Abstract
Genetic studies of midgut carcinoid cancer have exclusively focused on genomic changes of the tumor cells. We investigated the role of constitutional genetic polymorphisms in predisposing individuals to ileal carcinoids. In all, 239 cases and 110 controls were collected from three institutions: the Uppsala University Hospital; the Dana-Farber Cancer Institute; and the MD Anderson Cancer Center, and were genotyped using microarrays assaying >300 000 single nucleotide polymorphisms. Association with rs2208059 in KIF16B approached statistical significance (Mantel-Haenszel odds ratio=2.42, P=4.16×10−7) at a Bonferroni-corrected level (<1.62×10−7). Using two computational algorithms, four copy-number variants (CNVs) were identified in multiple cases that were absent in study controls and markedly less frequent in ~1500 population-based controls. Of these four constitutional CNVs identified in blood-derived DNA, a 40 kb heterozygous deletion in Chr18q22.1 corresponded with a region frequently showing loss of heterozygosity (LOH) in ileal carcinoid tumor cells based on our meta-analysis of previously published cytogenetic studies (69.7% LOH, 95% confidence interval=60.0–77.9%). We analyzed the constitutional 40 kb deletion on chr18 in our study samples with a real-time quantitative PCR assay; 14/226 cases (6.19%) and 2/97 controls (2.06%) carried the CNV, although the exact boundaries of each deletion have not been determined. Given the small sample size, our findings warrant an independent cohort for a replication study. Owing to the rarity of this disease, we believe these results will provide a valuable resource for future work on this serious condition by allowing others to make efficient use of their samples in targeted studies.
Introduction
Ileal carcinoids are well-differentiated and malignant neuroendocrine tumors of the small intestine. Ileal carcinoids are uncommon, with an age-adjusted annual incidence of <1 per 100 000 population per year in both the United States and Europe (Buchanan et al. 1986, Modlin et al. 2003, Yao et al. 2008). While some gastroenteropancreatic neuroendocrine tumors are associated with hereditary cancer syndromes (e.g. MEN1, MEN2A, and MEN2B), ileal carcinoids are a primarily sporadic neoplasm (D’adda et al. 2002). The genetic etiology of ileal carcinoids is largely unknown, but has been the focus of much recent research.
Numerous candidate genes have been studied, but no mutations have been consistently identified in TP53, KRAS, SMAD4, BRAF, APC, or CTNNB1 (Weckstrom et al. 1996, Younes et al. 1997, Lollgen et al. 2001, Wang et al. 2005, Su et al. 2006). Genome-wide cytogenetic studies of ileal carcinoids, which compare tumor tissue to matched healthy tissue, have sought to identify genomic alterations critical to ileal carcinoid tumorigenesis. Although ileal carcinoid tumors show genomic instability, such studies have had little success in narrowing down the list of genes potentially involved in disease pathogenesis (Oberg 2009).
While the genes involved remain unknown, cyto-genetic studies of ileal carcinoid tumors have begun to indicate that specific chromosomal regions are frequently altered (Kulke et al. 2008, Andersson et al. 2009). Recurrent chromosomal aberrations detected in multiple affected patients are believed to harbor dosage-sensitive genes, which influence ileal carcinoid tumorigenesis (Oberg 2009). Accumulating data on recurrent chromosomal deletions reveal that loss of one or more as yet unidentified tumor suppressor genes may be a critical step in the development of ileal carcinoids. Furthermore, recurrent chromosomal amplifications are believed to encompass a gene, or genes, which promote carcinoid tumor formation or survival (Zikusoka et al. 2005). While recurrent chromosomal aberrations are not a unique feature of ileal carcinoids, the specific chromosomal regions affected are largely particular, though not unique, to these tumors. Even among carcinoid tumors, ileal carcinoids show markedly different genomic profiles when compared with neuroendocrine tumors of the pancreas, lung, or another gastrointestinal site (Tonnies et al. 2001, Kim do et al. 2008). This suggests that neuroendocrine tumors develop through molecular pathways that are largely distinct across tumor sites, but common to tumors of a given site.
In an attempt to identify genes that are a component of the common pathway through which ileal carcinoids develop, we performed a genome-wide association study (GWAS) to identify constitutional genetic variants associated with development of ileal carcinoids using a multicenter case–control design comprising patients from the three centers. A total of 239 cases and 110 controls were genotyped at more than 300 000 single nucleotide polymorphisms (SNPs). As rare disorders may be associated with rare genetic variants (Galvan et al. 2010), and because chromosomal aberrations are a common feature of carcinoid tumors, SNP array data from the GWAS was used to identify copy-number variants (CNVs) potentially associated with case status. These CNVs were then subjected to verification by quantitative real-time PCR (RT-qPCR). Until now, genetic studies of ileal carcinoids have focused on genomic changes characteristic of carcinoid tumors, using matched healthy tissue as the reference. To our knowledge, this is the first study to investigate, genome-wide, the role of constitutional genetic polymorphisms in predisposing individuals to development of ileal carcinoids.
Materials and methods
Ethics statement
All participating institutions received the IRB approval, and appropriate informed consent was obtained from human subjects. DNA samples used in this study were obtained from archives wherein patients have already provided their informed consent to use specimens for research purposes.
Study population
A multicenter pilot case–control study was designed to include patients seen for ileal carcinoid cancer at the Dana-Farber Cancer Institute (82 cases and 35 controls), the University Hospital of Uppsala, Sweden (102 cases and 48 controls), and the MD Anderson Cancer Center (55 cases and 27 controls). All study participants have reported northern European ancestry. Clinical information was ascertained by the participating clinicians at each of the three sites. Tumor status and disease diagnosis were also performed by the participating clinicians. All cases obtained from the University Hospital of Uppsala, Sweden had metastatic disease, whereas cases obtained from the Dana-Farber Cancer Institute and the MD Anderson Cancer Center were at various stages of disease. Although cases were at various stages of disease, all tumors warranted surgical resection and all cases underwent surgery for carcinoid cancer as the primary indication.
Definition of cases and controls
All patients with ileal carcinoid tumors were eligible for inclusion, irrespective of age, gender, ethnicity, and tumor stage. Cancer-free controls were selected and frequency-matched to cases based on age, gender, and race, which have been shown to be associated with the incidence of intestinal carcinoid cancer (Modlin et al. 2003). To eliminate controls with indolent or subclinical carcinoid tumors, controls were, when possible, excluded if they reported symptoms associated with midgut carcinoids, including 1) anemia, 2) undiagnosed episodic serious stomach pains or possible partial obstruction, 3) any episode of blood loss from the upper or lower gastrointestinal tract, or 4) a family history of neuroendocrine cancer.
GWAS genotyping
DNA samples were extracted from peripheral blood, obtained by venipuncture. DNA samples were run on a 1% agarose gel to assess quality; all samples showed a single, high-molecular weight band and no sample was excluded based on this assay. Genotyping was performed at The Rockefeller University Genomics Resource Center using the Illumina 300K SNP Infinium platform (Sentrix HumanHap 300) according to the manufacturer’s recommended protocol. On average, the platform contains one SNP every 9 kb across the genome (median spacing=5 kb).
Statistical analysis
Fisher’s exact tests were used to test single SNP associations for both allele and genotype frequencies. This was carried out by constructing 2×2 tables of the allele counts and 2×3 tables of the genotype counts for each SNP in all cases and controls, stratified by site. To control for differences in allele frequency in patients recruited at different study sites, association statistics were calculated within single sites and then combined as follows: Z-scores for single SNP association at each site were weighted by sample size then summed across all the three sites to yield overall association statistics for the combined sample. The presence of population stratification among individuals from each study site was assessed using Eigenstrat (Price et al. 2006).
Systematic review and meta-analysis
A Medline search was performed on July 1, 2010 to identify all articles whose titles or abstracts contain the word ‘chromosome’ or ‘chromosomal’ and which were indexed with the MeSH terms ‘carcinoid’ or ‘neuro-endocrine’. Bibliographies of selected articles were scanned to identify pertinent publications, which the electronic search may have missed. Results were not filtered by language.
Studies that provided individual-level data were eligible for inclusion if the total number of ileal carcinoid tumors screened for chromosomal alterations was given, and the variants identified in each tumor were listed. Chromosomal alterations detected in other midgut carcinoid tumors (e.g. duodenal, jejunal, and appendiceal) were noted, but were excluded from the meta-analysis in order to minimize tumor heterogeneity.
From the selected publications, a list of the most commonly reported chromosomal alterations was compiled, and a meta-analysis of the prevalence of this alteration in patients with ileal carcinoids was performed. As different genetic alterations may distinguish localized ileal carcinoid tumor tissue from metastatic tumor tissue, a stratified analysis was performed to assess the prevalence in localized tumors separately from that in metastatic tumors. A fixed-effect model was used to combine studies within each subgroup, and the study-to-study variance was computed independently within the subgroups. Heterogeneity between the two subgroups was assessed using the Q value, which was compared with a χ2 distribution with one degree of freedom using a significance threshold of α = 0.10.
In the absence of substantial heterogeneity between local and metastatic tumors, the prevalence data may be combined to determine the prevalence of a given genetic aberration among all ileal carcinoid tumors. The study-to-study variance was not assumed to be the same for both the subgroups and was computed within the subgroups; 95% confidence intervals for this prevalence were calculated for primary tumors, metastatic tumors, and all carcinoids combined (excluding metastatic tumors of patients contributing a primary tumor, due to dependency of metastatic tumor genotype on primary tumor genotype). Study heterogeneity was assessed using the I2 statistic (Higgins et al. 2003). To assess the presence of reporting bias, a funnel plot graphing study precision against the logit event rate was created and Egger’s regression asymmetry test was performed (Egger et al. 1997). By abstracting data on chromosomal aberrations primarily from genome-wide scans, which are not hypothesis driven, reporting bias should be minimized. All statistics for the meta-analysis were calculated in Comprehensive Meta-Analysis, version 2 (Biostat Inc., Englewood, NJ, USA).
Detection of constitutional CNVs
CNVs were identified genome-wide by applying two distinct algorithms to the SNP array data. The first algorithm normalizes LogR and B allele frequency data to the larger sample pool to remove sample-specific noise. Based on the empirical distribution of LogR and B allele frequency values, the likelihoods of a given SNP being 0, 1, 2, and 3 or more copies were calculated respectively. If more than one SNP supporting deletion or duplication arose consecutively and the likelihood ratio of the SNPs exceeded threshold, they were called a deletion or duplication (Choi et al. 2009). The second algorithm, BioDiscovery’s SNPRank Segmentation algorithm (BioDiscovery Inc., El Segundo, CA, USA), is based on the circular binary segmentation algorithm of Venkatraman & Olshen (2007). This detection algorithm divides an individual’s genome into regions of equal copy-number and utilizes both the hybridization intensity and the B allele frequency to identify CNVs. Association tests were used on all individual CNVs detected, except those observed in only one case or in no controls.
CNVs were identified genome-wide and a limited number were confirmed on a per-sample basis using RT-qPCR. Real-time qPCR results were used to train the algorithms and adjusted calling thresholds. Following the training of the algorithms, a final list of CNVs detected with high confidence was assembled.
For those CNVs identified in several cases but absent in the study controls, we looked up the CNV frequency in the Database of Genomic Variants (http://projects.tcag.ca/variation/) to ensure their rarity in controls was not due to chance alone (Iafrate et al. 2004). We also assessed the frequency of these rare CNVs in a set of population-based controls, which were genotyped for another study using the Illumina 370K SNP array.
The most promising CNVs (those located in candidate regions from the systematic review) were assayed on the entire case–control dataset using a TaqMan copy-number genotyping assay.
TaqMan copy-number genotyping
Copy-number genotyping was performed using real-time quantitative PCR and commercially available reagents (Applied Biosystems, Foster City, CA, USA) following the manufacturer’s recommendations. Five nanograms of sample DNA was added to each of three replicate wells and dried overnight. Two calibrator samples available from Coriell Cell Repositories (NA15510 and NA10851) have been characterized extensively with respect to CNV content and were included in all plates analyzed to ensure genotyping consistency. A minimum of eight wells were left without DNA as ‘no template controls’ to set maximum limits for cycle threshold values. A copy-number reference assay containing two primers and a VIC and TAMRA dye-labeled probe assayed copy-number at the RNaseP locus, which is known to exist as one copy per haploid genome. This was amplified in a multiplex reaction with a copy-number target assay containing two primers and a FAM dye-labeled minor groove binder probe, which targeted a region in the candidate CNV. Plates were run on the ABI 7900HT machine using the manufacturer’s recommended PCR cycling conditions.
Cycle thresholds were calculated using the SDS v2.2.2 software with the autobaseline on and a manual CT threshold of 0.20. Wells with a cycle threshold exceeding 32.5 for either the target or the reference probe were excluded from analysis. Of the 346 samples that underwent SNP array genotyping, 323 samples had sufficient DNA available for RT-qPCR reactions and were included in the final CNV analysis. This included 226 cases and 97 controls.
CT values for the target and reference assay were imported into CopyCaller software version 1.0 (Applied Biosystems). A comparative CT (ΔΔCT) relative quantification analysis of the real-time data was performed. First, the difference between the target and reference CT value was determined for each well and then averaged across sample replicates. The ΔCT values of the samples are used to calculate copy-number at the target locus relative to that at the RNaseP control locus. These values were categorized into discrete copy-number classes using a maximum-likelihood algorithm in the CopyCaller software, which was conditioned to treat two copies as the likeliest outcome.
Results
SNP GWAS
A total of 349 samples (239 cases and 110 controls) were loaded to microarray chips for genotyping. Of these, 346 passed the default QC call rate threshold (90%) with an overall call rate range of 92.13–99.95%, leaving 239 cases and 107 controls for statistical analyses. Of the >317 000 SNPs genotyped, 308 330 autosomal SNPs with a call rate >95% in both cases and controls (stratified by site) and a Hardy–Weinberg equilibrium χ2 value of <50 in controls were included for analyses. The presence of population stratification was assessed within each study site using the Eigenstrat software (http://genepath.med.harvard.edu/~reich/Software.htm). The first two eigenvalues showed no significant population stratification was present, likely due to the self-reported northern European ancestry of all subjects and the careful matching that was performed.
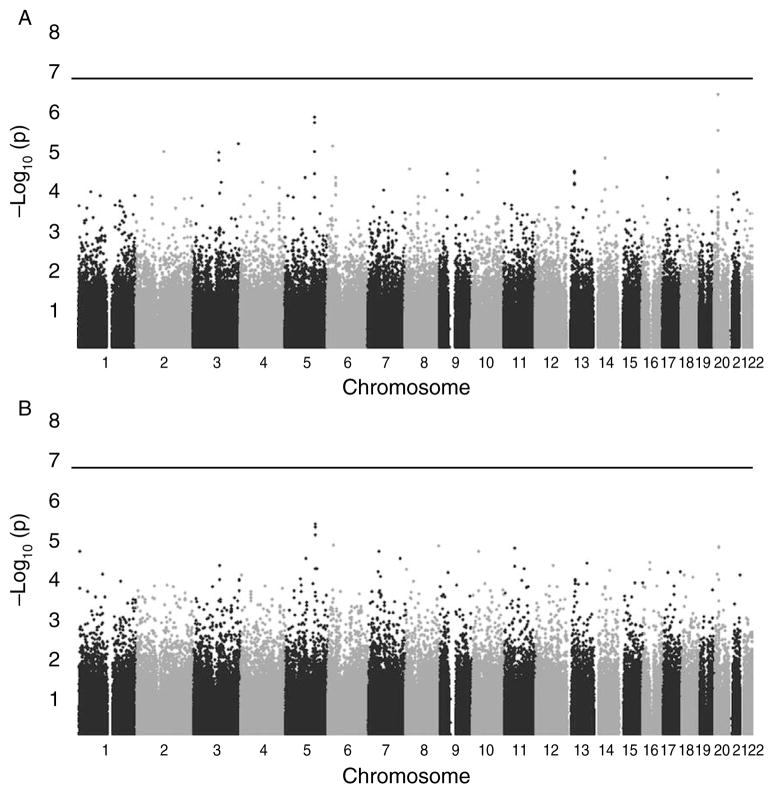
The GWAS did not identify any SNPs that were associated with case status at a strict Bonferroni-corrected level of significance (<1.62×10−7) using either the allelic or genotypic tests (Fig. 1A and B respectively). The most significant SNP identified was rs2208059 in KIF16B, which had a combined P value of 4.16×10−7 using the allelic test, and a Mantel–Haenszel odds ratio (adjusted for site) of 2.42 (95% confidence interval (CI)=1.72–3.42). Across study sites, no heterogeneity was detected in the odds ratio associated with this SNP using the Breslow–Day test. To facilitate further research, a list of the top 25 SNP associations is available from the authors upon request.
Figure 1.
Distribution of P values for the SNPs in a genome-wide association study of ileal carcinoid cancer. (A) P values for the 1 d.f. allelic test of association. (B) P values for the 2 d.f. genotypic test of association. P values are plotted as the −log10(P), with the SNPs in chromosomal order along the x-axis. The horizontal line indicates the Bonferroni-adjusted threshold for significant association at the 0.05 level.
CNV analyses from SNP array
A total of 2539 CNVs spanning five or more consecutive SNPs were detected in the 346 case–control patients from the carcinoid GWAS, ranging in size from 5.8 kb to 4.74 Mb (median=36.7 kb). A total of four regions contained recurrent rare CNVs, which both algorithms detected in multiple cases, but which were not detected in controls from the carcinoid GWAS (Table 1). The presence, among cases, of all CNVs appearing in Table 1 was successfully confirmed on a per-sample basis using RT-qPCR. Association tests were performed on all individual CNVs, but none returned P values <0.05. The rare CNVs presented in Table 1 are not amenable to traditional association testing due to the limited sample size of this pilot study, but provide promising candidates for future study.
Table 1.
Recurrent copy-number variant (CNV) regions identified in cases but absent in controlsa
| CNV Regiona | Case CNVs | Carcinoid Control CNVs | Additional population- based control CNVsb | CNV Type | Gene contents | Addn contrlsd | Addn controlse |
|---|---|---|---|---|---|---|---|
| Chr9: 137.3Mb-137.4 Mb | 4/239 | 0/107 | 11/1512 | Duplication | C9orf62 | 11/871 | 22/2621 |
| Chr10: 81.6-81.9 Mb | 3/239 | 0/107 | 7/1512 | Both§ | SFTPD, C10orf57, Plac9 | 1/871 | 46/2621 (7 duplications and 39 deletions) |
| Chr16: 15.4-16.8 Mb | 2/239 | 0/107 | 2/1512 | Duplication | C16orf45, KIAA0430, NDE1, MYH11, C16orf63, ABCC1, ABCC6 | 1/871 | 84/2621 |
| Chr18: 64.00-64.04 Mb | 5/239 | 0/107 | 0/1512 | Deletion | Intergenic | 9/871 | 24/2621 |
This is the minimum region of overlap for all case CNVs aligned to hg18/Build 36.
These are Caucasian samples genotyped on Illumina 370K chips for another study. Listed CNVs must have at least 50% overlap with the defined case CNV region (i.e. column 1).
Two cases showed duplication of this locus and one showed deletion. All seven population-based control CNVs were duplications
Controls from meningioma data genotyped on Illumina 610-Quad
Controls from the schizophrenia/bipolar dbGaP dataset on the Affymetrix 6.0 chip
Systematic review and meta-analysis
Overlap between constitutional CNVs and loci frequently altered in ileal carcinoid tumors may harbor shared dosage-sensitive genes, which predispose a subset of patients to future development of carcinoid cancer. In order to guide our selection of promising candidate CNVs for follow-up study, a systematic review and meta-analysis were conducted to determine the most common chromosomal aberration seen in ileal carcinoid tumors. Commonly amplified or deleted regions were considered loci of special interest in our study of constitutional CNVs associated with this disease.
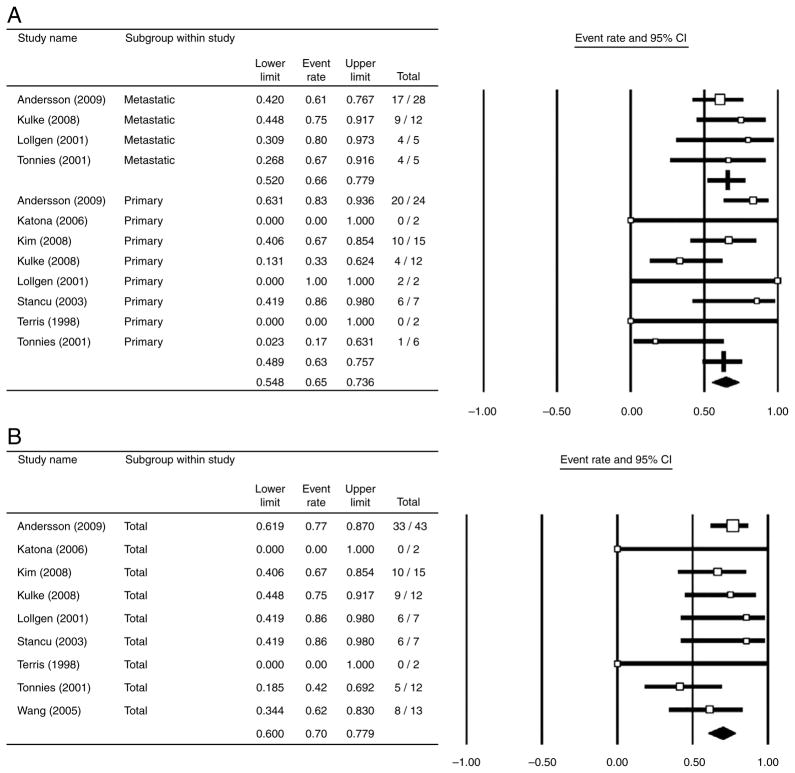
The Medline search returned 216 articles, of which 9 were included in the meta-analysis (Terris et al. 1998, Lollgen et al. 2001, Tonnies et al. 2001, Stancu et al. 2003, Wang et al. 2005, Katona et al. 2006, Kim do et al. 2008, Kulke et al. 2008, Andersson et al. 2009). Deletion of chromosome 18, especially the 18q22-qter region, was overwhelmingly the most commonly identified chromosomal aberration. In eight studies that included primary ileal carcinoid tumors, loss of chromosome 18q22-qter was observed in 63.3% of the primary tumors (95% CI=48.9–75.7%; Fig. 2A). Moderate study heterogeneity was observed in the primary tumor subgroup (I2=47.2%). In four studies that included metastatic ileal carcinoid tumors, loss of chromosome 18q22-qter was observed in 66.2% of the metastatic tumors (95% CI=52.0–77.9%; Fig. 2A). Low study heterogeneity was observed in the meta-static tumor subgroup (I2=0.0%). Significant heterogeneity between tumor subgroups (primary versus metastatic) was not detected (Q=0.085, P=0.771) and therefore the prevalence of 18q22-qter deletions could be analyzed jointly, combining all tumors.
Figure 2.
Meta-analysis results: forest plots of the prevalence of chromosome 18q22-qter deletions in ileal carcinoid tumors. (A) A forest plot of the prevalence of chromosome 18q22-qtrer deletions in ileal carcinoid tumors, stratified by tumor type (i.e. primary versus metastatic). (B) A cumulative forest plot of the prevalence of chromosome 18q22-qtrer deletions in ileal carcinoid tumors, regardless of tumor type (i.e. primary versus metastatic). The area of study symbols (white squares) is proportional to study weight.
The cumulative meta-analysis, pooling results from nine studies of primary and metastatic ileal carcinoid tumors, determined that 69.7% of these tumors showed loss of the 18q22-qter region (95% CI=60.0–77.9%; Fig. 2B), the majority of which have lost the entire chromosome. Low study heterogeneity was observed in the cumulative meta-analysis (I2=0.0%). The presence of reporting bias was assessed by visually inspecting a funnel plot, which graphed study precision against the logit event rate. The funnel plot showed only weak evidence of reporting bias, as there were no studies in the lower right corner. This region corresponds to studies with low precision (small sample size) and a large logit event rate. The Egger et al. (1997) regression asymmetry test did not suggest that publication bias was present (Intercept=−0.034, P=0.957), but due to the moderate number of studies in the analysis this result should be treated with caution.
Targeted analysis on the entire cohort of a candidate CNV on Chr18q22.1
Genome-wide detection of CNVs using SNP array data typically suffers from low sensitivity, thus it is desirable to use an accurate targeted assay to better establish the frequency of promising candidate CNVs in the entire case–control cohort (Scherer et al. 2007). As DNA for such experiments was extremely limited, it was decided a priori that any candidate CNV (Table 1) located within a highly recurrent chromosomal lesion identified by the systematic review would be assayed in the entire case–control sample using RT-qPCR.
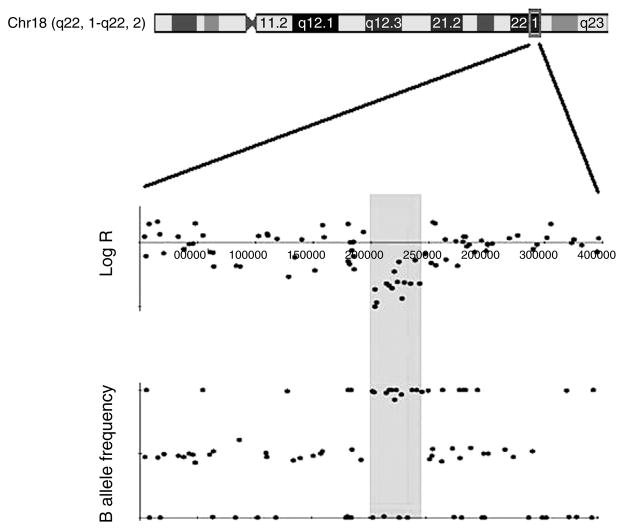
Of the four candidate CNVs returned from the genome-wide analysis, the one which was most enriched among cases is a ~40 kb deletion detected in five cases (2.1%) and zero controls. Although the exact CNV breakpoints have not yet been determined, the array data indicates that the breakpoints vary slightly across cases. This CNV is located at Chr18q22.1, within the chromosomal deletion returned from the systematic review (Fig. 3). The shared region of overlap for the five cases is from 64.00 to 64.042 Mb on chromosome 18 (genomic coordinates are from May 2004, Build 35). This deletion was not identified in any of the 1512 population-based controls, and has only been reported by two studies listed in the Database of Genomic Variants, both at very low frequencies (Itsara et al. 2009, Shaikh et al. 2009). One study found this region to be deleted in 7/1854 controls and another in 12/2026 controls (0.38 and 0.59% respectively), both of which are much lower than the 2.1% frequency detected in our cases using the SNP array data.
Figure 3.
LogR and B allele frequency from GWA data for a subject with a 40 kb deletion at 18q22.1. The LogR plot of this patient shows a clear decrease in the SNP hybridization intensity within a ~40 kb region of Chr18q22.1. The B allele frequency plot shows a stretch of homozygosity in this same region. Taken together, these indicate the patient has a constitutional deletion of the highlighted region, which was subsequently confirmed by real-time qPCR.
All samples from the carcinoid GWAS with DNA remaining were assayed for the presence of this deletion using RT-qPCR (ABI primer ID: Hs04003996_cn). The qPCR experiments determined that 14 of 226 cases (6.19%) and 2 of 97 controls (2.06%) had heterozygous copy-number deletions at 18q22.1. The B allele frequency data demonstrated that these 16 individuals had homozygous SNP calls throughout the 40 kb region, supporting the RT-qPCR results. These samples were found to have a copy-number deletion in this region regardless of whether Coriell control sample NA15510, Coriell control sample NA10851, or the maximum likelihood algorithm was used to set calling thresholds. These 16 samples include patients from all the three study sites and confirmed the presence of this deletion in the five samples predicted to have it using the SNP array data.
Discussion
A pilot GWAS assaying ~300 000 SNPs did not identify any variants that were significantly associated with ileal carcinoid cancer, although rs2208059 in KIF16B showed some evidence of an association. The lack of statistically significant results may be due to insufficient power to detect variants of small effect size, but the rarity of this disease severely limits the sample size that can be attained. Furthermore, rare diseases may be associated with rare variants not amenable to detection by GWAS (Galvan et al. 2010). Despite this, we have assembled a list of top SNP candidates, which will be useful in further studies of ileal carcinoid cancer and which highlight the promise of assembling larger case–control cohorts in the coming years. Until larger cohorts can be assembled for case–control analysis, sequencing studies of leukocytic and matched tumor genomes appear warranted.
Owing to the rarity of ileal carcinoid cancer, rare variants seem likely to be involved in the etiology of the disease. We therefore also sought to identify uncommon CNVs associated with case status. Four CNVs that were not found in controls from the carcinoid GWAS were recurrent among cases, all of which occurred at a higher frequency in cases than in the additional 1512 population-based controls. CNVs can alter gene dosage and cause numerous chronic disorders, such as lupus, osteoporosis, and neuropsy-chiatric disorders including autism, schizophrenia, and mental retardation (Yang et al. 2007, 2008, Itsara et al. 2009). Although constitutional CNVs have typically been associated with rare familial cancer syndromes (e.g. Von Hippel–Lindau disease (Richards et al. 1993), non-polyposis colorectal cancer (Charbonnier et al. 2005)), a ~120 kb deletion was recently shown to predispose children to neuroblastoma, serving as the first example of a non-syndromic cancer which is associated with constitutional CNVs (Diskin et al. 2009).
A systematic review and meta-analysis determined that nearly 70% of ileal carcinoids show loss of chromosome 18q22-qter or all of chromosome 18. This provides strong evidence that loss of a tumor suppressor gene in this region is a critical step in the tumorigenesis of most ileal carcinoids. As this deletion was detected at equal frequencies in primary tumors as in metastatic tumors, which typically harbor a greater number of chromosomal aberrations, this provides substantial evidence that loss of chromosome 18 is an early event in carcinoid tumorigenesis, as opposed to one that is acquired during progression toward metastatic disease.
In our genome-wide analysis of CNVs, the CNV that was most enriched among cases was located at Chr18q22.1. Although this deletion is located in a gene-poor region of the chromosome, it is nonetheless an intriguing observation that deserves further attention. Although most CNVs are believed to arise as a result of non-allelic homologous recombination (NAHR), this deletion is not flanked by any regions with high sequence homology, making NAHR an unlikely explanation for the formation of this CNV (Kidd et al. 2008). The center of the deleted region contains a palindromic AT-rich repeat, a structural element that can induce double-strand breaks and causes the most common known non-Robertsonian translocation (Kurahashi et al. 2010). While non-homologous end-joining can repair such DNA damage, it is an imperfect process capable of creating small CNVs (Conrad & Hurles 2007). Whether such a mechanism could be responsible for a deletion of 40 kb remains to be determined, but may underlie the genesis of this CNV.
Acknowledgments
Funding
Funding was provided by the Verto Institute.
Footnotes
Declaration of interest
Drs E Vosburgh and R S Sackler are affiliated with Purdue Pharma L P, which had no financial interests in the work reported here. No other potential conflict of interest, financial or otherwise, is reported.
References
- Andersson E, Sward C, Stenman G, Ahlman H, Nilsson O. High-resolution genomic profiling reveals gain of chromosome 14 as a predictor of poor outcome in ileal carcinoids. Endocrine-Related Cancer. 2009;16:953–966. doi: 10.1677/ERC-09-0052. [DOI] [PubMed] [Google Scholar]
- Buchanan KD, Johnston CF, O’Hare MM, Ardill JE, Shaw C, Collins JS, Watson RG, Atkinson AB, Hadden DR, Kennedy TL. Neuroendocrine tumors. A European view. American Journal of Medicine. 1986;81:14–22. doi: 10.1016/0002-9343(86)90581-4. [DOI] [PubMed] [Google Scholar]
- Charbonnier F, Baert-Desurmont S, Liang P, Di Fiore F, Martin C, Frerot S, Olschwang S, Wang Q, Buisine MP, Gilbert B, et al. The 5′ region of the MSH2 gene involved in hereditary non-polyposis colorectal cancer contains a high density of recombinogenic sequences. Human Mutation. 2005;26:255–261. doi: 10.1002/humu.20216. [DOI] [PubMed] [Google Scholar]
- Choi M, Scholl UI, Ji W, Liu T, Tikhonova IR, Zumbo P, Nayir A, Bakkaloglu A, Ozen S, Sanjad S, et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. PNAS. 2009;106:19096–19101. doi: 10.1073/pnas.0910672106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad DF, Hurles ME. The population genetics of structural variation. Nature Genetics. 2007;39:S30–S36. doi: 10.1038/ng2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’adda T, Pizzi S, Azzoni C, Bottarelli L, Crafa P, Pasquali C, Davoli C, Corleto VD, Delle Fave G, Bordi C. Different patterns of 11q allelic losses in digestive endocrine tumors. Human Pathology. 2002;33:322–329. doi: 10.1053/hupa.2002.32219. [DOI] [PubMed] [Google Scholar]
- Diskin SJ, Hou C, Glessner JT, Attiyeh EF, Laudenslager M, Bosse K, Cole K, Mosse YP, Wood A, Lynch JE, et al. Copy number variation at 1q21.1 associated with neuroblastoma. Nature. 2009;459:987–991. doi: 10.1038/nature08035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvan A, Ioannidis JP, Dragani TA. Beyond genome-wide association studies: genetic heterogeneity and individual predisposition to cancer. Trends in Genetics. 2010;26:132–141. doi: 10.1016/j.tig.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. Detection of large-scale variation in the human genome. Nature Genetics. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- Itsara A, Cooper GM, Baker C, Girirajan S, Li J, Absher D, Krauss RM, Myers RM, Ridker PM, Chasman DI, et al. Population analysis of large copy number variants and hotspots of human genetic disease. American Journal of Human Genetics. 2009;84:148–161. doi: 10.1016/j.ajhg.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katona TM, Jones TD, Wang M, Abdul-Karim FW, Cummings OW, Cheng L. Molecular evidence for independent origin of multifocal neuroendocrine tumors of the enteropancreatic axis. Cancer Research. 2006;66:4936–4942. doi: 10.1158/0008-5472.CAN-05-4184. [DOI] [PubMed] [Google Scholar]
- Kidd JM, Cooper GM, Donahue WF, Hayden HS, Sampas N, Graves T, Hansen N, Teague B, Alkan C, Antonacci F, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim do H, Nagano Y, Choi IS, White JA, Yao JC, Rashid A. Allelic alterations in well-differentiated neuro-endocrine tumors (carcinoid tumors) identified by genome-wide single nucleotide polymorphism analysis and comparison with pancreatic endocrine tumors. Genes, Chromosomes and Cancer. 2008;47:84–92. doi: 10.1002/gcc.20510. [DOI] [PubMed] [Google Scholar]
- Kulke MH, Freed E, Chiang DY, Philips J, Zahrieh D, Glickman JN, Shivdasani RA. High-resolution analysis of genetic alterations in small bowel carcinoid tumors reveals areas of recurrent amplification and loss. Genes, Chromosomes and Cancer. 2008;47:591–603. doi: 10.1002/gcc.20561. [DOI] [PubMed] [Google Scholar]
- Kurahashi H, Inagaki H, Ohye T, Kogo H, Tsutsumi M, Kato T, Tong M, Emanuel BS. The constitutional t(11;22): implications for a novel mechanism responsible for gross chromosomal rearrangements. Clinical Genetics. 2010;78:299–309. doi: 10.1111/j.1399-0004.2010.01445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lollgen RM, Hessman O, Szabo E, Westin G, Akerstrom G. Chromosome 18 deletions are common events in classical midgut carcinoid tumors. International Journal of Cancer. 2001;92:812–815. doi: 10.1002/ijc.1276. [DOI] [PubMed] [Google Scholar]
- Modlin IM, Lye KD, Kidd M. A 5-decade analysis of 13,715 carcinoid tumors. Cancer. 2003;97:934–959. doi: 10.1002/cncr.11105. [DOI] [PubMed] [Google Scholar]
- Oberg K. Genetics and molecular pathology of neuroendocrine gastrointestinal and pancreatic tumors (gastroenteropancreatic neuroendocrine tumors) Current Opinion in Endocrinology, Diabetes, and Obesity. 2009;16:72–78. doi: 10.1097/MED.0b013e328320d845. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nature Genetics. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Richards FM, Phipps ME, Latif F, Yao M, Crossey PA, Foster K, Linehan WM, Affara NA, Lerman MI, Zbar B. Mapping the Von Hippel–Lindau disease tumour suppressor gene: identification of germline deletions by pulsed field gel electrophoresis. Human Molecular Genetics. 1993;2:879–882. doi: 10.1093/hmg/2.7.879. [DOI] [PubMed] [Google Scholar]
- Scherer SW, Lee C, Birney E, Altshuler DM, Eichler EE, Carter NP, Hurles ME, Feuk L. Challenges and standards in integrating surveys of structural variation. Nature Genetics. 2007;39:S7–S15. doi: 10.1038/ng2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaikh TH, Gai X, Perin JC, Glessner JT, Xie H, Murphy K, O’Hara R, Casalunovo T, Conlin LK, D’Arcy M, et al. High-resolution mapping and analysis of copy number variations in the human genome: a data resource for clinical and research applications. Genome Research. 2009;19:1682–1690. doi: 10.1101/gr.083501.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stancu M, Wu TT, Wallace C, Houlihan PS, Hamilton SR, Rashid A. Genetic alterations in goblet cell carcinoids of the vermiform appendix and comparison with gastrointestinal carcinoid tumors. Modern Pathology. 2003;16:1189–1198. doi: 10.1097/01.MP.0000097362.10330.B1. [DOI] [PubMed] [Google Scholar]
- Su MC, Wang CC, Chen CC, Hu RH, Wang TH, Kao HL, Jeng YM, Yuan RH. Nuclear translocation of β-catenin protein but absence of β-catenin and APC mutation in gastrointestinal carcinoid tumor. Annals of Surgical Oncology. 2006;13:1604–1609. doi: 10.1245/s10434-006-9072-2. [DOI] [PubMed] [Google Scholar]
- Terris B, Meddeb M, Marchio A, Danglot G, Flejou JF, Belghiti J, Ruszniewski P, Bernheim A. Comparative genomic hybridization analysis of sporadic neuroendocrine tumors of the digestive system. Genes, Chromosomes and Cancer. 1998;22:50–56. doi: 10.1002/(SICI)1098-2264(199805)22:1 <50::AID-GCC7> 3.0. CO;2-6. [DOI] [PubMed] [Google Scholar]
- Tonnies H, Toliat MR, Ramel C, Pape UF, Neitzel H, Berger W, Wiedenmann B. Analysis of sporadic neuroendocrine tumours of the enteropancreatic system by comparative genomic hybridisation. Gut. 2001;48:536–541. doi: 10.1136/gut.48.4.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatraman ES, Olshen AB. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics. 2007;23:657–663. doi: 10.1093/bioinformatics/btl646. [DOI] [PubMed] [Google Scholar]
- Wang GG, Yao JC, Worah S, White JA, Luna R, Wu TT, Hamilton SR, Rashid A. Comparison of genetic alterations in neuroendocrine tumors: frequent loss of chromosome 18 in ileal carcinoid tumors. Modern Pathology. 2005;18:1079–1087. doi: 10.1038/modpathol.3800389. [DOI] [PubMed] [Google Scholar]
- Weckstrom P, Hedrum A, Makridis C, Akerstrom G, Rastad J, Scheibenpflug L, Uhlen M, Juhlin C, Wilander E. Midgut carcinoids and solid carcinomas of the intestine: differences in endocrine markers and p53 mutations. Endocrine Pathology. 1996;7:273–279. doi: 10.1007/BF02739834. [DOI] [PubMed] [Google Scholar]
- Yang Y, Chung EK, Wu YL, Savelli SL, Nagaraja HN, Zhou B, Hebert M, Jones KN, Shu Y, Kitzmiller K, et al. Gene copy-number variation and associated polymorphisms of complement component C4 in human systemic lupus erythematosus (SLE): low copy number is a risk factor for and high copy number is a protective factor against SLE susceptibility in European Americans. American Journal of Human Genetics. 2007;80:1037–1054. doi: 10.1086/518257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang TL, Chen XD, Guo Y, Lei SF, Wang JT, Zhou Q, Pan F, Chen Y, Zhang ZX, Dong SS, et al. Genome-wide copy-number-variation study identified a susceptibility gene, UGT2B17, for osteoporosis. American Journal of Human Genetics. 2008;83:663–674. doi: 10.1016/j.ajhg.2008. 10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao JC, Hassan M, Phan A, Dagohoy C, Leary C, Mares JE, Abdalla EK, Fleming JB, Vauthey JN, Rashid A, et al. One hundred years after “carcinoid”: epidemiology of and prognostic factors for neuroendocrine tumors in 35,825 cases in the United States. Journal of Clinical Oncology. 2008;26:3063–3072. doi: 10.1200/JCO.2007.15.4377. [DOI] [PubMed] [Google Scholar]
- Younes N, Fulton N, Tanaka R, Wayne J, Straus FH, II, Kaplan EL. The presence of K-12 ras mutations in duodenal adenocarcinomas and the absence of ras mutations in other small bowel adenocarcinomas and carcinoid tumors. Cancer. 1997;79:1804–1808. doi: 10.1002/(SICI)1097-0142(19970501)79:9<1804::AID-CNCR 24>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Zikusoka MN, Kidd M, Eick G, Latich I, Modlin IM. The molecular genetics of gastroenteropancreatic neuro-endocrine tumors. Cancer. 2005;104:2292–2309. doi: 10.1002/cncr.21451. [DOI] [PubMed] [Google Scholar]