Abstract
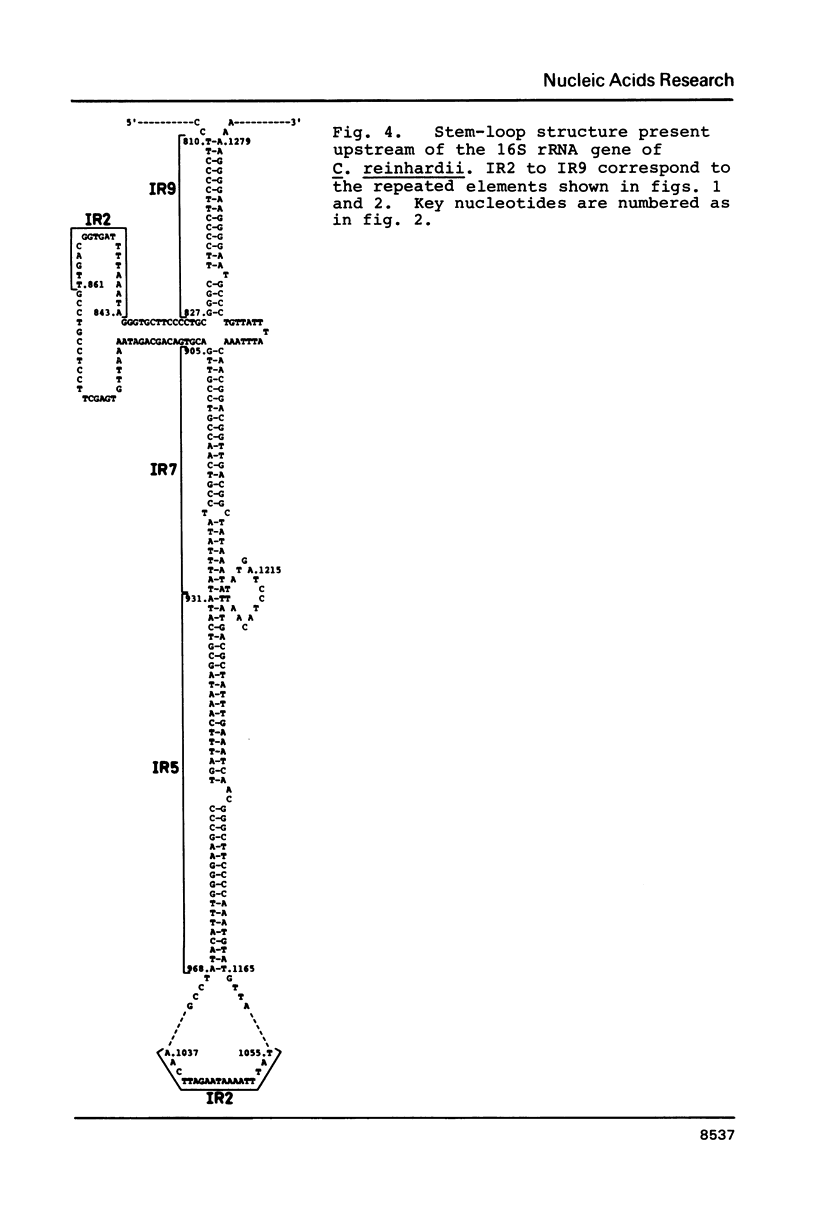
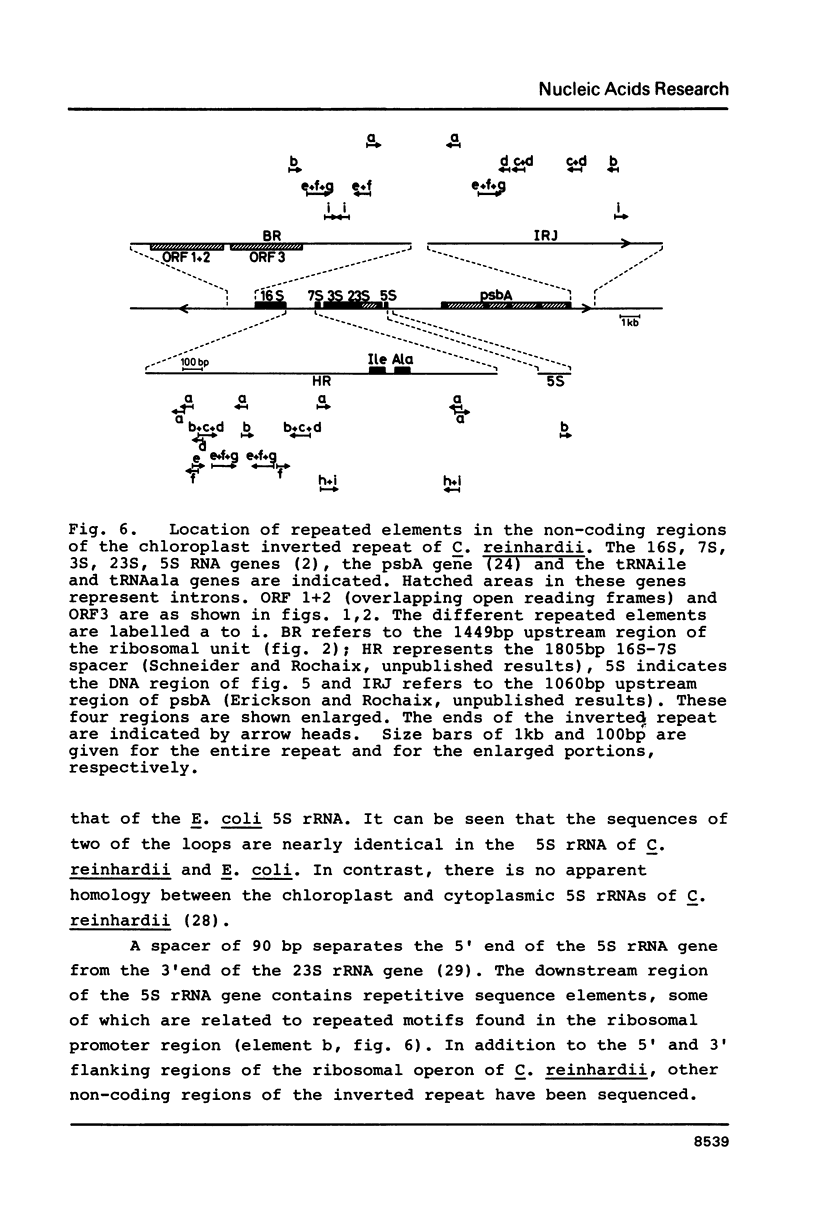
The flanking regions and the end of the chloroplast ribosomal unit of Chlamydomonas reinhardii have been sequenced. The upstream region of the ribosomal unit contains three open reading frames coding for 111, 117 and 124 amino acids, respectively. The latter polypeptide is partially related to the ribosomal protein L16 of E. coli. Two of the open reading frames overlap each other and are oriented in opposite direction. The region between these open reading frames and the 5' end of the 16S rRNA gene contains numerous short direct and inverted repeats which can be folded into large stem-loop structures. Sequence elements that resemble prokaryotic promoters are found in the same region. Several of the repeated elements are distributed throughout the non-coding regions of the chloroplast inverted repeat. Sequence comparison between the 5S rRNA and its gene does not reveal any significant sequence heterogeneity between the chloroplast 5S rRNA genes.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allet B., Rochaix J. D. Structure analysis at the ends of the intervening DNA sequences in the chloroplast 23S ribosomal genes of C. reinhardii. Cell. 1979 Sep;18(1):55–60. doi: 10.1016/0092-8674(79)90353-2. [DOI] [PubMed] [Google Scholar]
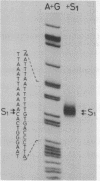
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Briat J. F., Dron M., Loiseaux S., Mache R. Structure and transcription of the spinach chloroplast rDNA leader region. Nucleic Acids Res. 1982 Nov 11;10(21):6865–6878. doi: 10.1093/nar/10.21.6865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J., Chen R. The primary structure of protein L16 located at the peptidyltransferase center of Escherichia coli ribosomes. FEBS Lett. 1976 Sep 15;68(1):105–109. doi: 10.1016/0014-5793(76)80415-2. [DOI] [PubMed] [Google Scholar]
- Darlix J. L., Rochaix J. D. Nucleotide sequence and structure of cytoplasmic 5S RNA and 5.8S RNA of Chlamydomonas reinhardii. Nucleic Acids Res. 1981 Mar 25;9(6):1291–1299. doi: 10.1093/nar/9.6.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donis-Keller H., Maxam A. M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977 Aug;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dron M., Rahire M., Rochaix J. D. Sequence of the chloroplast 16S rRNA gene and its surrounding regions of Chlamydomonas reinhardii. Nucleic Acids Res. 1982 Dec 11;10(23):7609–7620. doi: 10.1093/nar/10.23.7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dron M., Rahire M., Rochaix J. D. Sequence of the chloroplast DNA region of Chlamydomonas reinhardii containing the gene of the large subunit of ribulose bisphosphate carboxylase and parts of its flanking genes. J Mol Biol. 1982 Dec 25;162(4):775–793. doi: 10.1016/0022-2836(82)90547-2. [DOI] [PubMed] [Google Scholar]
- Erickson J. M., Rahire M., Rochaix J. D. Chlamydomonas reinhardii gene for the 32 000 mol. wt. protein of photosystem II contains four large introns and is located entirely within the chloroplast inverted repeat. EMBO J. 1984 Dec 1;3(12):2753–2762. doi: 10.1002/j.1460-2075.1984.tb02206.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz L., Kingsbury D. T., Helinski D. R. Stimulation by cyclic adenosine monophosphate of plasmid deoxyribonucleic acid replication and catabolite repression of the plasmid deoxyribonucleic acid-protein relaxation complex. J Bacteriol. 1973 May;114(2):577–591. doi: 10.1128/jb.114.2.577-591.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Malnoë P., Rochaix J. D. Localization of 4S RNA genes on the chloroplast genome of Chlamydomonas reinhardii. Mol Gen Genet. 1978 Nov 9;166(3):269–275. doi: 10.1007/BF00267618. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Orozco E. M., Jr, Rushlow K. E., Dodd J. R., Hallick R. B. Euglena gracilis chloroplast ribosomal RNA transcription units. II. Nucleotide sequence homology between the 16 S--23 S ribosomal RNA spacer and the 16 S ribosomal RNA leader regions. J Biol Chem. 1980 Nov 25;255(22):10997–11003. [PubMed] [Google Scholar]
- Rochaix J. D., Darlix J. L. Composite structure of the chloroplast 23 S ribosomal RNA genes of Chlamydomonas reinhardii. Evolutionary and functional implications. J Mol Biol. 1982 Aug 15;159(3):383–395. doi: 10.1016/0022-2836(82)90290-x. [DOI] [PubMed] [Google Scholar]
- Rochaix J. D., Malnoe P. Anatomy of the chloroplast ribosomal DNA of Chlamydomonas reinhardii. Cell. 1978 Oct;15(2):661–670. doi: 10.1016/0092-8674(78)90034-x. [DOI] [PubMed] [Google Scholar]
- Rochaix J. D., Rahire M., Michel F. The chloroplast ribosomal intron of Chlamydomonas reinhardii codes for a polypeptide related to mitochondrial maturases. Nucleic Acids Res. 1985 Feb 11;13(3):975–984. doi: 10.1093/nar/13.3.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochaix J. D., Rahire M., Michel F. The chloroplast ribosomal intron of Chlamydomonas reinhardii codes for a polypeptide related to mitochondrial maturases. Nucleic Acids Res. 1985 Feb 11;13(3):975–984. doi: 10.1093/nar/13.3.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochaix J. D. Restriction endonuclease map of the chloroplast DNA of Chlamydomonas reinhardii. J Mol Biol. 1978 Dec 25;126(4):597–617. doi: 10.1016/0022-2836(78)90011-6. [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Schmidt R. J., Richardson C. B., Gillham N. W., Boynton J. E. Sites of synthesis of chloroplast ribosomal proteins in Chlamydomonas. J Cell Biol. 1983 May;96(5):1451–1463. doi: 10.1083/jcb.96.5.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz Z., Kössel H., Schwarz E., Bogorad L. A gene coding for tRNA is located near 5' terminus of 16S rRNA gene in Zea mays chloroplast genome. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4748–4752. doi: 10.1073/pnas.78.8.4748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoncsits A., Brownlee G. G., Brown R. S., Rubin J. R., Guilley H. New rapid gel sequencing method for RNA. Nature. 1977 Oct 27;269(5631):833–836. doi: 10.1038/269833a0. [DOI] [PubMed] [Google Scholar]
- Strittmatter G., Gozdzicka-Jozefiak A., Kössel H. Identification of an rRNA operon promoter from Zea mays chloroplasts which excludes the proximal tRNAValGAC from the primary transcript. EMBO J. 1985 Mar;4(3):599–604. doi: 10.1002/j.1460-2075.1985.tb03672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohdoh N., Shinozaki K., Sugiura M. Sequence of a putative promoter region for the rRNA genes of tobacco chloroplast DNA. Nucleic Acids Res. 1981 Oct 24;9(20):5399–5406. doi: 10.1093/nar/9.20.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]