Abstract
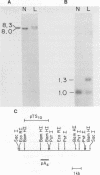
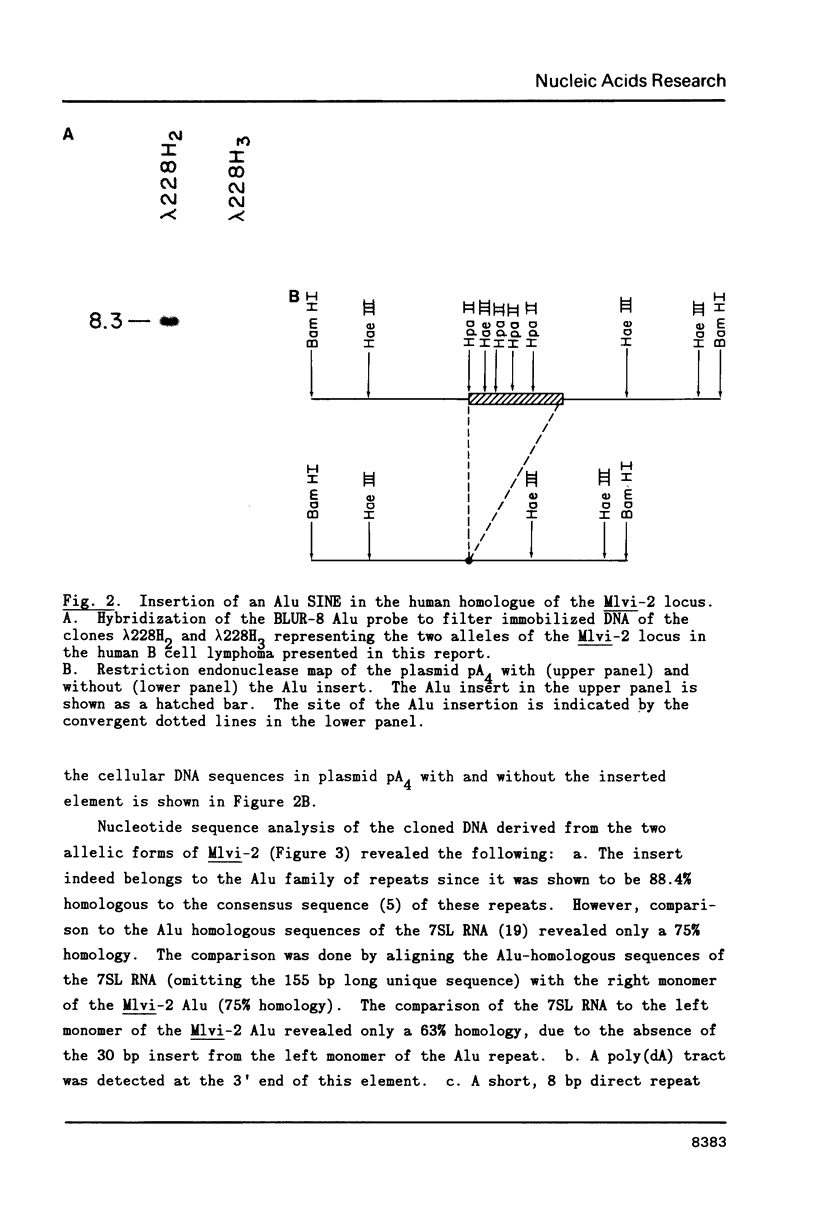
Fifty-nine human DNA samples derived from either normal tissues or hematopoietic neoplasias were examined for rearrangements in the Mlvi-2 locus, a putative oncogene. The rearranged Mlvi-2 sequences in one of them, a B cell lymphoma, were shown to result from the insertion of an approximately 300 bp DNA fragment that hybridized to a human Alu probe. DNA sequence analysis of both the rearranged and the nonrearranged allele around the site of the insertion revealed the following: a) the insert was 88.4% homologous to the consensus sequence of the Alu family of repeats and 75% homologous to the Alu related sequence in the human 7SL RNA; b) similar to other sequenced SINES, a poly(d.A) tract was present at the 3' end of this element; c) an 8 bp direct repeat was present at both ends of the inserted element; d) this repeat was present as a single copy in the unrearranged allele. We conclude from these findings that: Alu sequences can transpose and that the direct repeats flanking certain Alu SINES may be generated by the duplication of single copy cellular sequences at the site of the insertion. Furthermore the recent nature of the Alu insertion in the Mlvi-2 locus coupled to the low degree of homology of the inserted Alu to the Alu related sequence in the 7SL RNA suggest that this event did not occur via reverse transcription and reintegration of the 7SL RNA.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baltimore D. Retroviruses and retrotransposons: the role of reverse transcription in shaping the eukaryotic genome. Cell. 1985 Mar;40(3):481–482. doi: 10.1016/0092-8674(85)90190-4. [DOI] [PubMed] [Google Scholar]
- Barta A., Richards R. I., Baxter J. D., Shine J. Primary structure and evolution of rat growth hormone gene. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4867–4871. doi: 10.1073/pnas.78.8.4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten R. J., Kohne D. E. Repeated sequences in DNA. Hundreds of thousands of copies of DNA sequences have been incorporated into the genomes of higher organisms. Science. 1968 Aug 9;161(3841):529–540. doi: 10.1126/science.161.3841.529. [DOI] [PubMed] [Google Scholar]
- Chen P. J., Cywinski A., Taylor J. M. Reverse transcription of 7S L RNA by an avian retrovirus. J Virol. 1985 May;54(2):278–284. doi: 10.1128/jvi.54.2.278-284.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson E. H., Hough B. R., Amenson C. S., Britten R. J. General interspersion of repetitive with non-repetitive sequence elements in the DNA of Xenopus. J Mol Biol. 1973 Jun 15;77(1):1–23. doi: 10.1016/0022-2836(73)90359-8. [DOI] [PubMed] [Google Scholar]
- Deininger P. L., Jolly D. J., Rubin C. M., Friedmann T., Schmid C. W. Base sequence studies of 300 nucleotide renatured repeated human DNA clones. J Mol Biol. 1981 Sep 5;151(1):17–33. doi: 10.1016/0022-2836(81)90219-9. [DOI] [PubMed] [Google Scholar]
- Economou-Pachnis A., Lohse M. A., Furano A. V., Tsichlis P. N. Insertion of long interspersed repeated elements at the Igh (immunoglobulin heavy chain) and Mlvi-2 (Moloney leukemia virus integration 2) loci of rats. Proc Natl Acad Sci U S A. 1985 May;82(9):2857–2861. doi: 10.1073/pnas.82.9.2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enquist L., Tiemeier D., Leder P., Weisberg R., Sternberg N. Safer derivatives of bacteriophage lambdagt-lambdaC for use in cloning of recombinant DNA molecules. Nature. 1976 Feb 19;259(5544):596–598. doi: 10.1038/259596a0. [DOI] [PubMed] [Google Scholar]
- Grimaldi G., Queen C., Singer M. F. Interspersed repeated sequences in the African green monkey genome that are homologous to the human Alu family. Nucleic Acids Res. 1981 Nov 11;9(21):5553–5568. doi: 10.1093/nar/9.21.5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaldi G., Singer M. F. A monkey Alu sequence is flanked by 13-base pair direct repeats by an interrupted alpha-satellite DNA sequence. Proc Natl Acad Sci U S A. 1982 Mar;79(5):1497–1500. doi: 10.1073/pnas.79.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes S. R., Toomey T. P., Leinwand L., Jelinek W. R. The Chinese hamster Alu-equivalent sequence: a conserved highly repetitious, interspersed deoxyribonucleic acid sequence in mammals has a structure suggestive of a transposable element. Mol Cell Biol. 1981 Jul;1(7):573–583. doi: 10.1128/mcb.1.7.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollis G. F., Hieter P. A., McBride O. W., Swan D., Leder P. Processed genes: a dispersed human immunoglobulin gene bearing evidence of RNA-type processing. Nature. 1982 Mar 25;296(5855):321–325. doi: 10.1038/296321a0. [DOI] [PubMed] [Google Scholar]
- Houck C. M., Rinehart F. P., Schmid C. W. A ubiquitous family of repeated DNA sequences in the human genome. J Mol Biol. 1979 Aug 15;132(3):289–306. doi: 10.1016/0022-2836(79)90261-4. [DOI] [PubMed] [Google Scholar]
- Jagadeeswaran P., Forget B. G., Weissman S. M. Short interspersed repetitive DNA elements in eucaryotes: transposable DNA elements generated by reverse transcription of RNA pol III transcripts? Cell. 1981 Oct;26(2 Pt 2):141–142. doi: 10.1016/0092-8674(81)90296-8. [DOI] [PubMed] [Google Scholar]
- Kramerov D. A., Grigoryan A. A., Ryskov A. P., Georgiev G. P. Long double-stranded sequences (dsRNA-B) of nuclear pre-mRNA consist of a few highly abundant classes of sequences: evidence from DNA cloning experiments. Nucleic Acids Res. 1979 Feb;6(2):697–713. doi: 10.1093/nar/6.2.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krayev A. S., Kramerov D. A., Skryabin K. G., Ryskov A. P., Bayev A. A., Georgiev G. P. The nucleotide sequence of the ubiquitous repetitive DNA sequence B1 complementary to the most abundant class of mouse fold-back RNA. Nucleic Acids Res. 1980 Mar 25;8(6):1201–1215. doi: 10.1093/nar/8.6.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krayev A. S., Markusheva T. V., Kramerov D. A., Ryskov A. P., Skryabin K. G., Bayev A. A., Georgiev G. P. Ubiquitous transposon-like repeats B1 and B2 of the mouse genome: B2 sequencing. Nucleic Acids Res. 1982 Dec 11;10(23):7461–7475. doi: 10.1093/nar/10.23.7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemay G., Jolicoeur P. Rearrangement of a DNA sequence homologous to a cell-virus junction fragment in several Moloney murine leukemia virus-induced rat thymomas. Proc Natl Acad Sci U S A. 1984 Jan;81(1):38–42. doi: 10.1073/pnas.81.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemischka I., Sharp P. A. The sequences of an expressed rat alpha-tubulin gene and a pseudogene with an inserted repetitive element. Nature. 1982 Nov 25;300(5890):330–335. doi: 10.1038/300330a0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler L. A., Weber J. L., Gorski J. Polymorphism near the rat prolactin gene caused by insertion of an Alu-like element. Nature. 1983 Sep 8;305(5930):159–160. doi: 10.1038/305159a0. [DOI] [PubMed] [Google Scholar]
- Shapiro J. A. Molecular model for the transposition and replication of bacteriophage Mu and other transposable elements. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1933–1937. doi: 10.1073/pnas.76.4.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. A. Conversion of RNA to DNA in mammals: Alu-like elements and pseudogenes. Nature. 1983 Feb 10;301(5900):471–472. doi: 10.1038/301471a0. [DOI] [PubMed] [Google Scholar]
- Singer M. F. Highly repeated sequences in mammalian genomes. Int Rev Cytol. 1982;76:67–112. doi: 10.1016/s0074-7696(08)61789-1. [DOI] [PubMed] [Google Scholar]
- Steffen D. Proviruses are adjacent to c-myc in some murine leukemia virus-induced lymphomas. Proc Natl Acad Sci U S A. 1984 Apr;81(7):2097–2101. doi: 10.1073/pnas.81.7.2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J. G., Milner R. J., Bloom F. E., Lerner R. A. Common 82-nucleotide sequence unique to brain RNA. Proc Natl Acad Sci U S A. 1982 Aug;79(16):4942–4946. doi: 10.1073/pnas.79.16.4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temin H. M. Origin of retroviruses from cellular moveable genetic elements. Cell. 1980 Oct;21(3):599–600. doi: 10.1016/0092-8674(80)90420-1. [DOI] [PubMed] [Google Scholar]
- Tsichlis P. N., Strauss P. G., Hu L. F. A common region for proviral DNA integration in MoMuLV-induced rat thymic lymphomas. 1983 Mar 31-Apr 6Nature. 302(5907):445–449. doi: 10.1038/302445a0. [DOI] [PubMed] [Google Scholar]
- Ullu E., Murphy S., Melli M. Human 7SL RNA consists of a 140 nucleotide middle-repetitive sequence inserted in an alu sequence. Cell. 1982 May;29(1):195–202. doi: 10.1016/0092-8674(82)90103-9. [DOI] [PubMed] [Google Scholar]
- Ullu E., Tschudi C. Alu sequences are processed 7SL RNA genes. Nature. 1984 Nov 8;312(5990):171–172. doi: 10.1038/312171a0. [DOI] [PubMed] [Google Scholar]
- Weiner A. M. An abundant cytoplasmic 7S RNA is complementary to the dominant interspersed middle repetitive DNA sequence family in the human genome. Cell. 1980 Nov;22(1 Pt 1):209–218. doi: 10.1016/0092-8674(80)90169-5. [DOI] [PubMed] [Google Scholar]