Abstract
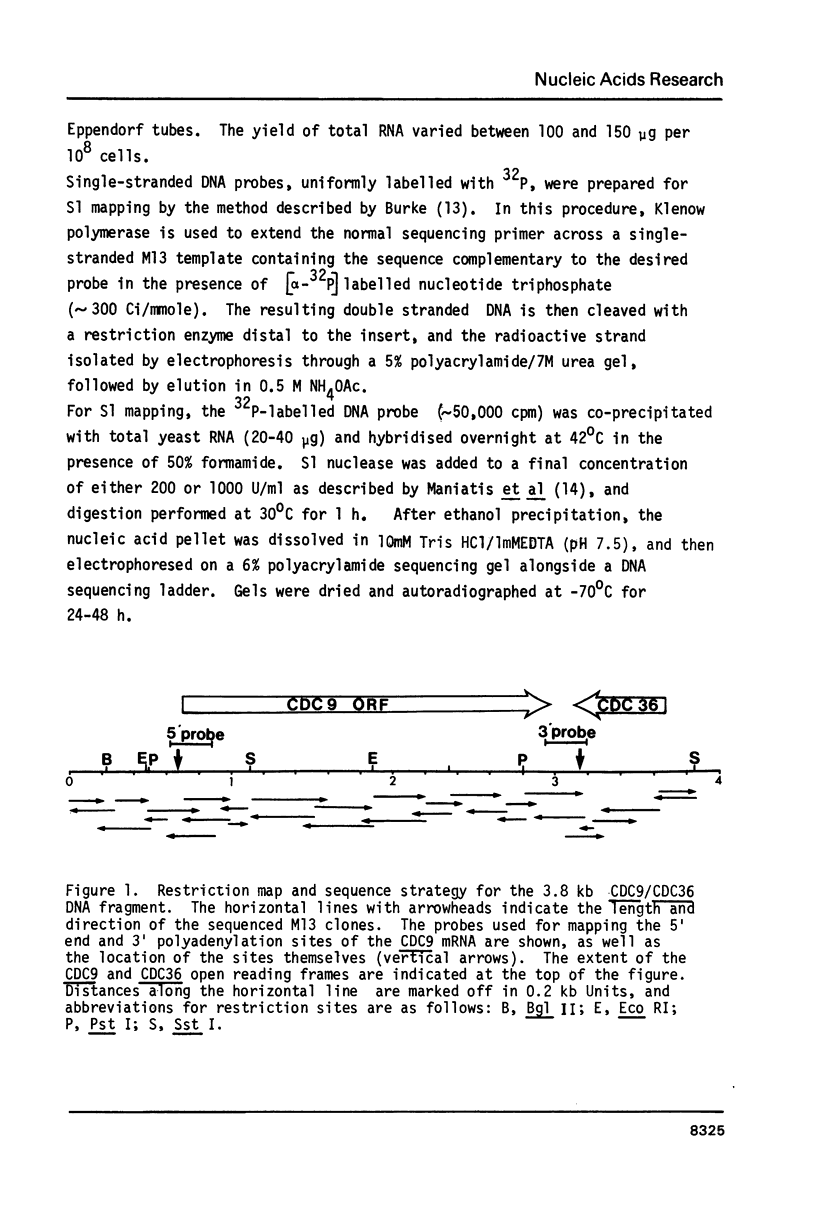
The CDC9 gene of Saccharomyces cerevisiae encodes a DNA ligase, and we have determined the nucleotide sequence of a 3.85 kb fragment of DNA which encompasses the convergently transcribed CDC9 and CDC36 genes. S1 nuclease mapping has revealed a major 5' end for the CDC9 mRNA, and one major and one minor site for 3' polyadenylation. These two sites lie within the C-terminal coding region of the CDC36 gene, implying that these two genes are transcribed from overlapping sequences. An interesting structural feature of the CDC9 gene is a series of 6 hexanucleotide repeats (ATGATT) which occur within the 650 bp immediately upstream from the site of transcription initiation. These repeat elements may be implicated in the cell division cycle regulated expression of CDC9. Comparison of the predicted amino acid sequence of the yeast DNA ligase (Mr 84,806) with the sequences of the T4 and T7 bacteriophage DNA ligases reveals little similarity except for a stretch of approximately 45 amino acids, comprising 3 short homologous segments. This region may represent an ATP-binding domain common to polynucleotide ligases.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Armstrong J., Brown R. S., Tsugita A. Primary structure and genetic organization of phage T4 DNA ligase. Nucleic Acids Res. 1983 Oct 25;11(20):7145–7156. doi: 10.1093/nar/11.20.7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aves S. J., Durkacz B. W., Carr A., Nurse P. Cloning, sequencing and transcriptional control of the Schizosaccharomyces pombe cdc10 'start' gene. EMBO J. 1985 Feb;4(2):457–463. doi: 10.1002/j.1460-2075.1985.tb03651.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker D. G., Johnson A. L., Johnston L. H. An improved assay for DNA ligase reveals temperature-sensitive activity in cdc9 mutants of Saccharomyces cerevisiae. Mol Gen Genet. 1985;200(3):458–462. doi: 10.1007/BF00425731. [DOI] [PubMed] [Google Scholar]
- Barker D. G., Johnston L. H. Saccharomyces cerevisiae cdc9, a structural gene for yeast DNA ligase which complements Schizosaccharomyces pombe cdc17. Eur J Biochem. 1983 Aug 1;134(2):315–319. doi: 10.1111/j.1432-1033.1983.tb07568.x. [DOI] [PubMed] [Google Scholar]
- Barker W. C., Dayhoff M. O. Viral src gene products are related to the catalytic chain of mammalian cAMP-dependent protein kinase. Proc Natl Acad Sci U S A. 1982 May;79(9):2836–2839. doi: 10.1073/pnas.79.9.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
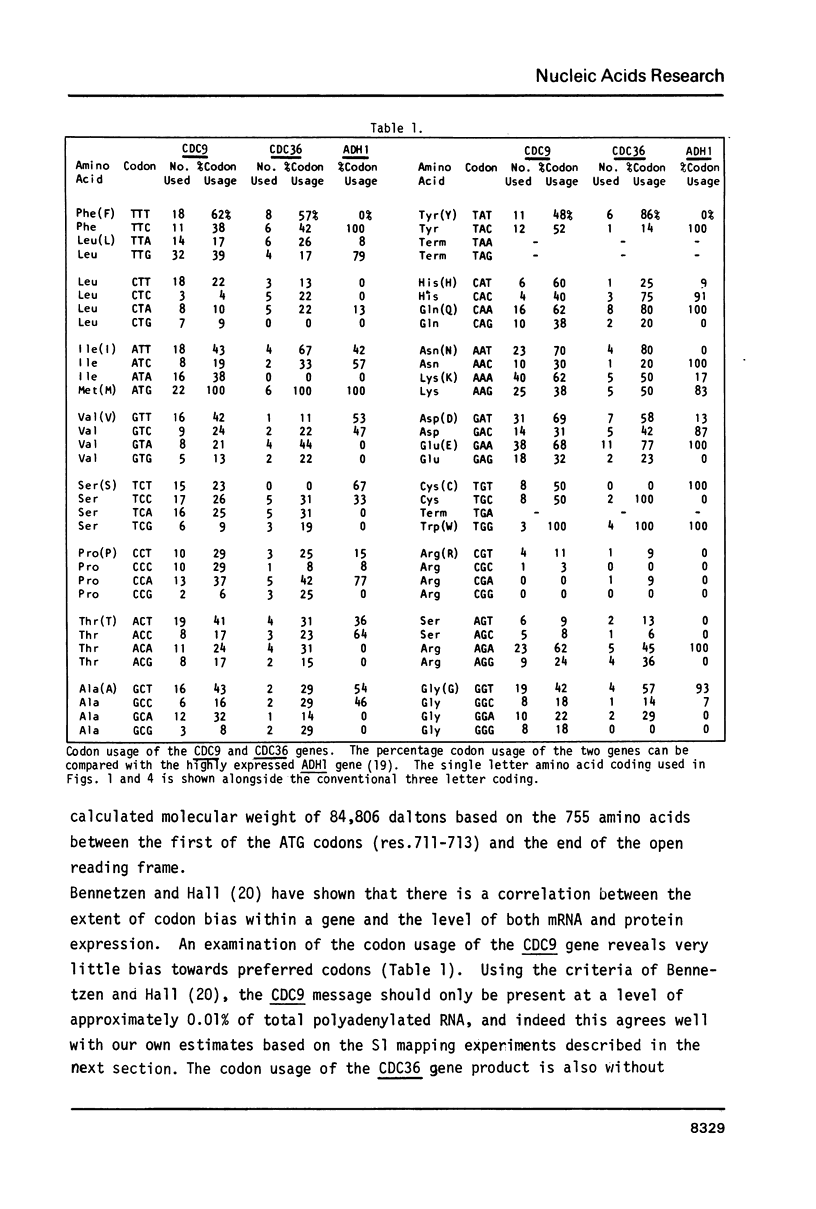
- Bennetzen J. L., Hall B. D. Codon selection in yeast. J Biol Chem. 1982 Mar 25;257(6):3026–3031. [PubMed] [Google Scholar]
- Bennetzen J. L., Hall B. D. The primary structure of the Saccharomyces cerevisiae gene for alcohol dehydrogenase. J Biol Chem. 1982 Mar 25;257(6):3018–3025. [PubMed] [Google Scholar]
- Benoist C., O'Hare K., Breathnach R., Chambon P. The ovalbumin gene-sequence of putative control regions. Nucleic Acids Res. 1980 Jan 11;8(1):127–142. doi: 10.1093/nar/8.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Breter H. J., Ferguson J., Peterson T. A., Reed S. I. Isolation and transcriptional characterization of three genes which function at start, the controlling event of the Saccharomyces cerevisiae cell division cycle: CDC36, CDC37, and CDC39. Mol Cell Biol. 1983 May;3(5):881–891. doi: 10.1128/mcb.3.5.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buehner M., Ford G. C., Olsen K. W., Moras D., Rossman M. G. Three-dimensional structure of D-glyceraldehyde-3-phosphate dehydrogenase. J Mol Biol. 1974 Nov 25;90(1):25–49. doi: 10.1016/0022-2836(74)90254-x. [DOI] [PubMed] [Google Scholar]
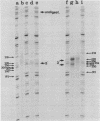
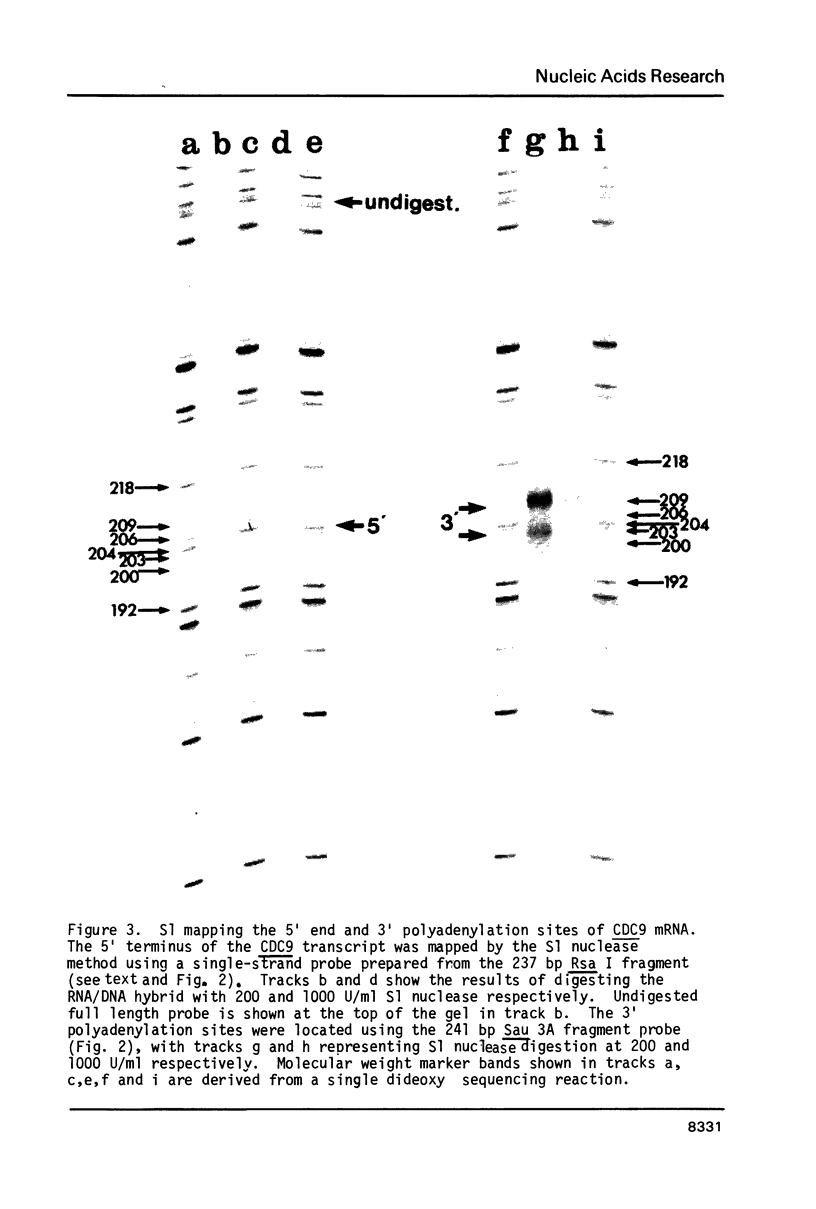
- Burke J. F. High-sensitivity S1 mapping with single-stranded [32P]DNA probes synthesized from bacteriophage M13mp templates. Gene. 1984 Oct;30(1-3):63–68. doi: 10.1016/0378-1119(84)90105-7. [DOI] [PubMed] [Google Scholar]
- Burke R. L., Tekamp-Olson P., Najarian R. The isolation, characterization, and sequence of the pyruvate kinase gene of Saccharomyces cerevisiae. J Biol Chem. 1983 Feb 25;258(4):2193–2201. [PubMed] [Google Scholar]
- Dunn J. J., Studier F. W. Nucleotide sequence from the genetic left end of bacteriophage T7 DNA to the beginning of gene 4. J Mol Biol. 1981 Jun 5;148(4):303–330. doi: 10.1016/0022-2836(81)90178-9. [DOI] [PubMed] [Google Scholar]
- Fabre F., Roman H. Evidence that a single DNA ligase is involved in replication and recombination in yeast. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4586–4588. doi: 10.1073/pnas.76.9.4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. H., Mortimer R. K., Culotti J., Culotti M. Genetic Control of the Cell Division Cycle in Yeast: V. Genetic Analysis of cdc Mutants. Genetics. 1973 Jun;74(2):267–286. doi: 10.1093/genetics/74.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hereford L. M., Osley M. A., Ludwig T. R., 2nd, McLaughlin C. S. Cell-cycle regulation of yeast histone mRNA. Cell. 1981 May;24(2):367–375. doi: 10.1016/0092-8674(81)90326-3. [DOI] [PubMed] [Google Scholar]
- Johnston L. H., Nasmyth K. A. Saccharomyces cerevisiae cell cycle mutant cdc9 is defective in DNA ligase. Nature. 1978 Aug 31;274(5674):891–893. doi: 10.1038/274891a0. [DOI] [PubMed] [Google Scholar]
- Johnston L. H. The DNA repair capability of cdc9, the Saccharomyces cerevisiae mutant defective in DNA ligase. Mol Gen Genet. 1979 Feb 16;170(1):89–92. doi: 10.1007/BF00268583. [DOI] [PubMed] [Google Scholar]
- Kozak M. How do eucaryotic ribosomes select initiation regions in messenger RNA? Cell. 1978 Dec;15(4):1109–1123. doi: 10.1016/0092-8674(78)90039-9. [DOI] [PubMed] [Google Scholar]
- Langford C. J., Gallwitz D. Evidence for an intron-contained sequence required for the splicing of yeast RNA polymerase II transcripts. Cell. 1983 Jun;33(2):519–527. doi: 10.1016/0092-8674(83)90433-6. [DOI] [PubMed] [Google Scholar]
- Laughon A., Gesteland R. F. Primary structure of the Saccharomyces cerevisiae GAL4 gene. Mol Cell Biol. 1984 Feb;4(2):260–267. doi: 10.1128/mcb.4.2.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman I. R. DNA ligase: structure, mechanism, and function. Science. 1974 Nov 29;186(4166):790–797. doi: 10.1126/science.186.4166.790. [DOI] [PubMed] [Google Scholar]
- Murray N. E., Bruce S. A., Murray K. Molecular cloning of the DNA ligase gene from bacteriophage T4. II. Amplification and preparation of the gene product. J Mol Biol. 1979 Aug 15;132(3):493–505. doi: 10.1016/0022-2836(79)90271-7. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. A repetitive DNA sequence that confers cell-cycle START (CDC28)-dependent transcription of the HO gene in yeast. Cell. 1985 Aug;42(1):225–235. doi: 10.1016/s0092-8674(85)80118-5. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. Molecular analysis of a cell lineage. Nature. 1983 Apr 21;302(5910):670–676. doi: 10.1038/302670a0. [DOI] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Peterson T. A., Prakash L., Prakash S., Osley M. A., Reed S. I. Regulation of CDC9, the Saccharomyces cerevisiae gene that encodes DNA ligase. Mol Cell Biol. 1985 Jan;5(1):226–235. doi: 10.1128/mcb.5.1.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N. J., Brownlee G. G. 3' non-coding region sequences in eukaryotic messenger RNA. Nature. 1976 Sep 16;263(5574):211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- Reed S. I. The selection of S. cerevisiae mutants defective in the start event of cell division. Genetics. 1980 Jul;95(3):561–577. doi: 10.1093/genetics/95.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds P., Higgins D. R., Prakash L., Prakash S. The nucleotide sequence of the RAD3 gene of Saccharomyces cerevisiae: a potential adenine nucleotide binding amino acid sequence and a nonessential acidic carboxyl terminal region. Nucleic Acids Res. 1985 Apr 11;13(7):2357–2372. doi: 10.1093/nar/13.7.2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. Identical 3'-terminal octanucleotide sequence in 18S ribosomal ribonucleic acid from different eukaryotes. A proposed role for this sequence in the recognition of terminator codons. Biochem J. 1974 Sep;141(3):609–615. doi: 10.1042/bj1410609a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuster J. R. Mating-defective ste mutations are suppressed by cell division cycle start mutations in Saccharomyces cerevisiae. Mol Cell Biol. 1982 Sep;2(9):1052–1063. doi: 10.1128/mcb.2.9.1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storms R. K., Ord R. W., Greenwood M. T., Mirdamadi B., Chu F. K., Belfort M. Cell cycle-dependent expression of thymidylate synthase in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Dec;4(12):2858–2864. doi: 10.1128/mcb.4.12.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M., Yamaguchi E., Uchida T. Thermophilic DNA ligase. Purification and properties of the enzyme from Thermus thermophilus HB8. J Biol Chem. 1984 Aug 25;259(16):10041–10047. [PubMed] [Google Scholar]
- Thøgersen H. C., Morris H. R., Rand K. N., Gait M. J. Location of the adenylylation site in T4 RNA ligase. Eur J Biochem. 1985 Mar 1;147(2):325–329. doi: 10.1111/j.1432-1033.1985.tb08753.x. [DOI] [PubMed] [Google Scholar]
- Walker J. E., Saraste M., Runswick M. J., Gay N. J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1(8):945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward D. F., Murray N. E. Convergent transcription in bacteriophage lambda: interference with gene expression. J Mol Biol. 1979 Sep 15;133(2):249–266. doi: 10.1016/0022-2836(79)90533-3. [DOI] [PubMed] [Google Scholar]
- Wierenga R. K., Hol W. G. Predicted nucleotide-binding properties of p21 protein and its cancer-associated variant. Nature. 1983 Apr 28;302(5911):842–844. doi: 10.1038/302842a0. [DOI] [PubMed] [Google Scholar]
- Zaret K. S., Sherman F. DNA sequence required for efficient transcription termination in yeast. Cell. 1982 Mar;28(3):563–573. doi: 10.1016/0092-8674(82)90211-2. [DOI] [PubMed] [Google Scholar]