Abstract
Relapse is a major challenge in the successful treatment of childhood acute lymphoblastic leukemia (ALL). Despite intensive research efforts, the mechanisms of ALL relapse are still not fully understood. An understanding of the molecular mechanisms underlying treatment outcome, therapy response and the biology of relapse is required. In this study, we carried out a genome-wide microRNA (miRNA) microarray analysis to determine the miRNA expression profiles and relapse-associated miRNA patterns in a panel of matched diagnosis–relapse or diagnosis–complete remission (CR) childhood ALL samples. A set of miRNAs differentially expressed either in relapsed patients or at diagnosis compared with CR was further validated by quantitative real-time polymerase chain reaction in an independent sample set. Analysis of the predicted functions of target genes based on gene ontology ‘biological process’ categories revealed that the abnormally expressed miRNAs are associated with oncogenesis, classical multidrug resistance pathways and leukemic stem cell self-renewal and differentiation pathways. Several targets of the miRNAs associated with ALL relapse were experimentally validated, including FOXO3, BMI1 and E2F1. We further investigated the association of these dysregulated miRNAs with clinical outcome and confirmed significant associations for miR-708, miR-223 and miR-27a with individual relapse-free survival. Notably, miR-708 was also found to be associated with the in vivo glucocorticoid therapy response and with disease risk stratification. These miRNAs and their targets might be used to optimize anti-leukemic therapy, and serve as novel targets for development of new countermeasures of leukemia. This fundamental study may also contribute to establish the mechanisms of relapse in other cancers.
INTRODUCTION
Childhood acute lymphoblastic leukemia (ALL) is the most common childhood malignancy worldwide (1,2). Despite substantial improvements in therapy, the number of cases in which relapse occurs is still higher than the number of newly diagnosed cases in other childhood cancers, and the outcome after relapse is generally poor (1,2). Therefore, there is a strong need to develop novel prognostic factors to predict relapse and therapeutic strategies. To this end, insight into the molecular mechanisms underlying treatment outcome, therapy response and the biology of relapse is required.
Several mechanisms for leukemia relapse have been reported in recent years. A number of studies have proposed that ALL relapse results from residual leukemic cells that have survived after therapy (3–6). Other groups have suggested that relapse could result from either the acquisition of a resistant phenotype in response to therapy and subsequent selection or the selection of an inherently resistant subclone initially undetected at diagnosis but present in low numbers (7). Recent work has also shown that ALL relapse is characterized by genomic alterations involving numerous genes, and molecular abnormalities have been identified in matched diagnosis–relapse pairs of childhood ALL samples through DNA microarray studies (8–11). These studies have provided novel biomarkers with potential use for diagnosis and tailored therapy in pediatric acute leukemia. However, it is clear that the pathways involved in ALL relapse are complicated, and the mechanisms that underlie relapse are largely elusive.
A class of small noncoding RNAs ranging from 19 to 25 nucleotides termed microRNAs (miRNAs) was shown to regulate gene expression at transcriptional or post-transcriptional levels (12). Widespread roles of miRNAs in diverse molecular processes driving the initiation and progression of various tumor types are known. The first evidence that miRNAs may be involved in the regulation of hematopoiesis came from a report that miRNAs modulate hematopoietic lineage differentiation (13), and subsequent studies have indicated that miRNAs indeed play a key role in cancer diagnosis and therapy (14). Altered miRNA expression has been observed in leukemia (15–20), and despite the link of miRNAs to childhood ALL development and progression and clinical therapy is revealed (18), little is known about the expression patterns and functions of miRNAs at relapse. Several studies have indicated that miRNAs respond to glucocorticoids (GC) (19) and play a role in multidrug resistance (21), suggesting a functional role for miRNAs associated with relapse in drug-resistant leukemic cells. Other groups have profiled miRNA expression in B-precursor ALL (B-ALL) and T-precursor ALL (T-ALL) (16,18); however, many of these studies focused primarily on the initial diagnosis. Dysregulated miRNAs from pairwise comparisons of matched diagnosis and relapse have not been reported, and their functional relevance in relapse has not been investigated.
In this study, we used genome-wide miRNA microarrays to analyze matched diagnosis–relapse samples in an attempt to gain insight into the biology of relapse in childhood ALL and investigate the possible contributions of miRNA deregulation. For comparison, matched diagnosis–complete remission (CR) samples were also studied. An additional 163 pediatric patients were used as a validation set to confirm the expression of differential miRNAs. A set of abnormally expressed miRNAs was found to be associated with oncogenesis, classical multidrug-resistance pathways and leukemic stem-cell self-renewal and differentiation pathways using gene ontology (GO) ‘biological process’ classifications and experimental validation. We detected the dysregulated expression of miRNAs, including miR-708, miR-223 and miR-27a, in childhood ALL relapse, and these miRNAs could be used to predict the relapse-free survival (RFS) of leukemia in a representative cohort of ALL patients with a 3-year follow-up. These results point to specific miRNA pathways that might be exploited for use as both prognostic indicators and novel targets for therapy.
RESULTS
miRNA expression analysis of paired samples indicates a relapse-associated miRNA pattern in childhood ALL
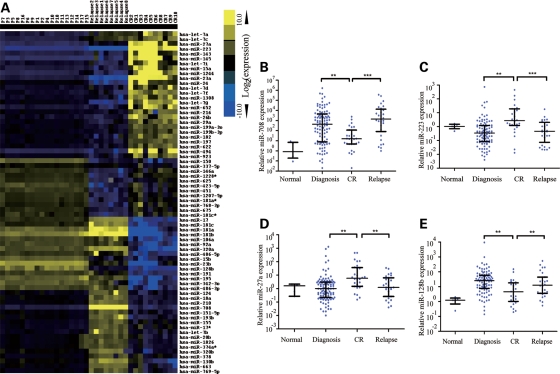
In an effort to identify the biological mechanisms of ALL relapse, we selected 18 matched-pair samples that had sufficient cell material available at diagnosis, relapse or CR and extracted enough total RNA to perform miRNA microarray-based gene-expression profiling. The data from 41 microarrays (8 pairs of diagnosis–relapse samples, 10 pairs of diagnosis–CR samples and 5 healthy controls) were summarized into probe set expression values and clustered. Unsupervised hierarchical clustering based on the expression of these miRNA sets identified three groups that coincided precisely with disease at diagnosis, at relapse and in CR when compared with healthy control; abnormally regulated miRNAs were presented in Supplementary Material, Figure S1.
Using SAM analysis, we identified probe sets that were differentially expressed at relapse compared with initial diagnosis or CR in the match-paired ALL cases. In total, 70 of 576 miRNAs were abnormally regulated in samples at relapse or CR compared with initial diagnosis of the same patients. For samples at relapse versus diagnosis, the most differentially expressed miRNAs included miR-223, miR-23a, let-7g, miR-181, miR-708 and miR-130b, while for the comparison of CR samples with diagnostic samples, miR-27a, miR-223, miR-23a, miR-181 and miR-128b have differential expression pattern. The results also showed that miRNA expression profile of the CR samples was significantly distinct from the relapse group (Fig. 1A and Supplementary Material, Table S1). Among these miRNAs, miR-223, miR-23a and let-7g were downregulated with higher fold changes in the relapse samples compared with the samples of the same patients at CR, whereas miR-181 family, miR-708 and miR-130b were upregulated with higher fold changes in the relapse samples. Moreover, miR-708 was the most upregulated miRNA in the relapse samples, whereas miR-223 was significantly downregulated at relapse, suggesting that miR-708 and miR-223 may have important roles in pediatric ALL relapse. Other miRNAs, such as miR-27a and miR-128b, whose expressions are downregulated in relapse compared with CR, may be involved in the progress of relapse caused by drug resistance after chemotherapy, whereas miR-23a whose expression is much lower in relapse than diagnosis when compared with CR and miR-181 whose expression is much higher in relapse than diagnosis when compared with CR might have a complicated role in leukemogenesis and relapse. To confirm the differential expression pattern, we performed a quantitative real-time polymerase chain reaction (qRT-PCR) assay to validate the expression of miR-23a, miR-223, let-7g, miR-181a, miR-181b, miR-128b, miR-27a, miR-708 and miR-130b in the samples at initial diagnosis and relapse as well as CR as shown in Supplementary Material, Figure S2. The qRT-PCR results largely confirmed the microarray data except for let-7g, whose expression did not show significantly different among three groups.
Figure 1.
Cluster analysis of miRNA expression of paired relapse and/or CR samples in childhood ALL and validation with qRT-PCR. (A) The 70 top-ranked differentially expressed miRNAs in pediatric ALL; the average miRNA level at diagnosis was set as 1. The fold changes of individual miRNA at diagnosis, relapse or CR, against average miRNA level at diagnosis were clustered in this tree distribution. (B–E) Differential expressions of miRNAs at diagnosis, relapse or CR were validated with qRT-PCR. (B) miR-708 (C) miR-223 (D) miR-27a and (E) miR-128b. Normal: healthy control; diagnosis: samples at initial diagnosis; CR: samples in first complete remission; relapse: samples at relapse.
The qRT-PCR results showed that miR-708, miR-223, miR-27a and miR-128b were differentially expressed in patients either in relapse or at original diagnosis compared with those in CR (P< 0.01). Notably, miR-708 and miR-223 have the most significant differences between the relapse and CR patients. Thus, we further detected their expression in a large sample set, which consisted of unmatched bone marrow samples at initial diagnosis (n= 104) and relapse (n= 29) as well as CR (n= 30) (Fig. 1B–E), suggesting that these molecules could have functional relevance in disease recurrence. For example, the expression of miR-708 was higher in relapse while lower in CR compared with diagnosis, implying that miR-708 might control a cancer-related factor in the progress of leukemia (Fig. 1B). In contrast, the expression of miR-223 was higher in CR, suggesting an oncogene-like factor or drug-resistant factor in the progress of leukemia was regulated by miR-223 (Fig. 1C). Result of qRT-PCR indicated that miR-27a presented a similar expression pattern as miR-223 (Fig. 1D). In a previous work, we have demonstrated that miR-27a could directly regulate expression of a drug-resistant factor, P-glycoprotein and overexpression of miR-27a increased sensitivity of leukemia cells to doxorubicin, indicating that downregulated miR-27a might be involved in relapse via inducing drug resistant of leukemia (21). MiR-128b was reported to promote drug resistance in many cancers, including ALL (22,23). Here, we also found that the expression of miR-128b was higher in relapse and at diagnosis compared with CR, implying that this miRNA might control a tumor suppressor in the progress of leukemia (Fig. 1E).
Dysregulated miRNAs target key factors involved in ALL progression and relapse
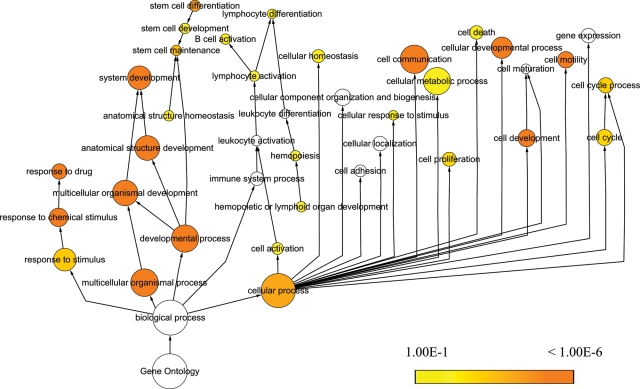
The temporal variation in miRNA expression levels at diagnosis and relapse suggested that miRNAs control the expression of key proteins underlying mechanisms leading to leukemogenesis and relapse. To elucidate the pathogenic role of the significantly differentially expressed miRNAs at diagnosis and relapse (fold change > 2.0 and P< 0.001), a total of 46 miRNAs were selected for targets analysis. The targets were predicted by PicTar, TargetScan and miRanda (24) and 395 genes were predicted as the targets of miRNAs output by GO analysis (Supplementary Material, Table S2). Among the 46 miRNAs, there are 5 miRNA families including miR-23, miR-181, let-7, miR-15 and miR-320. Because different members in a family share the same functions, thus only one member representing the family was shown in the cluster, as a result only 35 miRNAs presented (Fig. 2, on the top). By examining the significant GO ‘biological process’ classifications that were over-represented among the putative targets genes of the differentially expressed miRNAs, we analyzed the functional annotation for predicted target sets. Clustering of the over-represented GO classes in predicted targets of dysregulated miRNAs showed that the most significant GO terms were genes involved in macromolecular metabolic process, biosynthetic process and transcription regulation (Fig. 2). Notably, a number of miRNAs were related to drug transport, multidrug resistance or regulation of cellular biosynthetic process (Fig. 2, blue), and these annotated functions might be associated with miR-124, miR-145, miR-155 and miR-223 etc. Another cluster of significant GO terms was associated with stem-cell development and differentiation (Fig. 2, red), and these genes might be correlated with miR-27a, miR-128b, miR-29a, miR-18a etc. To evaluate the functional significance of the miRNA target genes, we also used BiNGO (Biological Networks Gene Ontology) (25) to construct a hierarchical ontology tree in Cytoscape (26), as shown in Figure 3. The color represents enrichment significance—the deeper the color on a color scale, the higher the enrichment significance. White color nodes are not enriched but show the hierarchical relationship among the enriched ontology branches. It was shown that the nodes representative genes response to chemical stimulus or drug as well as to stem-cell differentiation have a deeper color, implying that relapse could arise from activation of drug-resistance related genes or silent cancer stem-cell loci by miRNAs after diagnosis due to induction of resistance during chemotherapy.
Figure 2.
GO analysis. Clustering of over-represented GO classes in predicted targets of 46 differential miRNAs (fold change > 2.0 and P < 0.001). All genes with statistically overrepresented GO annotations were included (P < 0.001).
Figure 3.
Relapse-associated miRNA targets enrichment network based on GO molecular functions and biological processes. Significantly overrepresented GO terms based on GO molecular functions and biological processes were visualized in Cytoscape. The size of a node is proportional to the number of targets in the GO category. The color represents enrichment significance—the deeper the color on a color scale, the higher the enrichment significance. White color nodes are not enriched but show the hierarchical relationship among the enriched ontology branches.
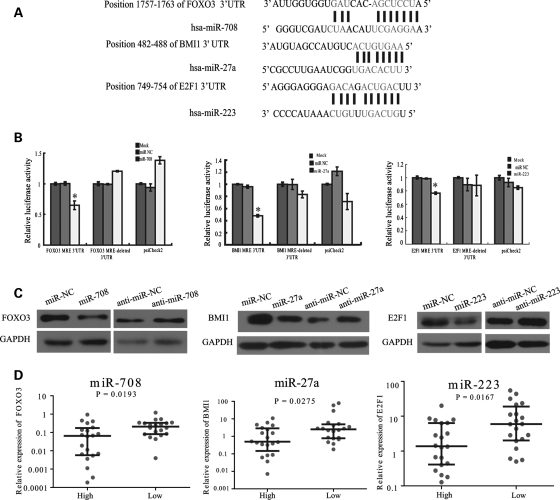
To further get insights into the biological pathways of the dysregulated miRNAs mentioned above, an mRNA–miRNA network by analysis of mRNA as opposed to miRNA was constructed to search the function of target genes involved in leukemogenesis and leukemia relapse. Dysregulated miRNAs and their targets or potential target mRNAs including the already known and our prediction were connected in Supplementary Material, Figure S3. Target prediction showed that BMI1, a transcription factor necessary for hematopoietic stem cell (HSC) and leukemia stem-cell self-renewal (27,28), is the putative target of both miR-27a and miR-128b, while E2F1, a master cell-cycle regulator, is the target of miR-223 (29). In addition, FOXO3, a forehead transcription factor that is critical for HSC self-renewal and mediates the initial apoptotic response (30–32), contained the conserved miR-708 response elements (MRE) in its 3′ untranslated terminal region (UTR).
To confirm the direct effects of these miRNAs, we fused the 3′UTR sequences of FOXO3, BMI1 and E2F1, each containing putative binding sites to their respective miRNA, to a luciferase reporter gene. By cotransfecting the miRNA mimics with the corresponding constructs, we found that all three miRNAs significantly repressed luciferase activity compared with miR-negative control (NC) (scrambled oligonucleotides) and mock (water) (Fig. 4A and B). Deletion of each seed sequence binding with the miRNAs showed that all contributed to the regulation of these transcripts, although not to the same degree (Fig. 4A and B). Cotransfection of the miR-708 mimics and the luciferase vector with the 3′UTR of FOXO3 into HEK-293T cells reduced luciferase activity to 40% compared with a miR-NC. Cotransfection of the BMI1 luciferase constructs with the precursor of miR-27a and cotransfection of the E2F1 luciferase constructs with the precursor of miR-223 reduced luciferase activity to 60 and 22%, respectively, compared with controls. To further determine whether these genes are regulated by their corresponding miRNAs, we performed both miRNA overexpression and knockdown experiments in the Jurkat cell, a T-ALL cell line and examined the expressions of FOXO3, BMI1 and E2F1, respectively. As shown in Figure 4C, when transfected with the miRNA mimics (small molecular mimics of mature miRNAs) into the Jurkat cell, miR-708, miR-27a and miR-223 significantly decreased the expression levels of the FOXO3, BMI1 and E2F1 proteins, respectively, compared with miRNA negative control (miR-NC, miRNA negative control, scrambled oligonucleotides). Alternatively, when the Jurkat cells were transfected with the miRNA inhibitors (antisenses, anti-miRs), the protein expression increased. We further examined the mRNA expression of FOXO3, BMI1 and E2F1 in patient samples. The results indicated that expressions of these miRNAs have an inverse correlation with the corresponding target gene expression (Fig. 4D). Taken together, these data support a direct effect of these miRNAs on their target at both transcription and post-transcription levels.
Figure 4.
miRNAs dysregulated specifically at relapse repress expression of key proteins. (A) Schematic representation of the interaction of miRNA with the 3′UTR of its corresponding targets. The MRE in the 3′UTR of human were predicted by TargetScan and miRBase target. Each predicted MRE 3′UTR was inserted into a psiCHECK-2 vector immediately downstream from the Renilla luciferase gene. (B) Luciferase reporter assays analyzing the putative targets of the three miRNAs. HEK-293T cells were co-transfected using Lipofectamine 2000 with psiCHECK-2 plasmids with the 3′UTR of target genes-wt or the target genes-mut and miRNA mimics or mimics negative control (mimics-NC). Firefly luciferase activity was normalized to renilla luciferase activity, and the results were expressed relative to the control (miRNA mimics/mock). (C) Jurkat cells were transfected with 100 nm miRNA inhibitor-NC or miRNA inhibitor and mimics-NC or miRNA mimics, respectively; miR-708 (left panel), miR-27a (middle panel) and miR-223 (right panel). Cells lysates were prepared for western blotting with antibodies against FOXO3, BMI1 or E2F1. GAPDH expression served as loading controls. (D) Differential expressions of FOXO3, BMI1 or E2F1 mRNA in diagnosis samples with different miRNA expressions were measured by qRT-PCR, GAPDH expression served as loading controls.
Specific miRNA expression is associated with the risk of relapse in childhood ALL
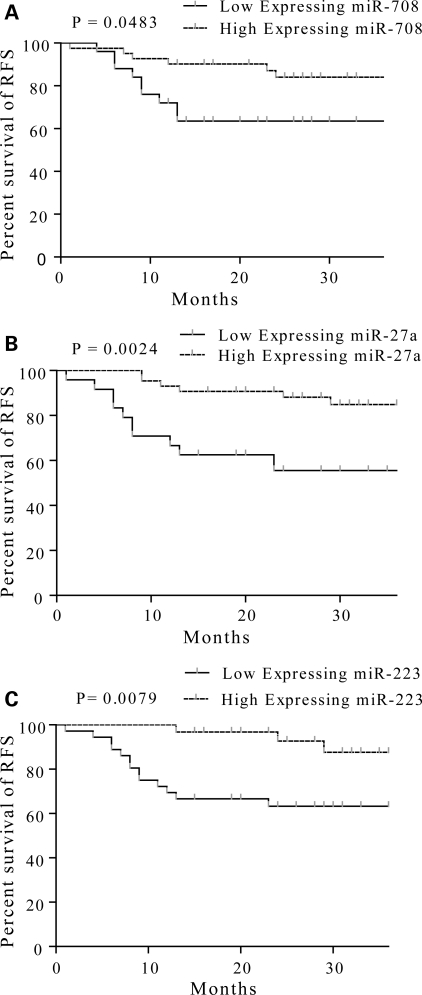
The above results indicated a specific pattern of miRNA expression and the biological pathway associated with leukemia relapse. We then asked whether these miRNAs could serve as the biomarkers for prediction of leukemia relapse? To address this issue, we analyzed the expressions of miR-708, miR-27a, miR-223 and miR-128b with qRT-PCR from a cohort of 70 newly diagnosed childhood ALL patients. These miRNAs were chosen because of their differential expression pattern and their targets involved in biological pathways related to drug resistance or leukemic stem-cell maintenance and differentiation. The patterns of these miRNAs expressions were compared with clinical outcome of childhood ALL in a 36-month follow-up survey. We found that the levels of miR-708 (P= 0.0483), miR-223 (P= 0.0079) and miR-27a (P= 0.0024) at diagnosis were correlated to RFS (Fig. 5A–C). To visualize the association of these miRNAs with outcome, the discrete variables of each miRNA at diagnosis were clustered into two groups, high and low expression according to the median expression in the full set of 70 samples. The Kaplan–Meier survival plot was generated based on these two groups of each miRNA. Patients with high miR-708 expression at diagnosis had a higher RFS rate (84.0%) compared with patients in the low expressional group, which had 66.2% of RFS. Similar correlations were observed in miR-223 of which a higher RFS rate was correlated with high expression cluster (87.6%) compared with 63.3% of RFS in low expression group, and miR-27a, where the RFS rate in the high expression group was 84.8% compared with 65.6% in the low expression group. No clear correlation between the level of miR-128b at diagnosis and RFS was identified (P= 0.7152) (data not shown).
Figure 5.
Dysregulated miRNAs at diagnosis can predict RFS. The representative RFS curves for miR-708 (A), miR-27a (B) and miR-223 (C) were measured by qRT-PCR. Statistical differences between curves were calculated using the log-rank test.
To determine further whether miRNA expression profiling could be used to predict ALL relapse, we validated the predictive values obtained from miRNA profiling. The specificities of miR-708, miR-223 and miR-27a were 67.9, 74.7 and 69.4%, respectively, and the sensitivities of these miRNAs were 55.9, 58.1 and 56.4%, respectively. Because the predictive objects of these three miRNAs did not completely overlap one another, we postulated that a combination of two or three miRNAs might improve the predictive value. As expected, the specificity of a combination of these three miRNAs reached as high as 91.4%, and the sensitivity of a combination of at least two of the three miRNAs reached 90.5%. Our results indicate that the use of a miRNA profile could yield a high accuracy of predicting pediatric ALL relapse.
Next, we used univariate Cox regression to evaluate the impact of these data. Both miR-708 and miR-223 were found to be univariate predictors (P= 0.011 and P= 0.004, respectively). However, miR-27a was failed in univariate Cox regression analysis due to high variance (P= 0.135). A multivariate Cox regression analysis was also assessed to evaluate the impact of these miRNAs at diagnosis on RFS. The frequently used clinical prognosis factors [age, immunophenotyping, prednisone treatment response, white blood cell (WBC) count before chemotherapy, remission status after 15-day and 33-day chemotherapy and the fusion gene BCR–ABL and MLL] were included in this analysis (Table 1). Patients with low miR-223 expression had a 24.22-fold higher risk of relapse (P= 0.001, 95% CI 3.73–157.28). Remission rate after 33-day chemotherapy, immunophenotyping, age and MLL positive were also significant predictors (P= 0.002, P= 0.002, P= 0.048 and P= 0.019, respectively). Results indicated that miR-223 expression at initial diagnosis is an independent and reliable predictor for relapse. Our analyses inferred that miR-708 expression at diagnosis is also a reliable predictor but not an independent one.
Table 1.
Univariate and multivariate Cox regression analysis
| Factor (categories) | Hazard ratio (95% CI) | P-value |
|---|---|---|
| Univariate Cox regression | ||
| Age (10-year increase) | 0.041 | |
| Immunophenotyping (T-ALL) | 0.340 | |
| Prednisone treatment response (PPR) | 0.021 | |
| WBC before chemotherapy (20 × 109 per liter increase) | 0.070 | |
| 15-day remission statusa (blast ≥ 25%) | 0.009 | |
| 33-day remission statusa (blast ≥ 5%) | <0.001 | |
| BCR–ABL (positive) | 0.002 | |
| MLL (positive) | 0.759 | |
| miR-708 (low expression) | 0.011 | |
| miR-223 (low expression) | 0.004 | |
| miR-27a (low expression) | 0.135 | |
| multivariate Cox regression | ||
| Age (10-year increase) | 3.30 (1.01 to 10.77) | 0.048 |
| Immunophenotyping (T-ALL) | 11.26 (2.45 to 51.69) | 0.002 |
| 33-day remission status (≥5%) | 6.22 (2.00 to 19.40) | 0.002 |
| miR-223 (low expression) | 24.22 (3.73 to 157.28) | 0.001 |
| MLL | 26.79 (1.71 to 420.28) | 0.019 |
aDuring induction treatment.
Upregulation of miR-708 is associated with in vivo GC response and disease risk aggressiveness in childhood ALL
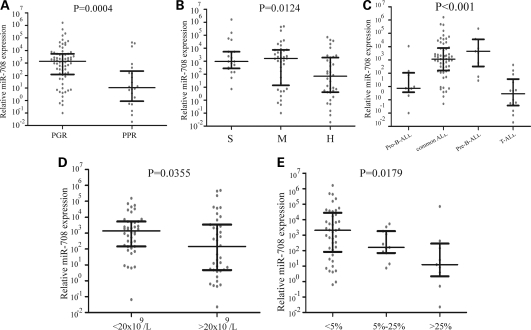
GCs are clinically used to treat childhood ALL and other lymphoid malignancies (33). Poor response to 7-day monotherapy with the GC prednisone is one of the strongest predictors of adverse outcomes in the treatment of childhood ALL (33,34). Despite rapid advances in biomedical technology facilitating the development of molecular diagnostic assays, more reliable predictor for GC resistance in ALL is still eagerly needed. As mentioned above, special miRNAs pattern could serve as the biomarkers to predict the relapse of childhood ALL. Can they also as a reliable stratification factor contribute to predict GC resistance? To address this question, we retrospectively analyzed the outcome of ALL patients with initial response to prednisone treatment data available. Therapeutic effect was defined as a reduction in leukemic blasts in the peripheral blood to below 1000 per µl after 7 days of treatment with prednisone. The 7-day window of prednisone monotherapy (before the combination chemotherapy) used to ascribe pediatric patients with ALL to either the non-high-risk (prednisone good response, PGR) or high-risk (prednisone poor response, PPR) categories and provided an opportunity to correlate the miRNA expression levels with the in vivo GC (prednisone) response. The miR-708, miR-223 and miR-27a were analyzed with the specimens at initial diagnosis through all the groups. The miR-708 expressions significantly distributed in different patterns between the prednisone response groups. Highly expressed miR-708 was detected from specimens in PGR, while lower miR-708 level appeared in PPR (P= 0.0004), indicating miR-708 level before chemotherapy was highly predictive of GC response (Fig. 6A). However, despite some patterns also obtained from miR-27a and miR-223, there were no statistical significances between the two groups (P= 0.2933 for miR-27a and P= 0.8963 for miR-223, Supplementary Material, Figs S4A and S5A).
Figure 6.
miR-708 expression is associated with prednisone response, risk group and relapse in ALL subtypes. (A) Differentially expressed miR-708 in ALL patients with varying responses to prednisone treatment. (B) miR-708 expression in different risk groups for ALL was confirmed using qRT-PCR. (C) miR-708 expression pattern in different subtypes of ALL. Lower expression was observed in T-ALL. (D) Differential expression of miR-708 in samples with different WBC counts before chemotherapy. (E) Differential expression of miR-708 at time of diagnosis in samples with different remission statuses (measured by percentage of blast cells) after 15 days of chemotherapy.
We then clustered patients into three risk stratification groups, standard risk (S), middle risk (M) and high risk (H), based on well-known risk factors, i.e. age, cytogenetics, remission status, response to induction therapy and survival outcomes according to the risk stratification system from ALLIC BFM 2002. We found that the patients in both standard risk and middle risk had higher expressed miR-708, while the high-risk patients displayed a significantly low level of miR-708 (H versus S: P= 0.0040; H versus M: P= 0.0280). There were no significant difference in miR-708 expression levels between standard risk and middle risk groups (P= 0.7444) (Fig. 6B). Similar pattern was not observed in either expression of miR-27a or miR-223 at patient groups (Supplementary Material, Figs S4B–E and S5B–E). In this study, we also found that the miR-708 expression at initial diagnosis differed among the four immunophenotypes, pro-B-ALL, pre-B-ALL, common ALL and T-ALL (Fig. 6C). The T-ALL phenotype, commonly recognized to correspond with higher risk of relapse, had the lowest miR-708 expression levels, implying lower miR-708 before therapy correlating to high relapse risk in patients received chemotherapy.
Correlation of miR-708 level before chemotherapy with other risk factors was also evaluated in this study. Patients with high miR-708 expression at diagnosis had a higher overall survival rate (P= 0.0174, Supplementary Material, Fig. S6) compared with patients in the low expressional group. WBC count at diagnosis of >20 × 109 per l is considered factor affecting 5-year survival rates of ALL patients. We therefore studied whether where was the negative correlation between miR-708 level and WBC count before chemotherapy. As shown in Figure 6D, miR-708 expression was lower in the higher WBC group (>20 × 109 per l) compared with the lower WBC group (P= 0.0355), giving another evidence of that patients whose miR-708 lower before therapy has higher relapse potential. We also noted that patients with the BCR–ABL fusion gene and patients older than 12 years, who are considered at higher risk for relapse, also have lower miR-708 expression (P= 0.0420 and P= 0.0030, respectively, data not shown). Notably, patients who exhibited a better remission status after 15-day chemotherapy (<5% blasts) had higher miR-708 expression than those in an unfavorable remission status (>25% blasts) (P= 0.0124) (Fig. 6E). In addition, patients with a better remission status after 33-day chemotherapy (<5% blasts) also had a higher miR-708 expression than those at a worse remission status (≥5% blasts) (P= 0.0177, data not shown), suggesting that low miR-708 expression levels could predict a higher risk for leukemogenesis or relapse.
DISCUSSION
Relapse following initial chemotherapy remains a barrier to survival in ∼20% of children suffering from ALL and is, therefore, of great interest to physicians and researchers. Despite intensive research efforts, the molecular mechanisms underlying ALL relapse are still not fully understood. In this study, we initiated a genome-wide miRNA microarray analysis in paired diagnosis–relapse and/or CR bone marrow samples of childhood ALL. There were two advances in this study. First, we identified a differential miRNA pattern between relapse and CR patients that included miR-708, miR-223 and miR-27a. In addition, miR-708 was upregulated in relapse, whereas both miR-223 and miR-27a were highly expressed in patients during CR, suggesting that these dysregulated miRNAs play important roles in controlling disease progression—namely, in the relapse of patients who had been in remission.
Secondly, to elucidate the fundamental roles of these relapse-associated miRNAs, we analyzed the putative gene targets of these dysregulated miRNAs using GO ‘biological process’ categories. This analysis revealed that a set of abnormally expressed miRNAs was associated with oncogenesis, classical multidrug-resistance pathways and leukemic stem-cell self-renewal and differentiation pathways. Our results provide evidence for a link between relapse and the leukemic stem-cell population; such a link has not previously been reported in the context of DNA microarray analyses in which significant differences have been found in the expression of genes involved in cell-cycle regulation, DNA repair and apoptosis (9–11).
Low miR-223 and miR-27a expression levels were detected in diagnostic specimens from patients who subsequently relapsed during the study. Furthermore, miR-223 had previously been identified as an essential factor in myeloid lineage development and as a putative tumor-suppressor gene in recurrent ovarian cancer (35). Previous gene-expression profiling identified a subset of T-cell ALL with myeloid-like genetic features and miR-223 overexpression (36). Also, miR-223 has been proposed to be an important factor in leukemogenesis (37); therefore, we propose that its downregulation contributes to the survival of leukemic cells.
We further determined that E2F1 was a target of miR-223 in leukemic cells. E2F1 has a controversial role in cell-cycle control, as it exhibits a dual behavior: it can act as either a tumor-suppressor or an oncogene. Therefore, we speculated that miR-223 could be used as an indicator of leukemia relapse mediated by E2F1. Another downregulated miRNA, miR-27a, is involved in the carcinogenesis of several cancer types. Interestingly, miR-27a exerts its oncogenic effects by regulating ZBTB10, which results in the overexpression of Sp proteins and Sp-dependent genes, which are important for cell survival and angiogenesis (38,39). We previously found that miR-27a is relevant to treatment outcome in vivo (21) and might be involved in the relapse of both lymphocytic leukemia and myeloid leukemia. This result suggests that downregulation of miR-27a might promote ALL relapse by regulating genes in classical drug or multi-drug resistance pathways.
The most notable finding from this study was that many of the identified miRNAs and their target genes are implicated in leukemic cell (including leukemic stem cell) development, differentiation and activation. For example, the polycomb gene BMI1 was identified as a bona fide target of miR-27a in this study. A number of studies have demonstrated that BMI1 plays an essential role in regulating self-renewing HSCs and leukemia stem cells in humans. Deletion of BMI1 can inhibit the self-renewal of tumor stem cells and prevent leukemia recurrence (40). Taken together with the finding that miR-27a targets the multi-drug resistance gene MDR1 (21), it seems possible that miR-27a downregulation leads to the accumulation of BMI1 or MDR1 protein, which, in turn, might promote leukemic cell proliferation or drug resistance, leading to leukemia recurrence. In addition, our results revealed that FOXO3 is a direct target of miR-708. FOXO3 has been reported to act as either an oncogene or a tumor suppressor in leukemia (41,42). It has also been shown that FOXO3 transcriptional activity is required to prevent B-chronic lymphocytic leukemia and chronic myelogenous leukemia (30,41). We, therefore, hypothesize that high miR-708 expression mediated by inhibition of FOXO3 is critical for switching from leukemia remission to relapse. Interestingly, miR-708 showed different expression patterns between patients who have relapse potential and those who are in relapse. The MiR-708 level is low in high relapse risk patients at diagnosis, while specimens of relapsed patients were measured the abundance of miR-708. One possible reason for this phenomenon is that miR-708 is critical for HSC self-renewal because it targets FOXO3. FOXO3 has an essential role in the maintenance of the leukemia-initiating cells (LICs) that drive the recurrence of chronic myeloid leukemia (41). Low miR-708 levels in a high-relapse risk patient might result in loss of control of FOXO3 stem-cell self-renewal functionality. As a result, LIC proliferation might be stimulated. The exact mechanism of this phenomenon has yet to be defined.
Another contribution of this study is the finding that some of these relapse-associated miRNAs (miR-708, miR-27a and miR-223) could potentially be used as biomarkers for predicting RFS. We found that the expression profiles of the three miRNA could yield highly accurate predictions of pediatric ALL outcomes, especially when two or three miRNAs were considered in combination. The sensitivity and specificity of these miRNAs are comparable to those of mRNAs, such as IKZF1 (the sensitivity and specificity of which are 90.1 and 65%, respectively) (43), IL-15 (56.7 and 55.5%) (44), CASP8AP2 (75 and 79.4%) (45) and a 21-gene classifier (71.6 and 77.4%) (46). Our data indicated that miRNA profiles could also be a novel and reliable indicator for the prediction of pediatric ALL relapses. However, because we did not measure mRNA expression and we do not have miRNA and mRNA data for the same samples, we could not address whether the miRNA data add anything to the mRNA data predictions. Further research will be needed to assess whether miRNA information adds predictive value or whether integrating information from both mRNA and miRNA profiles could improve the accuracy of predicting clinical ALL relapse.
In conclusion, our study reports the miRNA profiles from diagnosis, relapse and CR leukemia samples. To our knowledge, this report is the first to show that ALL relapse might be mediated by miRNA deregulation. The existence of ALL relapse-associated miRNAs provides new insight into the molecular mechanisms of leukemia relapse such as that relapse may be triggered as a consequence of therapy-induced miRNA dysregulation. We also demonstrated, for the first time, that some relapse-associated miRNAs that correlated with the RFS rate of childhood ALL, such as miR-708, miR-27a and miR-223, could be used to predict the risk of relapse before patients undergo therapy. These relapse-associated miRNAs and their targets might also be used to optimize anti-leukemia therapy and might be novel targets for the development of new leukemia countermeasures.
PATIENTS, MATERIALS AND METHODS
Patients and samples
Bone marrow samples were obtained from 18 patients where matched samples at initial diagnosis and first marrow relapse or CR were available. The majority of patients had a B-precursor phenotype (n= 11) and six patients had T-cell ALL (T-ALL; Table 2). These patients were treated according to the modified ALLIC BFM 2002 protocol. Patient characteristics are detailed in Table 2. An independent set of 104 marrow samples acquired at the time of initial diagnosis and 29 samples acquired at relapse were used to confirm the differential miRNA expression pattern and target gene expression by qRT-PCR. ALL samples were enrolled at the First and Second Affiliated Hospital of Sun Yat-sen University or Beijing Children's Hospital. Written informed consent was obtained from the parent/guardians. The study was approved by the ethics committee of the affiliated hospitals of Sun Yat-sen University.
Table 2.
Pediatric ALL patients' characteristics
| Type of sample | Characteristics | Median (range) | No. (%) |
|---|---|---|---|
| Initial diagnosed (N = 122) | Age at diagnosis, years | 6.0 (0.25–14.5) | |
| Sex | |||
| Male | 75 (61.5) | ||
| Female | 47 (38.5) | ||
| WBC count, ×109 per l | 21.7 (1.7–557.6) | ||
| Less than 20 | 60 (49.2) | ||
| 20 or higher | 62 (50.8) | ||
| FAB | |||
| L1 | 32 (26.2) | ||
| L2 | 64 (52.4) | ||
| L3 | 8 (6.6) | ||
| N/A | 18 (14.8) | ||
| Risk group | |||
| HR | 42 (34.4) | ||
| MR | 46 (37.7) | ||
| SR | 23 (18.9) | ||
| N/A | 11 (9.0) | ||
| Prednisone response | |||
| Good response | 77 (63.1) | ||
| Poor response | 26 (21.3) | ||
| N/A | 19 (15.6) | ||
| Immunophenotype | |||
| B | 92 (75.4) | ||
| T | 20 (16.4) | ||
| N/A | 10 (8.2) | ||
| Cytogenetics | |||
| BCR–ABL positive | 8 (6.6) | ||
| BCR–ABL negative | 114 (93.4) | ||
| MLL positive | 6 (4.9) | ||
| MLL negative | 116 (95.1) | ||
| After therapy (N = 77) | CRa | 40 | |
| Relapseb | 37 | ||
| Normal (N = 10) |
aCR samples are from 40 initial diagnosed patients with 3 years clinical follow-up.
bRelapse samples contain eight matched samples from initial diagnosis.
Cell lines and cell cultures
The human Jurkat leukemic T-cell line was cultured in RPMI-1640 medium (HyClone, UT, USA), and the HEK-293T cell line derived from human embryonic kidney was grown in DMEM medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (HyClone) at 37°C in a 5% CO2 atmosphere.
RNA isolation and miRNA microarray profiling
Total RNA was extracted from patient samples with TRIzol (Invitrogen) according to the manufacturer's instructions. miRNA microarray analysis was performed with the Affymetrix miRNA microarray platform (Capital Bio, Beijing, China), containing 743 probes in triplicate corresponding to 576 human (including 122 predicted miRNAs sequences from the published reference), 238 rat and 358 mouse mature miRNAs found in the miRNA Registry (http://microrna.sanger.ac.uk/sequences/; miRBase16; September 2010). Each sample was analyzed in triplicate.
The miRNA detection signal threshold was defined as twice the maximum background signal, and the maximum signal level of the background probes was 180. Normalization was performed using a cyclic LOWESS (locally weighted regression) method to remove the system-related variations. The data adjustments included data filtering, log2 transformation, gene centering and normalization. Diagnosis and relapse or CR samples were compared using t-test, and miRNAs with P< 0.05 were considered significant for cluster analysis. The cluster analysis was performed using a hierarchical method with average linkage and Euclidean distance metrics. All the data are MIAME compliant and the raw data are being deposited in a MIAME compliant database (ArrayExpress, GEO).
Quantitative real-time PCR analysis
qRT-PCR was performed to detect mature miRNAs. Briefly, 0.2 μg of small RNA extracted from patient samples was reverse transcribed to cDNA using M-MLV reverse transcriptase (Promega, WI, USA) and amplified with specific miRNA RT primers and PCR amplification primers (Sangon, Shanghai, China). All primer sequences are shown in Supplementary Material, Table S3. The expression level of each miRNA was determined using the 2−DeltaDeltaCt method. The results are presented as the fold change of each miRNA in the patient samples relative to the normal control samples.
Statistical analysis
Correlations were determined using the Pearson correlation coefficient (r), and a Fisher r-to-z transformation was performed to calculate a probability level (P-value). The mean time to first relapse was compared between groups using the rank sum test. Analysis of RFS—defined as the time from CR to relapse—was performed according to the Kaplan–Meier method, and comparisons of outcomes between subgroups were performed by using the log-rank test. Two-tailed tests were used for univariate comparisons. For univariate and multivariate analysis of prognostic factors, a Cox proportional hazard regression model was used. Accuracies, sensitivity, specificity, positive predictive values (the proportion of patients among those predicted to relapse who actually relapsed) and negative predictive values (the proportion of patients among those predicted to be non-relapsers who did not relapse within 3 years) were validated. In the combination of three miRNA models, patients with low expression of all three miRNAs were predicted to relapse, and in the combination of two of three miRNA models, patients with low expression of at least two miRNAs were predicted to relapse.
GO analysis of miRNA targets
Three complementary websites were used for GO analysis of miRNA targets. MiRNA predicted target was download from TargetScan website. For GO analysis, GO (47) terms and gene information were downloaded from the NCBI website (ftp://ftp.ncbi.nih.gov/gene/DATA) on November 20, 2010. The biological processes and molecular functions categories, as defined in the Gene Ontology Consortium database (http://www.geneontology.org) (47), were analyzed, and 395 genes (see Supplementary Material, Table S2) were predicted as the targets of miRNAs output by GO analysis. Significant overrepresentation of particular GO terms in the data set was determined using the software GeneMerge with corrections for multiple tests (48). Bonferroni-corrected P-values are reported, and a cutoff of 0.1 on the Bonferroni-corrected P-value was applied. Bonferroni-corrected P-values for overrepresented GO terms for each miRNA target were plotted on a negative log2 scale (e.g. P-values 20 = 1, 2−6 = 0.015625) (24). We performed two-way hierarchical clustering with the program MeV (49) using the Pearson correlation coefficient and average linkage clustering. To evaluate the functional significance of the miRNA target genes, we considered the GO distribution of all human genes as the background and then employed a functional enrichment analysis against GO biological process terms database. According to previous studies (50), over-represented functional themes present in background were mapped on the GO hierarchy using a Cytoscape's (26) plugin BINGO (25). A binomial distribution statistical testing method was selected and a significance level of 0.1 was applied. For multiple hypotheses testing, the Benjamini and Hochberg false discovery rate correction (51) was applied to reduce false negatives at the cost of a few more false positives. We only selected GO terms which are associated with our current study to draw figure.
Cell transfection and vector constructs
The miR-708, miR-223 and miR-27a mimics and scrambled oligonucleotides (named as miR-NC, the negative control) were purchased from GenePharm (Shanghai, China). The miR-708 antisense was purchased from RiboBio (Guangzhou, China). Jurkat cell were electrotransfected with mimics or miRNA antisense at a final concentration of 100 nm, and HEK-293T cell was transfected with mimics or miR-NC using Lipofectamine 2000 (Invitrogen). The transfected cells were collected to exact protein for western blot at 48 h after post-transfection.
The MRE in the 3′UTR segments of FOXO3, BMI1 and E2F1 containing putative binding sites to their respective miRNA were synthesized by Songon (Shanghai, China) and insert into the psiCHECK-2 vector (Promega). The reporter constructs, psiCHECK-2-FOXO3, psiCHECK-2-BMI1 and psiCHECK-2-E2F1, were used for miRNA functional analysis. The deletion mutant reporter constructs psiCHECK-2-FOXO3-M, psiCHECK-2-BMI1-M and psiCHECK-2-E2F1-M, in which the sites of perfect complementarity to miRNAs were deleted from parent constructs. Wild-type and mutant insertions were confirmed by DNA sequencing. All primers information is available in Supplementary Material, Table S3.
Target prediction and luciferase reporter assay
Three prediction algorithms PicTar (http://pictar.mdc-berlin.de/, New York University and Max Delbruck Centrum), miRanda (http://cbio.mskcc.org/mirnaviewer/, Memorial Sloan-Kettering Cancer Center) and TargetScan (http://www.targetscan.org/, Whitehead Institute for Biomedical Research) were used to identify possible targets of four miRNAs, miR-27a, miR-128b, miR-223 and miR-708. For luciferase reporter experiments, HEK-293T cells were transfected with each 3′UTR luciferase construct using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Each experiment was repeated at least three times. A statistical comparison of the luciferase results was performed with a two-tailed t test with the Bonferroni correction, and P< 0.05 was considered significant. Jurkat cell lines were transfected by Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions. Luciferase activities were measured at 48 and 72 h after transfection using dual-luciferase assays (Promega).
Western blot
Cells were collected 48 and 72 h after transfection. Total cellular extracts were prepared by homogenization of 5 × 106 cells. Lysates were incubated at 4°C for 30 min. followed by centrifugation at 15 700g at 4°C for 30 min. Equal amounts of proteins (2 µg) and 10 µl loading control (Fermentas) were separated by 8% SDS–PAGE and transferred to polyvinylidene difluoride membranes (GE Healthcare Biosciences). The membranes were incubated with antibodies against FOXO3 (Millipore, MA, USA). Antibody binding was assessed by incubation with horseradish peroxidase-conjugated antimouse secondary antibodies (Santa Cruz Biotechnology), BMI1 (Cell Signaling Technologies, Beverly, MA, USA) and E2F1 (Upstate, MA, USA). Chemiluminescence was detected using an ECL Plus immunoblotting detection system (GE Healthcare Biosciences).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
FUNDING
This work was supported by the funds from National Science and Technology Department (2011CBA01105, 2011CB811301 and 2009ZX09103-641), the National Natural Science Foundation of China (No. 30872784, and 30872297) and by ‘the Fundamental Research Funds for the Central Universities’. Funding to pay the Open Access publication charges for this article was provided by National Natural Science Foundation of China.
Supplementary Material
ACKNOWLEDGMENTS
We thank the following investigators and hospitals that provided samples for the analysis: Dr Hai-Xia Guo at the Second Affiliated Hospital of Sun Yat-sen University; Li-Bin Huang at the First Affiliated Hospital of Sun Yat-sen University.
REFERENCES
- 1.Pui C.H., Campana D., Evans W.E. Childhood acute lymphoblastic leukaemia—current status and future perspectives. Lancet Oncol. 2001;2:597–607. doi: 10.1016/S1470-2045(01)00516-2. [DOI] [PubMed] [Google Scholar]
- 2.Gaynon P.S. Childhood acute lymphoblastic leukaemia and relapse. Br. J. Haematol. 2005;131:579–587. doi: 10.1111/j.1365-2141.2005.05773.x. [DOI] [PubMed] [Google Scholar]
- 3.Borowitz M.J., Pullen D.J., Shuster J.J., Viswanatha D., Montgomery K., Willman C.L., Camitta B. Minimal residual disease detection in childhood precursor-B-cell acute lymphoblastic leukemia: relation to other risk factors. A Children's Oncology Group study. Leukemia. 2003;17:1566–1572. doi: 10.1038/sj.leu.2403001. [DOI] [PubMed] [Google Scholar]
- 4.Brisco M.J., Condon J., Hughes E., Neoh S.H., Sykes P.J., Seshadri R., Toogood I., Waters K., Tauro G., Ekert H., et al. Outcome prediction in childhood acute lymphoblastic leukaemia by molecular quantification of residual disease at the end of induction. Lancet. 1994;343:196–200. doi: 10.1016/s0140-6736(94)90988-1. [DOI] [PubMed] [Google Scholar]
- 5.Coustan-Smith E., Sancho J., Hancock M.L., Boyett J.M., Behm F.G., Raimondi S.C., Sandlund J.T., Rivera G.K., Rubnitz J.E., Ribeiro R.C., et al. Clinical importance of minimal residual disease in childhood acute lymphoblastic leukemia. Blood. 2000;96:2691–2696. [PubMed] [Google Scholar]
- 6.Cave H., van der Werff ten Bosch J., Suciu S., Guidal C., Waterkeyn C., Otten J., Bakkus M., Thielemans K., Grandchamp B., Vilmer E. Clinical significance of minimal residual disease in childhood acute lymphoblastic leukemia. European Organization for Research and Treatment of Cancer–Childhood Leukemia Cooperative Group. N. Engl. J. Med. 1998;339:591–598. doi: 10.1056/NEJM199808273390904. [DOI] [PubMed] [Google Scholar]
- 7.Choi S., Henderson M.J., Kwan E., Beesley A.H., Sutton R., Bahar A.Y., Giles J., Venn N.C., Pozza L.D., Baker D.L., et al. Relapse in children with acute lymphoblastic leukemia involving selection of a preexisting drug-resistant subclone. Blood. 2007;110:632–639. doi: 10.1182/blood-2007-01-067785. [DOI] [PubMed] [Google Scholar]
- 8.Zuna J., Ford A.M., Peham M., Patel N., Saha V., Eckert C., Kochling J., Panzer-Grumayer R., Trka J., Greaves M. TEL deletion analysis supports a novel view of relapse in childhood acute lymphoblastic leukemia. Clin. Cancer Res. 2004;10:5355–5360. doi: 10.1158/1078-0432.CCR-04-0584. [DOI] [PubMed] [Google Scholar]
- 9.Beesley A.H., Cummings A.J., Freitas J.R., Hoffmann K., Firth M.J., Ford J., de Klerk N.H., Kees U.R. The gene expression signature of relapse in paediatric acute lymphoblastic leukaemia: implications for mechanisms of therapy failure. Br. J. Haematol. 2005;131:447–456. doi: 10.1111/j.1365-2141.2005.05785.x. [DOI] [PubMed] [Google Scholar]
- 10.Bhojwani D., Kang H., Moskowitz N.P., Min D.J., Lee H., Potter J.W., Davidson G., Willman C.L., Borowitz M.J., Belitskaya-Levy I., et al. Biologic pathways associated with relapse in childhood acute lymphoblastic leukemia: a Children's Oncology Group study. Blood. 2006;108:711–717. doi: 10.1182/blood-2006-02-002824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Staal F.J., de Ridder D., Szczepanski T., Schonewille T., van der Linden E.C., van Wering E.R., van der Velden V.H., van Dongen J.J. Genome-wide expression analysis of paired diagnosis-relapse samples in ALL indicates involvement of pathways related to DNA replication, cell cycle and DNA repair, independent of immune phenotype. Leukemia. 2010;24:491–499. doi: 10.1038/leu.2009.286. [DOI] [PubMed] [Google Scholar]
- 12.Mattick J.S., Makunin I.V. Small regulatory RNAs in mammals. Hum. Mol. Genet. 2005;14(Suppl. 1):R121–R132. doi: 10.1093/hmg/ddi101. [DOI] [PubMed] [Google Scholar]
- 13.Chen C.Z., Li L., Lodish H.F., Bartel D.P. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 14.Blenkiron C., Miska E.A. miRNAs in cancer: approaches, aetiology, diagnostics and therapy. Hum. Mol. Genet. 2007;16(Suppl. 1):R106–R113. doi: 10.1093/hmg/ddm056. [DOI] [PubMed] [Google Scholar]
- 15.Marcucci G., Mrozek K., Radmacher M.D., Garzon R., Bloomfield C.D. The prognostic and functional role of microRNAs in acute myeloid leukemia. Blood. 2010;117:1121–1129. doi: 10.1182/blood-2010-09-191312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fulci V., Colombo T., Chiaretti S., Messina M., Citarella F., Tavolaro S., Guarini A., Foa R., Macino G. Characterization of B- and T-lineage acute lymphoblastic leukemia by integrated analysis of MicroRNA and mRNA expression profiles. Genes Chromosomes Cancer. 2009;48:1069–1082. doi: 10.1002/gcc.20709. [DOI] [PubMed] [Google Scholar]
- 17.Li Z., Lu J., Sun M., Mi S., Zhang H., Luo R.T., Chen P., Wang Y., Yan M., Qian Z., et al. Distinct microRNA expression profiles in acute myeloid leukemia with common translocations. Proc. Natl Acad. Sci. USA. 2008;105:15535–15540. doi: 10.1073/pnas.0808266105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mi S., Lu J., Sun M., Li Z., Zhang H., Neilly M.B., Wang Y., Qian Z., Jin J., Zhang Y., et al. MicroRNA expression signatures accurately discriminate acute lymphoblastic leukemia from acute myeloid leukemia. Proc. Natl Acad. Sci. USA. 2007;104:19971–19976. doi: 10.1073/pnas.0709313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rainer J., Ploner C., Jesacher S., Ploner A., Eduardoff M., Mansha M., Wasim M., Panzer-Grumayer R., Trajanoski Z., Niederegger H., et al. Glucocorticoid-regulated microRNAs and mirtrons in acute lymphoblastic leukemia. Leukemia. 2009;23:746–752. doi: 10.1038/leu.2008.370. [DOI] [PubMed] [Google Scholar]
- 20.Starczynowski D.T., Morin R., McPherson A., Lam J., Chari R., Wegrzyn J., Kuchenbauer F., Hirst M., Tohyama K., Humphries R.K., et al. Genome-wide identification of human microRNAs located in leukemia-associated genomic alterations. Blood. 2010;117:595–607. doi: 10.1182/blood-2010-03-277012. [DOI] [PubMed] [Google Scholar]
- 21.Feng D.D., Zhang H., Zhang P., Zheng Y.S., Zhang X.J., Han B.W., Luo X.Q., Xu L., Zhou H., Qu L.H., et al. Down-regulated miR-331–5p and miR-27a are associated with chemotherapy resistance and relapse in leukemia. J. Cell Mol. Med. 2010 doi: 10.1111/j.1582-4934.2010.01213.x. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kotani A., Ha D., Hsieh J., Rao P.K., Schotte D., den Boer M.L., Armstrong S.A., Lodish H.F. miR-128b is a potent glucocorticoid sensitizer in MLL-AF4 acute lymphocytic leukemia cells and exerts cooperative effects with miR-221. Blood. 2009;114:4169–4178. doi: 10.1182/blood-2008-12-191619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kotani A., Ha D., Schotte D., den Boer M.L., Armstrong S.A., Lodish H.F. A novel mutation in the miR-128b gene reduces miRNA processing and leads to glucocorticoid resistance of MLL-AF4 acute lymphocytic leukemia cells. Cell Cycle. 2010;9:1037–1042. doi: 10.4161/cc.9.6.11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lall S., Grun D., Krek A., Chen K., Wang Y.L., Dewey C.N., Sood P., Colombo T., Bray N., Macmenamin P., et al. A genome-wide map of conserved microRNA targets in C. elegans . Curr. Biol. 2006;16:460–471. doi: 10.1016/j.cub.2006.01.050. [DOI] [PubMed] [Google Scholar]
- 25.Maere S., Heymans K., Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 26.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raaphorst F.M. Self-renewal of hematopoietic and leukemic stem cells: a central role for the Polycomb-group gene Bmi-1. Trends Immunol. 2003;24:522–524. doi: 10.1016/s1471-4906(03)00241-2. [DOI] [PubMed] [Google Scholar]
- 28.Park I.K., Qian D., Kiel M., Becker M.W., Pihalja M., Weissman I.L., Morrison S.J., Clarke M.F. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423:302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- 29.Pulikkan J.A., Dengler V., Peramangalam P.S., Peer Zada A.A., Muller-Tidow C., Bohlander S.K., Tenen D.G., Behre G. Cell-cycle regulator E2F1 and microRNA-223 comprise an autoregulatory negative feedback loop in acute myeloid leukemia. Blood. 2010;115:1768–1778. doi: 10.1182/blood-2009-08-240101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hui R.C., Gomes A.R., Constantinidou D., Costa J.R., Karadedou C.T., Fernandez de Mattos S., Wymann M.P., Brosens J.J., Schulze A., Lam E.W. The forkhead transcription factor FOXO3a increases phosphoinositide-3 kinase/Akt activity in drug-resistant leukemic cells through induction of PIK3CA expression. Mol. Cell Biol. 2008;28:5886–5898. doi: 10.1128/MCB.01265-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miyamoto K., Araki K.Y., Naka K., Arai F., Takubo K., Yamazaki S., Matsuoka S., Miyamoto T., Ito K., Ohmura M., et al. Foxo3a is essential for maintenance of the hematopoietic stem cell pool. Cell Stem Cell. 2007;1:101–112. doi: 10.1016/j.stem.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 32.Tothova Z., Kollipara R., Huntly B.J., Lee B.H., Castrillon D.H., Cullen D.E., McDowell E.P., Lazo-Kallanian S., Williams I.R., Sears C., et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 2007;128:325–339. doi: 10.1016/j.cell.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 33.Inaba H., Pui C.H. Glucocorticoid use in acute lymphoblastic leukaemia. Lancet Oncol. 2010;11:1096–1106. doi: 10.1016/S1470-2045(10)70114-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang N., Koh G.S., Lim J.Y., Kham S.K., Ariffin H., Chew F.T., Yeoh A.E. BIM is a prognostic biomarker for early prednisolone response in pediatric acute lymphoblastic leukemia. Exp. Hematol. 2011;39:321–329. doi: 10.1016/j.exphem.2010.11.009. [DOI] [PubMed] [Google Scholar]
- 35.Laios A., O'Toole S., Flavin R., Martin C., Kelly L., Ring M., Finn S.P., Barrett C., Loda M., Gleeson N., et al. Potential role of miR-9 and miR-223 in recurrent ovarian cancer. Mol. Cancer. 2008;7:35. doi: 10.1186/1476-4598-7-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chiaretti S., Messina M., Tavolaro S., Zardo G., Elia L., Vitale A., Fatica A., Gorello P., Piciocchi A., Scappucci G., et al. Gene expression profiling identifies a subset of adult T-cell acute lymphoblastic leukemia with myeloid-like gene features and over-expression of miR-223. Haematologica. 2010;95:1114–1121. doi: 10.3324/haematol.2009.015099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stamatopoulos B., Meuleman N., Haibe-Kains B., Saussoy P., Van Den Neste E., Michaux L., Heimann P., Martiat P., Bron D., Lagneaux L. microRNA-29c and microRNA-223 down-regulation has in vivo significance in chronic lymphocytic leukemia and improves disease risk stratification. Blood. 2009;113:5237–5245. doi: 10.1182/blood-2008-11-189407. [DOI] [PubMed] [Google Scholar]
- 38.Mertens-Talcott S.U., Chintharlapalli S., Li X., Safe S. The oncogenic microRNA-27a targets genes that regulate specificity protein transcription factors and the G2-M checkpoint in MDA-MB-231 breast cancer cells. Cancer Res. 2007;67:11001–11011. doi: 10.1158/0008-5472.CAN-07-2416. [DOI] [PubMed] [Google Scholar]
- 39.Chintharlapalli S., Papineni S., Abdelrahim M., Abudayyeh A., Jutooru I., Chadalapaka G., Wu F., Mertens-Talcott S., Vanderlaag K., Cho S.D., et al. Oncogenic microRNA-27a is a target for anticancer agent methyl 2-cyano-3,11-dioxo-18beta-olean-1,12-dien-30-oate in colon cancer cells. Int. J. Cancer. 2009;125:1965–1974. doi: 10.1002/ijc.24530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lessard J., Sauvageau G. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature. 2003;423:255–260. doi: 10.1038/nature01572. [DOI] [PubMed] [Google Scholar]
- 41.Naka K., Hoshii T., Muraguchi T., Tadokoro Y., Ooshio T., Kondo Y., Nakao S., Motoyama N., Hirao A. TGF-beta-FOXO signalling maintains leukaemia-initiating cells in chronic myeloid leukaemia. Nature. 2010;463:676–680. doi: 10.1038/nature08734. [DOI] [PubMed] [Google Scholar]
- 42.Myatt S.S., Lam E.W. The emerging roles of forkhead box (Fox) proteins in cancer. Nat. Rev. Cancer. 2007;7:847–859. doi: 10.1038/nrc2223. [DOI] [PubMed] [Google Scholar]
- 43.Waanders E., van der Velden V.H., van der Schoot C.E., van Leeuwen F.N., van Reijmersdal S.V., de Haas V., Veerman A.J., van Kessel A.G., Hoogerbrugge P.M., Kuiper R.P., et al. Integrated use of minimal residual disease classification and IKZF1 alteration status accurately predicts 79% of relapses in pediatric acute lymphoblastic leukemia. Leukemia. 2011;25:254–258. doi: 10.1038/leu.2010.275. [DOI] [PubMed] [Google Scholar]
- 44.Wu S., Fischer L., Gökbuget N., Schwartz S., Burmeister T., Notter M., Hoelzer D., Fuchs H., Blau I.W., Hofmann W.K., et al. Expression of interleukin 15 in primary adult acute lymphoblastic leukemia. Cancer. 2010;116:387–392. doi: 10.1002/cncr.24729. [DOI] [PubMed] [Google Scholar]
- 45.Jiao Y., Cui L., Gao C., Li W., Zhao X., Liu S., Wu M., Deng G., Li Z. CASP8AP2 is a promising prognostic indicator in pediatric acute lymphoblastic leukemia. Leuk. Res. 2011 doi: 10.1016/j.leukres.2011.05.023. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 46.Kang H., Chen I.M., Wilson C.S., Bedrick E.J., Harvey R.C., Atlas S.R., Devidas M., Mullighan C.G., Wang X., Murphy M., et al. Gene expression classifiers for relapse-free survival and minimal residual disease improve risk classification and outcome prediction in pediatric B-precursor acute lymphoblastic leukemia. Blood. 2010;115:1394–1405. doi: 10.1182/blood-2009-05-218560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Castillo-Davis C.I., Hartl D.L. GeneMerge—post-genomic analysis, data mining, and hypothesis testing. Bioinformatics. 2003;19:891–892. doi: 10.1093/bioinformatics/btg114. [DOI] [PubMed] [Google Scholar]
- 49.Saeed A.I., Sharov V., White J., Li J., Liang W., Bhagabati N., Braisted J., Klapa M., Currier T., Thiagarajan M., et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 50.Zhang L., Chia J.M., Kumari S., Stein J.C., Liu Z., Narechania A., Maher C.A., Guill K., McMullen M.D., Ware D. A genome-wide characterization of microRNA genes in maize. PLoS Genet. 2009;5:e1000716. doi: 10.1371/journal.pgen.1000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Benjamini Y., Drai D., Elmer G., Kafkafi N., Golani I. Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 2001;125:279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.