Abstract
MicroRNAs (miRNAs) are a class of non-coding small RNAs with critical regulatory functions as post-transcriptional regulators. Due to the fundamental importance and broad impact of miRNAs on multiple genes and pathways, dysregulated miRNAs have been associated with human diseases, including cancer. Colorectal cancer (CRC) is among the most deadly diseases, and miRNAs offer a new frontier for target discovery and novel biomarkers for both diagnosis and prognosis. In this review, we summarize the recent advancement of miRNA research in CRC, in particular, the roles of miRNAs in CRC stem cells, epithelial-to-mesenchymal transition, chemoresistance, therapeutics, diagnosis, and prognosis.
Keywords: microRNA, colorectal cancer, EMT, biomarker, prognosis, cancer stem cell
Introduction
Colorectal cancer (CRC) is one of the most common cancer types and causes 655,000 deaths per year worldwide (Alwan, 2007). In the United States, CRC ranks second in cancer-related mortality (Hegde et al., 2008). It is estimated that over 141,000 new CRC cases will be diagnosed and more than 49,000 CRC patients will die in the year of 2011 according to the National Cancer Institute. Many early detection methods and therapeutic approaches have been explored to further improve the CRC treatment and prolong patient survival in the last 20 years (Hegde et al., 2008). CRC is a heterogeneous tumor containing different cell populations with distinct properties, which contribute to the disease complexity and lead to variable clinical patterns, including symptoms, tumor development, responses to treatments, and patients outcomes (Gutman and Fidler, 1995). Therefore, there is an urgent need to discover and develop reliable, highly sensitive, and specific novel comprehensive diagnostic biomarkers, which can predict the prognosis and provide the basis for individualized therapeutic strategies against CRC.
Recently, small regulatory RNAs have gained tremendous interest in cancer research. MicroRNAs (miRNAs) are non-coding RNA molecules, 18–25 nucleotide in length, that regulate the expression of their target genes by translational arrest or mRNA cleavage, most likely, through interaction mainly at the 3′-UTRs of the target mRNAs (Lee et al., 1993; Wightman et al., 1993; He and Hannon, 2004). Base pairing between at least six consecutive nucleotides within the 5′-seed of the miRNA with the target site on the mRNA is reported to be a minimum requirement for the miRNA–mRNA interaction (Lee et al., 1993; Wightman et al., 1993). miRNAs have been found to regulate many cellular processes including apoptosis (Brennecke et al., 2003; Chan et al., 2005; Hwang and Mendell, 2006; Ghodgaonkar et al., 2009), differentiation (Wightman et al., 1993; Navarro and Lieberman, 2010; Tang, 2010), and cell proliferation (Brennecke et al., 2003; He et al., 2005; Johnson et al., 2007; Navarro and Lieberman, 2010). Dysregulation of miRNAs have been associated with cancer development and progression, and miRNAs emerge as a new research frontier for understanding cancer development at the post-transcriptional and translational level (Ryan et al., 2010).
Genomics Profiling to Discover miRNAs Associated with Colorectal Cancer
The discovery of miRNAs associated with CRC began with high throughput systematic miRNA expression profiling. The first report of dysregulated miRNAs in CRC was published in 2003 (Michael et al., 2003), of which they discovered the down-regulation of miR-143 and miR-145 in CRC tumor tissues compared to the normal controls. Subsequently, several additional studies have been conducted to profile the expression levels of miRNAs in both CRC cell lines and tumor specimens, which provided more than 100 dysregulated miRNAs. Among these, several miRNAs were found to be consistently deregulated in CRC specimens, such as overexpression of miR-20, miR-21, miR-31, and miR-99b, and the reduction of miR-143, miR-145, and miR-192 (Bandres et al., 2006; Volinia et al., 2006; Slaby et al., 2007; Schetter et al., 2008; Chen et al., 2009). With regards to disease progression, miRNA levels were found to be associated with CRC disease stages, for example, the expression of miR-31 was significantly higher in stage IV tumors than that in stage II tumors, and miR-21 was found to be positively related to CRC metastasis and clinical stages (Slaby et al., 2007). Slattery et al. quantified the miRNA expression levels based on tumor location and mutation status of p53 and KRAS. There were 73 differentially expressed miRNAs between normal tissues from colon and rectum with at least twofold difference. The tumors were further categorized into several subtypes based on microsatellite instability, CpG Island Methylator Phenotype, KRAS2-mutation, and p53 mutation. Each subtype had a unique miRNA profile, indicating a potential of different genetic factors to influence miRNA expression levels (Slattery et al., 2011). More recently, a comprehensive genome wide analysis for the contribution of epigenetic mechanisms to miRNA dysregulation was conducted in CRC by high-resolution chromatin immunoprecipitation (ChIP)-sequencing approach. They have identified 47 miRNAs encoded in 37 primary transcription units as potential targets of epigenetic silencing. Among the miRNAs we found to be dysregulated, many of which are implicated in human cancer, miR-1-1 was methylated frequently in early and advanced CRC in which it may act as a tumor suppressor. These findings provide a new insight into the association between chromatin signatures and miRNA dysregulation in cancer (Suzuki et al., 2011). Given these findings, some of these miRNAs may be potential candidates as novel therapeutic targets and/or diagnostic and prognostic biomarkers.
Although currently there is still no experimental evidence that any dysregulated miRNAs directly cause the pathogenesis of CRC development, mounting evidence indicates that there is a close connection between inflammation and tumorigenesis (Williams et al., 1999; Hanada et al., 2006; Feagins et al., 2009; Dupaul-Chicoine et al., 2010). The pathogenesis of CRC is a rather complicated process and inflammation directly contributes to the development of CRC. In the case of CRC, the influence of miRNAs in inflammation and tumorigenesis was also investigated. It has been shown that expression levels of miRNAs are influenced by inflammation (Sonkoly et al., 2008). Necela et al. compared the miRNA expression levels in mouse CRC samples derived from chronic inflammation and genetic mutation. miR-215, miR-708, miR-31, and miR-135b were found consistently deregulated in both studies. miR-137, miR-133a, miR-467d, and miR-218 were uniquely differentially expressed in the mouse model (Necela et al., 2011).
p53 and miRNAs
It is well established that p53, a tumor suppressor gene, is among the most frequently deleted and mutated genes in many cancers, and the frequency in CRC is over 50% (Crawford et al., 1984; Vogelstein et al., 1988; Scott et al., 1991). p53 regulates its target gene expression at several levels. It functions either as a transcriptional activator or suppressor in certain cases in response to genotoxic and growth stresses respectively (Zhao et al., 2000). It can also act as an RNA-binding protein to modulate gene expression at the post-transcriptional level (Fu et al., 1996; Miller et al., 2000). We reasoned as a transcription factor, p53 may also regulate certain miRNA expression. In 2006, we first systematically analyzed the miRNA profiles in CRC cell lines HCT 116 with either wild-type p53 or null-p53, and identified putative p53-binding sites in nearly 40% of miRNA promoter regions (e.g., miR-34s, miR-192, miR-215, miR-200c, miR-26; Xi et al., 2006b). miR-26a was experimentally validated to be directly regulated by p53. Since then, this has now become a new frontier in cancer research. Several recent studies demonstrated a clear significance of such regulatory relationship in various cancer types (Shin et al., 2009a,b; Zhou et al., 2010).
Among p53-regulated miRNAs, miR-34a, miR-192, miR-215, and miR-26a have been extensively studied in CRC. miR-34a was shown to be directly regulated by p53 and suppress the proliferation of CRC cell lines HCT 116 and RKO in vitro and in vivo, and its expression level was reduced in 36% of human CRC specimens. The tumor suppressive function of miR-34a was a result of the suppression of key cell cycle regulators such as the E2F transcription factor pathway and activation of p53 and p21 (Chang et al., 2007; He et al., 2007; Raver-Shapira et al., 2007; Tazawa et al., 2007). Another important target of miR-34a is silent information regulator 1 (SIRT1), and the apoptosis of CRC cells induced by repression of SIRT1 was dependent on p53 expression (Yamakuchi et al., 2008). miR-26a has been demonstrated to impact colon cancer by influencing apoptotic pathway by targeting BAK1 (Kulshreshtha et al., 2007).
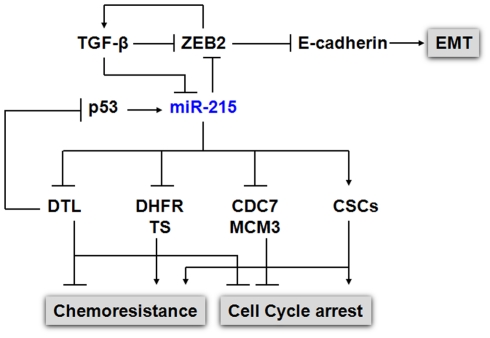
For other p53 mediated miRNAs such as miR-26a, miR-192, and miR-215, the expression levels were reduced in CRC. miR-192 and miR-215 can induce cell cycle arrest and enhance p53 mediated p21 expression when overexpressed in CRC cell lines (Xi et al., 2006b; Braun et al., 2008; Georges et al., 2008; Song et al., 2008; Karaayvaz et al., 2011). We further investigated the regulating mechanism of p53 level by miR-215 in CRC and discovered a key target, denticleless protein homolog (DTL). The suppression of DTL by miR-215 triggered an up-regulation of p53 and p21 (Song et al., 2010). DTL (RAMP, CDT2) is thought to play an essential role in DNA synthesis, cell cycle progression, proliferation, and differentiation (Pan et al., 2006). It has been reported that DTL controls cell cycle through many distinct mechanisms. First, it is a component of the proliferating cell nuclear antigen (PCNA) – coupled CUL4–DDB1 E3 ubiquitin ligase complex and is required for degrading Cdt1 during S phase (Higa et al., 2006), thereby preventing DNA replication (Sansam et al., 2006). Secondly, it has an important role in the early, radiation-induced G2/M checkpoint, possibly through degrading key cell cycle regulators in G2 phase by its ubiquitin ligase function (Sansam et al., 2006). Moreover, PCNA-coupled CUL4/DDB1/DTL complex has been reported to ubiquitinate and degrade key cell cycle proteins such as p53, MDM2, p21, and E2F1, further emphasizing the significance of this ubiquitination system in cell cycle (Banks et al., 2006; Nishitani et al., 2008; Shibutani et al., 2008). Although miR-192 has almost identical sequences as miR-215, it does not suppress DTL expression. miR-192 and miR-215 also localized at different chromosomes. These results suggest that not only can the functions of miRNAs be modulated by p53 status in CRC, the tumor suppressive function of p53 can also be modulated by the post-transcriptional controls of various miRNAs under different stress and/or physiological conditions, providing p53 with a greater flexibility to control cell cycle and cell death. The impact of miR-215 on several cellular processes [proliferation, cell cycle control, chemoresistance, epithelial–mesenchymal transition (EMT)] was summarized in Figure 1.
Figure 1.
Impact of miR-215 on cell proliferation, cell cycle, chemoresistance, and EMT. p53 and miR-215 form a positive feedback regulatory mechanism to regulate cell cycle control. The expression of p53 was induced by the suppression of DTL, a critical component of the e CUL4A–DDB1 E3 ubiquitin ligase complex, along with the p53 down stream gene p21. The elevated p53 in turn up-regulates the expression of miR-215. In addition, to DTL, miR-215 also suppresses the expression of TS, DHFR, CDC7, MCM3, MCM10. miR-215 triggers cell cycle arrest at the G2/M phase without triggering cell death, which contribute to chemoresistance to TS and DHFR inhibitors in CSCs. miR-215 also mediates EMT transition by targeting ZEB2 via TGFβ signaling pathway.
miRNAs and Epithelial–Mesenchymal Transition
Epithelial–mesenchymal transition is an evolutionarily conserved developmental process (Thiery, 2003), and it has been proposed as a key process for tumor invasion, metastasis, and carcinogenesis (Yang and Weinberg, 2008). EMT is characterized by a remarkable morphological change of epithelial cells from an epithelial cobblestone phenotype to an elongated fibroblastic phenotype (mesenchymal phenotype) accompanied by increased motility and invasive activity. During the acquisition of EMT characteristics, cells lose epithelial cell–cell junctions, undergo cytoskeleton reorganization, decrease the expression of proteins that promote cell–cell contact such as E-cadherin and γ-catenin, and increase the expression of mesenchymal markers such as vimentin, fibronectin, α-smooth muscle actin (SMA), and N-cadherin. Moreover, an increased activity of matrix metalloproteinases (MMPs) like MMP-2, MMP-3, and MMP-9 is observed during EMT, which can be associated with an invasive phenotype (Wang et al., 2010c).
Recently, studies have shown that miRNAs regulate EMT through the regulation of E-cadherin and its transcriptional repressors such as zinc finger E-box-binding factors (ZEB; Gregory et al., 2008b; Wang et al., 2010c). For example, miR-200 family and miR-205 have been identified as key regulators of EMT by repressing ZEB1 and ZEB2, which in turn repress the transcription of several members in miR-200 family, suggesting a double-negative feedback loop existed between these miRNAs and ZEBs to modulate the switch between epithelial and mesenchymal phenotype in cancer cells (Bracken et al., 2008; Burk et al., 2008; Gregory et al., 2008a, 2011). Moreover, it has been shown that TGF-β treatment was directly associated with phenotypic and molecular changes attributed to EMT, which led to the decreased expression of miR-200 with the up-regulation of ZEBs, indicating that TGF-β/ZEB/miR-200 signaling network can regulate the plasticity of EMT (Gregory et al., 2011). Similar functions were also found in miR-192/215, which was shown to regulate EMT in diabetic kidney cells (Kato et al., 2007; Wang et al., 2010a). Another p53 mediated miRNA, miR-194, also contributes directly to the EMT process by suppressing the expression of BMI1 in endometrial cancer (Dong et al., 2011).
Loboda et al. analyzed the gene expression profile from 326 CRC specimens by microarrays, and identified around 5000 differentially expressed genes, which were then divided into two subpopulations based on first principal component (PC1) analysis. Several gene signatures were also found to be tightly associated with PC1/EMT signature, such as TGF-β, RAS, proliferation, and MYC. TGF-β and RAS have been shown as key regulators for EMT process. They also profiled miRNAs in 70 CRC samples and found that miR-200 family was the most significant miRNA associated with PC1/EMT signature. Moreover, miR-200 overexpression was more abundant in tumors categorized as epithelial tumors, and its down-regulation was more evident in mesenchymal tumor category (Loboda et al., 2011).
In addition to the miR-200 family, other miRNAs were also involved in EMT process during CRC development. TGF-β treatment can trigger EMT transition in LIM 1863, a three-dimensional colon cancer organoid culture (Whitehead et al., 1987). Synergistic treatment of TNF-α with TGF-β can significantly accelerate the process of morphological changes, however, TNF-α alone had no effects (Bates and Mercurio, 2003). Cottonham et al. reported that the expression levels of miR-21 and miR-31 were significantly up-regulated during the EMT process in LIM 1863 induced by TGF-β/TNF-α treatment. Ectopic expression of miR-21 and miR-31 in LIM 1863 and other CRC cell lines increased their migratory and invasive activity, and had positive effects on TGF-β induced changes. Furthermore, the target of miR-21 and miR-31 mediating EMT was identified as T lymphoma invasion and metastasis 1 (TIAM1), a guanidine exchange factor for Rac GTPase. Overexpression of Rac GTPase antagonized the EMT process induced by TGF-β/TNF-α treatment and miR-21/miR-31 overexpression (Cottonham et al., 2010). These results suggest that certain miRNAs might act at the downstream of TGF-β signaling pathway to facilitate CRC invasion and metastasis.
Several other miRNAs such as miR-9 and miR-335 were reported to enhance cancer metastasis by inhibition of E-cadherin and SOX4 respectively (Tavazoie et al., 2008; Ma et al., 2010). Down-regulation of miR-31 has been shown to promote breast cancer cell migration, invasion, and metastasis, possibly through up-regulation of its targets, such as integrin α5, radixin, and Rho A (Valastyan et al., 2009, 2010). However, currently limited information was available on the impact of miRNAs in EMT in CRC development.
miRNAs and Colon Cancer Stem-Like Cells
Cancer stem-like cells (CSCs) are a small population of progenitor cells, which are distinct from other cells within a tumor mass based on morphology and function. The characteristics of CSCs are self-renewal, asymmetric division, slow proliferation, and differentiation into variable tumor cells (Clarke and Fuller, 2006). CSC-enriched populations have been isolated from CRCs based on their surface markers (CD44, CD133, Lgr5, aldehyde dehydrogenase; Ricci-Vitiani et al., 2007; Boman and Huang, 2008; Botchkina et al., 2009; Huang et al., 2009; Fan et al., 2011). This small population of cells exhibited several unique characteristics, such as formation of undifferentiated tumor spheres in serum-free media, differentiation into CRC cells in response to serum and growth factors and increased resistance to chemotherapeutic drugs in vitro, as well as rapid tumor formation in vivo, which contribute to the drug resistance, cancer metastasis, and tumor recurrence (Ricci-Vitiani et al., 2007). Due to the fundamental roles of miRNAs involved in the developmental process, we reasoned that certain miRNAs will contribute to the stem-like phenotype of CRC tumor cells and chemoresistance.
Previous studies from our laboratory have highlighted the association of miR-140 and miR-215 in colon CSCs (Song et al., 2009, 2010). Both of them can repress CRC cell proliferation and cause cell cycle arrest. CD133high/CD44high population displayed typical CSC characteristics, low proliferation, and highly resistant to chemotherapeutic drugs. After comparing miRNA levels between CRC stem cells and control parental cells, we demonstrated that miR-140 and miR-215 expression levels were increased by fourfold and threefold in CRC stem cells compared to control parental CRC cells respectively (Song et al., 2009, 2010). Differentially expressed miRNAs were also quantified in HT29 cells by isolating CRC stem-like cells with CD133 marker, which displayed a higher efficiency to form tumor spheres in vitro and tumors in nude mice. This population also showed elevated expression of stemness genes, such as Wnt2, BMI1, OCT3/4, and Notch 1. After profiling the miRNAs in CD133+ and CD133− population, 11 miRNAs were found to be up-regulated in CSC population, including miR-455-5p, miR-455-3p, miR-105, miR-1826, miR-155, miR-494, miR-16-2*, miR-744, miR-185, miR-423-5p, and miR-181b, and eight miRNAs were down-regulated, including miR-429, miR-548d-5p, miR-31, miR-151-5p, miR-151-3p, miR-221, miR-320, and miR-636 (Zhang et al., 2011). Among these, miR-429, a member of miR-200 family, and miR-155 has been reported to regulate CSC differentiation and EMT (Kong et al., 2008; Park et al., 2008).
Empirical evidence connecting EMT with the CSC phenotype has been previous demonstrated (Mani et al., 2008). It was reported that induction of EMT in immortalized human mammary epithelial cells (HMLEs) resulted in the acquisition of mesenchymal traits and the expression of stem-cell markers. Epithelial cells that have undergone EMT displayed stem-cell characteristics such as the abilities to form mammospheres, soft agar colonies, and tumors more efficiently. CSCs isolated from HMLE cultures express markers similar to HMLE cells that have undergone EMT. Thus, it has been suggested that EMT is able to generate cells with properties of stem cells. Furthermore, Brabletz et al. (2005) has reported that tumor cells at the tumor–host interface express EMT-associated genes as well as stemness-associated genes in CRC, suggesting a relationship between EMT and CSCs. It has also been demonstrated that many signaling pathways, such as Wnt, Notch, and Hedgehog are directly involved in regulating EMT and CSC self-renewal and maintenance (Wang et al., 2010c). Recent reports have described the involvement of several EMT-related miRNAs in the regulation of CSCs. One of the most prominent examples is miR-200c activated by p53, which suppresses the ability of normal mammary epithelial cells to form mammary ducts and breast cancer stem cells to form tumors in vivo (Shimono et al., 2009; Chang et al., 2011).
LIN28B, the repressor for let-7 biogenesis, was shown to be overexpressed in several cancer types (Guo et al., 2006; Viswanathan et al., 2009), and to induce pluripotency of somatic fibroblasts when co-expressed with OCT4, SOX2, and KLF4 (Yu et al., 2007b). In CRCs, let-7 level was decreased accompanied by increased level of LIN28B, which correlated with shorter patient survival and higher probability of tumor recurrence. Ectopic expression of LIN28B in immortalized intestinal epithelial cells (IEC-6) enhanced cell migration and invasion. More importantly, it switched normal epithelial cells to tumorigenic cells by soft agar colony formation assay. LIN28B also increased the migration and invasion of CRC cells, which was partially dependent on the inhibition of let-7 expression. Meanwhile, overexpression of LIN28B led to the increased expression of intestinal stem-cell marker LGR5 and PROM1. CRC cells with constitutive expression of LIN28B formed more tumors in vivo with higher metastasis as compared with parental cells, suggesting the dual roles of LIN28B in CRC development (King et al., 2011a,b).
miRNAs and CRC Diagnosis
The advantage of miRNAs as biomarkers is that they are highly stable in archival formalin fixed paraffin embedded (FFPE) tissues and body fluids (Xi et al., 2007; Chen et al., 2008; Mitchell et al., 2008; Hui et al., 2009). The common approaches for discovering miRNA based biomarkers is to quantify miRNAs from either fresh frozen or archival FFPE tumor tissues, and to calculate the miRNA ratio between normal and tumor tissue, and predict the patients’ prognosis and responses to chemotherapy. In addition, several new methods have been developed to detect miRNAs in plasma, serum and stool samples as non-invasive early screening approaches for CRC patients (Ng et al., 2009; Huang et al., 2010; Link et al., 2010; Cheng et al., 2011). The list of miRNAs as potential diagnostic, prognostic, and therapeutic candidates was summarized in Table 1.
Table 1.
Biomarker and therapeutic candidates of miRNAs in colorectal cancer.
| miRNAs | References |
|---|---|
| DIAGNOSTIC BIOMARKERS | |
| miR-106a | Link et al. (2010) |
| miR-141 | Cheng et al. (2011) |
| miR-17-3p | Ng et al. (2009) |
| miR-21 | Link et al. (2010), Cheng et al. (2011) |
| miR-29a | Huang et al. (2010) |
| miR-92 | Ng et al. (2009), Huang et al. (2010), Cheng et al. (2011) |
| PROGNOSTIC BIOMARKERS | |
| miR-106a | Diaz et al. (2008) |
| miR-141 | Cheng et al. (2011) |
| miR-143 | Kulda et al. (2010) |
| miR-200c | Xi et al. (2006a) |
| miR-21 | Slaby et al. (2007), Schetter et al. (2008), Kulda et al. (2010), Nielsen et al. (2011), Fassan et al. (2011) |
| miR-215 | Karaayvaz et al. (2011) |
| Let-7 and LIN28B | King et al. (2011a) |
| MODULATORS FOR CHEMOTHERAPY | |
| miR-140 | Song et al. (2009) |
| miR-143 | Borralho et al. (2009) |
| miR-192 | Song et al. (2008) |
| miR-215 | Song et al. (2010) |
| miR-22 | Li et al. (2011) |
| miR-31 | Wang et al. (2010b) |
| miR-34a | Akao et al. (2011) |
| miRNA REPLACEMENT THERAPY | |
| miR-143 | Akao et al. (2010) |
| miR-145 | Akao et al. (2010), Ibrahim et al. (2011) |
The ability to quantify miRNA expression from body fluids offers a potential huge advantage as a non-invasive approach for cancer diagnosis. Ng et al. (2009) first reported the quantification of miRNAs in the plasma from CRC patients by real-time qRT-PCR. The results showed that miR-135b, miR-95, miR-222, miR-17-3p, and miR-92 were increased in the plasma from CRC patients when compared with control normal plasma. The elevation of miR-17-3p and miR-92 was independently validated, and this elevation significantly decreased after surgical resection of primary tumors. Moreover, the elevation of miR-92 was able to distinguish CRCs from other diseases, including gastric cancer and inflammatory bowel disease. Similar results were obtained for miR-29a and miR-92a in CRC plasma (Huang et al., 2010), which highlighted these miRNAs as potential novel biomarkers for CRC early screening. However, before circulating miRNA based detection assay reaches to the clinical practice. A robust RNA extraction technique and normalization of miRNA expression from body fluids remained to be key factors to ensure the accuracy of the measurement.
Furthermore, the correlation between CRC stages and circulating plasma miRNAs was also analyzed. Cheng et al. quantified miR-21, miR-92, and miR-141 in two sets of CRC patients’ plasma cohorts by qRT-PCR, and found that only the miR-141 level was significantly correlated with CRC stages, although all of the three miRNAs showed differential expression in CRC plasma when compared to normal controls. Overexpression of miR-141 was highly associated with stage IV CRC with poor prognosis. It can distinguish stage IV CRC from other stages with 77.1% sensitivity and 89.7% specificity. Further, when a plasma test of miR-141 was combined with blood carcinoembryonic antigen (CEA) test, a common biomarker used for CRC diagnosis, the screening accuracy was further improved, providing a more accurate screening program for CRC patients (Cheng et al., 2011). The detailed mechanism of elevated miR-141 in plasma remained to be determined.
In addition to plasma tests, a recent study used stool samples from healthy person and CRC patients to analyze miRNA levels. They showed that the RNA extraction from stools was feasible and there were considerable amounts of miRNAs existing in the samples. The miRNA levels remained constant in fecal samples collected from the same person at different time. After comparing the miRNA profile from normal and CRC stool samples, miR-21 and miR-106a were found to be up-regulated in a CRC group when compared to normal controls, and their expression level decreased with advanced disease stages (Link et al., 2010). This study suggested that fecal miRNA detection could be a feasible method, and miR-21 and miR-106a are potential candidates for biomarkers in CRC screening. However, reliable control miRNAs have to be identified and fully validated. The stool samples are rather complex and a standard sample preparation needs to be established. Of note, none of these miRNA based biomarkers have yet made to clinical practice. Larger cohorts of CRC stool samples are needed for independent validation, especially due to the complex nature of the stool samples.
It has been observed that there was a lack of correlation between miRNAs that were up/down-regulated in tumor tissue with those identified in the serum or plasma CRC patients. The lack of correlation could be due to many reasons that need to be further investigated. The alterations of circulating miRNA levels may be an indication of pathological changes and therefore, might be possible as biomarkers. However, it can also be just an indirect indicator as one particular miRNA expression from tumor tissue can be indicative of disease while its expression in circulation is not relevant. The exact origin of miRNAs in circulation has not been thoroughly defined. One possibility is the release of miRNAs from lysed tumor cells packed in microvesicles (e.g., exosomes) against degradation. Recently it has been demonstrated that miRNAs can be transferred between cells through microvesicles (Valadi et al., 2007; Hunter et al., 2008). However, the exact mechanism of exosome formation and function of miRNAs in circulation still remained to be determined (Kosaka et al., 2010). Additional studies focusing on discovering tumor specific exosomes may contribute to our further understanding of the biological rules of circulating miRNAs.
miRNAs and CRC Prognosis
It is quite clear that miRNAs are involved in the tumorigenesis, disease progression, and fluoropyrimidine-based chemosensitivity of CRC. As a result, there is a great potential of miRNAs as prognostic biomarkers for CRC. The first report of miRNAs related to colon cancer prognosis was reported by Xi et al. as they compared expression levels of 10 miRNAs (let-7b, let-7g, miR-15b, miR-181b, miR-191, miR-200c, miR-26a, miR-27a, miR-30a-5p, and miR-30c) between 24 paired normal colon tissue and CRC specimens, and correlated with patient survival. Four miRNAs (miR-15b, miR-181b, miR-191, and miR-200c) were found to be up-regulated in CRC tumors, and miR-200c level was significantly correlated with CRC patient survival (Xi et al., 2006a). This study provides the clinical significance of miR-200c expression and CRC patient survival. Subsequent studies clearly demonstrated that miR-200c plays a key role in tumorigenesis and EMT transition by targeting key mRNA targets.
In addition to miR-200c, the miRNAs from miR-17-92 cluster (miR-17, miR-18a, miR-19a, miR-20a, miR-19b-1, miR-92a-1, and miR-106a) have been shown to be up-regulated in CRC tissue when compared with normal colon mucosa except miR-18a (Mendell, 2008; Diosdado et al., 2009). Moreover, the expression level of its target, E2F1, was inversely related to that of miR-17-92 cluster (O’Donnell et al., 2005; Woods et al., 2007). The expression level of miR-106a was associated with CRC stages, and its reduced expression was correlated with shorter survival. However, overexpression of miR-17-5p predicted shorter survival only at early stages (Diaz et al., 2008).
It has been proposed that miR-21 acts as a potential oncogene. The expression of miR-21 was up-regulated in CRC tissue with a significant association with CRC prognosis (Slaby et al., 2007; Schetter et al., 2008; Kulda et al., 2010; Fassan et al., 2011). Patients with higher miR-21 levels had shorter disease-free interval (DFI) when compared with those with lower levels (Kulda et al., 2010). Moreover, miR-21 was found be expressed mainly in fibroblast-like cells in the stroma of stage II CRC by in situ hybridization, and the overexpression of miR-21 correlated with shorter patient survival in stage II colon cancer but not in stage II rectal cancer (Nielsen et al., 2011). Accordingly, the target of miR-21, programmed cell death 4 (PDCD4), was inversely correlated with miR-21 levels with a lower nuclear staining of PDCD4 in CRC specimens than that of normal controls (Fassan et al., 2011). In contrast, miR-143, a potential tumor suppressor and down-regulated in CRC, was found to be inversely correlated with prognosis. Patients with higher level of miR-143 exhibited shorter DFI (Kulda et al., 2010). These results suggest that due to the impact of miRNA on many gene pathways and targets, the function of miRNA may be far more complex than simple definition of oncogene or tumor suppressor gene.
The repressors of miRNAs were also shown to be associated with CRC prognosis, such as LIN28B. King et al. (2011a) quantified LIN28B expression levels in CRC samples by immunohistochemistry, and discovered that stage I and II tumors had lower LIN28B staining intensity, which then was significantly correlated with longer patient survival and less chance of cancer recurrence, indicating the indirect role of miRNAs in the prediction of CRC prognosis.
It is established that more than 80% of stage II and 65% of stage III CRC patients can be cured by surgery alone without adjuvant therapy (McLornan et al., 2010). Moreover, the 5-year survival rates for stage II (78.5%) and stage III (54%) colon cancer patients are much higher than stage IV patients (8.1%; O’Connell et al., 2004). Therefore, it is crucial to investigate the prognosis potential of miRNAs in stage II and III colon cancer both to spare patients who can be cured by surgery alone from unnecessary chemotherapy treatment, and to target a population with long term survival with surgery and chemotherapy treatment. Previously our laboratory has shown the critical functions of miR-215 in CRC cell lines (Song et al., 2010). Recently, we further demonstrated the clinical significance of miR-215 mediated DTL expression in 34 paired specimens of stage II and stage III CRC patients prior to chemotherapy treatment. miR-215 levels were significantly decreased in colon cancer specimens compared to adjacent normal tissues, and its expression was associated with patient survival. Patients with low levels of miR-215 tended to have longer survival than patients with high levels of miR-215. Moreover, DTL, the target of miR-215, was found to be increasingly expressed in CRC specimens both on mRNA and protein levels, which was inversely correlated to miR-215 level, suggesting both molecules as potential novel biomarkers for CRC patient prognosis (Karaayvaz et al., 2011).
miRNAs and CRC Therapeutics
The major obstacle for the success of chemotherapy is due to the anti-cancer drug resistance (Longley et al., 2006). Many studies have shown that miRNAs can modulate the response of CRC cells to chemotherapeutic drugs.
To understand the underlying mechanisms responsible for drug resistance, we began to focus on the potential regulatory impact of miRNAs on two important genes, dihydrofolate reductase (DHFR) and thymidylate synthase (TS). DHFR catalyzes the reduction of dihydrofolate to tetrahydrofolate as the one-carbon donor essential for the de novo synthesis of thymidylate (dTMP), a precursor for DNA synthesis (Banerjee et al., 2002). Methotrexate (MTX) is used as antifolate-based chemotherapy to inhibit the activity of DHFR. TS catalyzes the reductive methylation of dUMP to dTMP (Carreras and Santi, 1995). 5-fluorouracil (5-FU) as fluoropyrimidine-based chemotherapy or tomudex (TDX) as antifolate-based chemotherapy is used to inhibit the activity of TS. Due to their critical functions, both DHFR and TS have been the major anti-cancer targets for the past 50 years (Salonga et al., 2000; Johnston et al., 2003; Wang et al., 2007). Recently, studies from our laboratory have shed light on miRNAs and chemoresistance mediated by DHFR and TS. miR-192 was shown to repress CRC cell proliferation and cell cycle arrest, which was dependent on wild-type p53 expression. Overexpression of miR-192 inhibited DHFR protein level by translational arrest, and induced increased chemosensitivity of CRC cells to MTX (Song et al., 2008). In the case of miR-215, we demonstrated that the expression of both TS and DHFR were suppressed by miR-215. However, despite this, the overexpression of miR-215 also counter-intuitively decreased chemosensitivity to MTX and TDX. This effect was due to reduced cell proliferation and increased G2-cell cycle arrest mediated by miR-215 by suppression of DTL, which rendered colon cancer cells less sensitive to MTX and TDX treatment (Song et al., 2010). Similar impact on chemoresistance was also found in miR-140, and its overexpression in CRC cells led to increased chemoresistance to MTX and 5-FU, which in part, was due to the reduced cell proliferation by the inhibition of histone deacetylase 4 (HDAC4) and the accumulation of CSC population. The knockdown of miR-140 expression in CRC stem cells restored their chemosensitivity to 5-FU (Song et al., 2009), indicating that miRNAs and their inhibitors can be potential candidates for adjuvant therapy in addition to chemotherapy. In another report, the inhibition of miR-31 in CRC cells was shown to decrease the chemoresistance to 5-FU (Wang et al., 2010b).
On the other hand, miRNAs with tumor suppressor like functions were also shown to modulate chemosensitivity in CRC. For example, the overexpression of miR-143 in CRC cell lines increased the chemosensitivity to 5-FU possibly through ERK5/NF-κB signaling pathway (Borralho et al., 2009). miR-34a level was found to be decreased in 5-FU-resistant DLD-1 CRC cells compared to the parental cells, while its target, SIRT1, was found to be elevated. The p53 and miR-34a positive feedback loop was silenced in the 5-FU-resistant cells. The overexpression of miR-34a in the resistant cells sensitized them to 5-FU treatment, which was mediated by inhibition of SIRT1 and E2F3 signaling pathways (Akao et al., 2011). This is quite different from the p53 mediated miR-215 on chemosensitivity as overexpressed miR-215 caused cell cycle arrest without triggering significant cell death as miR-34 does. Moreover, overexpression of miR-22 in p53-mutated CRC cells led to repression of cell proliferation, induction of apoptosis and increased chemosensitivity to paclitaxel. When miR-22 level was increased, phosphatase and tensin homolog (PTEN) expression was also elevated, and it resulted in the inhibition of Akt activation, reduction of metadherin (MTDH), and increased level of Bax and activated caspase-3, which contributed to higher apoptotic activity and lower chemoresistance to paclitaxel (Li et al., 2011).
In addition to the impact on chemosensitivity, miRNAs and their inhibitors have been tested in several in vivo animal models as a therapeutic agents based on their intrinsic tumor suppressive and oncogenic functions. For example, let-7 miRNAs have been shown to be reduced in many cancer types, and proposed as tumor suppressors since they inhibited multiple oncogenes (RAS, MYC, HMGA2) and key regulators of cell cycle control genes (CDC25A, CDK6, cyclin D2; Takamizawa et al., 2004; Johnson et al., 2005, 2007; Lee and Dutta, 2007; Mayr et al., 2007; Park et al., 2007; Sampson et al., 2007; Yu et al., 2007a). Ectopic expression of let-7 miRNAs in lung cancer cells and breast cancer cells has been shown to prevent or reduce the tumor formation in mouse models (Johnson et al., 2007; Yu et al., 2007a; Esquela-Kerscher et al., 2008; Kumar et al., 2008), and administration of synthetic let-7 precursor to the established tumors in mice led to the tumor regression (Trang et al., 2010). The latter study suggested miRNA replacement as a novel therapeutic strategy for cancer patients. However, the miRNA based anti-cancer therapy will likely face the some of the same technical hurdles of siRNA based therapy.
miR-143 and miR-145 have been extensively studied in CRCs, and as potential candidates for CRC treatment. Both miRNAs have been shown to be down-regulated in many cancer types (Akao et al., 2006), especially in CRCs (Michael et al., 2003; Akao et al., 2007; Slaby et al., 2007), and ectopic expression of miR-143 or miR-145 suppressed tumor cell proliferation and triggered apoptosis of tumor cells in vitro and in vivo (Shi et al., 2007; Akao et al., 2009, 2010; Sachdeva et al., 2009). Among the identified targets for both miRNAs in CRCs, there are several important oncogenes and transcription factors such as ERK5 and KRAS for miR-143 (Akao et al., 2006, 2009, 2010; Chen et al., 2009; Gao et al., 2011), c-myc, insulin receptor substrate 1 (IRS1), YES, and STAT1 for miR-145 (Shi et al., 2007; La Rocca et al., 2009; Sachdeva et al., 2009; Gregersen et al., 2010). In mouse CRC xenograft models, miR-145 was delivered systemically or locally to the tumors mediated by polyethylenimine (PEI), and both methods led to decreased tumor burden, up-regulation of apoptosis and repression of miR-145 targets, c-myc, and ERK5 (Ibrahim et al., 2011). To increase the resistance to nucleases, chemically modified miR-143 was locally delivered to human CRC xenografts via cationic liposomes. The synthetic miR-143 had the addition of aromatic benzene-pyridine like structure to its 3′-overhang region and sequence changes in the passenger strand in miR-143 duplex, which showed higher stability against nuclease digestion and greater anti-tumor activity when compared with endogenous miR-143 (Akao et al., 2010; Kitade and Akao, 2010). These results suggest that synthetic miRNAs may be potential candidates for the development of CRC RNA replacement therapies. These results establish a foundation for the pursuit of therapeutic potentials for both miR-143 and miR-145 in CRCs. With the continued advancement of in vivo delivery of miRNA and its mimics, the therapeutic application of miRNAs for cancer treatment is on the horizon.
Conclusion
Given the significant role of miRNAs in many aspects of CRC development such as proliferation, cell cycle control, invasion, EMT, and maintained tumor stem-cell phenotype, we remain hopeful that miRNA based therapeutics, diagnosis, and prognosis may emerge in the near future to benefit patients. Multi-center, large patient cohort studies are clearly needed to fully validate the clinical values of miRNA as biomarkers for CRCs. With the progress in technology of miRNA detection and quantification such as RNA-Seq, personalized medicine based on miRNA biomarkers will eventually emerge as new tools for the CRC disease management due to the superior stability, broad impact on gene pathways and targets.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We apologize to our colleagues whose research was not cited in this review due to the focus, space limitations and timing. We appreciate the critical review from Ms. Sonya Lorrain and suggestions from our reviewers. This study was supported in part by Stony Brook University Translational Research Laboratory Start-up fund (J. Ju), R01CA155019 (J. Ju) and R33CA147966 (J. Ju).
References
- Akao Y., Nakagawa Y., Hirata I., Iio A., Itoh T., Kojima K., Nakashima R., Kitade Y., Naoe T. (2010). Role of anti-oncomirs miR-143 and -145 in human colorectal tumors. Cancer Gene Ther. 17, 398–408 10.1038/cgt.2009.88 [DOI] [PubMed] [Google Scholar]
- Akao Y., Nakagawa Y., Iio A., Naoe T. (2009). Role of microRNA-143 in Fas-mediated apoptosis in human T-cell leukemia Jurkat cells. Leuk. Res. 33, 1530–1538 10.1016/j.leukres.2009.04.019 [DOI] [PubMed] [Google Scholar]
- Akao Y., Nakagawa Y., Naoe T. (2006). MicroRNAs 143 and 145 are possible common onco-microRNAs in human cancers. Oncol. Rep. 16, 845–850 [PubMed] [Google Scholar]
- Akao Y., Nakagawa Y., Naoe T. (2007). MicroRNA-143 and -145 in colon cancer. DNA Cell Biol. 26, 311–320 10.1089/dna.2006.0550 [DOI] [PubMed] [Google Scholar]
- Akao Y., Noguchi S., Iio A., Kojima K., Takagi T., Naoe T. (2011). Dysregulation of microRNA-34a expression causes drug-resistance to 5-FU in human colon cancer DLD-1 cells. Cancer Lett. 300, 197–204 10.1016/j.canlet.2010.10.006 [DOI] [PubMed] [Google Scholar]
- Alwan A. (2007). World Health Organization. Disaster Med. Public. Health Prep. 1, 7–8 10.1097/DMP.0b013e3181514969 [DOI] [PubMed] [Google Scholar]
- Bandres E., Cubedo E., Agirre X., Malumbres R., Zarate R., Ramirez N., Abajo A., Navarro A., Moreno I., Monzo M., Garcia-Foncillas J. (2006). Identification by real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol. Cancer 5, 29. 10.1186/1476-4598-5-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee D., Mayer-Kuckuk P., Capiaux G., Budak-Alpdogan T., Gorlick R., Bertino J. R. (2002). Novel aspects of resistance to drugs targeted to dihydrofolate reductase and thymidylate synthase. Biochim. Biophys. Acta 1587, 164–173 [DOI] [PubMed] [Google Scholar]
- Banks D., Wu M., Higa L. A., Gavrilova N., Quan J., Ye T., Kobayashi R., Sun H., Zhang H. (2006). L2DTL/CDT2 and PCNA interact with p53 and regulate p53 polyubiquitination and protein stability through MDM2 and CUL4A/DDB1 complexes. Cell Cycle 5, 1719–1729 10.4161/cc.5.15.3150 [DOI] [PubMed] [Google Scholar]
- Bates R. C., Mercurio A. M. (2003). Tumor necrosis factor-alpha stimulates the epithelial-to-mesenchymal transition of human colonic organoids. Mol. Biol. Cell 14, 1790–1800 10.1091/mbc.E02-09-0583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boman B. M., Huang E. (2008). Human colon cancer stem cells: a new paradigm in gastrointestinal oncology. J. Clin. Oncol. 26, 2828–2838 10.1200/JCO.2008.17.7436 [DOI] [PubMed] [Google Scholar]
- Borralho P. M., Kren B. T., Castro R. E., Da Silva I. B., Steer C. J., Rodrigues C. M. (2009). MicroRNA-143 reduces viability and increases sensitivity to 5-fluorouracil in HCT116 human colorectal cancer cells. FEBS J. 276, 6689–6700 10.1111/j.1742-4658.2009.07383.x [DOI] [PubMed] [Google Scholar]
- Botchkina I. L., Rowehl R. A., Rivadeneira D. E., Karpeh M. S., Jr., Crawford H., Dufour A., Ju J., Wang Y., Leyfman Y., Botchkina G. I. (2009). Phenotypic subpopulations of metastatic colon cancer stem cells: genomic analysis. Cancer Genomics Proteomics 6, 19–29 [PubMed] [Google Scholar]
- Brabletz T., Jung A., Spaderna S., Hlubek F., Kirchner T. (2005). Opinion: migrating cancer stem cells – an integrated concept of malignant tumour progression. Nat. Rev. Cancer 5, 744–749 10.1038/nrc1694 [DOI] [PubMed] [Google Scholar]
- Bracken C. P., Gregory P. A., Kolesnikoff N., Bert A. G., Wang J., Shannon M. F., Goodall G. J. (2008). A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 68, 7846–7854 10.1158/0008-5472.CAN-08-1942 [DOI] [PubMed] [Google Scholar]
- Braun C. J., Zhang X., Savelyeva I., Wolff S., Moll U. M., Schepeler T., Orntoft T. F., Andersen C. L., Dobbelstein M. (2008). p53-Responsive micrornas 192 and 215 are capable of inducing cell cycle arrest. Cancer Res. 68, 10094–10104 10.1158/0008-5472.CAN-08-1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J., Hipfner D. R., Stark A., Russell R. B., Cohen S. M. (2003). Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113, 25–36 10.1016/S0092-8674(03)00231-9 [DOI] [PubMed] [Google Scholar]
- Burk U., Schubert J., Wellner U., Schmalhofer O., Vincan E., Spaderna S., Brabletz T. (2008). A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 9, 582–589 10.1038/embor.2008.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreras C. W., Santi D. V. (1995). The catalytic mechanism and structure of thymidylate synthase. Annu. Rev. Biochem. 64, 721–762 10.1146/annurev.bi.64.070195.003445 [DOI] [PubMed] [Google Scholar]
- Chan J. A., Krichevsky A. M., Kosik K. S. (2005). MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 65, 6029–6033 10.1158/0008-5472.CAN-05-0137 [DOI] [PubMed] [Google Scholar]
- Chang C. J., Chao C. H., Xia W., Yang J. Y., Xiong Y., Li C. W., Yu W. H., Rehman S. K., Hsu J. L., Lee H. H., Liu M., Chen C. T., Yu D., Hung M. C. (2011). p53 Regulates epithelial-mesenchymal transition and stem cell properties through modulating miRNAs. Nat. Cell Biol. 13, 317–323 10.1038/ncb2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang T. C., Wentzel E. A., Kent O. A., Ramachandran K., Mullendore M., Lee K. H., Feldmann G., Yamakuchi M., Ferlito M., Lowenstein C. J., Arking D. E., Beer M. A., Maitra A., Mendell J. T. (2007). Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol. Cell 26, 745–752 10.1016/j.molcel.2007.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Ba Y., Ma L., Cai X., Yin Y., Wang K., Guo J., Zhang Y., Chen J., Guo X., Li Q., Li X., Wang W., Wang J., Jiang X., Xiang Y., Xu C., Zheng P., Zhang J., Li R., Zhang H., Shang X., Gong T., Ning G., Zen K., Zhang C. Y. (2008). Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 18, 997–1006 10.1038/cr.2008.66 [DOI] [PubMed] [Google Scholar]
- Chen X., Guo X., Zhang H., Xiang Y., Chen J., Yin Y., Cai X., Wang K., Wang G., Ba Y., Zhu L., Wang J., Yang R., Zhang Y., Ren Z., Zen K., Zhang J., Zhang C. Y. (2009). Role of miR-143 targeting KRAS in colorectal tumorigenesis. Oncogene 28, 1385–1392 10.1038/onc.2009.55 [DOI] [PubMed] [Google Scholar]
- Cheng H., Zhang L., Cogdell D. E., Zheng H., Schetter A. J., Nykter M., Harris C. C., Chen K., Hamilton S. R., Zhang W. (2011). Circulating plasma MiR-141 is a novel biomarker for metastatic colon cancer and predicts poor prognosis. PLoS ONE 6, e17745. 10.1371/journal.pone.0017745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke M. F., Fuller M. (2006). Stem cells and cancer: two faces of eve. Cell 124, 1111–1115 10.1016/j.cell.2006.03.011 [DOI] [PubMed] [Google Scholar]
- Cottonham C. L., Kaneko S., Xu L. (2010). miR-21 and miR-31 converge on TIAM1 to regulate migration and invasion of colon carcinoma cells. J. Biol. Chem. 285, 35293–35302 10.1074/jbc.M110.160069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford L. V., Pim D. C., Lamb P. (1984). The cellular protein p53 in human tumours. Mol. Biol. Med. 2, 261–272 [PubMed] [Google Scholar]
- Diaz R., Silva J., Garcia J. M., Lorenzo Y., Garcia V., Pena C., Rodriguez R., Munoz C., Garcia F., Bonilla F., Dominguez G. (2008). Deregulated expression of miR-106a predicts survival in human colon cancer patients. Genes Chromosomes Cancer 47, 794–802 10.1002/gcc.20580 [DOI] [PubMed] [Google Scholar]
- Diosdado B., Van De Wiel M. A., Terhaar Sive Droste J. S., Mongera S., Postma C., Meijerink W. J., Carvalho B., Meijer G. A. (2009). MiR-17-92 cluster is associated with 13q gain and c-myc expression during colorectal adenoma to adenocarcinoma progression. Br. J. Cancer 101, 707–714 10.1038/sj.bjc.6605037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong P., Kaneuchi M., Watari H., Hamada J., Sudo S., Ju J., Sakuragi N. (2011). MicroRNA-194 inhibits epithelial to mesenchymal transition of endometrial cancer cells by targeting oncogene BMI-1. Mol. Cancer 10, 99. 10.1186/1476-4598-10-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupaul-Chicoine J., Yeretssian G., Doiron K., Bergstrom K. S., Mcintire C. R., Leblanc P. M., Meunier C., Turbide C., Gros P., Beauchemin N., Vallance B. A., Saleh M. (2010). Control of intestinal homeostasis, colitis, and colitis-associated colorectal cancer by the inflammatory caspases. Immunity 32, 367–378 10.1016/j.immuni.2010.02.012 [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A., Trang P., Wiggins J. F., Patrawala L., Cheng A., Ford L., Weidhaas J. B., Brown D., Bader A. G., Slack F. J. (2008). The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle 7, 759–764 10.4161/cc.7.6.5834 [DOI] [PubMed] [Google Scholar]
- Fan X., Ouyang N., Teng H., Yao H. (2011). Isolation and characterization of spheroid cells from the HT29 colon cancer cell line. Int. J. Colorectal Dis. 26, 1279–1285 10.1007/s00384-011-1248-y [DOI] [PubMed] [Google Scholar]
- Fassan M., Pizzi M., Giacomelli L., Mescoli C., Ludwig K., Pucciarelli S., Rugge M. (2011). PDCD4 nuclear loss inversely correlates with miR-21 levels in colon carcinogenesis. Virchows Arch. 458, 413–419 10.1007/s00428-011-1046-5 [DOI] [PubMed] [Google Scholar]
- Feagins L. A., Souza R. F., Spechler S. J. (2009). Carcinogenesis in IBD: potential targets for the prevention of colorectal cancer. Nat. Rev. Gastroenterol. Hepatol. 6, 297–305 10.1038/nrgastro.2009.44 [DOI] [PubMed] [Google Scholar]
- Fu L., Minden M. D., Benchimol S. (1996). Translational regulation of human p53 gene expression. EMBO J. 15, 4392–4401 [PMC free article] [PubMed] [Google Scholar]
- Gao J. S., Zhang Y., Tang X., Tucker L. D., Tarwater P. M., Quesenberry P. J., Rigoutsos I., Ramratnam B. (2011). The Evi1, microRNA-143, K-Ras axis in colon cancer. FEBS Lett. 585, 693–699 10.1016/j.febslet.2011.01.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georges S. A., Biery M. C., Kim S. Y., Schelter J. M., Guo J., Chang A. N., Jackson A. L., Carleton M. O., Linsley P. S., Cleary M. A., Chau B. N. (2008). Coordinated regulation of cell cycle transcripts by p53-Inducible microRNAs, miR-192 and miR-215. Cancer Res. 68, 10105–10112 10.1158/0008-5472.CAN-08-1846 [DOI] [PubMed] [Google Scholar]
- Ghodgaonkar M. M., Shah R. G., Kandan-Kulangara F., Affar E. B., Qi H. H., Wiemer E., Shah G. M. (2009). Abrogation of DNA vector-based RNAi during apoptosis in mammalian cells due to caspase-mediated cleavage and inactivation of Dicer-1. Cell Death Differ. 16, 858–868 10.1038/cdd.2009.15 [DOI] [PubMed] [Google Scholar]
- Gregersen L. H., Jacobsen A. B., Frankel L. B., Wen J., Krogh A., Lund A. H. (2010). MicroRNA-145 targets YES and STAT1 in colon cancer cells. PLoS ONE 5, e8836. 10.1371/journal.pone.0008836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory P. A., Bert A. G., Paterson E. L., Barry S. C., Tsykin A., Farshid G., Vadas M. A., Khew-Goodall Y., Goodall G. J. (2008a). The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 10, 593–601 10.1038/ncb1722 [DOI] [PubMed] [Google Scholar]
- Gregory P. A., Bracken C. P., Bert A. G., Goodall G. J. (2008b). MicroRNAs as regulators of epithelial-mesenchymal transition. Cell Cycle 7, 3112–3118 10.4161/cc.7.20.6851 [DOI] [PubMed] [Google Scholar]
- Gregory P. A., Bracken C. P., Smith E., Bert A. G., Wright J. A., Roslan S., Morris M., Wyatt L., Farshid G., Lim Y. Y., Lindeman G. J., Shannon M. F., Drew P. A., Khew-Goodall Y., Goodall G. J. (2011). An autocrine TGF-beta/ZEB/miR-200 signaling network regulates establishment and maintenance of epithelial-mesenchymal transition. Mol. Biol. Cell 22, 1686–1698 10.1091/mbc.E11-02-0103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Chen Y., Ito H., Watanabe A., Ge X., Kodama T., Aburatani H. (2006). Identification and characterization of lin-28 homolog B (LIN28B) in human hepatocellular carcinoma. Gene 384, 51–61 10.1016/j.gene.2006.07.011 [DOI] [PubMed] [Google Scholar]
- Gutman M., Fidler I. J. (1995). Biology of human colon cancer metastasis. World J. Surg. 19, 226–234 10.1007/BF00308631 [DOI] [PubMed] [Google Scholar]
- Hanada T., Kobayashi T., Chinen T., Saeki K., Takaki H., Koga K., Minoda Y., Sanada T., Yoshioka T., Mimata H., Kato S., Yoshimura A. (2006). IFNgamma-dependent, spontaneous development of colorectal carcinomas in SOCS1-deficient mice. J. Exp. Med. 203, 1391–1397 10.1084/jem.20060436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L., Hannon G. J. (2004). MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 5, 522–531 10.1038/nrg1379 [DOI] [PubMed] [Google Scholar]
- He L., He X., Lim L. P., De Stanchina E., Xuan Z., Liang Y., Xue W., Zender L., Magnus J., Ridzon D., Jackson A. L., Linsley P. S., Chen C., Lowe S. W., Cleary M. A., Hannon G. J. (2007). A microRNA component of the p53 tumour suppressor network. Nature 447, 1130–1134 10.1038/nature05939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L., Thomson J. M., Hemann M. T., Hernando-Monge E., Mu D., Goodson S., Powers S., Cordon-Cardo C., Lowe S. W., Hannon G. J., Hammond S. M. (2005). A microRNA polycistron as a potential human oncogene. Nature 435, 828–833 10.1038/nature03552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde S. R., Sun W., Lynch J. P. (2008). Systemic and targeted therapy for advanced colon cancer. Expert Rev. Gastroenterol. Hepatol. 2, 135–149 10.1586/17474124.2.1.135 [DOI] [PubMed] [Google Scholar]
- Higa L. A., Banks D., Wu M., Kobayashi R., Sun H., Zhang H. (2006). L2DTL/CDT2 interacts with the CUL4/DDB1 complex and PCNA and regulates CDT1 proteolysis in response to DNA damage. Cell Cycle 5, 1675–1680 10.4161/cc.5.1.2266 [DOI] [PubMed] [Google Scholar]
- Huang E. H., Hynes M. J., Zhang T., Ginestier C., Dontu G., Appelman H., Fields J. Z., Wicha M. S., Boman B. M. (2009). Aldehyde dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (SC) and tracks SC overpopulation during colon tumorigenesis. Cancer Res. 69, 3382–3389 10.1158/0008-5472.CAN-08-3413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z., Huang D., Ni S., Peng Z., Sheng W., Du X. (2010). Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int. J. Cancer 127, 118–126 10.1002/ijc.25007 [DOI] [PubMed] [Google Scholar]
- Hui A. B., Shi W., Boutros P. C., Miller N., Pintilie M., Fyles T., Mccready D., Wong D., Gerster K., Waldron L., Jurisica I., Penn L. Z., Liu F. F. (2009). Robust global micro-RNA profiling with formalin-fixed paraffin-embedded breast cancer tissues. Lab. Invest. 89, 597–606 10.1038/labinvest.2009.46 [DOI] [PubMed] [Google Scholar]
- Hunter M. P., Ismail N., Zhang X., Aguda B. D., Lee E. J., Yu L., Xiao T., Schafer J., Lee M. L., Schmittgen T. D., Nana-Sinkam S. P., Jarjoura D., Marsh C. B. (2008). Detection of microRNA expression in human peripheral blood microvesicles. PLoS ONE 3, e3694. 10.1371/journal.pone.0003694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang H. W., Mendell J. T. (2006). MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br. J. Cancer 94, 776–780 10.1038/sj.bjc.6603023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim A. F., Weirauch U., Thomas M., Grunweller A., Hartmann R. K., Aigner A. (2011). MicroRNA replacement therapy for miR-145 and miR-33a is efficacious in a model of colon carcinoma. Cancer Res. 71, 5214–5224 10.1158/0008-5472.CAN-10-4645 [DOI] [PubMed] [Google Scholar]
- Johnson C. D., Esquela-Kerscher A., Stefani G., Byrom M., Kelnar K., Ovcharenko D., Wilson M., Wang X., Shelton J., Shingara J., Chin L., Brown D., Slack F. J. (2007). The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 67, 7713–7722 10.1158/0008-5472.CAN-07-1195 [DOI] [PubMed] [Google Scholar]
- Johnson S. M., Grosshans H., Shingara J., Byrom M., Jarvis R., Cheng A., Labourier E., Reinert K. L., Brown D., Slack F. J. (2005). RAS is regulated by the let-7 microRNA family. Cell 120, 635–647 10.1016/j.cell.2005.01.014 [DOI] [PubMed] [Google Scholar]
- Johnston P. G., Benson A. B., III, Catalano P., Rao M. S., O’Dwyer P. J., Allegra C. J. (2003). Thymidylate synthase protein expression in primary colorectal cancer: lack of correlation with outcome and response to fluorouracil in metastatic disease sites. J. Clin. Oncol. 21, 815–819 10.1200/JCO.2003.10.064 [DOI] [PubMed] [Google Scholar]
- Karaayvaz M., Pal T., Song B., Zhang C., Georgakopoulos P., Mehmood S., Burke S., Shroyer K., Ju J. (2011). Prognostic significance of miR-215 in colon cancer. Clin. Colorectal Cancer. [Epub ahead of print]. 10.1016/j.clcc.2011.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M., Zhang J., Wang M., Lanting L., Yuan H., Rossi J. J., Natarajan R. (2007). MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of E-box repressors. Proc. Natl. Acad. Sci. U.S.A. 104, 3432–3437 10.1073/pnas.0704462104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King C. E., Cuatrecasas M., Castells A., Sepulveda A. R., Lee J. S., Rustgi A. K. (2011a). LIN28B promotes colon cancer progression and metastasis. Cancer Res. 71, 4260–4268 10.1158/0008-5472.CAN-10-4637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King C. E., Wang L., Winograd R., Madison B. B., Mongroo P. S., Johnstone C. N., Rustgi A. K. (2011b). LIN28B fosters colon cancer migration, invasion and transformation through let-7-dependent and -independent mechanisms. Oncogene 30, 4185–4193 10.1038/onc.2011.131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitade Y., Akao Y. (2010). MicroRNAs and their therapeutic potential for human diseases: microRNAs, miR-143 and -145, function as anti-oncomirs and the application of chemically modified miR-143 as an anti-cancer drug. J. Pharmacol. Sci. 114, 276–280 10.1254/jphs.10R12FM [DOI] [PubMed] [Google Scholar]
- Kong W., Yang H., He L., Zhao J. J., Coppola D., Dalton W. S., Cheng J. Q. (2008). MicroRNA-155 is regulated by the transforming growth factor beta/Smad pathway and contributes to epithelial cell plasticity by targeting RhoA. Mol. Cell. Biol. 28, 6773–6784 10.1128/MCB.00941-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaka N., Iguchi H., Yoshioka Y., Takeshita F., Matsuki Y., Ochiya T. (2010). Secretory mechanisms and intercellular transfer of microRNAs in living cells. J. Biol. Chem. 285, 17442–17452 10.1074/jbc.M110.107821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulda V., Pesta M., Topolcan O., Liska V., Treska V., Sutnar A., Rupert K., Ludvikova M., Babuska V., Holubec L., Jr., Cerny R. (2010). Relevance of miR-21 and miR-143 expression in tissue samples of colorectal carcinoma and its liver metastases. Cancer Genet. Cytogenet. 200, 154–160 10.1016/j.cancergencyto.2010.04.015 [DOI] [PubMed] [Google Scholar]
- Kulshreshtha R., Ferracin M., Wojcik S. E., Garzon R., Alder H., Agosto-Perez F. J., Davuluri R., Liu C. G., Croce C. M., Negrini M., Calin G. A., Ivan M. (2007). A microRNA signature of hypoxia. Mol. Cell. Biol. 27, 1859–1867 10.1128/MCB.01395-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M. S., Erkeland S. J., Pester R. E., Chen C. Y., Ebert M. S., Sharp P. A., Jacks T. (2008). Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc. Natl. Acad. Sci. U.S.A. 105, 3903–3908 10.1073/pnas.0711480105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rocca G., Badin M., Shi B., Xu S. Q., Deangelis T., Sepp-Lorenzinoi L., Baserga R. (2009). Mechanism of growth inhibition by MicroRNA 145: the role of the IGF-I receptor signaling pathway. J. Cell. Physiol. 220, 485–491 10.1002/jcp.21796 [DOI] [PubMed] [Google Scholar]
- Lee R. C., Feinbaum R. L., Ambros V. (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854 10.1016/0092-8674(93)90529-Y [DOI] [PubMed] [Google Scholar]
- Lee Y. S., Dutta A. (2007). The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 21, 1025–1030 10.1101/gad.1540407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Zhang Y., Zhao J., Kong F., Chen Y. (2011). Overexpression of miR-22 reverses paclitaxel-induced chemoresistance through activation of PTEN signaling in p53-mutated colon cancer cells. Mol. Cell. Biochem. 357, 31–38 10.1007/s11010-011-0916-0 [DOI] [PubMed] [Google Scholar]
- Link A., Balaguer F., Shen Y., Nagasaka T., Lozano J. J., Boland C. R., Goel A. (2010). Fecal MicroRNAs as novel biomarkers for colon cancer screening. Cancer Epidemiol. Biomarkers Prev. 19, 1766–1774 10.1158/1055-9965.EPI-10-0027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loboda A., Nebozhyn M. V., Watters J. W., Buser C. A., Shaw P. M., Huang P. S., Van’t Veer L., Tollenaar R. A., Jackson D. B., Agrawal D., Dai H., Yeatman T. J. (2011). EMT is the dominant program in human colon cancer. BMC Med. Genomics 4, 9. 10.1186/1755-8794-4-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longley D. B., Allen W. L., Johnston P. G. (2006). Drug resistance, predictive markers and pharmacogenomics in colorectal cancer. Biochim. Biophys. Acta 1766, 184–196 [DOI] [PubMed] [Google Scholar]
- Ma L., Young J., Prabhala H., Pan E., Mestdagh P., Muth D., Teruya-Feldstein J., Reinhardt F., Onder T. T., Valastyan S., Westermann F., Speleman F., Vandesompele J., Weinberg R. A. (2010). miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat. Cell Biol. 12, 247–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani S. A., Guo W., Liao M. J., Eaton E. N., Ayyanan A., Zhou A. Y., Brooks M., Reinhard F., Zhang C. C., Shipitsin M., Campbell L. L., Polyak K., Brisken C., Yang J., Weinberg R. A. (2008). The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 133, 704–715 10.1016/j.cell.2008.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C., Hemann M. T., Bartel D. P. (2007). Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science 315, 1576–1579 10.1126/science.1137999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLornan D. P., Barrett H. L., Cummins R., Mcdermott U., Mcdowell C., Conlon S. J., Coyle V. M., Van Schaeybroeck S., Wilson R., Kay E. W., Longley D. B., Johnston P. G. (2010). Prognostic significance of TRAIL signaling molecules in stage II and III colorectal cancer. Clin. Cancer Res. 16, 3442–3451 10.1158/1078-0432.CCR-10-0052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell J. T. (2008). miRiad roles for the miR-17-92 cluster in development and disease. Cell 133, 217–222 10.1016/j.cell.2008.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael M. Z., Sm O. C., Van Holst Pellekaan N. G., Young G. P., James R. J. (2003). Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol. Cancer Res. 1, 882–891 [PubMed] [Google Scholar]
- Miller S. J., Suthiphongchai T., Zambetti G. P., Ewen M. E. (2000). p53 binds selectively to the 5′ untranslated region of cdk4, an RNA element necessary and sufficient for transforming growth factor beta- and p53-mediated translational inhibition of cdk4. Mol. Cell. Biol. 20, 8420–8431 10.1128/MCB.20.2.542-555.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P. S., Parkin R. K., Kroh E. M., Fritz B. R., Wyman S. K., Pogosova-Agadjanyan E. L., Peterson A., Noteboom J., O’briant K. C., Allen A., Lin D. W., Urban N., Drescher C. W., Knudsen B. S., Stirewalt D. L., Gentleman R., Vessella R. L., Nelson P. S., Martin D. B., Tewari M. (2008). Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. U.S.A. 105, 10513–10518 10.1073/pnas.0803384105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro F., Lieberman J. (2010). Small RNAs guide hematopoietic cell differentiation and function. J. Immunol. 184, 5939–5947 10.4049/jimmunol.0902567 [DOI] [PubMed] [Google Scholar]
- Necela B. M., Carr J. M., Asmann Y. W., Thompson E. A. (2011). Differential expression of microRNAs in tumors from chronically inflamed or genetic (APC(Min/+) models of colon cancer. PLoS ONE 6, e18501. 10.1371/journal.pone.0018501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng E. K., Chong W. W., Jin H., Lam E. K., Shin V. Y., Yu J., Poon T. C., Ng S. S., Sung J. J. (2009). Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut 58, 1375–1381 10.1136/gut.2008.167817 [DOI] [PubMed] [Google Scholar]
- Nielsen B. S., Jorgensen S., Fog J. U., Sokilde R., Christensen I. J., Hansen U., Brunner N., Baker A., Moller S., Nielsen H. J. (2011). High levels of microRNA-21 in the stroma of colorectal cancers predict short disease-free survival in stage II colon cancer patients. Clin. Exp. Metastasis 28, 27–38 10.1007/s10585-010-9355-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani H., Shiomi Y., Iida H., Michishita M., Takami T., Tsurimoto T. (2008). CDK inhibitor p21 is degraded by a proliferating cell nuclear antigen-coupled Cul4-DDB1Cdt2 pathway during S phase and after UV irradiation. J. Biol. Chem. 283, 29045–29052 10.1074/jbc.M806045200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell J. B., Maggard M. A., Ko C. Y. (2004). Colon cancer survival rates with the new American Joint Committee on Cancer sixth edition staging. J. Natl. Cancer Inst. 96, 1420–1425 10.1093/jnci/djh275 [DOI] [PubMed] [Google Scholar]
- O’Donnell K. A., Wentzel E. A., Zeller K. I., Dang C. V., Mendell J. T. (2005). c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435, 839–843 10.1038/nature03677 [DOI] [PubMed] [Google Scholar]
- Pan H. W., Chou H. Y., Liu S. H., Peng S. Y., Liu C. L., Hsu H. C. (2006). Role of L2DTL, cell cycle-regulated nuclear and centrosome protein, in aggressive hepatocellular carcinoma. Cell Cycle 5, 2676–2687 10.4161/cc.5.22.3500 [DOI] [PubMed] [Google Scholar]
- Park S. M., Gaur A. B., Lengyel E., Peter M. E. (2008). The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 22, 894–907 10.1101/gad.1640608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S. M., Shell S., Radjabi A. R., Schickel R., Feig C., Boyerinas B., Dinulescu D. M., Lengyel E., Peter M. E. (2007). Let-7 prevents early cancer progression by suppressing expression of the embryonic gene HMGA2. Cell Cycle 6, 2585–2590 10.4161/cc.6.21.4845 [DOI] [PubMed] [Google Scholar]
- Raver-Shapira N., Marciano E., Meiri E., Spector Y., Rosenfeld N., Moskovits N., Bentwich Z., Oren M. (2007). Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol. Cell 26, 731–743 10.1016/j.molcel.2007.05.017 [DOI] [PubMed] [Google Scholar]
- Ricci-Vitiani L., Lombardi D. G., Pilozzi E., Biffoni M., Todaro M., Peschle C., De Maria R. (2007). Identification and expansion of human colon-cancer-initiating cells. Nature 445, 111–115 10.1038/nature05384 [DOI] [PubMed] [Google Scholar]
- Ryan B. M., Robles A. I., Harris C. C. (2010). Genetic variation in microRNA networks: the implications for cancer research. Nat. Rev. Cancer 10, 389–402 10.1038/nrc2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachdeva M., Zhu S., Wu F., Wu H., Walia V., Kumar S., Elble R., Watabe K., Mo Y. Y. (2009). p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc. Natl. Acad. Sci. U.S.A. 106, 3207–3212 10.1073/pnas.0808042106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salonga D., Danenberg K. D., Johnson M., Metzger R., Groshen S., Tsao-Wei D. D., Lenz H. J., Leichman C. G., Leichman L., Diasio R. B., Danenberg P. V. (2000). Colorectal tumors responding to 5-fluorouracil have low gene expression levels of dihydropyrimidine dehydrogenase, thymidylate synthase, and thymidine phosphorylase. Clin. Cancer Res. 6, 1322–1327 [PubMed] [Google Scholar]
- Sampson V. B., Rong N. H., Han J., Yang Q., Aris V., Soteropoulos P., Petrelli N. J., Dunn S. P., Krueger L. J. (2007). MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res. 67, 9762–9770 10.1158/0008-5472.CAN-07-2462 [DOI] [PubMed] [Google Scholar]
- Sansam C. L., Shepard J. L., Lai K., Ianari A., Danielian P. S., Amsterdam A., Hopkins N., Lees J. A. (2006). DTL/CDT2 is essential for both CDT1 regulation and the early G2/M checkpoint. Genes Dev. 20, 3117–3129 10.1101/gad.1482106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schetter A. J., Leung S. Y., Sohn J. J., Zanetti K. A., Bowman E. D., Yanaihara N., Yuen S. T., Chan T. L., Kwong D. L., Au G. K., Liu C. G., Calin G. A., Croce C. M., Harris C. C. (2008). MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA 299, 425–436 10.1001/jama.299.4.425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott N., Sagar P., Stewart J., Blair G. E., Dixon M. F., Quirke P. (1991). p53 in colorectal cancer: clinicopathological correlation and prognostic significance. Br. J. Cancer 63, 317–319 10.1038/bjc.1991.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi B., Sepp-Lorenzino L., Prisco M., Linsley P., Deangelis T., Baserga R. (2007). Micro RNA 145 targets the insulin receptor substrate-1 and inhibits the growth of colon cancer cells. J. Biol. Chem. 282, 32582–32590 10.1074/jbc.M611680200 [DOI] [PubMed] [Google Scholar]
- Shibutani S. T., De La Cruz A. F., Tran V., Turbyfill W. J., III, Reis T., Edgar B. A., Duronio R. J. (2008). Intrinsic negative cell cycle regulation provided by PIP box- and Cul4Cdt2-mediated destruction of E2f1 during S phase. Dev. Cell 15, 890–900 10.1016/j.devcel.2008.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimono Y., Zabala M., Cho R. W., Lobo N., Dalerba P., Qian D., Diehn M., Liu H., Panula S. P., Chiao E., Dirbas F. M., Somlo G., Pera R. A., Lao K., Clarke M. F. (2009). Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell 138, 592–603 10.1016/j.cell.2009.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S., Cha H. J., Lee E. M., Jung J. H., Lee S. J., Park I. C., Jin Y. W., An S. (2009a). MicroRNAs are significantly influenced by p53 and radiation in HCT116 human colon carcinoma cells. Int. J. Oncol. 34, 1645–1652 [DOI] [PubMed] [Google Scholar]
- Shin S., Lee E. M., Cha H. J., Bae S., Jung J. H., Lee S. M., Yoon Y., Lee H., Kim S., Kim H., Lee S. J., Park I. C., Jin Y. W., An S. (2009b). MicroRNAs that respond to histone deacetylase inhibitor SAHA and p53 in HCT116 human colon carcinoma cells. Int. J. Oncol. 35, 1343–1352 [DOI] [PubMed] [Google Scholar]
- Slaby O., Svoboda M., Fabian P., Smerdova T., Knoflickova D., Bednarikova M., Nenutil R., Vyzula R. (2007). Altered expression of miR-21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology 72, 397–402 10.1159/000113489 [DOI] [PubMed] [Google Scholar]
- Slattery M. L., Wolff E., Hoffman M. D., Pellatt D. F., Milash B., Wolff R. K. (2011). MicroRNAs and colon and rectal cancer: differential expression by tumor location and subtype. Genes Chromosomes Cancer 50, 196–206 10.1002/gcc.20844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song B., Wang Y., Kudo K., Gavin E. J., Xi Y., Ju J. (2008). miR-192 Regulates dihydrofolate reductase and cellular proliferation through the p53-microRNA circuit. Clin. Cancer Res. 14, 8080–8086 10.1158/1078-0432.CCR-07-4672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song B., Wang Y., Titmus M. A., Botchkina G., Formentini A., Kornmann M., Ju J. (2010). Molecular mechanism of chemoresistance by miR-215 in osteosarcoma and colon cancer cells. Mol. Cancer 9, 96. 10.1186/1476-4598-9-96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song B., Wang Y., Xi Y., Kudo K., Bruheim S., Botchkina G. I., Gavin E., Wan Y., Formentini A., Kornmann M., Fodstad O., Ju J. (2009). Mechanism of chemoresistance mediated by miR-140 in human osteosarcoma and colon cancer cells. Oncogene 28, 4065–4074 10.1038/onc.2009.181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonkoly E., Stahle M., Pivarcsi A. (2008). MicroRNAs: novel regulators in skin inflammation. Clin. Exp. Dermatol. 33, 312–315 10.1111/j.1365-2230.2008.02804.x [DOI] [PubMed] [Google Scholar]
- Suzuki H., Takatsuka S., Akashi H., Yamamoto E., Nojima M., Maruyama R., Kai M., Yamano H. O., Sasaki Y., Tokino T., Shinomura Y., Imai K., Toyota M. (2011). Genome-wide profiling of chromatin signatures reveals epigenetic regulation of microRNA genes in colorectal cancer. Cancer Res. 71, 5646–5658 10.1158/0008-5472.CAN-11-1076 [DOI] [PubMed] [Google Scholar]
- Takamizawa J., Konishi H., Yanagisawa K., Tomida S., Osada H., Endoh H., Harano T., Yatabe Y., Nagino M., Nimura Y., Mitsudomi T., Takahashi T. (2004). Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 64, 3753–3756 10.1158/0008-5472.CAN-04-0637 [DOI] [PubMed] [Google Scholar]
- Tang F. (2010). Small RNAs in mammalian germline: tiny for immortal. Differentiation 79, 141–146 10.1016/j.diff.2009.11.002 [DOI] [PubMed] [Google Scholar]
- Tavazoie S. F., Alarcon C., Oskarsson T., Padua D., Wang Q., Bos P. D., Gerald W. L., Massague J. (2008). Endogenous human microRNAs that suppress breast cancer metastasis. Nature 451, 147–152 10.1038/nature06487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tazawa H., Tsuchiya N., Izumiya M., Nakagama H. (2007). Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc. Natl. Acad. Sci. U.S.A. 104, 15472–15477 10.1073/pnas.0707351104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery J. P. (2003). Epithelial-mesenchymal transitions in development and pathologies. Curr. Opin. Cell Biol. 15, 740–746 10.1016/j.ceb.2003.10.006 [DOI] [PubMed] [Google Scholar]
- Trang P., Medina P. P., Wiggins J. F., Ruffino L., Kelnar K., Omotola M., Homer R., Brown D., Bader A. G., Weidhaas J. B., Slack F. J. (2010). Regression of murine lung tumors by the let-7 microRNA. Oncogene 29, 1580–1587 10.1038/onc.2009.445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadi H., Ekstrom K., Bossios A., Sjostrand M., Lee J. J., Lotvall J. O. (2007). Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9, 654–659 10.1038/ncb1596 [DOI] [PubMed] [Google Scholar]
- Valastyan S., Chang A., Benaich N., Reinhardt F., Weinberg R. A. (2010). Concurrent suppression of integrin alpha5, radixin, and RhoA phenocopies the effects of miR-31 on metastasis. Cancer Res. 70, 5147–5154 10.1158/0008-5472.CAN-10-0410 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Valastyan S., Reinhardt F., Benaich N., Calogrias D., Szasz A. M., Wang Z. C., Brock J. E., Richardson A. L., Weinberg R. A. (2009). A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell 137, 1032–1046 10.1016/j.cell.2009.03.047 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Viswanathan S. R., Powers J. T., Einhorn W., Hoshida Y., Ng T. L., Toffanin S., O’Sullivan M., Lu J., Phillips L. A., Lockhart V. L., Shah S. P., Tanwar P. S., Mermel C. H., Beroukhim R., Azam M., Teixeira J., Meyerson M., Hughes T. P., Llovet J. M., Radich J., Mullighan C. G., Golub T. R., Sorensen P. H., Daley G. Q. (2009). Lin28 promotes transformation and is associated with advanced human malignancies. Nat. Genet. 41, 843–848 10.1038/ng.392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B., Fearon E. R., Hamilton S. R., Kern S. E., Preisinger A. C., Leppert M., Nakamura Y., White R., Smits A. M., Bos J. L. (1988). Genetic alterations during colorectal-tumor development. N. Engl. J. Med. 319, 525–532 10.1056/NEJM198809013190901 [DOI] [PubMed] [Google Scholar]
- Volinia S., Calin G. A., Liu C. G., Ambs S., Cimmino A., Petrocca F., Visone R., Iorio M., Roldo C., Ferracin M., Prueitt R. L., Yanaihara N., Lanza G., Scarpa A., Vecchione A., Negrini M., Harris C. C., Croce C. M. (2006). A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. U.S.A. 103, 2257–2261 10.1073/pnas.0510565103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Herman-Edelstein M., Koh P., Burns W., Jandeleit-Dahm K., Watson A., Saleem M., Goodall G. J., Twigg S. M., Cooper M. E., Kantharidis P. (2010a). E-cadherin expression is regulated by miR-192/215 by a mechanism that is independent of the profibrotic effects of transforming growth factor-beta. Diabetes 59, 1794–1802 10.2337/db10-0192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C. J., Stratmann J., Zhou Z. G., Sun X. F. (2010b). Suppression of microRNA-31 increases sensitivity to 5-FU at an early stage, and affects cell migration and invasion in HCT-116 colon cancer cells. BMC Cancer 10, 616. 10.1186/1471-2407-10-616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Li Y., Ahmad A., Azmi A. S., Kong D., Banerjee S., Sarkar F. H. (2010c). Targeting miRNAs involved in cancer stem cell and EMT regulation: an emerging concept in overcoming drug resistance. Drug Resist. Updat. 13, 109–118 10.1016/j.drup.2010.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Mcleod H. L., Cassidy J., Collie-Duguid E. S. (2007). Mechanisms of acquired chemoresistance to 5-fluorouracil and tomudex: thymidylate synthase dependent and independent networks. Cancer Chemother. Pharmacol. 59, 839–845 10.1007/s00280-006-0300-z [DOI] [PubMed] [Google Scholar]
- Whitehead R. H., Jones J. K., Gabriel A., Lukies R. E. (1987). A new colon carcinoma cell line (LIM1863) that grows as organoids with spontaneous differentiation into crypt-like structures in vitro. Cancer Res. 47, 2683–2689 [PubMed] [Google Scholar]
- Wightman B., Ha I., Ruvkun G. (1993). Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75, 855–862 10.1016/0092-8674(93)90530-4 [DOI] [PubMed] [Google Scholar]
- Williams C. S., Mann M., Dubois R. N. (1999). The role of cyclooxygenases in inflammation, cancer, and development. Oncogene 18, 7908–7916 10.1038/sj.onc.1203286 [DOI] [PubMed] [Google Scholar]
- Woods K., Thomson J. M., Hammond S. M. (2007). Direct regulation of an oncogenic micro-RNA cluster by E2F transcription factors. J. Biol. Chem. 282, 2130–2134 10.1074/jbc.C600252200 [DOI] [PubMed] [Google Scholar]
- Xi Y., Edwards J. R., Ju J. (2007). Investigation of miRNA biology by bioinformatic tools and impact of miRNAs in colorectal cancer – regulatory relationship of c-Myc and p53 with miRNAs. Cancer Inform. 3, 245–253 [PMC free article] [PubMed] [Google Scholar]
- Xi Y., Formentini A., Chien M., Weir D. B., Russo J. J., Ju J., Kornmann M. (2006a). Prognostic values of microRNAs in colorectal cancer. Biomark. Insights 2, 113–121 [PMC free article] [PubMed] [Google Scholar]
- Xi Y., Shalgi R., Fodstad O., Pilpel Y., Ju J. (2006b). Differentially regulated micro-RNAs and actively translated messenger RNA transcripts by tumor suppressor p53 in colon cancer. Clin. Cancer Res. 12, 2014–2024 10.1158/1078-0432.CCR-05-1853 [DOI] [PubMed] [Google Scholar]
- Yamakuchi M., Ferlito M., Lowenstein C. J. (2008). miR-34a repression of SIRT1 regulates apoptosis. Proc. Natl. Acad. Sci. U.S.A. 105, 13421–13426 10.1073/pnas.0801613105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Weinberg R. A. (2008). Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev. Cell 14, 818–829 10.1016/j.devcel.2007.12.004 [DOI] [PubMed] [Google Scholar]
- Yu F., Yao H., Zhu P., Zhang X., Pan Q., Gong C., Huang Y., Hu X., Su F., Lieberman J., Song E. (2007a). let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell 131, 1109–1123 10.1016/j.cell.2007.10.054 [DOI] [PubMed] [Google Scholar]
- Yu J., Vodyanik M. A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J. L., Tian S., Nie J., Jonsdottir G. A., Ruotti V., Stewart R., Slukvin Ii., Thomson J. A. (2007b). Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917–1920 10.1126/science.1151526 [DOI] [PubMed] [Google Scholar]
- Zhang H., Li W., Nan F., Ren F., Wang H., Xu Y., Zhang F. (2011). MicroRNA expression profile of colon cancer stem-like cells in HT29 adenocarcinoma cell line. Biochem. Biophys. Res. Commun. 404, 273–278 10.1016/j.bbrc.2010.12.109 [DOI] [PubMed] [Google Scholar]
- Zhao R., Gish K., Murphy M., Yin Y., Notterman D., Hoffman W. H., Tom E., Mack D. H., Levine A. J. (2000). Analysis of p53-regulated gene expression patterns using oligonucleotide arrays. Genes Dev. 14, 981–993 10.1101/gad.827700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Zhou Y., Yin B., Hao W., Zhao L., Ju W., Bai C. (2010). 5-Fluorouracil and oxaliplatin modify the expression profiles of microRNAs in human colon cancer cells in vitro. Oncol. Rep. 23, 121–128 [PubMed] [Google Scholar]