Abstract
As part of an ongoing study of the response of the Streptococcus pneumoniae population to conjugate vaccination, we applied multi-locus sequence typing (MLST) to 291 isolates sampled from nasopharyngeal carriage in Massachusetts children. We found 94 distinct sequence types (STs), including 19 that had not been previously recorded, and a xpt allele containing a large insertion. Comparison with a similar sample collected in 2007 revealed no significant overall difference in the ST composition (p=0.51) suggesting that the population has reached a new equilibrium following the introduction of 7 valent vaccination in 2000. Within serotypes, a large and statistically significant increase (p=0.014 Fisher’s Exact test) was noted in the prevalence of the major multiresistant clone ST 320, which is apparently outcompeting ST 199 among serotype 19A strains. This sample will be used as a baseline to study the future evolution of the pneumococcal population in Massachusetts following introduction of vaccines with higher valency.
Keywords: MLST, serotype replacement, carriage
Introduction
The implementation of a conjugate vaccine against seven pneumococcal serotypes (PCV-7) has been followed by the near disappearance of vaccine types (VTs) as causes of invasive disease and asymptomatic carriage [1–3]. However, non-vaccine types (NVTs) have become more common in carriage, such that the overall carriage prevalence of pneumococcus has remained constant, a phenomenon known as serotype replacement. The increased exposure to NVTs in carriage has been followed by a small but significant increase in invasive pneumococcal disease (IPD) due to these serotypes [4], in particular 19A [2]. A new 13 valent vaccine (PCV-13) extends coverage to some of the NVTs which have been documented in IPD both in the US and elsewhere, including 19A. PCV-13 has been introduced into the routine immunisation schedule in the US. It is anticipated that this will address some of the continued morbidity due to pneumococcal serotypes not covered by PCV-7, as well as leading to another round of serotype replacement in carriage and IPD the consequences of which are unknown.
Pneumococcal serotypes are typically made up of several lineages or clones, which are readily distinguished using multi-locus sequence typing (MLST)[5]. MLST characterises strains based on the combinations of alleles at seven different housekeeping loci scattered around the genome, which together determine the sequence type (ST). MLST is a useful molecular tool for the characterization of pneumococcal clones because, in most cases, features such as antibiotic resistance or serotype, as well as the presence of accessory loci such as pilus [6], are clonal properties, meaning that two strains with the same ST are likely to share them as a result of common ancestry. ST can therefore be used to characterise clones and their properties (for a review of such clonal analyses, see [7]). The pneumococcus is, however, naturally transformable; it is able to take up homologous DNA from other strains and incorporate it into its genome. As a result of such recombination events, genes including those encoding serotype or antimicrobial resistance may be transferred between clones, and over the longer term this diversification will reach the point at which isolates identical by MLST exhibit multiple serotypes or resistance profiles and the association between ST and phenotype breaks down. The diversification of one such lineage (ST 81, or PMEN23F-1) was recently explored by whole genome sequencing, and showed substantial variation arising over the period of a few decades [8]. Serotype switching refers to the process by which the loci determining serotype are transferred between STs, leading to organisms with the same ST expressing more than one capsular type.
We have previously used MLST to study the effect of vaccination with PCV-7 on the carried pneumococcal population among Massachusetts children, through longitudinal samples collected in the community in 2001, 2004 and 2007 [9, 10]. By 2007, the clonal composition of the pneumococcal population had changed dramatically in response to immunization. Nevertheless, by this time, the relative prevalence of different serotypes appeared similar to that observed in pre-vaccine samples, interpreted as evidence that replacement in carriage has led to a new equilibrium in which 19A is the most common serotype in carriage and disease [11]. Between 2001 and 2007, we have been able to document the emergence in carriage, and expansion of important STs in the post vaccine era Some of these have been documented to be increasingly important in resistance and invasive disease [12, 13], and have apparently been generated by serotype switching [14]. We now report the further evolution of the pneumococcal population as determined by sampling from the same communities in 2009. This allows us to document clonal changes within serotypes, and also provides a baseline for continued study following introduction of PCV-13.
Materials and methods
Data collection
Carried pneumococcal strains were collected by nasopharyngeal swab from children aged from 3 months to 7 years of age, between October 2008 and April 2009, from children attending pediatric and family medicine practices for well-child or acute illness visits in 8 Massachusetts communities. Parental consent was obtained by study staff and nasopharyngeal swabs were obtained by trained nurses [9]. All study procedures were approved by the Harvard Pilgrim Health Care institutional review board. Samples were processed for S. pneumoniae growth, antibiotic susceptibility, and serotype using the quellung reaction as previously described [15]. Strains were maintained as glycerol stocks at −80°C and DNA was purified using DNeasy tissue kits (Qiagen, Valencia, CA). We consider 15B and 15C to be a single, rapidly interconverting serotype, 15B/C. The results from the 2008–9 sample were compared with samples from the same communities in the 2006–7 season which have previously been described in detail [9].
Multilocus Sequence Typing (MLST)
Sequence types (STs) of isolates were determined by MLST as previously described [5]. Sequences of each of the seven gene fragments used in the pneumococcal MLST scheme were obtained using an ABI 3730xl DNA analyzer. The sequences were aligned and trimmed to defined start and end positions using MEGA version 4 [16]. Allele and ST assignments were made using the MLST website (www.spneumoniae.mlst.net). All alleles not already present in the pneumococcal MLST database were verified by re-sequencing the gene fragment on both strands. In one isolate, a large insertion was found in the xpt locus. For this isolate the usual forward and reverse primers for xpt were used together with an additional pair of primers located within the insert to amplify and sequence the entire locus (xpt Insert Forward:CAGAGAACTGGCAGACACGA; xpt Insert Reverse:AAAAACAGGGCAATACTACCTCA). For STs found among isolates of more than one serotype the MLST loci were resequenced and serotypes were confirmed by Quellung reaction.
Analysis
Differences in the ST composition of the two populations were estimated using the classification index, with significance assessed using a permutation method as previously described [10, 17]. eBURSTv3 software was used to compare the population structures in 2007 and 2009 [18]. This program groups related STs into clonal complexes (CCs), identifies the probable ancestor of each clonal complex and outputs a graphical representation of these relationships. This enabled us to assess the frequency of CCs of related strains in the 2009 sample and compare them with the sample collected in 2007 [10]. Fisher’s exact test was used to assess the significance of ST changes in each serotype. For individual STs and CCs Fisher’s exact test was used to test for the significance of changes in frequency compared to all other STs and clonal complexes in the serotype. The R statistical package was used in the analysis [19].
Results
Summary
94 distinct STs were found, 19 of which had not been previously recorded. Those present 5 or more times in the sample, and the serotypes associated with them, are shown in Table 1 and the complete results are presented in the supplementary results (SOM Table 1). Several alleles were recorded that were new to the MLST database; 2 gdh, 2 gki, 1 recP, 1 spi, 2 xpt and 2 ddl. One of the novel xpt alleles was found to contain a large ~1.7kb insertion starting at position 358 within the locus. The ancestral allele was xpt 31. Sequencing of a total of 1kb of the insert using normal MLST primers allowed us to design additional primers located within it, which were sufficient to obtain full sequence. Comparison with BLAST results suggested it was a Spn1 transposase (>99% sequence identity). This xpt allele is deposited at the MLST website (www.spneumoniae.mlst.net) in the non-standard length alleles database as allele 324.
Table 1. Most common STs in the 2009 sample.
For those STs found 5 or more times in the sample, the total number of isolates with that ST is shown (N) along with the percentage of the sample comprising isolates with that ST. The serotypes associated with the indicated ST are reported. In some cases different isolates with the same ST may express different capsules. Where this is the case, the numbers of each serotype are shown in parentheses. Full details of all isolates are provided in SOM Table 1.
| ST | N (%) | Serotypes (N) |
|---|---|---|
| 199 | 24 (8.2) | 15B/C (19), 19A (5) |
| 320 | 19 (6.5) | 19A (19) |
| 62 | 16 (5.5) | 11A (14), 11D (1), 36 (1) |
| 558 | 15 (5.2) | 35B (15) |
| 439 | 10 (3.4) | 23B (10) |
| 63 | 9 (3.1) | 15A (7), 15F (1), 19A (1) |
| 191 | 9 (3.1) | 7F (9) |
| 1390 | 9 (3.1) | 6C (8), 6B (1) |
| 3280 | 9 (3.1) | 15B/C (8), 15F (1) |
| 338 | 8 (2.7) | 23A (7) 15B/C (1) |
| 433 | 8 (2.7) | 22F (8) |
| 695 | 8 (2.7) | 19A (7), 6A (1) |
| 816 | 8 (2.7) | 10A (8) |
| 498 | 7 (2.4) | 35F (6), 35B (1) |
| 1379 | 7 (2.4) | 6C (5), 23B (2) |
| 42 | 5 (1.7) | 23A (5) |
| 138 | 5 (1.7) | 6C (4), 6A (1) |
| 547 | 5 (1.7) | 34 (5) |
| 1847 | 5 (1.7) | 23B (5) |
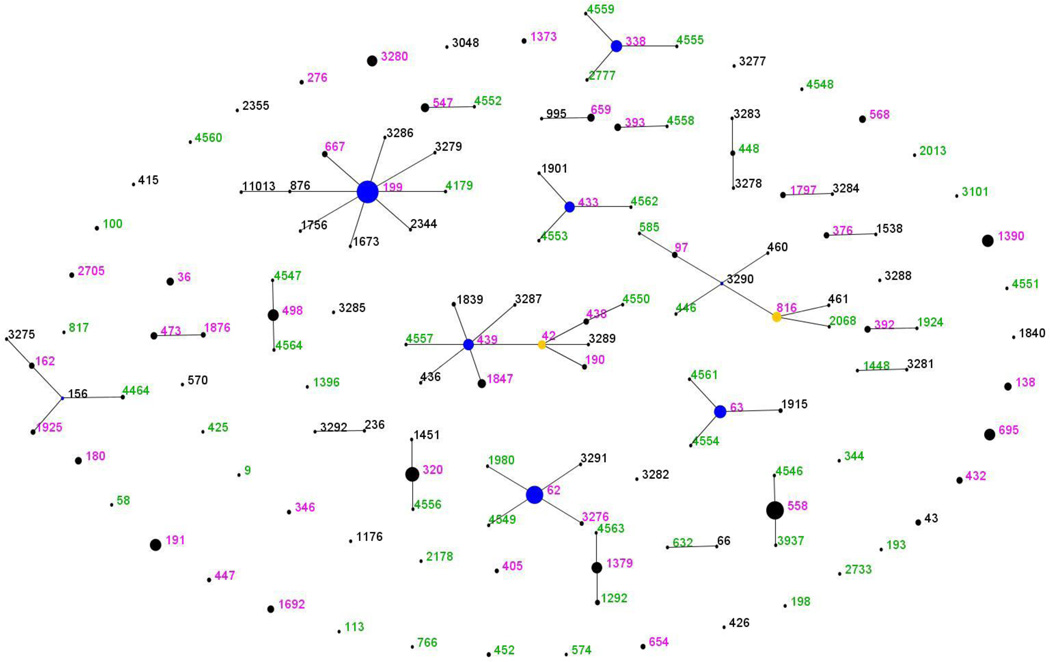
We asked whether the 2007 and 2009 samples were significantly different in their ST composition with the classification index [17]. This test calculates a statistic for the similarity between two populations, and then randomly permutes the pooled populations into two samples of the same size to estimate how the test statistic is distributed under the null hypothesis of the two samples being from the same population. We found no significant evidence of difference between the 2007 and 2009 samples (p=0.507 from 1000 permutations). To allow a broad comparison of the latest results with those obtained in 2007, we generated a comparative eBURST population snapshot (Figure 1). This shows the clonal relationships among STs found at the two time points. STs are represented as circles, with larger circles representing more isolates, with those differing at one of the seven MLST loci linked by solid lines. Groups of related STs are referred to as clonal complexes. STs shown in pink are found in both samples, with those found only in 2007 shown in black. STs that were only recorded in the most recent sample are shown in green. The only novel combinations of ST and serotype in comparison with previous samples were a single isolate of ST 137923B, which is usually associated with a 6C capsule, and a single isolate of ST 33815B/C, which is normally found with a 23A capsule. In both cases serotype was confirmed by repeat Quellung reaction. Note that ST 6319A was recorded in both the 2007 and 2004 samples.
Figure 1. Comparative eBURST diagram showing changes between the 2007 and 2009 samples.
Each ST is represented by a point, the size of which is determined by the number of isolates with that ST in the combined dataset. STs differing at a single MLST locus are shown linked by a straight line. A clonal complex (CC) is a group of STs sharing 6 of 7 alleles with at least one other member of the group. STs which are the putative founders of their CC are shown in blue, with those which are thought to have given rise to subgroups shown in yellow. Those STs which cannot be linked to any other in the sample are termed singletons and appear as unlinked points. For more information see http://spneumoniae.mlst.net/eburst/. The ST numbers are colored according to whether they were found only in 2007 (black), only in 2009 (green) or in both (pink).
Changes within individual serotypes
Changes in the 19A and 6C populations between 2007 and 2009 are shown in Figure 2. The only significant difference in ST composition as assessed by Fisher’s exact test is in the 19A sample (p=0.014). This is due to a decline in ST 199, which is paralleled by the increase in ST 320 and related strains. No significant differences were observed in the populations within serotypes 23A, 15B/C, or 6C, although the marked increase in ST 138 in the 6C population should be noted.
Figure 2. Changes between the 2007 and 2009 samples within individual serotypes.
a) 19A and b) 6C. Results for 2007 are shown in blue, and 2009 in red. STs are shown as the proportion of isolates within each serotype, arranged by clonal complex according to eBURST analysis. The ancestral ST of each clonal complex in eBURST analysis is shown below the panel. Other indicates STs for which ancestry could not be determined by eBURST.
Discussion
We have previously used MLST to characterise samples of carried pneumococci from MA children collected in 2001, 2004 and 2007. In this paper, we report the results of the MLST analysis of the most recent sample, collected in 2009. In marked contrast with all the previous samples, the 2009 sample was not found to be significantly different in terms of overall ST composition from the 2007 sample (CI; p=0.51). This is in contrast with all previous consecutive samples [9, 10], which showed a continuous process of change in the STs composing the population, reflecting the process of serotype replacement that we have described previously. It has previously been shown that by 2007 the population structure of carried serotypes, in terms of diversity, was similar to that observed in the absence of vaccination [11], an observation which was interpreted as a result of the serotype distribution reaching a new equilibrium in the presence of vaccination. We interpret the similarity between the ST composition of the 2009 and 2007 samples as reflecting this new equilibrium.
Our finding of a xpt allele harbouring a large insertion should be understood in the context of other alleles found at this locus. The MLST database contains, at the time of writing, 15 alleles at the xpt locus that contain insertions or deletions (including allele 324, which we report here. See www.spneumoniae.mlst.net). No alleles of the other MLST loci have been found containing insertions or deletions. This may indicate that the xpt locus can be lost or severely mutated with little or no selective consequence. In further support of this, 12 of the recorded xpt alleles at www.spneumoniae.mlst.net contain premature stop codons.
Despite the overall similarity of the 2007 and 2009 samples, this does not preclude considerable changes between 2007 and 2009 in the STs that make up individual serotypes. Since 2007, ST 320 has replaced ST 199 as the most common ST in the 19A population. In parallel, ST 695 has continued to be an increasingly important ST in 19A. Both these STs were previously associated with vaccine serotypes, and demonstrate the ability of successful pneumococcal clones to respond to selective pressures [8]. This capacity will continue to be important as we observe the implementation of new vaccines covering an expanded repertoire of serotypes.
The expansion of STs 320 and 695 in 19A appear to be at the expense of ST 199 (Figure 2a). The reasons for the success of these STs cannot be definitively ascertained from the data we present here, though the publication of a complete genome sequence for ST 320 has highlighted a number of features that could contribute to fitness [20]. We can be reasonably sure that this clone has enjoyed a number of selective advantages dating back to the pre-vaccine era, when it was an important 19F lineage. It was known prior to vaccination that ST 320 and its relatives were associated with resistance to multiple classes of antibiotics. After acquiring a 19A serotype, allowing vaccine evasion [14], this resistance profile has been maintained. We also note the continued presence of ST 695, another example of serotype switching from a vaccine serotype. In fact, ST 695 may be more common with a 19A capsule than it was in the pre-vaccine era, when it was associated with serotype 4 (typically a rather rare serotype in carriage, though important in IPD [21]). Both ST 320 and ST 695 are associated with a type 1 pilus, which has been documented as increasing in the pneumococcal population in Massachusetts [22], and may contribute to fitness.
Despite these changes in the 19A population, ST 199 remains the single most common ST in the 2009 population among all serotypes considered together, as it was in 2007 and 2004 because ST 199 remains common among isolates with a 15B/C capsule. However it has shown a marked decline: 31/222 (14% of all isolates) in 2004, 40/294 (14%) in 2007 to 24/293 (8%) in 2009. Another example of serotype switching from a previously vaccine type strain is the emergence of ST 138 with a 6C capsule, which was previously a 6B strain, and has been increasing since it was recorded as a single isolate in the 2007 sample (see figure 2b). The increase is not significant in this case (Fisher’s exact p = 0.06 comparing ST 138 with all other 6C STs), but the persistence of this clone suggests some variants arising from serotype switching are likely to continue to contribute to the population. It should be noted that clones such as ST 1386C will not be covered by PCV-13, and are expected to derive an advantage from the implementation of this vaccine. Whether serotype switching will enable ST 320 to evade vaccination a second time remains to be seen.
Highlights.
Long-term ongoing study of PCV-7 impact on pneumococcal population structure using MLST.
Multiply resistant 19A clone ST 320 replacing previously dominant ST 199.
Emergence of ST 138 (previously 6B capsule) with a 6C capsule
Evidence population has reached a new equilibrium following serotype replacement
Essential baseline data for studying PCV-13 impact
Supplementary Material
Table 1. Allelic profiles, sequence types and serotypes of 291 isolates of pneumococcus from pediatric carriage in MA in 2009
Acknowledgments
Funding statement
This study was funded by the National Institutes of Health (RO1 AI066304, Finkelstein). WPH was a Royal Society University Research Fellow during this study. The funders had no input into the presentation of the data, the writing of the manuscript or the decision to publish. WPH has received honoraria for advisory board participation for GSK. ML reports consulting and/or honoraria from Pfizer, Novartis, AIR Worldwide, and the Avian/Pandemic Flu Registry (Outcome Sciences) funded by Roche. SIP has been awarded investigator initiated research grants from Pfizer, GSK, Novartis and Intercell, and received honoraria for advisory board participation and/or participation in symposium for GSK, Pfizer and Merck.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
conflicts of interest
The other authors have no conflict of interest to report.
References
- 1.Huang SS, Hinrichsen VL, Stevenson AE, Rifas-Shiman SL, Kleinman K, Pelton SI, et al. Continued impact of pneumococcal conjugate vaccine on carriage in young children. Pediatrics. 2009 Jul;124(1):e1–e11. doi: 10.1542/peds.2008-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pilishvili T, Lexau C, Farley MM, Hadler J, Harrison LH, Bennett NM, et al. Sustained reductions in invasive pneumococcal disease in the era of conjugate vaccine. J Infect Dis. Jan 1;201(1):32–41. doi: 10.1086/648593. [DOI] [PubMed] [Google Scholar]
- 3.Sa-Leao R, Nunes S, Brito-Avo A, Frazao N, Simoes AS, Crisostomo MI, et al. Changes in pneumococcal serotypes and antibiotypes carried by vaccinated and unvaccinated day-care centre attendees in Portugal, a country with widespread use of the seven-valent pneumococcal conjugate vaccine. Clin Microbiol Infect. 2009 Nov;15(11):1002–1007. doi: 10.1111/j.1469-0691.2009.02775.x. [DOI] [PubMed] [Google Scholar]
- 4.Hicks LA, Harrison LH, Flannery B, Hadler JL, Schaffner W, Craig AS, et al. Increase in Non-vaccine-Type Pneumococcal Disease in the Era of Widespread Pneumococcal Conjugate Vaccination, United States, 1998–2004. Journal of Infectious Diseases. 2007;196:1346–1354. doi: 10.1086/521626. [DOI] [PubMed] [Google Scholar]
- 5.Enright MC, Sprattt BG. A multilocus sequence typing scheme for Streptococcus pneumoniae identification of clones associated with serious invasive disease. Microbiology. 1998 Nov;144(Pt 11):3049–3060. doi: 10.1099/00221287-144-11-3049. [DOI] [PubMed] [Google Scholar]
- 6.Moschioni M, Donati C, Muzzi A, Masignani V, Censini S, Hanage WP, et al. Streptococcus pneumoniae contains 3 rlrA pilus variants that are clonally related. J Infect Dis. 2008 Mar 15;197(6):888–896. doi: 10.1086/528375. [DOI] [PubMed] [Google Scholar]
- 7.Henriques-Normark B, Blomberg C, Dagerhamn J, Battig P, Normark S. The rise and fall of bacterial clones:Streptococcus pneumoniae. Nat Rev Microbiol. 2008 Nov;6(11):827–837. doi: 10.1038/nrmicro2011. [DOI] [PubMed] [Google Scholar]
- 8.Croucher NJ, Harris SR, Fraser C, Quail MA, Burton J, van der Linden M, et al. Rapid pneumococcal evolution in response to clinical interventions. Science. 2011 Jan 28;331(6016):430–434. doi: 10.1126/science.1198545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanage WP, Bishop CJ, Huang SS, Stevenson AE, Pelton SI, Lipsitch M, et al. Carried pneumococci in Massachusetts children: the contribution of clonal expansion and serotype switching. Pediatr Infect Dis J. 2011 Apr;30(4):302–308. doi: 10.1097/INF.0b013e318201a154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanage WP, Huang SS, Lipsitch M, Bishop CJ, Godoy D, Pelton SI, et al. Diversity and antibiotic resistance among nonvaccine serotypes of Streptococcus pneumoniae carriage isolates in the post-heptavalent conjugate vaccine era. J Infect Dis. 2007 Feb 1;195(3):347–352. doi: 10.1086/510249. [DOI] [PubMed] [Google Scholar]
- 11.Hanage WP, Finkelstein JA, Huang SS, Pelton SI, Stevenson AE, Kleinman K, et al. Evidence that pneumococcal serotype replacement in Massachusetts following conjugate vaccination is now complete. Epidemics. 2010 doi: 10.1016/j.epidem.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beall B, McEllistrem MC, Gertz RE, Jr., Wedel S, Boxrud DJ, Gonzalez AL, et al. Pre- and postvaccination clonal compositions of invasive pneumococcal serotypes for isolates collected in the United States in 1999, 2001, and 2002. J Clin Microbiol. 2006 Mar;44(3):999–1017. doi: 10.1128/JCM.44.3.999-1017.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pelton SI, Huot H, Finkelstein JA, Bishop CJ, Hsu KK, Kellenberg J, et al. Emergence of 19A as virulent and multidrug resistant Pneumococcus in Massachusetts following universal immunization of infants with pneumococcal conjugate vaccine. Pediatr Infect Dis J. 2007 Jun;26(6):468–472. doi: 10.1097/INF.0b013e31803df9ca. [DOI] [PubMed] [Google Scholar]
- 14.Brueggemann AB, Pai R, Crook DW, Beall B. Vaccine escape recombinants emerge after pneumococcal vaccination in the United States. PLoS Pathog. 2007 Nov;3(11):e168. doi: 10.1371/journal.ppat.0030168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang SS, Platt R, Rifas-Shiman SL, Pelton SI, Goldmann D, Finkelstein JA. Post-PCV7 changes in colonizing pneumococcal serotypes in 16 Massachusetts communities, 2001 and 2004. Pediatrics. 2005 Sep;116(3):e408–e413. doi: 10.1542/peds.2004-2338. [DOI] [PubMed] [Google Scholar]
- 16.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007 Aug;24(8):1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 17.Jolley KA, Wilson DJ, Kriz P, McVean G, Maiden MC. The influence of mutation, recombination, population history, and selection on patterns of genetic diversity in Neisseria meningitidis. Mol Biol Evol. 2005 Mar;22(3):562–569. doi: 10.1093/molbev/msi041. [DOI] [PubMed] [Google Scholar]
- 18.Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol. 2004 Mar;186(5):1518–1530. doi: 10.1128/JB.186.5.1518-1530.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Team RDC. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2005. [Google Scholar]
- 20.Pillai DR, Shahinas D, Buzina A, Pollock RA, Lau R, Khairnar K, et al. Genome-wide dissection of globally emergent multi-drug resistant serotype 19A Streptococcus pneumoniae. BMC Genomics. 2009;10:642. doi: 10.1186/1471-2164-10-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brueggemann AB, Peto TE, Crook DW, Butler JC, Kristinsson KG, Spratt BG. Temporal and geographic stability of the serogroup-specific invasive disease potential of Streptococcus pneumoniae in children. J Infect Dis. 2004 Oct 1;190(7):1203–1211. doi: 10.1086/423820. [DOI] [PubMed] [Google Scholar]
- 22.Regev-Yochay G, Hanage WP, Trzcinski K, Rifas-Shiman SL, Lee G, Bessolo A, et al. Re-emergence of the type 1 pilus among Streptococcus pneumoniae isolates in Massachusetts, USA. Vaccine. 2010 Jul 5;28(30):4842–4846. doi: 10.1016/j.vaccine.2010.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table 1. Allelic profiles, sequence types and serotypes of 291 isolates of pneumococcus from pediatric carriage in MA in 2009