Abstract
Studies of the major psychoses, schizophrenia (SZ) and bipolar disorder (BD), have traditionally focused on genetic and environmental risk factors, although more recent work has highlighted an additional role for epigenetic processes in mediating susceptibility. Since monozygotic (MZ) twins share a common DNA sequence, their study represents an ideal design for investigating the contribution of epigenetic factors to disease etiology. We performed a genome-wide analysis of DNA methylation on peripheral blood DNA samples obtained from a unique sample of MZ twin pairs discordant for major psychosis. Numerous loci demonstrated disease-associated DNA methylation differences between twins discordant for SZ and BD individually, and together as a combined major psychosis group. Pathway analysis of our top loci highlighted a significant enrichment of epigenetic changes in biological networks and pathways directly relevant to psychiatric disorder and neurodevelopment. The top psychosis-associated, differentially methylated region, significantly hypomethylated in affected twins, was located in the promoter of ST6GALNAC1 overlapping a previously reported rare genomic duplication observed in SZ. The mean DNA methylation difference at this locus was 6%, but there was considerable heterogeneity between families, with some twin pairs showing a 20% difference in methylation. We subsequently assessed this region in an independent sample of postmortem brain tissue from affected individuals and controls, finding marked hypomethylation (>25%) in a subset of psychosis patients. Overall, our data provide further evidence to support a role for DNA methylation differences in mediating phenotypic differences between MZ twins and in the etiology of both SZ and BD.
INTRODUCTION
Schizophrenia (SZ) and bipolar disorder (BD) are two related psychiatric conditions that together contribute significantly to the global burden of disease (1). SZ is defined primarily by the presence of psychotic symptoms, and is also characterized by dysfunctional affective responses and altered cognition. BD presents with episodic swings in mood, potentially ranging from extreme mania to severe depression, and is often accompanied by psychotic symptoms and impaired cognition. This overlap between the symptoms of SZ and BD, which can be classified together as ‘major psychosis’, suggests there may be etiological factors common to both disorders (2,3).
SZ and BD clearly aggregate in families, and quantitative genetic analyses highlight a strong inherited component to both. However, although heritability is estimated to be ∼70% (4,5), disease concordance within monozygotic (MZ) twin pairs is far from 100% (6,7), indicating that non-genetic factors are also important in the etiology of the diseases. Recently, increasing emphasis has been placed on the potential role of epigenetic dysfunction in the etiology of psychosis (8).
Epigenetic mechanisms mediate mitotically heritable, but reversible, changes in gene expression without changing the genomic DNA sequence, principally through alterations in DNA methylation and chromatin structure (9). Epigenetic changes in the brain have been associated with a range of biological and cognitive processes including neurogenesis (10), brain growth and development (11), learning and memory (12), drug addiction (13), neurodegeneration (14) and circadian rhythm (15). It has been widely speculated that epigenetic dysfunction in the brain may be involved in a spectrum of psychiatric disorders including psychosis (16,17). Although epigenetic studies in psychiatric disorders are in their infancy, a recent postmortem brain study of patients with psychosis and healthy controls uncovered significant epigenetic changes associated with both SZ and BD across the genome (18).
Investigating discordant MZ twin pairs is a powerful strategy for uncovering disease-associated epigenetic changes because it allows an assessment of the epigenome independent of any underlying genomic sequence variation (19). Recent studies have uncovered a large amount of DNA methylation variability between MZ twins (20,21), supporting the notion that epigenetic variation may contribute to phenotypic discordance between genetically identical individuals. The utility of disease-discordant twins in epigenetic epidemiological studies of complex disease was highlighted by a recent analysis of DNA methylation differences between MZ twins discordant for systemic lupus erythematosus (22). To date, few studies have empirically tested for epigenetic differences between MZ twins discordant for neuropsychiatric disorders and none has looked genome-wide across a large number of twin pairs. Petronis et al. (23) found evidence for DNA methylation differences in the promoter of DRD2 in one SZ-discordant MZ twin pair. Kuratomi et al. (24) examined two pairs discordant for BD and found increased methylation in affected twins upstream of the spermine synthase gene (SMS) and decreased methylation upstream of the peptidylprolyl isomerase E-like gene (PPIEL). Rosa et al. (25) assessed skewed X-chromosome inactivation in 63 female MZ twin pairs with mixed concordance for BD or SZ, reporting that discordant female BD twins showed greater differences in the methylation of the maternal and paternal chromosome X alleles than concordant twin pairs.
In this study, we report the first systematic genome-wide analysis of DNA methylation differences between MZ twins discordant for SZ and BD, using whole-blood DNA obtained from a unique collection of twins. We found numerous disease-associated differences in DNA methylation, many located in the vicinity of genes previously implicated in psychosis. Our data support the hypothesis that epigenetic alterations play an important role in the etiology of SZ and BD.
RESULTS
Overview of experimental strategy
Genome-wide DNA methylation was measured in a unique collection of 22 twin pairs (44 individuals) discordant for SZ or BD, using the Illumina Infinium HumanMethylation27 BeadChip. Demographic data for the samples included in this study are given in Table 1 and Supplementary Material, Table S1. We used an analytical approach that took advantage of the discordant MZ twin design and tested both the magnitude of differences and significance to maximize our chances of identifying real within-twin differences in DNA methylation associated with SZ, BD and a combined ‘psychosis’ diagnosis. Furthermore, given the known phenotypic heterogeneity of both SZ and BD, we screened for large DNA methylation differences within each discordant MZ twin pair to examine the possibility that disease-associated epigenetic changes are not consistent across all patients. The top-ranked disease-associated probe was verified in the same samples, using the Sequenom EpiTYPER platform, and then tested in a set of 45 postmortem brain samples from patients and unaffected controls. Full experimental details are given in Materials and Methods.
Table 1.
Demographic details for the discordant MZ twin-pair samples utilized in this study
| SZ-discordant twin pairs | BD-discordant twin pairs | Psychosis (SZ and BD)-discordant twin pairs | |
|---|---|---|---|
| Sex (males:females) | 8:3 | 2:9 | 10:12 |
| Ethnicity | 10 Caucasian, 1 unknown | 10 Caucasian, 1 Afro-Caribbean | 20 Caucasian, 1 unknown, 1 Afro-Caribbean |
| Time discordant (years) | 10.4 ± 10.6 | 14.6 ± 10.7 | 12.6 ± 10.6 |
| Age of onset (years) | 20.0 ± 4.6 | 21.7 ± 12.3 | 20.9 ± 9.3 |
Values shown are mean ± standard deviation.
Genome-wide DNA methylation analysis
Genome-wide DNA methylation was measured in 22 twin pairs (44 individuals) discordant for SZ or BD, using the Illumina Infinium HumanMethylation27 BeadChip. Demographic data for the samples included in this study are given in Table 1 and Supplementary Material, Table S1. All arrays passed standard quality control metrics and were included in our analyses. As expected, there was a high correlation in genome-wide DNA methylation within each MZ twin pair (Supplementary Material, Fig. S1), indicating no systemic changes in genome-wide epigenetic programming. The mean within-pair correlation (r2) was 0.99 across all analyzed probes on the array and 0.96 across all ‘variable’ probes (i.e. excluding those with a probe-wise standard deviation smaller than the estimated standard deviation across all probes on the array). This concurred with the results of our LINE-1 element pyrosequencing assay (Supplementary Material, Fig. S2), which identified no significant differences in global DNA methylation between affected and unaffected twins for either SZ or BD (SZ-discordant twins: affected-twin mean = 85%, unaffected-twin mean = 86%, P= 0.28; BD-discordant twins: affected-twin mean = 85%, unaffected-twin mean = 86%, P= 0.38).
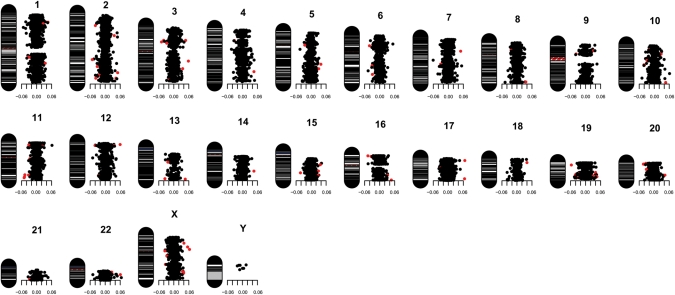
In contrast, DNA methylation at individual CpG sites showed considerable variability within MZ twin pairs (Fig. 1 and Supplementary Material, Figs S3 and S4). Using an analysis method designed to identify the largest and most significant differences in DNA methylation at specific CpG sites, we identified numerous sites throughout the genome exhibiting disease-associated differential DNA methylation (Figs 1, 2 and Supplementary Material, Fig. S4). Table 2 shows the genes located nearest to the eight top-ranking disease-associated, differentially methylated CpG sites in each of the three diagnostic analysis groups: SZ, BD and a combined psychosis group. Supplementary Material, Tables S2–S4 show the top 100 differentially methylated CpG sites for each group. The full methylation data set for all CpG sites analyzed in this study is available for download from our laboratory website (http://epigenetics.iop.kcl.ac.uk/psychosis). Of note, analysis of the location of the 100 top-ranked psychosis-associated, differentially methylated CpG sites demonstrates a significant under-representation of CpG sites located within CpG islands (62% versus 76% for all probes on the Illumina 27K array, χ2 P< 0.001).
Figure 1.
Idiogram illustrating the mean DNA methylation difference (Δβ-value) (well-twin minus ill-twin) for each CpG site included in our analysis across all 22 pairs of psychosis-discordant MZ twins. The red-colored dots correspond to the 100 top-ranked CpG sites (Supplementary Material, Table S3). Similar idiograms for SZ- and BD-discordant twins-pairs are shown in Supplementary Material, Figure S4.
Figure 2.
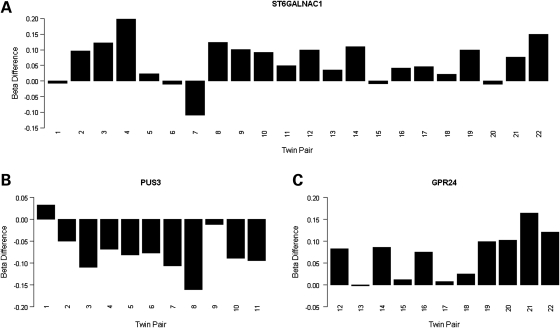
DNA methylation differences (Δβ-value) (well-twin minus ill-twin) for the top-ranked probes from (A) the combined psychosis-discordant analysis group: ST6GALNAC1 (cg13015534), (B) SZ-discordant analysis group: PUS3 (cg02659232) and (C) the BD-discordant analysis group: GPR24 (cg21342728). Psychosis = twin pairs 1–22, SZ = twin pairs 1–11, BD = twin pairs 12–22.
Table 2.
The top eight differentially methylated CpG sites for each of the three diagnostic analysis groups, ranked by a combination of both mean Δβ (well-twin minus ill-twin) and statistical significance
| Analysis group | Rank | Gene name | Chromosome | Paired t-test P-value | Mean Δβ (minimum–maximum) | Weighted Δβ P-value | Weighted q-value |
|---|---|---|---|---|---|---|---|
| Psychosis | 1 | ST6GALNAC1 | 17q25.1 | 4.03E − 04 | 0.06 (−0.11–0.20) | 1.19E − 07 | 7.97E − 04 |
| 2 | ACADL | 2q34 | 2.49E − 04 | 0.05 (−0.05–0.17) | 1.59E − 06 | 3.19E − 03 | |
| 3 | TBC1D10A | 22q12.2 | 8.56E − 04 | 0.06 (−0.05–0.23) | 9.40E − 08 | 7.97E − 04 | |
| 4 | PUS3 | 11q24.2 | 7.66E − 04 | −0.05 (−0.16–0.06) | 1.71E − 06 | 3.19E − 03 | |
| 5 | FXR2 | 17p13.1 | 1.74E − 03 | 0.06 (−0.09–0.26) | 9.03E − 08 | 7.97E − 04 | |
| 6 | TSP50 | 3p21.31 | 4.92E − 04 | 0.04 (−0.05–0.12) | 2.26E − 05 | 1.12E − 02 | |
| 7 | PCOLN3 | 16q24.3 | 1.27E − 03 | 0.04 (−0.06–0.21) | 1.79E − 05 | 1.03E − 02 | |
| 8 | SOX1 | 13q34 | 1.04E − 03 | 0.04 (−0.04–0.13) | 2.64E − 05 | 1.18E − 02 | |
| SZ | 1 | PUS3 | 11q24.2 | 7.66E − 04 | −0.07 (−0.16–0.03) | 5.16E − 05 | 0.10 |
| 2 | SYNGR2 | 17q25.3 | 8.29E − 04 | 0.07 (0.01–0.13) | 9.82E − 05 | 0.14 | |
| 3 | KDELR1 | 19q13.3 | 1.25E − 03 | −0.06 (−0.14–0.01) | 3.07E − 04 | 0.18 | |
| 4 | PDK3 | Xp22.11 | 7.54E − 04 | 0.06 (0.00–0.14) | 3.67E − 04 | 0.18 | |
| 5 | PPARGC1A | 4p15.1 | 1.85E − 03 | 0.06 (−0.02–0.12) | 2.89E − 04 | 0.18 | |
| 6 | ACADL | 2q34 | 3.74E − 03 | 0.07 (0.00–0.17) | 7.81E − 05 | 0.12 | |
| 7 | FLJ90650 | 5q23.1 | 4.19E − 04 | 0.05 (−0.01–0.09) | 6.98E − 04 | 0.19 | |
| 8 | TUBB6 | 18p11.21 | 3.54E − 03 | 0.06 (−0.01–0.18) | 1.78E − 04 | 0.16 | |
| BD | 1 | GPR24 | 22q13.2 | 1.30E − 03 | 0.07 (0.00–0.16) | 7.59E − 05 | 0.17 |
| 2 | TLE6 | 19p13.3 | 1.97E − 03 | −0.09 (−0.21–0.01) | 1.54E − 05 | 0.12 | |
| 3 | STAB1 | 3p21.1 | 1.63E − 03 | −0.07 (−0.18–0.02) | 8.11E − 05 | 0.17 | |
| 4 | PPYR1 | 10q11.2 | 5.13E − 04 | −0.06 (−0.12–0.01) | 3.44E − 04 | 0.25 | |
| 5 | CTNNA2 | 2p12 | 3.59E − 03 | 0.09 (0.00–0.21) | 1.56E − 05 | 0.12 | |
| 6 | ST6GALNAC1 | 17q25.1 | 3.06E − 03 | 0.06 (−0.01–0.15) | 2.82E − 04 | 0.23 | |
| 7 | C1orf35 | 1q42.13 | 5.30E − 03 | 0.06 (−0.01–0.18) | 1.88E − 04 | 0.23 | |
| 8 | IQCH | 15q23 | 3.94E − 03 | 0.05 (−0.06–0.10) | 7.81E − 04 | 0.30 |
The top 100 ranked differentially methylated CpG sites for each diagnostic analysis group are given in Supplementary Material, Tables S2–S4, and data for all CpG sites included in our analyses are available for download from our laboratory website (http://epigenetics.iop.kcl.ac.uk/psychosis).
Overall, the top differentially methylated site across all 22 psychosis-discordant MZ twin pairs was located in the promoter region of the gene encoding alpha-N-acetylgalacto saminide alpha-2,6-sialyltransferase 1 (ST6GALNAC1), which was hypomethylated in affected individuals compared with their unaffected co-twins (mean Δβ = 0.06, P= 4.03E − 04). The top differentially methylated CpG site across the discordant SZ twin pairs was located upstream of the gene encoding pseudouridylate synthase 3 (PUS3), being hypermethylated in affected twins (mean Δβ = −0.07, P= 7.66E − 04). The top-ranked CpG site across the discordant BD MZ twin pairs resided upstream of the gene encoding melanin-concentrating hormone receptor 1 (GPR24), which was hypomethylated in affected twins (mean Δβ = 0.07, P = 1.3E − 03). Twin-pair-specific DNA methylation differences for the top-ranked CpG sites for each analysis group are shown in Figure 2 and Supplementary Material, Figure S5.
Although the direction of effect for the top-ranked loci is consistent across individual pairs within each diagnostic group, some loci show variation in the degree of methylation differences between affected and unaffected twins, indicating that there is inter-family heterogeneity even within our top-ranked loci. Our top-ranked psychosis differentially methylated site in the promoter of ST6GALNAC1, for instance, showed a spectrum of within-twin Δβ-values up to 0.20 (Fig. 2A). Given the clinical heterogeneity of psychosis, it is widely hypothesized that there are rare etiological factors of large effect (e.g. copy-number variation) (26) underlying some cases, and it is plausible that individual twin pairs may similarly harbor large methylation differences at specific disease-associated loci. We therefore assessed DNA methylation differences separately within individual twin pairs to look for the largest family-specific alterations. An example of the differences within one representative discordant MZ twin pair is given in Supplementary Material, Figure S3, with data for the full set of twins available for download from our laboratory webpage (http://epigenetics.iop.kcl.ac.uk/psychosis). A number of novel and known psychiatric candidate genes showed large DNA methylation differences between members of one or a few twin pairs, with some loci showing a Δβ-value >0.60 between the affected and unaffected twin. A full list of CpG probes showing a Δβ > 0.30 within each individual twin pair is given in Supplementary Material, Table S5.
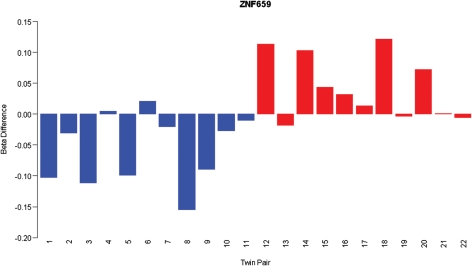
Intriguingly, some CpG sites ranked highly in both the analysis of the SZ-discordant and BD-discordant twins, but with opposite changes in DNA methylation between disorders, indicating that some epigenetic changes may be disorder-specific. Of note, a CpG site in the promoter of ZNF659 (cg18267381), ranking in the 100 top-ranked loci in both the discordant SZ and BD MZ twin pairs, was hypomethylated in the BD twins [average Δβ = 0.04, (P< 0.005)] but hypermethylated [average Δβ = −0.06 (P< 0.01)] in the SZ twins (Fig. 3). Interestingly, a number of other probes in the 100 top-ranked SZ and BD loci show nominally-significant β differences in the opposite direction in the alternative diagnostic group (Supplementary Material, Fig. S6).
Figure 3.
DNA methylation differences (Δβ-value) (well-twin minus ill-twin) for a CpG site located at ZNF659 (cg18267381), which shows disease-associated hypermethylation in SZ (twin pairs 1–11, blue bars) but disease-associated hypomethylation in BD (twin pairs 12–22, red bars).
Analyses were performed to assess correlation between DNA methylation and ‘time discordant’, with no significantly correlated loci suggesting that the twins are not becoming more epigenetically different over the course of their illness. Given recent reports of a correlation between age and DNA methylation at specific loci (27,28), we also investigated the effects of age on the data. Including age as a covariate in our analyses made little change to the list of top-ranked loci. Intriguingly, however, we did replicate several of the previously reported correlations between age and DNA methylation in our twin data (Pidsley et al., in preparation), supporting the notion that DNA methylation changes dynamically through life and affirming the reproducibility of data generated using the Illumina array platform across studies.
Verification and replication of disease-associated hypomethylation at ST6GALNAC1
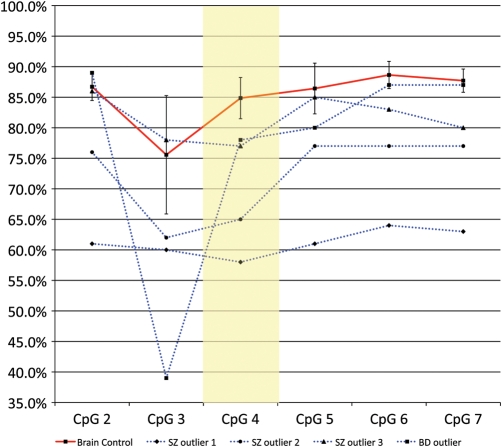
Our Sequenom EpiTYPER data exactly replicated the array data at the specific CpG site represented on the Illumina platform (cg13015534), demonstrating hypomethylation in affected psychosis twins (psychosis twin average = 35% methylation, unaffected twin = 41% methylation). Interestingly, the verification data on the SZ twins highlighted an even larger difference (average 15% hypomethylation) in DNA methylation between the affected and unaffected twins than that was detected on the array at this site. Further validation of disease-associated DNA methylation at ST6GALNAC1 was performed on postmortem brain samples from the Stanley Foundation Neuropathology Consortium. Overall levels of DNA methylation in the brain were higher (∼85%) than those observed in the blood (∼40%, from both raw Illumina array data and Sequenom EpiTYPER data). Although no overall significant difference in DNA methylation at the array-nominated site (CpG4 in the Sequenom assay) was observed between psychosis cases and controls, it was striking that 4 out of the 30 (13.3%) psychosis cases tested showed marked (up to 27%) hypomethylation and were clear outliers at this CpG, and several adjacent CpG sites (Fig. 4). No such hypomethylation of the ST6GALNAC1 promoter was observed in any control samples, which showed a consistently tight methylation profile across the region. These brain data reflect the significant heterogeneity between twin pairs seen in the blood data (Fig. 1), where some affected siblings demonstrated up to 20% hypomethylation but other twin pairs showed much smaller differences. Together, these data indicate that large disease-associated changes at this locus may affect a small number of patients.
Figure 4.
DNA methylation across multiple CpG sites in the promoter of ST6GALNAC1 in postmortem brain tissue from psychosis patients and controls. Postmortem brain tissue from affected individuals showed no overall significant hypomethylation, but 4 out of the 30 (13.3%) psychosis cases tested showed marked (up to 27%) hypomethylation at the same CpG site nominated from the array analysis, with hypomethylation extending across several adjacent CpG sites. No such hypomethylation was observed in any unaffected control samples which all demonstrated very similar methylation values (shown is mean control sample methylation; error bars denote standard deviation).
Ingenuity pathway analysis
We performed ingenuity pathway analysis (IPA) on the 100 top-ranked loci from the analysis of MZ twins discordant for SZ and BD (listed in Supplementary Material, Tables S2–S4) to examine whether common networks or pathways were over-represented in the list of genes associated with the top differentially methylated CpG sites for each disorder. Strikingly, core IPA analysis revealed strong enrichment for phenotypically relevant gene networks: the top scoring functional network in the SZ gene list was ‘nervous system development and function’, and in the BD gene list the top network was ‘developmental, genetic and neurological disorder’ followed by a network related to ‘psychological disorders’ (Table 3). The top biological function pathway in the SZ gene list was ‘psychological disorders’, comprising nine genes directly implicated in SZ (CCND2, CHRNA2, FXR2, FXYD6, HRH3, MUC1, PFN2, SLC31A2, SLC6A3) (P= 3.64E − 03). The first and third canonical pathways were dopamine receptor signaling (P= 6.08E − 03) and glutamate receptor signaling (P= 3.31E − 02), respectively. Furthermore ‘nervous system development and function’ was the top ranking function within ‘physiological system development and function’ from the BD gene list (P= 1.07E − 03–4.85E − 0.2) and third highest from the SZ gene list (P= 7.83E − 0.5–3.45E − 0.2). These results are presented in Supplementary Material, Tables S6–S9.
Table 3.
The top-scoring functional networks of genes nominated by analysis of SZ-discordant twin pairs and BD-discordant twin pairs
| Associated network functions | Network score | |
|---|---|---|
| Top SZ networks | ||
| 1 | Nervous system development and function, cellular development, reproductive system development and function | 73 |
| 2 | Cell signaling, nucleic acid metabolism, small-molecule biochemistry | 39 |
| 3 | Embryonic development, organ development, organ morphology | 30 |
| Top BD networks | ||
| 1 | Neurological disease, developmental disorder, genetic disorder | 67 |
| 2 | Psychological disorders, genetic disorder, nutritional disease | 48 |
| 3 | Gene expression, RNA post-transcriptional modification, organismal development | 18 |
The network score is a numerical value used to rank networks according to their degree of relevance to the network eligible molecules in the gene list. A number of functional networks directly related to psychosis were highlighted by these analyses.
DISCUSSION
This study represents the first comprehensive analysis of disease-associated DNA methylation differences in MZ twins discordant for SZ and BD, using a genome-wide approach. We found no alterations in global DNA methylation between affected and unaffected twins, but considerable disease-associated between-twin differences at specific loci across the genome. Some differences were consistently altered across a combined discordant psychosis group, whereas others appear to be specific to either SZ or BD. Furthermore, although many differences were identified across all discordant twin pairs for each diagnostic category, others were specific to one or a couple of pairs. Our hypothesis-free experimental design allowed us to identify disease-associated DNA methylation differences at loci not previously implicated in psychiatric disorders, but we also found evidence for DNA methylation differences at genes previously implicated in psychosis, such as GPR24 and CTNNA2 in BD. Pathway analysis of our top loci highlighted a significant enrichment of epigenetic disruption to biological networks and pathways relevant to psychiatric disease and neurodevelopment. Overall, our data provide further evidence to support a role for DNA methylation differences in the etiology of both SZ and BD.
The overall top-ranked psychosis-associated methylation difference identified in this study was at a CpG site in the promoter of ST6GALNAC1, located at 17q25.1, which we found was hypomethylated in affected twins. Interestingly, we also found that >13% of postmortem brain samples from SZ and BD patients tested showed marked hypomethylation over an extended region encompassing the nominated CpG site, suggesting that epigenetic changes at this region may be present in a subset of psychosis patients. The psychosis-associated CpG site does not reside in a CpG island, reflecting the observation that our top disease-associated, differentially methylated loci are significantly under-represented for classical CpG-rich promoters and concurring with data from recent epigenomic profiling studies showing that phenotypically relevant variation in DNA methylation often occurs outside of such regions (29).
ST6GALNAC1 is a member of the sialyltransferease family of molecules, involved in protein glycosylation, with a key role in mediating cell–cell interactions. ST6GALNAC1 is expressed in the brain and other CNS tissue and is differentially regulated during key periods of neurodevelopment (30). We performed in silico analysis of the specific CpG site (cg13015534) nominated in our study, using publicly available ENCODE ChIP-seq data in the UCSC genome browser (http://genome.ucsc.edu/) (Supplementary Material, Fig. S7). The site lies directly proximal to a peak of Pol2 occupancy and within a region able to bind neuron restrictive silencer factor/RE1-silencing transcription factor (NSRF/REST), a key regulator of neuronal differentiation that acts to silence neuronal genes in non-neuronal tissue (31). ENCODE DNA methylation data generated using the Illumina 27K array shows that DNA methylation at cg13015534 is inversely correlated with Pol2 binding and ST6GALNAC1 expression, suggesting that variation in methylation at this locus is directly associated with gene transcription and function. Interestingly, a rare inherited genomic duplication spanning the gene, including the disease-associated CpG identified in this study, was reported recently in a single case of SZ (32), suggesting that over-expression of the gene may be implicated in pathogenesis. Given that promoter hypomethylation is typically associated with increased gene expression, a disease-associated duplication of ST6GALNAC1 is consistent with our data. Hypermethylation at this locus has also been associated with estrogen and progesterone receptor-positive breast cancers (33), highlighting that epigenetic variation in the ST6GALNAC1 promoter has important phenotypic consequences that may well be pleiotropic. Finally, another member of the sialyltransferease family of molecules, sialyltransferease 8B (SIAT8B), has been previously associated with SZ (34) and animal studies indicate that ablation of this gene leads to incomplete polysialylation of the neuronal cell adhesion molecule causing severe defects in the anatomical organization of the forebrain (35), which potentially provides a direct mechanism for the role of this gene in the predisposition to SZ.
A number of the other top-ranked genes nominated by our study have been previously implicated in SZ and BD. For example, animal models and human genetic studies suggest that GPR24 (22q13.2), encoding G protein-coupled receptor 24, is involved in the susceptibility to BD. GPR24 knock-out animals show dysregulation of the mesolimbic dopamine system (36), a key system in psychosis. Furthermore, GPR24 antagonists have antidepressant and anxiolytic effects, implicating this gene in the regulation of mood (37,38). There is also genetic evidence to suggest a direct involvement of GPR24 in psychosis; it lies within one of the strongest linkage peaks nominated by a comprehensive meta-analysis (39), and polymorphisms in the gene have been associated with both BD and SZ (40). Interestingly, our significant CpG site is located only 21 bp from rs133073 (although not overlapping the probe), one of the SNPs strongly associated with psychosis.
The alpha catenin gene (CTNNA2) is located in an SZ linkage region 2p11 (41), and encodes a neuronal cadherin-associated protein that plays a major role in folding and lamination of the cerebral cortex (42). This gene has been previously identified as a susceptibility gene in a genome-wide association study and meta-analysis of BD comprising 3683 cases and 14 507 controls (43). Furthermore, polymorphisms in LRRTM1, a gene residing in an intron of CTNNA2, have been associated with psychosis in a parental-origin-specific manner (44). Schalkwyk et al. (45) found evidence for allele-specific methylation in this region supporting the theory that this region could be imprinted and mediating risk via an interaction between genetic and epigenetic factors.
Previous studies investigating MZ-discordant pairs for psychosis have been limited to one or a few twin pairs. One strength of our study is that by using DNA samples from 22 discordant MZ twin pairs, representing one of the largest twin studies performed for any complex disease phenotype to date, we were also able to identify DNA methylation differences that are consistently changed across multiple discordant MZ twin pairs in addition to identifying family-specific disease-associated epigenetic changes. The only other large-scale genome-wide study of DNA methylation changes associated with psychosis investigated postmortem brains from patients and controls and found a number of highly significant alterations in DNA methylation (18). Interestingly five of our top 100 ranked genes overlap with those found to be significantly associated with psychosis in that study—GGN, SLC117A, SMUG1, SOX1 and TCF7L2 (Supplementary Material, Table S10)—suggesting that epigenetic variation at these loci deserves further investigation. Individual twin pair analysis in our study also highlighted considerable familial heterogeneity, with some pairs showing much greater disparity in DNA methylation at certain loci. Furthermore, we observed some large methylation changes that were specific to one or a few twin pairs (Supplementary Material, Table S5), suggesting that some rare epigenetic alterations of large effect may be associated with psychosis.
Our study has a number of limitations that should be considered when interpreting the data presented here. First, although this represents the largest disease-discordant MZ twin study to date for any complex psychiatric disorder, our analysis was limited to only 22 pairs of twins. A power calculation based on an estimate of the standard deviation of the whole data set from the Illumina array data shows that, at a stringent Bonferroni-corrected P-value cut-off (1.86E − 06), our sample of 22 discordant MZ pairs gives good power (>80%) to detect a Δβ of 0.06, although the power to detect smaller differences at this level of significance is more limited, as are analyses in the diagnostic subgroups. It is noteworthy that using our weighted t-test based on the magnitude of change observed between affected and unaffected twins at each locus, many of our top-ranked loci are highly significant, with FDR values (q-values) <0.05. The significant over-enrichment of biologically relevant pathways in our IPA network analyses of top-ranked loci adds further weight to the power of our study to uncover valid differences. Because the discordant MZ-twin design is such a powerful tool for identifying epigenetic variation associated with phenotype (19), a notion affirmed by our success in identifying disease-associated epigenetic changes, future efforts should focus on collecting biological material from additional twin pairs for use in methylomic analyses.
Second, by necessity, this study was performed on DNA samples extracted from peripheral blood rather than from the brain. To our knowledge, there is no archived collection of postmortem MZ twin brains discordant for psychosis. Although the epigenome clearly shows tissue-specific patterns, there is mounting evidence from other disorders that disease-associated epimutations may be detectable across tissues (46,47). Our own methylomic profiling across the brain and blood from multiple individuals shows that although tissue-specific variation far outweighs between-individual variation, many between-individual differences are observed across tissues (unpublished data, see http://epigenetics.iop.kcl.ac.uk/brain). In this regard, it is reassuring that we find evidence for disease-associated hypomethylation of the ST6GALNAC1 promoter in postmortem brain tissue from affected patients, and that some of the genes nominated from a previous study on brain samples were also in our list of top-ranked loci (Supplementary Material, Table S10). Furthermore, our IPA data demonstrates that we can clearly identify epigenetic changes to disease-relevant pathways from peripheral samples. A recent study by Kaminsky et al. (48) found BD-associated methylation changes in the promoter of HCG9 in both brain and peripheral samples, and other reports have also successfully used peripheral tissues to identify disease-associated epigenetic changes in psychosis (24,49). Finally, the use of peripheral samples may have some advantages over the use of the brain; sample collection is more standardized between twins and is not subject to potential confounding issues such as postmortem delay (8).
Third, it is hard to draw conclusions about causality for any of the differentially methylated regions identified in our study, in part because we do not have DNA samples from the twins taken before they became discordant for disease. It is plausible that many of the changes we identify have occurred downstream of the disease, resulting from exposure to antipsychotic medication, for example. There is mounting evidence that many of the drugs used to treat both SZ and BD induce epigenetic changes (50). Such changes are interesting, however, and understanding the pathways via which these drugs work may provide information about the neurobiological processes involved in disease. The ideal study design would be to longitudinally assess DNA methylation changes in the brain during an individual's' transition into disease, although such a study is clearly unfeasible at present.
Finally, although the microarray platform utilized in this study (the Illumina 27K Methylation Array) is reliable and has been previously used to identify quantitative changes in DNA methylation (51), it is relatively limited in the density of probe coverage, interrogating only a couple of CpG sites for the majority of genes with a bias to loci implicated in cancer. This actually makes the results of our pathway analysis more striking, given the clear over-representation of DMRs related to neurobiological processes. Future work should build on the recent technological advances in methylomic profiling and examine DNA methylation differences between discordant twins at much higher resolution.
To conclude, we have undertaken the first large-scale analysis of disease-associated epigenetic changes in MZ twins discordant for major psychosis identifying numerous DNA methylation differences associated with both SZ and BD. These findings further support the notion that epigenetic processes probably play an important role in neuropsychiatric disease and highlight the power of the discordant MZ twin design for epigenomic studies of complex disease.
MATERIALS AND METHODS
Samples
Genomic DNA was extracted using a standard protocol from whole blood obtained from 22 discordant MZ twin pairs (n= 44 individuals) enrolled in the Maudsley Twin Studies of Bipolar Disorder and Schizophrenia. All subjects underwent extensive clinical assessment by two psychiatrists and two psychologists, as detailed previously to confirm discordance for psychotic illness (52). At the time of blood collection, an average of 12.6 (±10.6) years had elapsed since the onset of the illness in the affected twin member (Table 1). Full demographic data pertaining to each of the samples used in this study are given in Table 1 and Supplementary Material, Table S1.
Genome-wide DNA methylation assay
Five hundred nanograms of genomic DNA from each individual was treated with sodium bisulfite in duplicate, using the EZ 96-DNA methylation kit (Zymo Research, CA, USA), following the manufacturer's standard protocol. Genome-wide DNA methylation was assessed using the Illumina Infinium HumanMethylation27 BeadChip (Illumina, Inc., CA, USA), which interrogates 27 578 CpG sites associated with approximately 14 000 genes. Arrays were processed using a standard protocol as described elsewhere (53), with both members of a twin pair being hybridized to the same microarray to control against batch effects. The Illumina GenomeStudio software was used to extract the signal intensities of each probe and to perform initial quality control checks, with all data sets being considered to be high quality. Probes with a detection P-value >0.05 in any of the samples were removed across all individuals (n= 733 probes) to stringently control for poor-quality probes.
Microarray data analysis
All computations and statistical analyses were performed within the R statistical analysis environment (http://www.r-project.org), and all analysis scripts are available on request from the authors. Briefly, signal intensities for each probe were normalized using quantile normalization to reduce unwanted inter-array variation. The relative methylation level of each interrogated CpG site was calculated as the ratio of the normalized signal from the methylated probe to the sum of the normalized signals of the methylated and unmethylated probes. This gave an average β-value for each CpG site, ranging from 0 (unmethylated) to 1 (fully methylated). A density plot of β-values for every sample revealed that, as expected, the data followed a bimodal distribution. An empirical variance stabilizing transformation was used to account for the bimodal distribution of the data (Supplementary Material, Fig. S8).
We developed an analytical approach that took advantage of the discordant MZ twin design and incorporated both magnitude and significance to maximize our chances of identifying real within-twin differences in DNA methylation. For the genome-wide correlation analyses, ‘variable probes’ were identified by calculating an estimate of the standard deviation of the whole data set and then filtering out probes that had a probe-wise standard deviation smaller than the estimated standard deviation. Our ranking analysis comprised two separate tests. The first was a standard paired t-test to assess the significance of DNA methylation differences between the affected and unaffected member in each twin pair. The second assessed the size of methylation differences: a delta-β (Δβ) value was calculated representing the mean difference in methylation between the affected and unaffected member in each twin pair (values stated refer to unaffected minus affected twin). Results from both tests were ranked, by P-value and size, respectively, and then the two ranked lists were combined to give a final ranked list of the CpG sites demonstrating the largest and most consistent differences in DNA methylation across all twin pairs. To generate statistics based on the magnitude of change observed between affected and unaffected twins at each locus, we developed a custom-weighted t-test that adds weight to bigger twin differences by using a constant standard deviation as the denominator in the t-test (estimated from array-wide distribution). This analysis approach was conducted for (i) all twin pairs discordant for major psychosis (n= 22 pairs, 44 individuals), then separately for (ii) the SZ-discordant twins (n= 11 pairs, 22 individuals) and (iii) the BD-discordant twins (n= 11 pairs, 22 individuals). A summary of our analysis strategy is presented in Supplementary Material, Figure S9. Furthermore, given the known phenotypic heterogeneity of both SZ and BD, we screened for large Δβ-values within each discordant MZ twin pair to examine the possibility that disease-associated epigenetic changes are not consistent across all patients.
Global DNA methylation assay
The heavily methylated, abundant, long interspersed nucleotide element-1 (LINE-1) is routinely assayed to give a proxy for global DNA methylation levels (54). We used the commercially available Pyrosequencing LINE-1 assay (Qiagen, Hilden, Germany) and followed the manufacturers' instructions using the Pyromark Q24 (Qiagen) platform. DNA from each individual was treated with sodium bisulfite in duplicate, using the EZ 96-DNA methylation kit and positive controls—both artificially methylated and artificially unmethylated samples were included in all experimental procedures to ensure accuracy in quantification. LINE-1 methylation was calculated as an average across the three interrogated CpG sites across both replicates after bisulfite-PCR-pyrosequencing, and a standard paired t-test was used to assess the significance of global DNA methylation differences between the affected and unaffected member in each twin pair.
Pathway and network analysis
IPA software (version 9.0) (Ingenuity Systems, Redwood City, CA, USA; www.ingenuity.com) was used to assess the top-ranked disease-associated, differentially methylated genes for the enrichment of gene networks, canonical pathways and biological processes relevant to the pathogenesis of psychosis. Lists of the top 100 ranked genes from the analysis of twin pairs discordant for SZ and twin pairs discordant for BD were uploaded to IPA and each gene was mapped to its corresponding gene object in the Ingenuity Knowledge Base (IKB), a repository of biological interactions and functional annotations created from millions of individually modeled relationships between proteins, genes, complexes, cells, tissues, drugs and diseases (see www.ingenuity.com). Enrichment for specific pathways and biological functions was determined relative to the IKB database, using a right-tailed Fisher's test at a significance level of P< 0.05.
Verification of ST6GALNAC1
For the top-ranked psychosis CpG site, in ST6GALNAC1, we performed verification analysis using the Sequenom EpiTYPER platform. Our assay spanned six CpG sites, including the specific CpG interrogated on the Illumina 27 K array. Five hundred nanograms of DNA from each individual (n= 22 pairs, 44 individuals) was independently treated with sodium bisulfite, using the EZ 96-DNA methylation kit. Bisulfite-PCR amplification was performed in duplicate, using the primers and assay conditions given in Supplementary Material, Table S11. Quantitative DNA methylation analysis was conducted using the Sequenom EpiTYPER system (Sequenom, Inc., CA, USA), which utilizes base-specific cleavage followed by MALDI-TOF mass spectrometry (55). Positive controls, including both artificially methylated and artificially unmethylated samples, were included in all experimental procedures to ensure accuracy in quantification. A standard paired t-test was used to assess the significance of DNA methylation differences between the affected and unaffected member in each twin pair.
Replication in postmortem brain samples
Forty-five postmortem brain samples were obtained from the Stanley Medical Neuropathology Consortium (n= 15 SZ, n= 15 BD, n= 15 control) (56). DNA was isolated using a standard phenol–chloroform extraction method and bisulfite treated using the EZ 96 DNA methylation kit. DNA methylation across six CpG sites in the ST6GALNAC1 promoter was assessed using bisulfite-PCR and the Sequenom EpiTYPER system, as described above.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
FUNDING
This work was funded by a NARSAD Young Investigator Award, a London University Research Grant and NIH grant AG036039 to J.M. R.P. is funded by an MRC PhD studentship. M.P. was funded by a Research Training Fellowship from the Wellcome Trust. This work was supported by the European Community's Sixth Framework Programme through a Marie Curie Training Network MRTN-CT-2006-035987 called the European Twin Study Network on Schizophrenia (EUTwinsS). Postmortem brains were donated by The Stanley Medical Research Institute courtesy of Michael B. Knable, E. Fuller Torrey, Maree J. Webster and Robert H. Yolken. Funding to pay the Open Access publication charges for this article was provided by departmental research funds.
Supplementary Material
REFERENCES
- 1.Patel V., Prince M. Global mental health: a new global health field comes of age. JAMA. 2010;303:1976–1977. doi: 10.1001/jama.2010.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Craddock N., Owen M.J. Rethinking psychosis: the disadvantages of a dichotomous classification now outweigh the advantages. World Psychiatry. 2007;6:84–91. [PMC free article] [PubMed] [Google Scholar]
- 3.Cardno A.G., Rijsdijk F.V., Sham P.C., Murray R.M., McGuffin P. A twin study of genetic relationships between psychotic symptoms. Am. J. Psychiatry. 2002;159:539–545. doi: 10.1176/appi.ajp.159.4.539. [DOI] [PubMed] [Google Scholar]
- 4.Craddock N., O'Donovan M.C., Owen M.J. The genetics of schizophrenia and bipolar disorder: dissecting psychosis. J. Med. Genet. 2005;42:193–204. doi: 10.1136/jmg.2005.030718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sullivan P.F., Kendler K.S., Neale M.C. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch. Gen. Psychiatry. 2003;60:1187–1192. doi: 10.1001/archpsyc.60.12.1187. [DOI] [PubMed] [Google Scholar]
- 6.Cardno A.G., Gottesman I.I. Twin studies of schizophrenia: from bow-and-arrow concordances to star wars Mx and functional genomics. Am. J. Med. Genet. 2000;97:12–17. [PubMed] [Google Scholar]
- 7.Craddock N., Jones I. Genetics of bipolar disorder. J. Med. Genet. 1999;36:585–594. doi: 10.1136/jmg.36.8.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pidsley R., Mill J. Epigenetic studies of psychosis: current findings, methodological approaches, and implications for postmortem research. Biol. Psychiatry. 2011;69:146–156. doi: 10.1016/j.biopsych.2010.03.029. [DOI] [PubMed] [Google Scholar]
- 9.Henikoff S., Matzke M.A. Exploring and explaining epigenetic effects. Trends Genet. 1997;13:293–295. doi: 10.1016/s0168-9525(97)01219-5. [DOI] [PubMed] [Google Scholar]
- 10.Ma D.K., Marchetto M.C., Guo J.U., Ming G.L., Gage F.H., Song H. Epigenetic choreographers of neurogenesis in the adult mammalian brain. Nat. Neurosci. 2010;13:1338–1344. doi: 10.1038/nn.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pidsley R., Dempster E.L., Mill J. Brain weight in males is correlated with DNA methylation at IGF2. Mol. Psychiatry. 2010;15:880–881. doi: 10.1038/mp.2009.138. [DOI] [PubMed] [Google Scholar]
- 12.Lubin F.D., Roth T.L., Sweatt J.D. Epigenetic regulation of BDNF gene transcription in the consolidation of fear memory. J. Neurosci. 2008;28:10576–10586. doi: 10.1523/JNEUROSCI.1786-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Renthal W., Nestler E.J. Histone acetylation in drug addiction. Semin. Cell Dev. Biol. 2009;20:387–394. doi: 10.1016/j.semcdb.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Migliore L., Coppede F. Genetics, environmental factors and the emerging role of epigenetics in neurodegenerative diseases. Mutat. Res. 2009;667:82–97. doi: 10.1016/j.mrfmmm.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 15.Nakahata Y., Grimaldi B., Sahar S., Hirayama J., Sassone-Corsi P. Signaling to the circadian clock: plasticity by chromatin remodeling. Curr. Opin. Cell Biol. 2007;19:230–237. doi: 10.1016/j.ceb.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 16.Iwamoto K., Kato T. Epigenetic profiling in schizophrenia and major mental disorders. Neuropsychobiology. 2009;60:5–11. doi: 10.1159/000234811. [DOI] [PubMed] [Google Scholar]
- 17.Tsankova N., Renthal W., Kumar A., Nestler E.J. Epigenetic regulation in psychiatric disorders. Nat. Rev. Neurosci. 2007;8:355–367. doi: 10.1038/nrn2132. [DOI] [PubMed] [Google Scholar]
- 18.Mill J., Tang T., Kaminsky Z., Khare T., Yazdanpanah S., Bouchard L., Jia P., Assadzadeh A., Flanagan J., Schumacher A., et al. Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. Am. J. Hum. Genet. 2008;82:696–711. doi: 10.1016/j.ajhg.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bell J.T., Spector T.D. A twin approach to unraveling epigenetics. Trends Genet. 2011;27:116–125. doi: 10.1016/j.tig.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaminsky Z.A., Tang T., Wang S.C., Ptak C., Oh G.H., Wong A.H., Feldcamp L.A., Virtanen C., Halfvarson J., Tysk C., et al. DNA methylation profiles in monozygotic and dizygotic twins. Nat. Genet. 2009;41:240–245. doi: 10.1038/ng.286. [DOI] [PubMed] [Google Scholar]
- 21.Wong C.C., Caspi A., Williams B., Craig I.W., Houts R., Ambler A., Moffitt T.E., Mill J. A longitudinal study of epigenetic variation in twins. Epigenetics. 2010;5:516–526. doi: 10.4161/epi.5.6.12226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Javierre B.M., Fernandez A.F., Richter J., Al-Shahrour F., Martin-Subero J.I., Rodriguez-Ubreva J., Berdasco M., Fraga M.F., O'Hanlon T.P., Rider L.G., et al. Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Res. 2010;20:170–179. doi: 10.1101/gr.100289.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petronis A., Gottesman I.I., Kan P., Kennedy J.L., Basile V.S., Paterson A.D., Popendikyte V. Monozygotic twins exhibit numerous epigenetic differences: clues to twin discordance? Schizophr. Bull. 2003;29:169–178. doi: 10.1093/oxfordjournals.schbul.a006988. [DOI] [PubMed] [Google Scholar]
- 24.Kuratomi G., Iwamoto K., Bundo M., Kusumi I., Kato N., Iwata N., Ozaki N., Kato T. Aberrant DNA methylation associated with bipolar disorder identified from discordant monozygotic twins. Mol. Psychiatry. 2008;13:429–441. doi: 10.1038/sj.mp.4002001. [DOI] [PubMed] [Google Scholar]
- 25.Rosa A., Picchioni M.M., Kalidindi S., Loat C.S., Knight J., Toulopoulou T., Vonk R., van der Schot A.C., Nolen W., Kahn R.S., et al. Differential methylation of the X-chromosome is a possible source of discordance for bipolar disorder female monozygotic twins. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2008;147B:459–462. doi: 10.1002/ajmg.b.30616. [DOI] [PubMed] [Google Scholar]
- 26.Merikangas A.K., Corvin A.P., Gallagher L. Copy-number variants in neurodevelopmental disorders: promises and challenges. Trends Genet. 2009;25:536–544. doi: 10.1016/j.tig.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 27.Rakyan V.K., Down T.A., Maslau S., Andrew T., Yang T.P., Beyan H., Whittaker P., McCann O.T., Finer S., Valdes A.M., et al. Human aging-associated DNA hypermethylation occurs preferentially at bivalent chromatin domains. Genome Res. 2010;20:434–439. doi: 10.1101/gr.103101.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bocklandt S., Lin W., Sehl M.E., Sanchez F.J., Sinsheimer J.S., Horvath S., Vilain E. Epigenetic predictor of age. PLoS One. 2011;6:e14821. doi: 10.1371/journal.pone.0014821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weber M., Hellmann I., Stadler M.B., Ramos L., Paabo S., Rebhan M., Schubeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat. Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 30.Smith F.I., Qu Q., Hong S.J., Kim K.S., Gilmartin T.J., Head S.R. Gene expression profiling of mouse postnatal cerebellar development using oligonucleotide microarrays designed to detect differences in glycoconjugate expression. Gene Expr. Patterns. 2005;5:740–749. doi: 10.1016/j.modgep.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 31.Ballas N., Grunseich C., Lu D.D., Speh J.C., Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 32.Xu B., Roos J.L., Levy S., van Rensburg E.J., Gogos J.A., Karayiorgou M. Strong association of de novo copy number mutations with sporadic schizophrenia. Nat. Genet. 2008;40:880–885. doi: 10.1038/ng.162. [DOI] [PubMed] [Google Scholar]
- 33.Li L., Lee K.M., Han W., Choi J.Y., Lee J.Y., Kang G.H., Park S.K., Noh D.Y., Yoo K.Y., Kang D. Estrogen and progesterone receptor status affect genome-wide DNA methylation profile in breast cancer. Hum. Mol. Genet. 2010;19:4273–4277. doi: 10.1093/hmg/ddq351. [DOI] [PubMed] [Google Scholar]
- 34.Tao R., Li C., Zheng Y., Qin W., Zhang J., Li X., Xu Y., Shi Y.Y., Feng G., He L. Positive association between SIAT8B and schizophrenia in the Chinese Han population. Schizophr. Res. 2007;90:108–114. doi: 10.1016/j.schres.2006.09.029. [DOI] [PubMed] [Google Scholar]
- 35.Angata K., Huckaby V., Ranscht B., Terskikh A., Marth J.D., Fukuda M. Polysialic acid-directed migration and differentiation of neural precursors are essential for mouse brain development. Mol. Cell. Biol. 2007;27:6659–6668. doi: 10.1128/MCB.00205-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pissios P., Frank L., Kennedy A.R., Porter D.R., Marino F.E., Liu F.F., Pothos E.N., Maratos-Flier E. Dysregulation of the mesolimbic dopamine system and reward in MCH−/− mice. Biol. Psychiatry. 2008;64:184–191. doi: 10.1016/j.biopsych.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 37.Borowsky B., Durkin M.M., Ogozalek K., Marzabadi M.R., DeLeon J., Lagu B., Heurich R., Lichtblau H., Shaposhnik Z., Daniewska I., et al. Antidepressant, anxiolytic and anorectic effects of a melanin-concentrating hormone-1 receptor antagonist. Nat. Med. 2002;8:825–830. doi: 10.1038/nm741. [DOI] [PubMed] [Google Scholar]
- 38.Chaki S., Funakoshi T., Hirota-Okuno S., Nishiguchi M., Shimazaki T., Iijima M., Grottick A.J., Kanuma K., Omodera K., Sekiguchi Y., et al. Anxiolytic- and antidepressant-like profile of ATC0065 and ATC0175: nonpeptidic and orally active melanin-concentrating hormone receptor 1 antagonists. J. Pharmacol. Exp. Ther. 2005;313:831–839. doi: 10.1124/jpet.104.081711. [DOI] [PubMed] [Google Scholar]
- 39.Badner J.A., Gershon E.S. Meta-analysis of whole-genome linkage scans of bipolar disorder and schizophrenia. Mol. Psychiatry. 2002;7:405–411. doi: 10.1038/sj.mp.4001012. [DOI] [PubMed] [Google Scholar]
- 40.Severinsen J.E., Als T.D., Binderup H., Kruse T.A., Wang A.G., Vang M., Muir W.J., Blackwood D.H., Mors O., Borglum A.D. Association analyses suggest GPR24 as a shared susceptibility gene for bipolar affective disorder and schizophrenia. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2006;141B:524–533. doi: 10.1002/ajmg.b.30335. [DOI] [PubMed] [Google Scholar]
- 41.DeLisi L.E., Shaw S.H., Crow T.J., Shields G., Smith A.B., Larach V.W., Wellman N., Loftus J., Nanthakumar B., Razi K., et al. A genome-wide scan for linkage to chromosomal regions in 382 sibling pairs with schizophrenia or schizoaffective disorder. Am. J. Psychiatry. 2002;159:803–812. doi: 10.1176/appi.ajp.159.5.803. [DOI] [PubMed] [Google Scholar]
- 42.Smith A., Bourdeau I., Wang J., Bondy C.A. Expression of Catenin family members CTNNA1, CTNNA2, CTNNB1 and JUP in the primate prefrontal cortex and hippocampus. Brain Res. Mol. Brain Res. 2005;135:225–231. doi: 10.1016/j.molbrainres.2004.12.025. [DOI] [PubMed] [Google Scholar]
- 43.Scott L.J., Muglia P., Kong X.Q., Guan W., Flickinger M., Upmanyu R., Tozzi F., Li J.Z., Burmeister M., Absher D., et al. Genome-wide association and meta-analysis of bipolar disorder in individuals of European ancestry. Proc. Natl Acad. Sci. USA. 2009;106:7501–7506. doi: 10.1073/pnas.0813386106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Francks C., Maegawa S., Lauren J., Abrahams B.S., Velayos-Baeza A., Medland S.E., Colella S., Groszer M., McAuley E.Z., Caffrey T.M., et al. LRRTM1 on chromosome 2p12 is a maternally suppressed gene that is associated paternally with handedness and schizophrenia. Mol. Psychiatry. 2007;12:1129–1139. doi: 10.1038/sj.mp.4002053. 1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schalkwyk L.C., Meaburn E.L., Smith R., Dempster E.L., Jeffries A.R., Davies M.N., Plomin R., Mill J. Allelic skewing of DNA methylation is widespread across the genome. Am. J. Hum. Genet. 2010;86:196–212. doi: 10.1016/j.ajhg.2010.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pedersen K.S., Bamlet W.R., Oberg A.L., de Andrade M., Matsumoto M.E., Tang H., Thibodeau S.N., Petersen G.M., Wang L. Leukocyte DNA methylation signature differentiates pancreatic cancer patients from healthy controls. PLoS One. 2011;6:e18223. doi: 10.1371/journal.pone.0018223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sapienza C., Lee J., Powell J., Erinle O., Yafai F., Reichert J., Siraj E.S., Madaio M. DNA methylation profiling identifies epigenetic differences between diabetes patients with ESRD and diabetes patients without nephropathy. Epigenetics. 2011;6:20–28. doi: 10.4161/epi.6.1.13362. [DOI] [PubMed] [Google Scholar]
- 48.Kaminsky Z., Tochigi M., Jia P., Pal M., Mill J., Kwan A., Ioshikhes I., Vincent J.B., Kennedy J.L., Strauss J., et al. A multi-tissue analysis identifies HLA complex group 9 gene methylation differences in bipolar disorder. Mol Psychiatry. 2011 doi: 10.1038/mp.2011.64. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 49.Ghadirivasfi M., Nohesara S., Ahmadkhaniha H.R., Eskandari M.R., Mostafavi S., Thiagalingam S., Abdolmaleky H.M. Hypomethylation of the serotonin receptor type-2A Gene (HTR2A) at T102C polymorphic site in DNA derived from the saliva of patients with schizophrenia and bipolar disorder. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2011;156:536–545. doi: 10.1002/ajmg.b.31192. [DOI] [PubMed] [Google Scholar]
- 50.Dong E., Nelson M., Grayson D.R., Costa E., Guidotti A. Clozapine and sulpiride but not haloperidol or olanzapine activate brain DNA demethylation. Proc. Natl Acad. Sci. USA. 2008;105:13614–13619. doi: 10.1073/pnas.0805493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Breitling L.P., Yang R., Korn B., Burwinkel B., Brenner H. Tobacco-smoking-related differential DNA methylation: 27K discovery and replication. Am. J. Hum. Genet. 2011;88:450–457. doi: 10.1016/j.ajhg.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Picchioni M.M., Toulopoulou T., Landau S., Davies N., Ribchester T., Murray R.M. Neurological abnormalities in schizophrenic twins. Biol. Psychiatry. 2006;59:341–348. doi: 10.1016/j.biopsych.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 53.Bibikova M., Le J., Barners B., Saedinia-Melnyk S., Shen R., Gunderson K. Genome-wide DNA methylation profiling using Infinium assay. Epigenomics. 2009;1:177–200. doi: 10.2217/epi.09.14. [DOI] [PubMed] [Google Scholar]
- 54.Yang A.S., Estecio M.R., Doshi K., Kondo Y., Tajara E.H., Issa J.P. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res. 2004;32:e38. doi: 10.1093/nar/gnh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Coolen M.W., Statham A.L., Gardiner-Garden M., Clark S.J. Genomic profiling of CpG methylation and allelic specificity using quantitative high-throughput mass spectrometry: critical evaluation and improvements. Nucleic Acids Res. 2007;35:e119. doi: 10.1093/nar/gkm662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Torrey E.F., Webster M., Knable M., Johnston N., Yolken R.H. The Stanley Foundation Brain Collection and Neuropathology Consortium. Schizophr Res. 2000;44:151–155. doi: 10.1016/S0920-9964(99)00192-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.