (Lehmann et al., 2009). Prior to this the presence of the nuclear-encoded mitochondrial protein, ETFQO, which accepts electrons from the ETF localized in the mitochondrial matrix and reduces ubiquinone had exclusively been characterized in mammalian systems (Ruzicka and Beinert, 1977; Beckmann and Frerman, 1985). In contrast, in mammals ETF is the physiological electron acceptor for at least nine different mitochondrial FAD-containing acyl-CoA dehydrogenases of fatty acid β-oxidation and amino acid catabolism (Frerman, 1988; Frerman and Goodman, 2001; Ghisla and Thorpe, 2004; He et al., 2007). The same pathway also accepts electrons from sarcosine dehydrogenase and dimethyl-Gly dehydrogenase, two enzymes of mitochondrial one-carbon metabolism (Hoskins and MacKenzie, 1961; Watmough and Frerman, 2010) while in plants only two major electron donors—isovaleryl-CoA dehydrogenase and 2-hydroxyglutarate dehydrogenase—have been characterized, to date (Engqvist et al., 2009; Araújo et al., 2010) and accordingly the dramatic conditional phenotypes observed in plants with mutations in either of these genes demonstrates the importance of this pathway in plants. Nevertheless, when taken together with the functional characterization of the role of Arabidopsis ETFQO and ETF proteins themselves (Ishizaki et al., 2005, 2006), these findings suggest that the Arabidopsis ETF/ETFQO complex, like that of its mammalian counterpart, can be regarded as a branch of the mitochondrial electron transport chain.
In plants, the ETF/ETFQO complex appears to participate in an important mechanism by which the plant cell can sustain respiration under conditions in which carbon supply is severely compromised. These conditions include those induced by growth in extended darkness mentioned above and also those induced under much milder conditions of stress such as continuous light (24 h light/0 h dark), short days (8 h light/16 h dark), and chilling conditions (13°C, 16 h light/8 h dark; Araújo et al., 2010). Here we have extended our earlier extensive studies of the role of the ETF/ETFQO system in dark-induced senescence to investigate whether phytanoyl-CoA is a major substrate for this important and complex system. In our previous study we were able to demonstrate, by the characterization of Arabidopsis knockout mutants of isovaleryl-CoA dehydrogenase and 2-hydroxyglutarate dehydrogenase, that during dark-induced senescence both enzymes supplied electrons and carbon skeletons to the tricarboxylic acid (TCA) cycle, under these conditions. We were additionally able to ascertain that isovaleryl-CoA dehydrogenase was involved in degradation of the branched-chain amino acids, phytol, and Lys while in contrast 2-hydroxyglutarate dehydrogenase was exclusively involved in Lys degradation (Araújo et al., 2010). In agreement with this, it has been recently demonstrated that this alternative pathway and particularly 2-hydroxyglutarate dehydrogenase, is operational during the dark period in Arabidopsis grown under regular diurnal light cycles and indicated Lys as the most suitable substrate generating 2-hydroxyglutarate in plants (Engqvist et al., 2011).
In humans, phytol is converted into phytanoyl-CoA that is further degraded by α-oxidation in a pathway containing phytanoyl-CoA 2-hydroxylase (PAHX) and 2-hydroxyphytanoyl-CoA lyase (HPCL) that have been shown to be the key enzymes of the phytol degradation pathway in mammals (Mukherji et al., 2003; Wanders et al., 2003). There are unique homologs of both PAHX (At2g01490) and HPCL (At5g17380) in the Arabidopsis genome, suggesting that the phytol degradation pathway in plants is similar to that of mammals.
Given our previous demonstration of the dramatic accumulation of phytanoyl-CoA both in knockout mutants of the ETF/ETFQO complex (Ishizaki et al., 2005, 2006) and of isovaleryl-CoA dehydrogenase (Araújo et al., 2010) following growth in extended dark periods we wished to establish if degradation via the isovaleryl-CoA dehydrogenase is the major (or only) route directly linking phytol degradation to the mitochondrial electron transport chain. This question is particularly intriguing in light of recent data from other studies of the well-characterized mobilization of chlorophyll and fatty acids during dark-induced senescence (Kunz et al., 2009; Schelbert et al., 2009). Therefore, to address this question, we here characterized three independent Arabidopsis knockout lines of PAHX (At2g01490), following dark-induced senescence, and compared their response at the morphological and biochemical levels to that of previously studied knockouts of key enzymes of the Arabidopsis ETF/ETFQO complex. Our analyses showed that these mutants were not compromised in their ability to withstand significant extension of the dark period but do accumulate phytanoyl-CoA and to a lesser extent 2-hydroxyglutarate as well as sharing some of the other metabolic features of mutants of the ETF/ETFQO complex following this treatment. These findings suggest that the breakdown of phytanoyl does not significantly contribute, if at all, to electron donation to the ubiquinol pool via the ETF system. This conclusion is further supported by the finding that isovaleryl-CoA dehydrogenase/2-hydroxyglutarate dehydrogenase double mutants phenocopy mutants of the ETF/ETFQO complex. These combined results are discussed in the context of current models of reserve mobilization during dark-induced senescence in leaves.
RESULTS
Isolation of T-DNA Insertional Mutants of PAHX
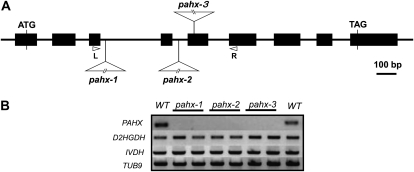
To investigate the in vivo functions of the PAHX protein (which is single copy in Arabidopsis), three independent Arabidopsis lines were isolated that contained T-DNA elements inserted into the PAHX gene. Segregation analysis of the encoded antibiotic resistance markers in the three insertional elements was in good agreement with the 3:1 (resistant:susceptible) ratio, suggesting insertion at a single Mendelian locus. Homozygous lines for each mutant were characterized by genomic PCR and designated pahx-1, pahx-2, and pahx-3, respectively (Fig. 1). The respective sites of insertion of these mutants were confirmed by sequencing of PCR products amplified from each mutant. Reverse transcription (RT)-PCR using primer pairs designed to span the T-DNA insertion sites of the three mutant loci was used to investigate transcription of the PAHX gene. The Arabidopsis β-tubulin gene, TUB9, ISOVALERYL-COA DEHYDROGENASE (IVDH), and 2-HYDROXYGLUTARATE DEHYDROGENASE (D2HGDH) genes were used as controls to demonstrate the integrity of the RNA preparation. PAHX mRNAs were detected in the wild type (Columbia-0) using the primer set L1/R1 and L2/R2, respectively; however, no amplification products were observed for the transcripts in pahx-1, pahx-2, and pahx-3 (Fig. 1). Thus, these results confirm that transcripts spanning the T-DNA insertion site are absent in these mutant lines.
Figure 1.
A schematic representation of the sites of T-DNA insertion in the pahx mutants. A, Genomic structure of PAHX (At2g01490). Arrowheads represent positions of primers used for genotyping of wild-type and mutant lines; black boxes indicate exons. The T-DNAs are inserted in intron 3, intron 4, and exon 5 of PAHX, in pahx-1, pahx-2, and pahx-3, respectively. B, RT-PCR analysis on total RNA (two biological replicates) from the wild type (WT) and the pahx-1, pahx-2, and pahx-3 mutant lines, with primer sets for the genes indicated on the left. The positions of the primers (R and L) in the PAHX genomic locus are represented in A.
Activities in Extracts of Arabidopsis Wild-Type and Knockout Plants
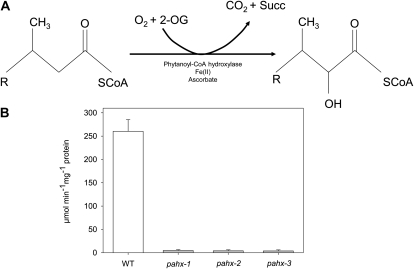
Phytanoyl-CoA hydroxylase, an Fe(II) and 2-oxoglutarate-dependent oxygenase, catalyzes the α-hydroxylation of phytanoyl-CoA to α-hydroxyphytanoyl-CoA, the first step in α-oxidation with the concomitant release of CO2 (Fig. 2A). Arabidopsis leaf crude extracts were analyzed for the activity of PAHX by using a 14CO2 turnover assay (Searls et al., 2005). To determine the conditions under which the reaction rate is linear, the effects of increasing times of incubation, substrate, and enzyme protein concentrations were examined. As expected the T-DNA insertion resulted in effectively complete loss of latent activity in mutant lines (i.e. no conversion of 2-oxoglutarate and phytanoyl-CoA was observed) with CO2 release observed been similar to background values while wild-type plants have shown a substrate and time-dependent CO2 release (Fig. 2B). When considered together with the lack of expression (described above) these results indicate that the pahx mutant lines described here represent a useful resource for investigating the role of the AtPAHX protein.
Figure 2.
Conversion of phytanoyl-CoA to α-hydroxyphytanoyl-CoA by the Fe(II)- and 2-oxoglutarate-dependent enzyme phytanoyl-coenzyme A α-hydroxylase (PAHX), with coevolution of carbon dioxide and succinic acid. A, Schematic reaction catalyzed by PAHX and enzyme activity of pahx mutant lines compared to the wild-type (WT) control (Col 0; B).The activity was determined in leaves of 4-week-old, short-day-grown, Arabidopsis plants. Values are means ± se of five independent samplings.
Phenotypes of the pahx Lines
Following the molecular characterization of the T-DNA insertional mutants, they were grown in soil under short-day (8 h light/16 h dark) conditions alongside wild-type controls. Under these conditions, there were no visible aberrant phenotypes in the mutants during vegetative growth. The whole-plant phenotypes were observed in both short- and long-day growth conditions, as outlined in “Materials and Methods.” Given that several previous studies have implicated phytanoyl-CoA degradation in supporting respiration during dark-induced senescence (Ishizaki et al., 2005, 2006; Engqvist et al., 2009; Araújo et al., 2010), we next transferred 4-week-old pahx and corresponding wild-type controls grown under short-day (8 h light/16 h dark) conditions to extended dark conditions that had previously been used to prove the metabolic importance of the metabolic reactions catalyzed by isovaleryl-CoA dehydrogenase and 2-hydroxyglutarate dehydrogenase as well as the electron transfer reactions mediated by the ETF/ETFQO complex. The pahx mutants were identical to the wild type grown under these conditions both in terms of visible phenotype (Supplemental Fig. S1A), total chlorophyll content (Supplemental Fig. S1B), chlorophyll a/b ratio (Supplemental Fig. S1C), and the maximum variable fluorescence/maximum yield of fluorescence (Fv/Fm ratio; Supplemental Fig. S1D) throughout the time period of the experiment. Moreover, the survival rate of the pahx mutants following reversion to either short or long days was similar to that of the wild-type plants. Thus while the mutants display the typical decrease in phytochemical efficiency and chlorophyll breakdown that are diagnostic of leaf senescence (Oh et al., 1996; Pruzinská et al., 2005), they do not displayed an accelerated senescence such as that we had previously documented for mutants in key enzymes intimately associated with the ETF/ETFQO complex. It should also be noted that all genotypes used in this study showed similar levels of starch, nitrate, amino acids, and proteins in samples harvested immediately prior to the start of the dark treatment (Supplemental Table S1).
Fatty Acid Composition of the pahx Lines
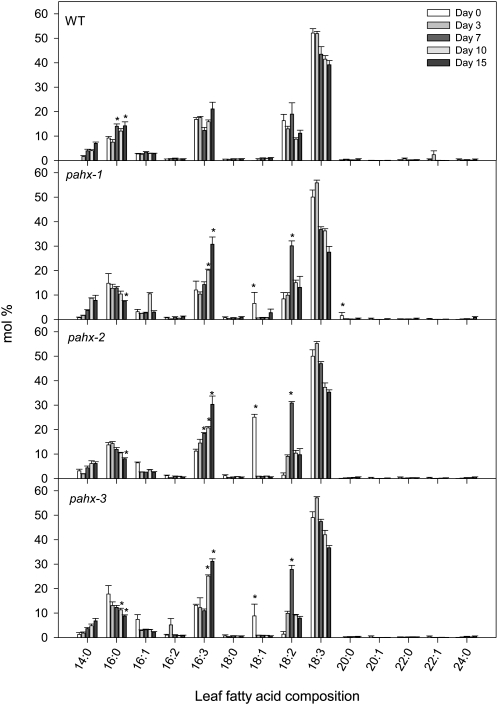
Cumulative evidence has demonstrated that one of the major functions of the ETF/ETFQO system in mammals is to allow respiration of substrates other than Glc. Therefore we first analyzed the changes in the fatty acid composition of leaf glycolipids during the extended dark treatment. As we have previously observed (Ishizaki et al., 2005; Araújo et al., 2010) and reported by Yang and Ohlrogge (2009) a typical distribution of leaf fatty acids was found with the major fatty acids in Arabidopsis being palmitic acid (16:0), hexadecatrienoic acid (16:3), linoleic acid (18:2), and linolenic acid (18:3) while hexadecenoic acid (16:1), hexadecadienoic acid (16:2), stearic acid (18:0), and oleic acid (18:1) are minor components. Interestingly we observed several changes in fatty acid composition of the mutants during the extended dark treatment, when compared to wild-type plants (Fig. 3). Thus although all fatty acids detected were present in leaves during the extended dark treatment, their relative amounts varied considerably and in some instances in a different manner than that observed in wild-type leaves. For instance the levels of palmitic acid increased in the wild-type plants whereas they decreased in the pahx mutants during the course of the dark treatment (Fig. 3). Another conspicuous feature is the substantial increase in hexadecatrienoic acid (16:3) observed in the mutants while no significant changes were detected in wild-type plants. It is also worth mentioning that the minor component oleic acid (18:1) is significantly higher in the mutants before the dark-induced senescence and reaches values similar to wild-type plants after only 3 d of darkness-induced treatment.
Figure 3.
Leaf fatty acid composition in pahx Arabidopsis mutants plants during growth under extended dark conditions. Data (in % mol) represent means ± se for six independent samplings of the ninth or 10th leaves of 4-week-old, short-day-grown, Arabidopsis plants after further treatment for 0, 3, 7, 10, and 15 d in extended darkness. Fatty acid composition was analyzed by GC of fatty acid methyl esters. An asterisk indicates values that were determined by the Student’s t test to be significantly different (P < 0.05) from the wild type at the same time point.
Acyl-CoA and 2-Hydroxyglutarate Profiles of the pahx Lines
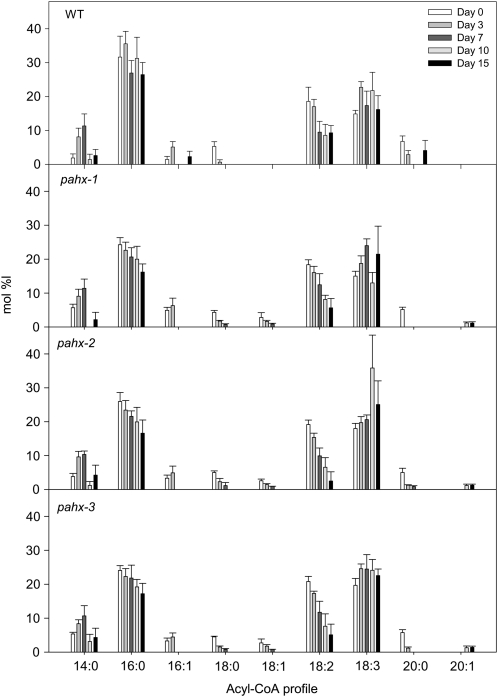
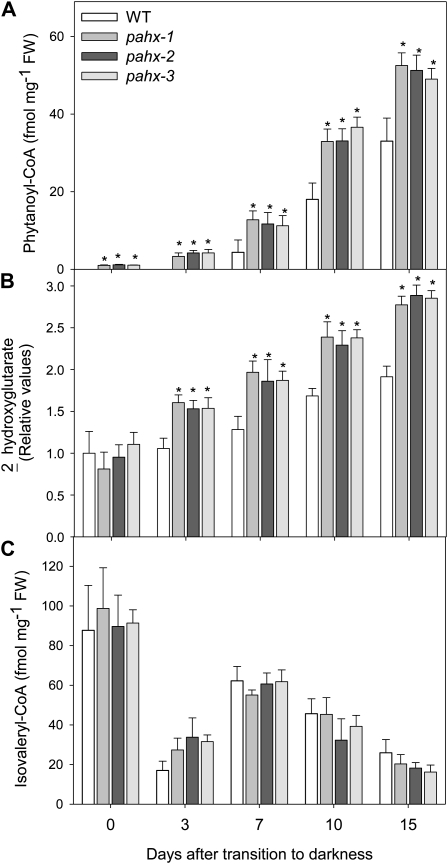
Disruption of the electron transfer function in Arabidopsis presumably compromises dehydrogenase activity and leads to accumulation of the isovaleryl-CoA substrate. In mammals, defects in ETFQO result in a functional deficiency of mitochondrial flavoprotein dehydrogenases and accumulation of their substrates (Frerman and Goodman, 2001). To investigate the functional linkage between the ETF/ETFQO system and phytanoyl-CoA, which in mammals catalyzes an essential step in the α-oxidation pathway (Wierzbicki et al., 2002), the well-established method of Larson and Graham (2001) was used to analyze changes in acyl-CoAs profile in the mutants and wild-type plants during the extended dark treatment. Interestingly the results shown in Figure 4 generally demonstrate no obvious differences in amounts of the typical spectrum of medium- to long-chain acyl-CoAs between wild-type and mutants plants. However, some intriguing changes were observed during the extended dark treatment. For instance while an obvious reduction of 16:0-CoA was observed in all mutant lines no significant changes were observed in wild-type leaves. It is also interesting to note that while the 18:1-CoA was present in all mutant lines with a clear trend leading to a reduction during the dark-induced treatment and finally disappearing after 10 d of darkness it was not detected in wild-type leaves. Another pertinent feature observed in the acyl-CoA profile was a substantial decrease in 18:0-CoA observed in wild type after 3 d in darkness with it finally being undetectable after 7 d while in contrast the rate of decrease in this acyl-CoA was significantly slower in all mutant lines finally becoming undetectable after 10 d of darkness-induced treatment. Surprising measurable levels of the very-long-chain acyl-CoA 20:1-CoA were only detectable after 10 d of darkness in the mutants and not detectable at all in the wild type (Fig. 4). We reported a similar pattern of changes in acyl-CoA for the etfqo-1, etfqo-2, and ivdh-1 mutants but not for d2hgdh-2 mutant in Araújo et al. (2010). Of further note is the significantly increased accumulation of phytanoyl-CoA in the pahx mutants when compared to wild-type plants (Fig. 5A). Additionally an increased accumulation of 2-hydroxyglutarate was observed in all knockout lines tested here (Fig. 5B) with respect to the observed increment in wild type, confirming a linkage between 2-hydroxyglutarate metabolism and the ETF/ETFQO system. However the levels of isovaleryl-CoA were essentially unaltered in all mutants when compared to wild-type levels (Fig. 5C). It should be noted however that the relatively increased values observed here are by no means similar to those observed in our previous work (Araújo et al., 2010) nor to those observed by Engqvist et al. (2009).
Figure 4.
Acyl-CoA profiles in pahx Arabidopsis mutants plants during growth under extended dark conditions. Data (in % mol) represent means ± se for six independent samplings of the ninth or 10th leaves of 4-week-old, short-day-grown, Arabidopsis plants after further growth for 0, 3, 7, 10, and 15 d in extended darkness. Acyl-CoAs in samples of 10 mg (fresh weight) each were derivatized to their acyl-etheno-CoA esters, separated by HPLC, and detected fluorometrically.
Figure 5.
Metabolic phenotype of pahx Arabidopsis mutants plants during growth under extended dark conditions. Phytanoyl-CoA (A), 2-hydroxyglutarate (B), and isovaleryl-CoA (C) profiles in pahx Arabidopsis mutants under extended dark treatment. Samples were taken from the ninth or 10th leaves of 4-week-old, short-day-grown, Arabidopsis plants after further growth for 0, 3, 7, 10, and 15 d in extended darkness. Values are means ± se of six independent samplings; an asterisk indicates values that were determined by the Student’s t test to be significantly different (P < 0.05) from the wild type.
Sugar and Organic Acid Levels in the pahx Lines
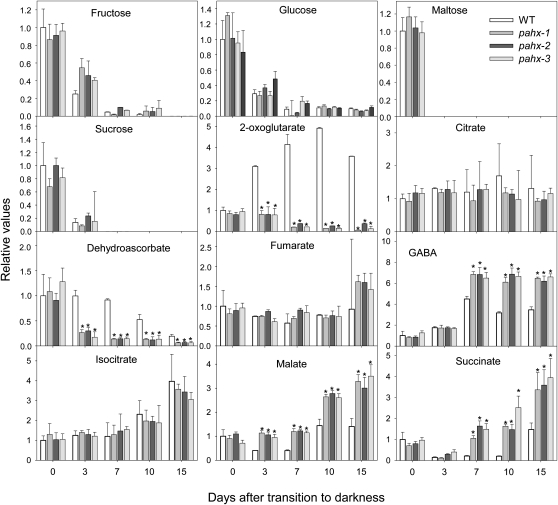
Further metabolic characterization of the pahx mutants and wild type during dark-induced senescence was performed using an established gas chromatography (GC)-MS metabolic profiling protocol (Lisec et al., 2006). Growth of plants for an extended dark period results in a rapid decline in Suc and other sugars in all genotypes analyzed (Fig. 6). Within the TCA cycle intermediates only malate and succinate significantly increased in all mutants at the end of dark treatment (Fig. 6). In sharp contrast the levels of 2-oxoglutarate and dehydroascorbate were dramatically reduced, declining to as low as 10% of the level measured at the start of the dark treatment. While these changes are striking, the exact mechanism underlying this phenomenon cannot be elucidated from the results in this study. In addition the levels of γ-amino butyric acid (GABA) were significantly increased in all genotypes analyzed (Fig. 6). Interestingly, both GABA and succinate have been previously demonstrated to increase as a strategy to bypass blocks in the TCA cycle and thus augment respiration (Studart-Guimarães et al., 2007). When taken together these data are likely indicative of small up-regulation of the GABA shunt as an alternative source of mitochondrial succinate (Studart-Guimarães et al., 2007).
Figure 6.
Relative levels of sugars and organic acids in Arabidopsis mutant plants during growth under extended dark conditions as measured by GC-MS. The y axis values represent the metabolite level relative to wild type. Data were normalized to the mean response calculated for the 0-d dark-treated leaves of the wild type (in case no response was detected at 0 d, normalization was performed against 3-d dark-treated leaves of the wild type). Values presented are means ± se of determinations on six independent samplings; an asterisk indicates values that were determined by the Student’s t test to be significantly different (P < 0.05) from the wild type.
Amino Acid Levels in the pahx Lines
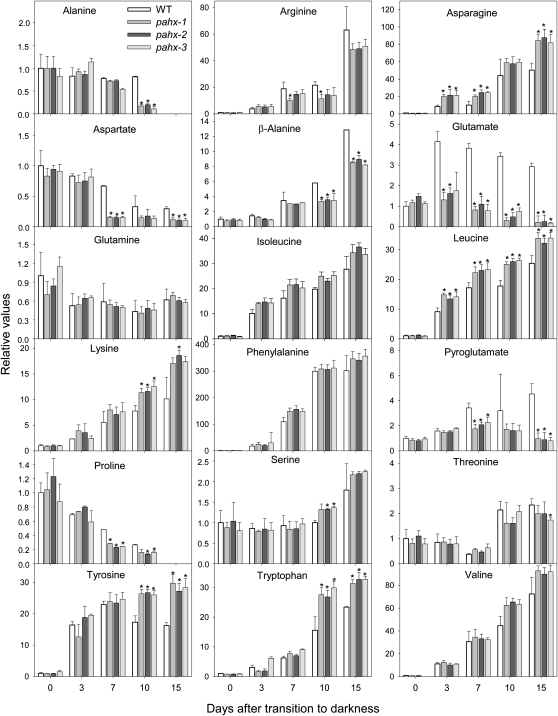
Analyses of the levels of free amino acids in wild-type and pahx mutants revealed that Arg, Asn, Leu, Lys, Tyr, β-Ala, and Trp significantly increased in all genotypes during the extended dark period while in contrast the levels of Ala, Asp, and Pro significantly decreased (Fig. 7). It is worth mentioning that some metabolites showed the same trend of relative change with respect to the wild type (i.e. increase or decrease) but with different intensity, as in the case of Arg and β-Ala that increased less in knockout plants than in wild-type plants. In addition, the levels of Glu and pyro-Glu (Fig. 7) as well as 2-oxoglutarate (Fig. 6) declined in all mutants at 7 to 10 d, whereas they increased in wild type (Figs. 6 and 7). These results are likely indicative of relatively similar pattern of protein degradation in both wild-type and the pahx mutants in contrast to our previous studies (Ishizaki et al., 2005; Araújo et al., 2010) since here there were relatively few changes in free amino acids content (Fig. 7). The elevated level of some amino acids, especially the branched-chain amino acids, aromatic amino acids, and Lys, in the mutants indicates the involvement of the ETF/ETFQO complex in their degradation. However it is important to note that in our previous work this increase was significantly higher, suggesting a small, if any, contribution of PAHX in the donation of electrons to the mitochondrial electron transport chain.
Figure 7.
Relative levels of amino acids in Arabidopsis mutants plants during growth under extended dark conditions as measured by GC-MS. Levels of the indicated amino acids are presented as in Figure 6.
Redistribution of 13C Label Following Feeding of Wild-Type and Mutant Plants with U-([U-13C]-Val and Lys
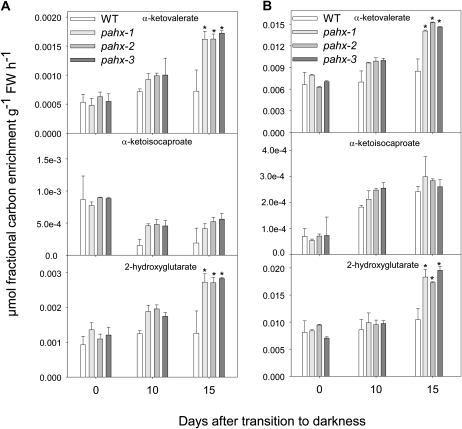
To further investigate changes in primary metabolism following reduction in the expression of PAHX and the connection between ETF/ETFQO complex and PAHX-mediated metabolism we fed isotopically labeled Val and Lys to leaves excised from wild-type and mutant plants and followed isotope redistribution at various time points during extended dark treatment. For this purpose we used a combination of feeding 13C-labeled substrate ([U-13C]-Val and [U-13C]-Lys) to the leaf via the transpiration stream and an adapted GC-MS protocol that facilitates isotope tracing (Roessner-Tunali et al., 2004). Interestingly, but not surprisingly, the pattern of changes in redistribution of label were essentially conserved across the mutants and wild-type plants, with results from this experiment in close agreement with the observed alteration in the steady-state levels of sugars, organic acids, and amino acids (Figs. 6 and 7). Nevertheless following feeding of ([U-13C]-Val it is clear that the branched-chain amino acid α-ketovalerate but not α-ketoisocaproate significantly increase in all pahx mutants after 15 d of darkness (Fig. 8). The same is true following for [U-13C]-Lys feeding. Similarly following feeding of either Val or Lys, a relatively small increase in label accumulated as 2-hydroxyglutarate in all pahx mutants was observed (Fig. 8). Again, in contrast to our previous work these changes were relatively small (Ishizaki et al., 2005; Araújo et al., 2010).
Figure 8.
Redistribution of 13C label following incubation of Arabidopsis mutants and wild-type leaves. The incubation was followed via the transpiration stream in [U-13C] Val (A) or [U-13C] Lys (B) solution. The leaf material was harvested from the ninth or 10th leaves of 4-week-old, short-day-grown, Arabidopsis plants after growth for 0, 10, and 15 d in extended darkness. Values represent absolute redistribution and are given as means ± se of determinations on six independent samplings; an asterisk indicates values that were determined by the Student’s t test to be significantly different (P < 0.05) from the wild type.
Comparative Response of pahx Mutants and d2hgdh and ivdh Double Mutants to Growth in Extended Darkness
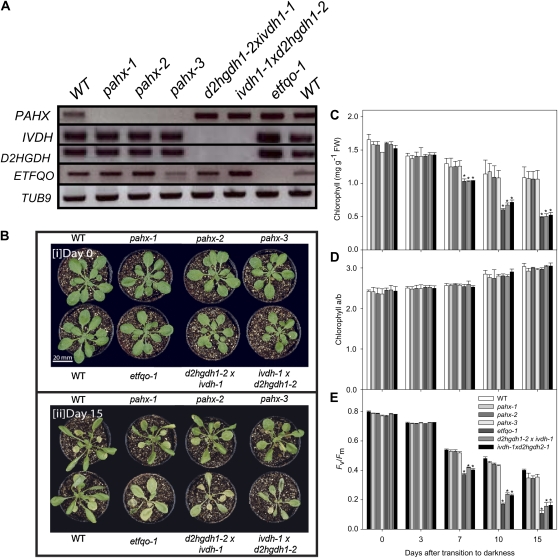
The fact that the PAHX mutants do not show phenotypical differences with respect to wild type following growth in extended darkness compared to the significant phenotypic changes we reported in previously characterized mutants in the ETF/ETFQO complex (Ishizaki et al., 2005; Araújo et al., 2010) and that the metabolic changes observed were not so evident it is reasonable to hypothesize that PAHX plays a small role in the electron donation to the ETF/ETFQO pathway and that both D2HGDH and IVDH account for the largest part of the electron input via the ETF complex. To further address this hypothesis we first generated two isovaleryl-CoA dehydrogenase/2-hydroxyglutarate dehydrogenase double mutants (d2hgdh1-2 × ivdh1-1 and ivdh1-1 × d2hgdh1-2). Here it is important to stress that these mutants lines germinated and grew normally without the addition of an external carbon source, which showed that the homozygous mutants are viable as well as exhibiting no aberrant phenotype during vegetative growth. Secondly using primers described here and previously (Araújo et al., 2010) we were able to confirm the molecular identity of all pahx mutants, the ivdh/d2hgdh double mutant, and the previously characterized etfqo mutants (Ishizaki et al., 2005). As expected we confirmed that the transcripts spanning the T-DNA insertion site, and downstream of the T-DNA insertion, are absent in all these mutant lines (Fig. 9A). That said probes for the Arabidopsis genes IVDH, D2HGDH, and ETFQO were also used as controls and our results indicated that there was no effect of the T-DNA in any of the other genes of interest, on transcript levels, confirming the specificity of our results (Fig. 9A). The Arabidopsis β-tubulin gene, TUB9 was also used as a control to analyze transcript levels and demonstrate the integrity of the RNA preparations.
Figure 9.
Phenotype of ETF/ETFQO-related Arabidopsis mutants during growth under extended dark conditions. A, RT-PCR analysis on total RNA from the wild type (WT) and the pahx-1, pahx-2, and pahx-3 mutant lines, as well as two double knockout lines ivdhh1-1 × d2hgdh1-2 and etfqo-1 with primer sets for the genes indicated on the left. B, Images of 4-week-old, short-day-grown Arabidopsis plants immediately (0 d) and after further growth for up to 15 d in darkness conditions. The leaves of the pahx mutant lines were only partially yellowed and dehydrated following 15 d growth in darkness compared to the wild-type (WT) control (Col0). Chlorophyll content (C), chlorophyll a/b ratio (D), and Fv/Fm (E), the maximum quantum yield of PSII electron transport, of leaves of 4-week-old, short-day-grown, Arabidopsis plants after further growth for 0, 3, 7, 10, and 15 d in extended darkness. Values are means ± se of six independent samplings; an asterisk indicates values that were determined by the Student’s t test to be significantly different from the wild type (P < 0.05).
We next transferred 4-week-old mutant lines and corresponding wild-type controls grown under short-day (8 h light/16 h dark) conditions to extended dark conditions for periods of up to 15 d. Interestingly we observed that the ivdh/d2hgdh double mutant showed a very similar phenotype to the previously characterized etfqo mutants (Ishizaki et al., 2005). Those homozygous mutant lines started to wilt and show signs of senescence after 10 d of continuous darkness and were apparently dead after 15 d of continuous darkness, whereas both wild-type and pahx mutants were still alive after 15 d of continuous darkness and showed only limited signs of senescence (Fig. 9B). To further confirm this accelerated senescence in the mutants, we measured two parameters related to the function of chloroplasts, chlorophyll content and photochemical efficiency (maximum variable fluorescence/maximum yield of fluorescence [Fv/Fm]), as diagnostics for leaf senescence (Oh et al., 1996). During extended dark conditions, chlorophyll content declined at the same rate in the etfqo mutant and the ivdh/d2hgdh double mutant (Fig. 9C), and was accompanied with a minor increase in the chlorophyll a/b ratio, a feature typical of senescence-related chlorophyll breakdown in Arabidopsis (Pruzinská et al., 2005; Fig. 9D). Accordingly these results were consistent with a rapid decline in the photochemical efficiency of PSII (Fv/Fm; Fig. 9E). Additionally both wild-type and pahx mutants display similar decrease in phytochemical efficiency and chlorophyll breakdown that are diagnostic of leaf senescence (Oh et al., 1996; Pruzinská et al., 2005). When taken together these results suggest that the phytanoyl-CoA does not donate electrons to the ubiquinol pool via the ETF/ETFQO complex and support the view that both the isovaleryl-CoA dehydrogenase and 2-hydroxyglutarate dehydrogenase, previously documented to be intimately associated with the ETF complex, are the major contributors to the electron input into the ETF/ETFQO complex during extended darkness and the associated induction of senescence.
DISCUSSION
In the case of 3-methyl fatty acids, such as phytanic acid, the presence of a methyl group at the 3 position prevents direct β-oxidation, leaving α- and the much less well characterized ω- (Wanders and Komen, 2007) oxidation as the only remaining option(s) for its catabolism. In mammals α-oxidation is the preferred mechanism of phytanic acid oxidation and two key enzymes for α-oxidation, PAHX and HPCL have been identified and relatively well characterized to date (for review, see Wanders et al., 2003). In brief, studies in mammals have shown that PAHX is localized in the peroxisome (Jansen et al., 1999) and that defects in the corresponding gene cause the rare inborn error of metabolism Refsum’s disease (a neurological syndrome characterized by adult-onset retinitis pigmentosa, anosmia, sensory neuropathy, and phytanic acidemia; Mukherji et al., 2001). Details underlying the biochemical nature of the reaction catalyzed by this protein (Mihalik et al., 1995, 1997) as well as the mutations underlying this syndrome have now been fairly well described with a reasonable x-ray crystallographic structure of human PAHX being available (McDonough et al., 2005). In plants, the significant accumulation of phytanoyl-CoA, which is the first-step intermediate of phytanic acid α-oxidation, during dark-induced senescence in etf/etfqo mutants (Ishizaki et al., 2005, 2006; Araújo et al., 2010) strongly suggests the existence of a similar operational pathway of α-oxidation in Arabidopsis. Interestingly, however, there ;is only a single homolog for PAHX (At2g01490) in the Arabidopsis genome, and surprisingly, the AtPAHX shares greater homology with human Phytanoyl-CoA Hydroxylase Domain Containing1 (HsPHYHD1) gene than to human PAHX itself (Supplemental Fig. S2). Nevertheless the close homologs of mammalian PAHX are exclusively found in animals whereas those of mammalian PHYHD1 homologs are distributed to a large variety of eukaryotes including both plants and animals. It is important to note that the existence of an HPCL (At5g17380) homolog in the Arabidopsis genome (Supplemental Fig. S3) additionally provides circumstantial support for PAHX activity in Arabidopsis and as such for the involvement of AtPAHX in the α-oxidation pathway of phytanoyl degradation. These bioinformatic analyses were closely supported by metabolite analysis of three independent knockout lines of this gene in Arabidopsis that showed elevated levels of the metabolite phytanoyl-CoA. Furthermore, we were able to provide evidence of its enzyme activity in Arabidopsis and this largely helps us to answer the main question we approached here that was namely whether phytanoyl-CoA was an important physiological substrate that fuels the ETF complex in Arabidopsis. The suggested role of phytanoyl-CoA in such a pathway remains an open question since despite the accumulation of phytanoyl-CoA in etf, etfqo, and ivdh mutants it was unaltered in d2hgdh mutants (Araújo et al., 2010). Surprisingly although the levels of phytanoyl-CoA and 2-hydroxyglutarate determined here are significantly higher than their wild-type counterparts, no senescence phenotype could be observed in all pahx mutants, indicating that the phenotype observed in this study as well as in previous work (Ishizaki et al., 2005, 2006; Araújo et al., 2010) cannot be associated exclusively with only one single metabolite, but rather with a complex metabolic interaction. Moreover, quantitative analysis of the mutant phenotypes, as assessed for instance from Fv/Fm values (Fig. 9), suggests that the IVDH and D2HGDH may be sufficient to satisfy the entire electron input into the mitochondrial electron transport chain via the ETF/ETFQO complex (Araújo et al., 2010). It is noteworthy that two independent coexpression analyses revealed strong correlation in gene expression between phytol, fatty acid, and branched-chain amino acid degradation genes and the expression of the D2HGDH gene (Engqvist et al., 2009; Araújo et al., 2010). Furthermore, the relationship between changes in chlorophyll fluorescence and senescence is well documented (Hörtensteiner and Kräutler, 2011) while the role of lipid oxidation as well as β-oxidation in dark-induced senescence in Arabidopsis has been underscored by a range of elegant experimental studies (Kunz et al., 2009). To address this question we assessed the direct consequences of phytanoyl-CoA accumulation by investigating and characterizing knockouts of a putative key enzyme PAHX involved in its catabolism as well as generating and characterizing reciprocal crosses of the ivdh and d2hgdh mutants (Fig. 9). Such studies should additionally help to clarify the currently poorly understood pathways of phytanoyl-CoA degradation in particular and for that matter latter stage of chlorophyll degradation in general.
Perhaps the most striking observation made in this work was the lack of an apparent phenotype since none of the three independent pahx mutants display a highly chlorotic or even necrotic phenotype following 15 d of darkness (Supplemental Fig. S1; Fig. 9). The above-mentioned phenotype being characteristically displayed by mutants of the ETF/ETFQO complex (Ishizaki et al., 2005, 2006) and indeed, albeit to a lesser extent, of the ivdh-1 and d2hgdh1-2 mutants (Araújo et al., 2010). This could be seen at both a visual phenotypic level and more quantitatively when comparing diagnostic chlorophyll and Fv/Fm ratios (Oh et al., 1996; Pruzinská et al., 2005), with those of the wild type (Supplemental Fig. S1). This result, in itself, suggests that phytanoyl-CoA degradation is unlikely to represent a major source of electrons to the alternative input into the ubiquinone pool mediated by the ETF/ETFQO complex. We were, however, also interested to see if the metabolic consequences of pahx deficiency supported this hypothesis and hence implied the existence of alternative routes of phytanoyl-CoA degradation that were able to compensate for the presumed loss of the ability to degrade phytanoyl-CoA to 2-hydroxyphytanoyl-CoA. To investigate this further we carried out a broad range of analyses for metabolic profiling to assess the levels of primary metabolites, fatty acids, and acyl-CoAs among which we have previously identified as clear biomarkers of dark-induced senescence (Ishizaki et al., 2005, 2006; Araújo et al., 2010).
As would be anticipated from the proposed function of PAHX under study the levels of phytanoyl-CoA were consistently increased in the knockout mutants (Fig. 5). However, intriguingly they were only about 1.3-fold higher than the wild-type values, whereas those in the etf, etfqo, ivdh, and d2gdh mutants were up to 30-fold higher (Ishizaki et al., 2005, 2006; Araújo et al., 2010; the significance of this observation is discussed in detail below). Consistent with the observation of the lack of an apparent phenotype there was no change in the levels of isovaleryl-CoA and a relatively minor increase in the levels of 2-hydroxyglutarate in all pahx mutants (Fig. 5). This finding implies that PAHX plays a negligible role, if any in supporting the function of the ETF/ETFQO complex—a fact that was confirmed following our finding that reciprocal crosses between the ivdh-1 and d2gdh1-2 were able to phenocopy both etf and etfqo mutants at the visual and functional level (Fig. 9). Furthermore, although the levels of the branched-chain amino acids, Lys, and aromatic acids increased they were only elevated to about 1.2-fold of the wild-type level (Fig. 7) whereas in the etf, etfqo, ivdh, and d2gdh mutants they were between 2- and 15-fold higher (Ishizaki et al., 2005, 2006; Araújo et al., 2010). Despite these changes being relatively minor the magnitude of several other changes were consistent with those mutants we had previously studied (Figs. 6 and 7). Such changes, including those of sugars and the Glc derivative, ascorbate (which decreased), TCA cycle intermediates (which, with the exception of 2-oxoglutarate that massively decreased, mainly increased), while GABA and Ala also displayed very similar changes in their levels. These findings suggest that although the PAHX is not of critical importance in support of the ETF/ETFQO complex it does play somewhat of an adaptive role in metabolism under conditions of dark-induced senescence since the changes observed are reminiscent, although relatively minor, of those previously observed (Ishizaki et al., 2005, 2006; Araújo et al., 2010). Recent studies have however illustrated that although it is clear that alternative pathways are not constitutively active (Araújo et al., 2010) they are induced under several environmentally and developmentally associated stress conditions that lead to a restriction in the provision of carbon for mitochondrial respiration and are coordinated with a general increase in protein degradation and amino acid metabolism (Araújo et al., 2011) as partially observed in this study.
Analysis of the relative levels of phytanoyl-CoA accumulation of the etf, etfqo, ivdh d2gdh, and pahx mutants lines revealed the surprising result that the levels of the substrate of PAHX accumulate less in the pahx mutants than they do in the etf, etfqo, ivdh, and d2gdh mutants when the opposite would have been anticipated. One possible explanation for this is that a downstream metabolite of the reaction catalyzed by PAHX, such as 2-hydroxyphytanoyl-CoA, acts as a signal to promote chlorophyll degradation. Support for this theory comes from the observation that etf, etfqo, ivdh, and d2gdh mutants, but not pahx mutants, do indeed contain less chlorophyll and lower Fv/Fm ratios than the wild type following exposure to continuous darkness (Fig. 9; Araújo et al., 2010).
This hypothesis is illustrated in Supplemental Figure S4 and would hence explain why phytanoyl-CoA is accumulating in the etf, etfqo, ivdh, and d2gdh mutants—which could be anticipated to be continuously producing such a signal metabolite; but to a much lesser extent in the pahx mutants—which could not. Acceptance of this model suggests that a downstream product of phytanoyl-CoA catabolism by PAHX induces chlorophyll degradation. The role of chlorophyll catabolites as signaling molecules is not without precedence. Indeed, Mur et al. (2010) have recently identified that accumulation of chlorophyll catabolites photosensitizes the hypersensitive response elicited by Pseudomonas syringae in Arabidopsis. Furthermore, Hörtensteiner and Kräutler (2011) postulate that there may be several other chlorophyll-derived signaling molecules with importance in determining antioxidant potential, fruit coloration, and color changes in autumn/fall leaves. It is also important to note that possible components of such a signaling pathway are still unknown (Hörtensteiner and Kräutler, 2011). Given the vast literature on the signaling function of pigment precursors (Pogson et al., 2008; Woodson and Chory, 2008; Kleine et al., 2009; Cazzonelli and Pogson, 2010; Chen et al., 2010), it seems reasonable to speculate that such roles may also be played by their breakdown products. Nevertheless it should be noted that our understanding of intraorganellar communication is currently hampered since the molecular nature and physiological function of transporters at the plastid and mitochondrial membranes remains incomplete (Bräutigam and Weber, 2011; Palmieri et al., 2011). Although we have provided circumstantial evidence for the presence of an important downstream metabolite able to initiate chlorophyll degradation we cannot rule out the presence of an alternative pathway of phytol degradation in plants. We also are not able to ascertain whether this pathway is independent or not of PAHX. It is thus tempting to speculate the presence a plant-specific enzyme or an enzyme with lack of specificity in plants. While the exact mechanism by which this phenotype seems to be unclear from this study, it remains to be tested if the blockage of the phytanoyl-CoA degradation is able to avoid the chlorophyll degradation, not only in ETF/ETFQO-related mutants but also in other mutants involved in the chlorophyll breakdown.
In summary, in this study we have been able to refine our understanding of upstream events of the ETF/ETFQO system during dark-induced senescence. The data presented here suggests that PAHX provides negligible, if any, support to the donation of electrons to the EFT/ETFQO complex under these conditions. This fact aside, intriguingly our data suggest that a signal emanating from a reaction downstream of the PAHX reaction has a role as an initiator of chlorophyll degradation. Confirmation and identification of the exact nature of this signal and indeed of the precise biological function of PAHX itself in the regulation of metabolism remain as important questions that should be addressed in future studies. However, results from this study show it clearly has an important role in determining fatty acid and acyl-CoA profiles in unstressed plants as well as being a seemingly important component of non-ETF-related aspects of the modulation of metabolism in response to dark-induced senescence.
MATERIALS AND METHODS
Plant Material
All Arabidopsis (Arabidopsis thaliana) plants used in this study were of the Columbia-0 ecotype. Arabidopsis seeds were handled exactly as described previously (Ishizaki et al., 2005, 2006). The T-DNA mutant lines SALK065006 (pahx-1), GK010F09 (pahx-2), and GK220D06 (pahx-3) were obtained from Nottingham Arabidopsis Stock Centre. The T-DNA mutant lines GK756G02 (ivdh-1) and SAIL844G06 (d2hgdh1-2, as described in Engqvist et al., 2009) were obtained as described in Araújo et al. (2010).
Isolation of T-DNA Insertion Mutants and Genotype Characterization
Homozygous mutant lines were isolated by PCR using primers specific for PAHX open reading frame (PAHX_L, 5′-GCGCTGAGAAAATCTCCTTC-3′ and PAHX_R, 5′-CACAACTTCGCCTCCAATTC-3′) in combination with the T-DNA left border primer (GABI-LB8409, 5′-ATATTGACCATCATACTCATTGC-3′). Crosses were generated by using both the ivdh-1 and d2hgdh1-2 mutants previously described to generate ivdh-1 × d2hgdh1-2 and d2hgdh1-2 × ivdh-1 double knockout.
Analysis of Phytanol-CoA 2-Hydrolase mRNA Expression by RT-PCR
Total RNA was isolated from rosette leaves of 3-d dark-treated plants using TRIzol reagent. First-strand cDNA was synthesized from 10 μg of total RNA with Superscript II Rnase H− reverse transcriptase (Invitrogen) and oligo(dT) primer. PCR amplification of cDNA-specific sequence was performed using primers specific for the open reading frame, L and R, described above. PCR amplification of the cDNA encoding the β-tubulin of Arabidopsis (TUB9) with a forward primer (5′-GATATCTGTTTCCGTACCTTGAAGC-3′) and a reverse primer (5′-CCGACTGTAGCATCTTGATATTGC-3′) served as a control.
Enzyme Assays
Enzymatic assays for 2-oxoglutarate conversion were performed using the 14CO2 turnover assay described previously (Searls et al., 2005). Radiochemical [U14C] 2-oxoglutarate was purchased from Moravek Biochemicals and Radiochemicals (http://www.moravek.com/). Phytanoyl-CoA, for use as an enzyme substrate, was enzymatically synthesized from phytanic acid using Pseudomonas acyl-CoA synthetase from Sigma-Aldrich (Taylor et al., 1990). Additional confirmation of phytanoyl-CoA identity was obtained exactly as described in Ishizaki et al. (2005). Enzyme extracts were prepared as described previously (Gibon et al., 2004), except that Triton X-100 was used at a concentration of 1% and glycerol at 20% and dithiothreitol was omitted. In brief, phytanoyl-CoA hydroyxylase activity was measured by determining 4CO2 release in a reaction medium containing 150 mm phytanoyl-CoA (substrate), 50 mm Tris-HCl (pH 7.5), 5 mm 2-oxoglutarate, 10 mm l-ascorbate, 0.1 mm FeCl2, 5 mm sodium acetate, 50 mm imidazole, 100 mm KCl, 0.25% (w/v) bovine serum albumin, 1 mm EDTA, 5 mm ATP, 5 mm MgCl2, 0.5 mm CoA, and 50 mg of protein extract. The reaction was started by adding 0.5 μCi of [U-14C] 2-oxoglutarate (specific activity of 2.22 MBq mmol−1). The 14CO2 liberated was captured after 1 h incubation in a KOH trap and the amount of radiolabel was subsequently quantified by liquid scintillation counting.
Dark Treatment
For dark treatments, 7- to 10-d-old seedlings were transferred to soil and then grown at 22°C under short-day conditions (8 h light/16 h dark) for 4 weeks. Following bolting, plants were grown at 22°C in the dark in the same growth cabinet. The fully expanded ninth to 12th rosette leaves were harvested at intervals of 0, 3, 7, 10, and 15 d from control and dark-grown plants for subsequent analysis. Additionally, the experiment was repeated at least five times (three in the case of the double knockout mutants—even in different growth facilities) with similar phenotypes observed each time. For phenotype characterization plants were also grown under a long-day photoperiod (16 h of light/8 h of dark) and similar responses were generally observed.
Measurement of Senescence Parameters
Chlorophyll content was determined photometrically as described in the literature (Porra et al., 1989) and the protein content as in Bradford (1976). The ratio of Fv to Fm, which corresponds to the potential quantum yield of the photochemical reactions of PSII, was measured as previously described (Oh et al., 1996) as a measure of the photochemical efficiency. Starch, nitrate, and total amino acids were determined as in Sienkiewicz-Porzucek et al. (2010).
Profiling of Acyl-CoAs, Fatty Acids, and Polar Primary Metabolites
Acyl-CoAs, fatty acids, and polar primary metabolites were extracted and evaluated exactly as previously described (Ishizaki et al., 2005), with the exception that additional peaks were looked for in the GC-MS chromatograms of the polar primary metabolites using the libraries housed in the Golm Metabolome Database (Kopka et al., 2005; Schauer et al., 2005).
Extraction, Derivatization, and Analysis of Arabidopsis Leaf Metabolites Using GC-MS
Metabolite extraction for GC-MS was performed by a method modified from that described by Roessner-Tunali et al. (2003). Arabidopsis leaf tissue (approximately 180 mg) was homogenized using a ball mill precooled with liquid nitrogen and extracted in 1,400 μL of methanol, and 60 μL of internal standard (0.2 mg ribitol mL−1 water) was subsequently added as a quantification standard. The extraction, derivatization, standard addition, and sample injection were exactly as described previously (Lisec et al., 2006). Both chromatograms and mass spectra were evaluated using either TAGFINDER (Luedemann et al., 2008) or the MASSLAB program (ThermoQuest), and the resulting data were prepared and presented as described by Roessner et al. (2001).
Analysis of [U-13C]-Val and [U-13C]-Lys Labeled Samples
Arabidopsis leaves of similar size (the fully expanded ninth to 10th rosette leaves), but from different genotypes and following varying lengths of darkness treatment were fed via the petiole with 20 mm [U-13C]-Lys or 20 mm [U-13C]-Val (both from Cambridge Isotope Laboratories) by incubation in buffered solution (10 mm MES-KOH; pH 6.5) for a period of 6 h. At the end of the incubation leaves were snap frozen in liquid nitrogen. They were subsequently extracted in 100% methanol and processed exactly as described in Timm et al. (2008). The metabolic fate of these substrates was subsequently assessed exactly as described previously (Tieman et al., 2006).
The Arabidopsis Genome Initiative locus numbers for the major genes discussed in this article are: PAHX, At2g01490; IVDH, At3g45300; D2HGDH, At4g364000; ETFQO, At2g43400.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phenotype of pahx Arabidopsis mutants plants during growth under extended dark conditions.
Supplemental Figure S2. Sequence comparison of the Arabidopsis homolog of PAHX with human PHYD1 and human PAHX.
Supplemental Figure S3. Sequence comparison of the Arabidopsis homolog of 2-HPCL and human 2-HPCL.
Supplemental Figure S4. Metabolic model showing the involvement of phytanoyl-CoA catabolism in the chlorophyll degradation pathway.
Supplemental Table S1. Key primary metabolite levels in pahx Arabidopsis mutant plants.
Acknowledgments
We thank Mark Stitt, Ralph Bock, and Joost T. van Dongen (Max-Planck-Institut für Molekulare Pflanzenphysiologie) for helpful discussions as well as Josef Bergstein (Max-Planck-Institut für Molekulare Pflanzenphysiologie) for excellent photographic work.
References
- Araújo WL, Ishizaki K, Nunes-Nesi A, Larson TR, Tohge T, Krahnert I, Witt S, Obata T, Schauer N, Graham IA, et al. (2010) Identification of the 2-hydroxyglutarate and isovaleryl-CoA dehydrogenases as alternative electron donors linking lysine catabolism to the electron transport chain of Arabidopsis mitochondria. Plant Cell 22: 1549–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo WL, Tohge T, Ishizaki K, Leaver CJ, Fernie AR. (2011) Protein degradation—an alternative respiratory substrate for stressed plants. Trends Plant Sci (in press) [DOI] [PubMed] [Google Scholar]
- Beckmann JD, Frerman FE. (1985) Electron-transfer flavoprotein-ubiquinone oxidoreductase from pig liver: purification and molecular, redox, and catalytic properties. Biochemistry 24: 3913–3921 [DOI] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Bräutigam A, Weber AP. (2011) Do metabolite transport processes limit photosynthesis? Plant Physiol 155: 43–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan-Wollaston V, Page T, Harrison E, Breeze E, Lim PO, Nam HG, Lin J-F, Wu S-H, Swidzinski J, Ishizaki K, et al. (2005) Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J 42: 567–585 [DOI] [PubMed] [Google Scholar]
- Cazzonelli CI, Pogson BJ. (2010) Source to sink: regulation of carotenoid biosynthesis in plants. Trends Plant Sci 15: 266–274 [DOI] [PubMed] [Google Scholar]
- Chen M, Galvão RM, Li M, Burger B, Bugea J, Bolado J, Chory J. (2010) Arabidopsis HEMERA/pTAC12 initiates photomorphogenesis by phytochromes. Cell 141: 1230–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engqvist M, Drincovich MF, Flügge UI, Maurino VG. (2009) Two D-2-hydroxy-acid dehydrogenases in Arabidopsis thaliana with catalytic capacities to participate in the last reactions of the methylglyoxal and beta-oxidation pathways. J Biol Chem 284: 25026–25037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engqvist MK, Kuhn A, Wienstroer J, Weber K, Jansen EEW, Jakobs C, Weber APM, Maurino VG. (2011) Plant D-2-hydroxyglutarate dehydrogenase participates in the catabolism of lysine especially during senescence. J Biol Chem 286: 11382–11390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frerman FE. (1988) Acyl-CoA dehydrogenases, electron transfer flavoprotein and electron transfer flavoprotein dehydrogenase. Biochem Soc Trans 16: 416–418 [DOI] [PubMed] [Google Scholar]
- Frerman FE, Goodman SI. (2001) Defects of electron transfer flavoprotein and electron transfer flavoprotein-ubiquinone oxidoreductase: glutaric acidemia type II. Scriver CR, Sly WS, Childs B, Beaudet AL, Valle D, , The Metabolic and Molecular Bases of Inherited Disease. McGraw-Hill, New York, pp 2357–2365 [Google Scholar]
- Ghisla S, Thorpe C. (2004) Acyl-CoA dehydrogenases: a mechanistic overview. Eur J Biochem 271: 494–508 [DOI] [PubMed] [Google Scholar]
- Gibon Y, Blaesing OE, Hannemann J, Carillo P, Höhne M, Hendriks JHM, Palacios N, Cross J, Selbig J, Stitt M. (2004) A robot-based platform to measure multiple enzyme activities in Arabidopsis using a set of cycling assays: comparison of changes of enzyme activities and transcript levels during diurnal cycles and in prolonged darkness. Plant Cell 16: 3304–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M, Rutledge SL, Kelly DR, Palmer CA, Murdoch G, Majumder N, Nicholls RD, Pei Z, Watkins PA, Vockley J. (2007) A new genetic disorder in mitochondrial fatty acid beta-oxidation: ACAD9 deficiency. Am J Hum Genet 81: 87–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heazlewood JL, Tonti-Filippini JS, Gout AM, Day DA, Whelan J, Millar AH. (2004) Experimental analysis of the Arabidopsis mitochondrial proteome highlights signaling and regulatory components, provides assessment of targeting prediction programs, and indicates plant-specific mitochondrial proteins. Plant Cell 16: 241–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörtensteiner S, Kräutler B. (2011) Chlorophyll breakdown in higher plants. Biochim Biophys Acta 1807: 977–988 [DOI] [PubMed] [Google Scholar]
- Hoskins DD, MacKenzie CG. (1961) Solubilization and electron transfer flavoprtein requirement of mitochondrial sarcosine dehydrogenase and dimethylglycine dehydrogenase. J Biol Chem 236: 177–183 [PubMed] [Google Scholar]
- Ishizaki K, Larson TR, Schauer N, Fernie AR, Graham IA, Leaver CJ. (2005) The critical role of Arabidopsis electron-transfer flavoprotein:ubiquinone oxidoreductase during dark-induced starvation. Plant Cell 17: 2587–2600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki K, Schauer N, Larson TR, Graham IA, Fernie AR, Leaver CJ. (2006) The mitochondrial electron transfer flavoprotein complex is essential for survival of Arabidopsis in extended darkness. Plant J 47: 751–760 [DOI] [PubMed] [Google Scholar]
- Jansen GA, Ofman R, Denis S, Ferdinandusse S, Hogenhout EM, Jakobs C, Wanders RJA. (1999) Phytanoyl-CoA hydroxylase from rat liver: protein purification and cDNA cloning with implications for the subcellular localization of phytanic acid α-oxidation. J Lipid Res 40: 2244–2254 [PubMed] [Google Scholar]
- Kleine T, Voigt C, Leister D. (2009) Plastid signalling to the nucleus: messengers still lost in the mists? Trends Genet 25: 185–192 [DOI] [PubMed] [Google Scholar]
- Kopka J, Schauer N, Krueger S, Birkemeyer C, Usadel B, Bergmüller E, Dörmann P, Weckwerth W, Gibon Y, Stitt M, et al. (2005) GMD@CSBDB: the Golm Metabolome Database. Bioinformatics 21: 1635–1638 [DOI] [PubMed] [Google Scholar]
- Kunz HH, Scharnewski M, Feussner K, Feussner I, Flügge UI, Fulda M, Gierth M. (2009) The ABC transporter PXA1 and peroxisomal β-oxidation are vital for metabolism in mature leaves of Arabidopsis during extended darkness. Plant Cell 21: 2733–2749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson TR, Graham IA. (2001) Technical advance: a novel technique for the sensitive quantification of acyl CoA esters from plant tissues. Plant J 25: 115–125 [DOI] [PubMed] [Google Scholar]
- Lehmann M, Schwarzländer M, Obata T, Sirikantaramas S, Burow M, Olsen CE, Tohge T, Fricker MD, Møller BL, Fernie AR, et al. (2009) The metabolic response of Arabidopsis roots to oxidative stress is distinct from that of heterotrophic cells in culture and highlights a complex relationship between the levels of transcripts, metabolites, and flux. Mol Plant 2: 390–406 [DOI] [PubMed] [Google Scholar]
- Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR. (2006) Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat Protoc 1: 387–396 [DOI] [PubMed] [Google Scholar]
- Luedemann A, Strassburg K, Erban A, Kopka J. (2008) TagFinder for the quantitative analysis of gas chromatography—mass spectrometry (GC-MS)-based metabolite profiling experiments. Bioinformatics 24: 732–737 [DOI] [PubMed] [Google Scholar]
- McDonough MA, Kavanagh KL, Butler D, Searls T, Oppermann U, Schofield CJ. (2005) Structure of human phytanoyl-CoA 2-hydroxylase identifies molecular mechanisms of Refsum disease. J Biol Chem 280: 41101–41110 [DOI] [PubMed] [Google Scholar]
- Mihalik SJ, Morrell JC, Kim DO, Sacksteder KA, Watkins PA, Gould SJ. (1997) Identification of PAHX, a Refsum disease gene. Nat Genet 17: 185–189 [DOI] [PubMed] [Google Scholar]
- Mihalik SJ, Rainville AM, Watkins PA. (1995) Phytanic acid α-oxidation in rat liver peroxisomes: production of α-hydroxyphytanoyl-CoA and formate is enhanced by dioxygenase cofactors. Eur J Biochem 232: 545–551 [DOI] [PubMed] [Google Scholar]
- Mukherji M, Chien W, Kershaw NJ, Clifton IJ, Schofield CJ, Wierzbicki AS, Lloyd MD. (2001) Structure-function analysis of phytanoyl-CoA 2-hydroxylase mutations causing Refsum's disease. Hum Mol Genet 10: 1971–1982 [DOI] [PubMed] [Google Scholar]
- Mukherji M, Schofield CJ, Wierzbicki AS, Jansen GA, Wanders RJ, Lloyd MD. (2003) The chemical biology of branched-chain lipid metabolism. Prog Lipid Res 42: 359–376 [DOI] [PubMed] [Google Scholar]
- Mur LA, Aubry S, Mondhe M, Kingston-Smith A, Gallagher J, Timms-Taravella E, James C, Papp I, Hörtensteiner S, Thomas H, et al. (2010) Accumulation of chlorophyll catabolites photosensitizes the hypersensitive response elicited by Pseudomonas syringae in Arabidopsis. New Phytol 188: 161–174 [DOI] [PubMed] [Google Scholar]
- Oh SA, Lee SY, Chung IK, Lee CH, Nam HG. (1996) A senescence-associated gene of Arabidopsis thaliana is distinctively regulated during natural and artificially induced leaf senescence. Plant Mol Biol 30: 739–754 [DOI] [PubMed] [Google Scholar]
- Palmieri F, Pierri CL, De Grassi A, Nunes-Nesi A, Fernie AR. (2011) Evolution, structure and function of mitochondrial carriers: a review with new insights. Plant J 66: 161–181 [DOI] [PubMed] [Google Scholar]
- Pogson BJ, Woo NS, Förster B, Small ID. (2008) Plastid signalling to the nucleus and beyond. Trends Plant Sci 13: 602–609 [DOI] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE. (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta 975: 384–394 [Google Scholar]
- Pruzinská A, Tanner G, Aubry S, Anders I, Moser S, Müller T, Ongania K-H, Kräutler B, Youn J-Y, Liljegren SJ, et al. (2005) Chlorophyll breakdown in senescent Arabidopsis leaves: characterization of chlorophyll catabolic enzymes involved in the degreening reaction. Plant Physiol 139: 52–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner U, Luedemann A, Brust D, Fiehn O, Linke T, Willmitzer L, Fernie AR. (2001) Metabolic profiling allows comprehensive phenotyping of genetically or environmentally modified plant systems. Plant Cell 13: 11–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner-Tunali U, Hegemann B, Lytovchenko A, Carrari F, Bruedigam C, Granot D, Fernie AR. (2003) Metabolic profiling of transgenic tomato plants overexpressing hexokinase reveals that the influence of hexose phosphorylation diminishes during fruit development. Plant Physiol 133: 84–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner-Tunali U, Liu J, Leisse A, Balbo I, Perez-Melis A, Willmitzer L, Fernie AR. (2004) Kinetics of labelling of organic and amino acids in potato tubers by gas-chromatography mass-spectrometry following incubation in 13C labelled isotopes. Plant J 39: 669–679 [DOI] [PubMed] [Google Scholar]
- Ruzicka FJ, Beinert H. (1977) A new iron-sulfur flavoprotein of the respiratory chain: a component of the fatty acid beta oxidation pathway. J Biol Chem 252: 8440–8445 [PubMed] [Google Scholar]
- Schauer N, Steinhauser D, Strelkov S, Schomburg D, Allison G, Moritz T, Lundgren K, Roessner-Tunali U, Forbes MG, Willmitzer L, et al. (2005) GC-MS libraries for the rapid identification of metabolites in complex biological samples. FEBS Lett 579: 1332–1337 [DOI] [PubMed] [Google Scholar]
- Schelbert S, Aubry S, Burla B, Agne B, Kessler F, Krupinska K, Hörtensteiner S. (2009) Pheophytin pheophorbide hydrolase (pheophytinase) is involved in chlorophyll breakdown during leaf senescence in Arabidopsis. Plant Cell 21: 767–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searls T, Butler D, Chien W, Mukherji M, Lloyd MD, Schofield CJ. (2005) Studies on the specificity of unprocessed and mature forms of phytanoyl-CoA 2-hydroxylase and mutation of the iron binding ligands. J Lipid Res 46: 1660–1667 [DOI] [PubMed] [Google Scholar]
- Sienkiewicz-Porzucek A, Sulpice R, Osorio S, Krahnert I, Leisse A, Urbanczyk-Wochniak E, Hodges M, Fernie AR, Nunes-Nesi A. (2010) Mild reductions in mitochondrial NAD-dependent isocitrate dehydrogenase activity result in altered nitrate assimilation and pigmentation but do not impact growth. Mol Plant 3: 156–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studart-Guimarães C, Fait A, Nunes-Nesi A, Carrari F, Usadel B, Fernie AR. (2007) Reduced expression of succinyl-coenzyme A ligase can be compensated for by up-regulation of the gamma-aminobutyrate shunt in illuminated tomato leaves. Plant Physiol 145: 626–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DC, Weber N, Hogge LR, Underhill EW. (1990) A simple enzymatic method for the preparation of radiolabeled erucoyl-CoA and other long-chain fatty acyl-CoAs and their characterization by mass spectrometry. Anal Biochem 184: 311–316 [DOI] [PubMed] [Google Scholar]
- Tieman D, Taylor M, Schauer N, Fernie AR, Hanson AD, Klee HJ. (2006) Tomato aromatic amino acid decarboxylases participate in synthesis of the flavor volatiles 2-phenylethanol and 2-phenylacetaldehyde. Proc Natl Acad Sci USA 103: 8287–8292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timm S, Nunes-Nesi A, Pärnik T, Morgenthal K, Wienkoop S, Keerberg O, Weckwerth W, Kleczkowski LA, Fernie AR, Bauwe H. (2008) A cytosolic pathway for the conversion of hydroxypyruvate to glycerate during photorespiration in Arabidopsis. Plant Cell 20: 2848–2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanders RJ, Jansen GA, Lloyd MD. (2003) Phytanic acid alpha-oxidation, new insights into an old problem: a review. Biochim Biophys Acta 1631: 119–135 [DOI] [PubMed] [Google Scholar]
- Wanders RJ, Komen JC. (2007) Peroxisomes, Refsum’s disease and the α- and ω-oxidation of phytanic acid. Biochem Soc Trans 35: 865–869 [DOI] [PubMed] [Google Scholar]
- Watmough NJ, Frerman FE. (2010) The electron transfer flavoprotein: ubiquinone oxidoreductases. Biochim Biophys Acta 1797: 1910–1916 [DOI] [PubMed] [Google Scholar]
- Wierzbicki AS, Lloyd MD, Schofield CJ, Feher MD, Gibberd FB. (2002) Refsum’s disease: a peroxisomal disorder affecting phytanic acid alpha-oxidation. J Neurochem 80: 727–735 [DOI] [PubMed] [Google Scholar]
- Woodson JD, Chory J. (2008) Coordination of gene expression between organellar and nuclear genomes. Nat Rev Genet 9: 383–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Ohlrogge JB. (2009) Turnover of fatty acids during natural senescence of Arabidopsis, Brachypodium, and switchgrass and in Arabidopsis β-oxidation mutants. Plant Physiol 150: 1981–1989 [DOI] [PMC free article] [PubMed] [Google Scholar]