Abstract
Spinocerebellar ataxia type 3 is one of the polyglutamine (polyQ) diseases, which are caused by a CAG-repeat expansion within the coding region of the associated genes. The CAG repeat specifies glutamine, and the expanded polyQ domain mutation confers dominant toxicity on the protein. Traditionally, studies have focused on protein toxicity in polyQ disease mechanisms. Recent findings, however, demonstrate that the CAG-repeat RNA, which encodes the toxic polyQ protein, also contributes to the disease in Drosophila. To provide insights into the nature of the RNA toxicity, we extracted brain-enriched RNA from flies expressing a toxic CAG-repeat mRNA (CAG100) and a non-toxic interrupted CAA/G mRNA repeat (CAA/G105) for microarray analysis. This approach identified 160 genes that are differentially expressed specifically in CAG100 flies. Functional annotation clustering analysis revealed several broad ontologies enriched in the CAG100 gene list, including iron ion binding and nucleotide binding. Intriguingly, transcripts for the Hsp70 genes, a powerful suppressor of polyQ and other human neurodegenerative diseases, were also upregulated. We therefore tested and showed that upregulation of heat shock protein 70 mitigates CAG-repeat RNA toxicity. We then assessed whether other modifiers of the pathogenic, expanded Ataxin-3 polyQ protein could also modify the CAG-repeat RNA toxicity. This approach identified the co-chaperone Tpr2, the transcriptional regulator Dpld, and the RNA-binding protein Orb2 as modifiers of both polyQ protein toxicity and CAG-repeat RNA-based toxicity. These findings suggest an overlap in the mechanisms of RNA and protein-based toxicity, providing insights into the pathogenicity of the RNA in polyQ disease.
INTRODUCTION
A large number of neurodegenerative diseases are caused by unstable trinucleotide repeat expansions in the associated genes (1). Based on pathogenic mechanisms, these disorders are categorized into different groups, including loss of protein function, RNA toxicity and dominant protein-based toxicity. Myotonic dystrophy type 1 (DM1) and myotonic dystrophy type 2 (DM2) are thought to be caused primarily by a gain-of-function RNA toxicity. CUG and CCUG RNA expansions in the non-coding regions of the dystrophia myotonica-protein kinase (DMPK) gene and the zinc finger 9 (ZNF9) gene cause DM1 and DM2, respectively (2). Polyglutamine (polyQ) diseases are dominantly inherited disorders expanded CAG repeats within the coding region of the associated genes, such as Huntington's disease (HD) and the spinocerebellar ataxias (SCA1, 2, 3, 6, 7 and 17) (3). A pathological hallmark of these diseases is ubiquitinated nuclear inclusions (NIs) containing chaperones and subunits of the proteasome (3).
The prevailing hypothesis for polyQ disease mechanisms is a protein effect due to the polyQ. Upregulating chaperones suppresses toxicity and supports the hypothesis that protein misfolding or aberrant interactions of the mutant protein cause disease (4). Therefore, studies have traditionally focused on protein-based mechanisms of toxicity conferred by the expanded polyQ domain. Recent studies, however, suggest that RNA toxicity is a component of degeneration (5). The toxicity of a pathogenic, expanded polyQ Ataxin-3 protein in Drosophila is significantly reduced by interrupting the typically pure CAG-repeat sequence encoding the polyQ domain, with a CAA-interrupted repeat which also encodes glutamine. CAG-repeat RNA forms a hairpin structure which is thought to confer toxicity (6,7). Interruptions of the CAG-repeat RNA by CAA codons are predicted to disrupt the hairpin structure (8). Furthermore, evidence suggests that pure or interrupted CAG-repeat expansions in ATAXIN-2 (ATXN2) are associated with distinct effects (9–12). Expanded pure CAG repeat in ATXN2 typifies SCA2. CAA interruptions in ATXN2 are found in CAG-repeat expansions that present with parkinsonism or amyotrophic lateral sclerosis (ALS). Taken together, these findings suggest a role of the CAG-repeat RNA in toxicity and effects. Additional studies indicate that simply a non-coding RNA bearing only an abnormally long CAG repeat but not coding for a protein can induce progressive neuronal dysfunction, including shortened lifespan and locomotor defects (5). Interestingly, muscleblind (mbl), an RNA-binding protein implicated as a modifier in DM1, can dramatically enhance the toxicity of both the Ataxin-3 protein and non-coding CAG-repeat RNA in Drosophila (13,14). These findings suggest that the CAG-repeat RNA, in the absence of coding for a protein, may be a component of toxicity in polyQ disease. However, mechanisms of CAG-repeat RNA toxicity remain unclear.
To further understand mechanisms of CAG-repeat RNA toxicity, we assessed transcriptional changes in flies expressing toxic expanded CAG-repeat RNA using microarray analysis in Drosophila. This approach identified transcriptional changes in 160 genes specifically in the brain of flies expressing toxic CAG repeats, suggesting a robust component of transcriptional alterations. These findings led us to test the overlap between modifiers defined for protein-based polyQ toxicity and the toxicity of the non-coding RNA. Our findings highlight an overlap between the two situations, suggesting a component of shared mechanism.
RESULTS
Defining gene expression changes in response to a toxic non-coding CAG-repeat RNA
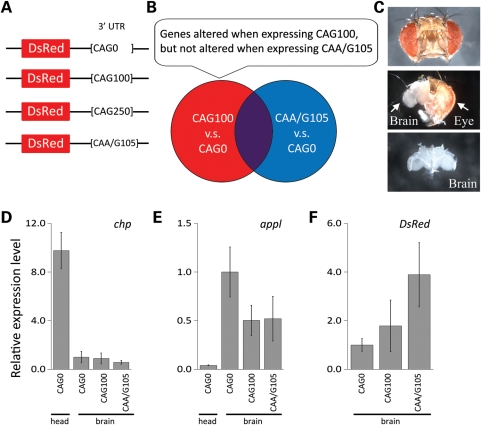
To identify gene pathways involved in response to a toxic non-coding CAG-repeat RNA, we performed microarray analysis on Drosophila brains using the Affymetrix Drosophila Genome 2.0 Array. In order to assess early transcriptional changes and to avoid detecting secondary responses, pre-symptomatic adult flies expressing a CAG repeat in the 3′ untranslated region (3′-UTR) of a transgene encoding the control protein DsRed were used (Fig. 1A). The control CAG0 flies express a DsRed reporter gene with no CAG repeat in the 3′-UTR. The CAG100 and CAG250 flies have a CAG repeat of about 100 and 250 CAG trinucleotides in the 3′-UTR, as previously described (5). The CAA/G105 flies are a non-toxic trinucleotide control and express DsRed with an interrupted CAA/CAG-repeat sequence in the 3′-UTR. The experimental design is illustrated in Figure 1B. To define genes whose expression is altered in response to a toxic CAG-repeat RNA, we compared CAG100 flies with age-matched flies expressing CAG0. To exclude transcriptional changes in response to a non-toxic trinucleotide repeat, a second gene list was generated by comparing CAA/G105 flies with age-matched CAG0 flies. To then focus on the list of genes specific to the toxic CAG-repeat RNA, we eliminated genes that were differentially expressed in common to both CAG100 and CAA/G105 flies. At 3 days, flies expressing CAG100 in all neurons of the brain with elav-GAL4 driver show normal climbing ability and normal survival compared with age-matched control flies expressing CAG0 (5). Fly brains were thus dissected from 3-day animals, and RNA was isolated for microarray analysis (Fig. 1C). To assess whether the brain dissection enriched for brain RNAs, the dissection quality was assessed by determining the relative level of expression of an eye gene, chaoptin (chp), which is selectively expressed in photoreceptor cells, and the neural gene marker amyloid precursor protein like (appl), which is selectively expressed in neurons thus enriched in brain compared with eye tissue (15,16). Quantitative real-time polymerase chain reaction (qRT-PCR) analysis indicated that the relative expression level of chp was significantly low (less than 10-fold compared to head RNA) in all samples, whereas the level of appl was enriched more than 12-fold compared with head RNA (Fig. 1D and E). This analysis thus indicated that dissection of the brain from the eyes successfully enriched for RNA expressed in the brain among the four independent replicates of the microarray analysis.
Figure 1.
Microarray analysis with brain-enriched RNA. (A) DNA constructs with untranslated CAG or CAA/G repeats placed in the 3′-UTR of a control protein DsRed. The control CAG0 flies express the control DsRed reporter with no CAG repeat in the 3′-UTR. The CAG100 and CAG250 flies express the DsRed reporter with repeats of about 100 and 250 CAGs in the 3′-UTR (5). CAA/G105 flies, a non-toxic trinucleotide control, express the DsRed reporter with a trinucleotide sequence of CAA and CAG of 105 units (5). (B) Venn diagram illustrates the rationale to generate lists of genes differentially expressed in CAG100 animals selectively. (C) Illustration of brain dissection process, to enrich for RNA expressed in the brain tissue. The dissection removes the eyes and the outer retinal lamina from the head capsule. (D and E) To verify the brain dissection quality, we quantified relative expression levels of (D) chaoptin (chp) and (E) appl by qRT-PCR. (D) The gene chp is expressed in the retina. Quantifying its expression in brain, compared with head in all the preparations, indicated a lack of eye tissue in the brain preparations. (D) The gene appl is selectively expressed in neurons and showed an enrichment in the brain preparations. (F) The expression levels of each trinucleotide repeat transgene were also controlled. Transcript levels were determined by assessing the level of the DsRed portion of the transgenes by qRT-PCR. Expression levels in (D–F) are relative to CAG0 (brain CAG0 was set to 1) and are presented as mean ± SD (n= 4 biological replicates used in the microarray analysis). Genotypes of flies from left to right: elav-GAL4 in trans to (CAG0) UAS-DsRed-CAG0, (CAG100) UAS-DsRed-CAG100 (5×), (CAA/G105) is UAS-DsRed-CAA/G105 (2×).
The expression levels of each trinucleotide repeat transgene were also controlled. Transcript levels were determined by assessing the level of the DsRed portion of the transgenes by qRT-PCR. The transgenes were adjusted to match at the mRNA expression level, such that CAG0 flies had one copy of the transgene and the CAG100 flies had five copies. The CAA/G105 line expressed the transgene at levels lower than those of the CAG100. Thus, we used two copies of this transgene in order to express this RNA at a level at least as high as that of the CAG100 flies (Fig. 1F).
Global analysis of genes differentially expressed in flies expressing a toxic CAG RNA
Genes differentially expressed in CAG100 and/or CAA/G105 were defined by comparing RNA from brains of flies expressing CAG100 versus CAG0 and CAA/G105 versus CAG0. Pearson's correlation coefficient showed a strong correlation between the biological replicates of any one transgene (r> 0.99). We predicted that the transcriptional profile of CAA/G105 flies would be more similar to CAG0 flies than to CAG100 flies, because the interrupted CAA/G105 repeat is not toxic. Hierarchical clustering confirmed a higher similarity between the transcriptional profiles of CAG0- and CAA/G105-expressing flies than between CAG0- and CAG100-expressing flies (Fig. 2A). In addition, it further supported our experimental design to exclude genes differentially expressed in response to a long trinucleotide repeat by using the interrupted CAA/G105 RNA.
Figure 2.
Transcriptional profiles of CAG100 and CAA/G105 flies. (A) A total of four independent biological replicates were examined by microarray analysis. Hierarchical clustering of all elements on the array by Euclidean distance showed a higher similarity between transcriptional profiles of the same genotypes, regardless of the replicate. Hierarchical clustering also showed a higher similarity between the transcriptional profiles of CAG0 and CAA/G105 than between CAG0 and CAG100. (B) Venn diagram highlighting the number of differentially expressed genes that have at least a 2-fold change in CAG100- and/or CAA/G105-expressing flies (step-up P < 0.05).
About 129 transcripts were identified that were 2-fold or greater upregulated and 81 that were 2-fold or greater downregulated in CAG100 flies compared with CAG0 flies (Fig. 2B). Among these, 19% (25 transcripts) upregulated and 31% (25 transcripts) downregulated transcripts were also differentially up- or downregulated in CAA/G105 flies (Supplementary Material, Table S1a). These transcripts were not pursued at this time; these transcripts might reflect a general response to expressing a long trinucleotide repeat RNA, whether toxic or not. About 160 transcripts (104 up and 56 down) were altered selectively in CAG100 flies (Supplementary Material, Table S1b). About 122 transcripts (48 up and 74 down) were altered in CAA/G105 flies, although not in CAG100 flies (Supplementary Material, Table S1c). To verify the microarray analysis, we selected 12 genes of a range of gene expression alterations to assess for expression changes by qRT-PCR in CAG100 flies (Supplementary Material, Table S2). Five out of 12 genes expression changes, or 42%, were positively validated in CAG100 (P < 0.05). Genes for which the expression level was not changed in a significant manner still showed a trend consistent with the microarray analysis. Eleven out of 12 genes expression changes, or 92%, were positively correlated with the change detected in the microarray.
Gene Ontology (GO) analysis based on the Database of Annotation, Visualization and Integrated Discovery was performed to provide insights into the biological processes affected by the genes differentially expressed selectively in CAG100 flies (Supplementary Material, Table S1b) (17,18). We used the GO_FAT category which filters out very broad GO terms to identify statistically enriched functional groups. Several broad GO terms were enriched, including iron ion binding, nucleotide binding and oxidation reduction (Fig. 3). Genes related to ion channels and receptors were also affected in response to CAG100. Trpgamma of the transient receptor potential (TRP) classic channel family and CG34123 of the TRP melastatin-like channel family were upregulated in CAG100 flies (2.4-fold change). TRP channels are essential for the control of extracellular magnesium levels (19).
Figure 3.
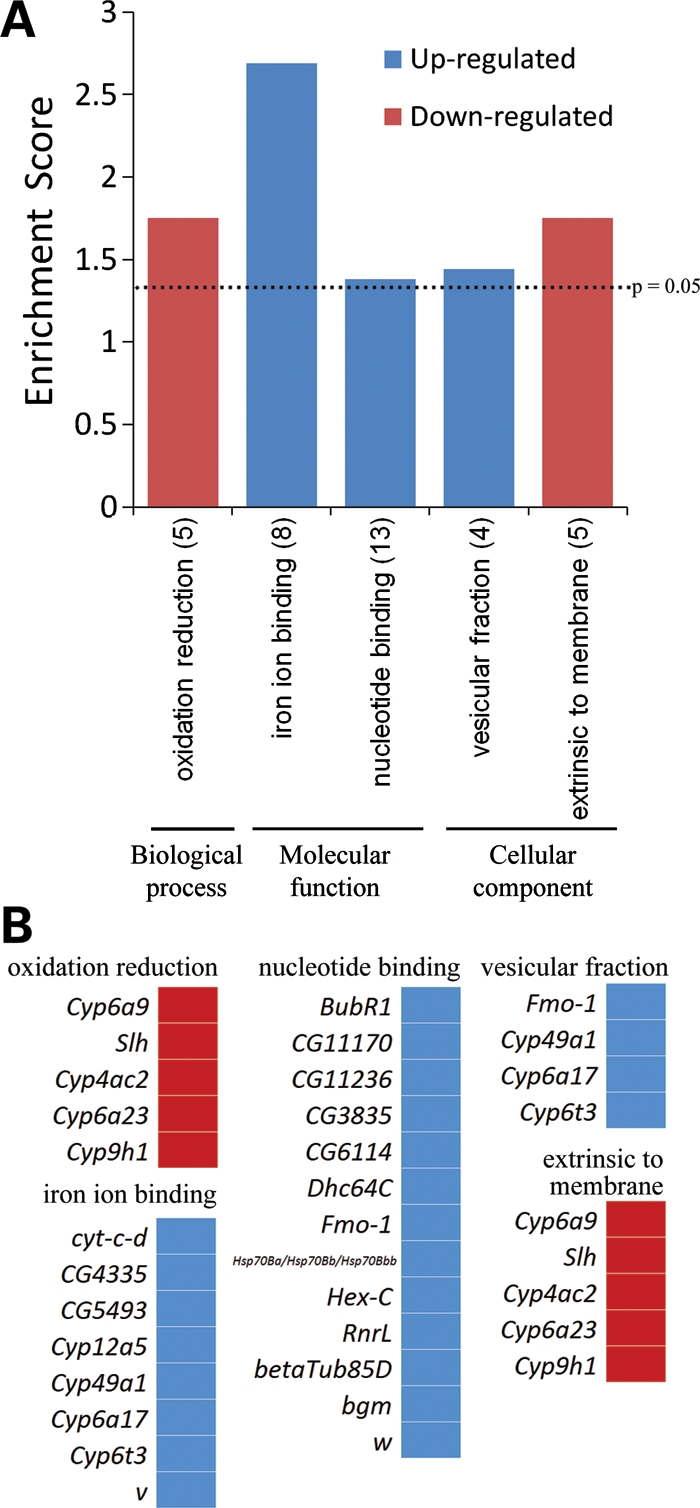
Analysis of GO categories for genes differentially expressed in CAG100. (A) Bar graph shows the GO terms and the number of genes in each category that are significantly enriched in the CAG100 gene list. The enrichment score (E) is the P-value in the -log scale. The P-value shows whether the gene list is specifically enriched in the corresponding annotation cluster compared with random chance. The enrichment score is ≥1.3 (dashed line) when the P-value is ≤0.05. (B) Lists of genes in each enriched category in (A).
RNA-binding proteins are especially interesting targets for potential mechanisms of CAG-repeat RNA toxicity. The transcripts of two RNA-binding protein genes were downregulated in CAG100 flies: RNA-binding protein 4 (−2.4-fold change), which has an RNA recognition motif (RRM), and CHKov1 (−2.3-fold change), predicted to have an RNA-binding motif and a domain of RNA-directed DNA polymerase activity. In the list of upregulated transcripts, expression of the RNA-binding protein eIF4E-5 was upregulated 4.9-fold in CAG100 flies.
The cytochrome P450 superfamily (Cyp) is a large and diverse group of enzymes that catalyse oxidation. Of these, four cytochrome P450-related genes were downregulated and four were upregulated. Their differential expression might reflect a detoxification response in CAG100 flies at the pre-symptomatic stage. Significant upregulation of cytochrome P450-related genes has also been observed in the microarray analysis of a Drosophila model of Parkinson's disease (PD) (20).
Hsp70, a genetic modifier of polyQ toxicity, is upregulated in microarray analysis
Intriguingly, in the microarray analysis, we found that some of the Hsp70 genes were significantly upregulated in brains of animals expressing the toxic CAG100 RNA (Supplementary Material, Table S1b). Heat shock protein 70 (Hsp70) is a powerful modulator of the pathogenic polyQ protein, such that in Drosophila models for SCA3, Hsp70 becomes localized to the NIs (4). Furthermore, upregulation of Hsp70 suppresses polyQ-mediated neurodegeneration. There are multiple copies of the Hsp70 genes in Drosophila at distinct loci, including Hsp70Aa and Hsp70Ab at chromosomal position 87A and Hsp70Ba, Hsp70Bb, Hsp70Bbb and Hsp70Bc at 87B. We found that probe sets of transcripts for Hsp70Ba, Hsp70Bb and Hsp70Bbb genes were upregulated 4.06-fold.
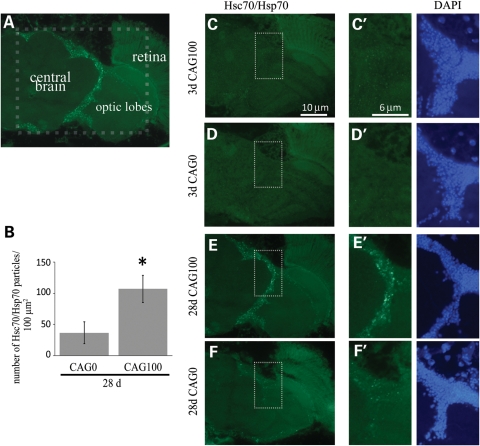
Given this upregulation of Hsp70 transcripts, we used a second technique to determine whether abnormal localization or levels of Hsp70 protein were detectable in flies expressing CAG100. Tissue immunostaining of fly brains using an antibody that detects the Hsc70/Hsp70 protein family was performed on flies expressing the toxic RNA repeat (Fig. 4A). At 3 days, when an Hsp70 transcriptional change was detectable by the microarray analysis, we did not see an alteration of the Hsc70/Hsp70 protein in CAG100 flies compared with controls (Fig. 4C and D). By 28 days, the brain normally shows a modest degree of Hsc70/Hsp70 accumulations (Fig. 4F). Strikingly, immunostaining of CAG100-expressing animals revealed abundant Hsc70/Hsp70 accumulations in the cortical region of the brain at 28 days (Fig. 4E). Quantification of the accumulations (Fig. 4E′ and F′) showed that the number of Hsc70/Hsp70 accumulations was significantly increased (P < 0.05) in CAG100-expressing flies compared with control animals (Fig. 4B). Thus, the microarray analysis revealed a transcriptional increase in Hsp70 genes in CAG100 flies in 3-day pre-symptomatic animals and a protein translational response in Hsc70/Hsp70 in 28-day animals. Furthermore, these data suggested an overlap in the biological response of the animal to the toxic expanded polyQ protein and toxic expanded CAG-repeat RNA, namely an Hsc70/Hsp70 response.
Figure 4.
Hsc70/Hsp70 accumulated in aged CAG100 flies. (A) Cryosection of a CAG100 fly head at 28 days, immunostained for Hsc70/Hsp70. The dashed outline represents the area enlarged in (C–F). (B) Graph of the mean number of Hsc70/Hsc70 particles in CAG0- and CAG100-expressing animals at 28 days. The average number of Hsc70/Hsc70-positive particles within the area shown in (E′) and (F′) was quantified. Error bars represent SD (*P< 0.05, Student's t-test; n= 3–4). (C and D) At 3 days, Hsc70/Hsp70 accumulation is not detectable in CAG0 and CAG100 fly brains. Enlarged images of the dashed outline area are shown in (C′) and (D′) (green: anti-Hsc70/Hsp70, blue: DAPI). (E and F) At 28 days, there are prominent Hsc70/Hsp70 accumulations in CAG100 flies. Enlarged images of the dashed outline area are shown in (E′) and (F′). Genotypes of flies: elav-GAL4 in trans to (CAG0) UAS-DsRed-CAG0 and (CAG100) UAS-DsRed-CAG100 (5×). DAPI images are shown next to (C′–F′) for orientation.
Hsp70 is a modifier of Ataxin-3 protein-based neurodegeneration and of CAG-repeat RNA-based toxicity
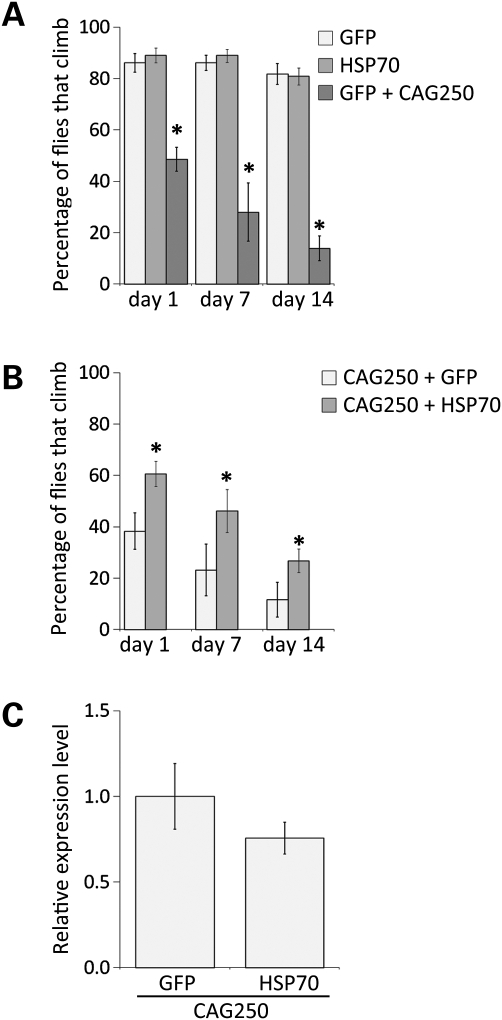
To determine whether upregulation of Hsp70 could modify the toxicity of the CAG-repeat RNA, we co-expressed human Hsp70 together with a non-coding CAG250 RNA ubiquitously in the animal with daughterless-GAL4 (da-GAL4). CAG250 flies express a sequence of 250 CAG trinucleotide repeats in the 3′-UTR of the DsRed reporter. Flies expressing CAG250 showed a stronger toxicity than animals with tissue-specific expression of the RNA, thus facilitating the analysis of modifier genes. When expressed in this manner, the CAG250 repeat caused compromised climbing ability at day 1 that progressively deteriorated over 14 days (Fig. 5A). Upregulation of Hsp70 mitigated the toxic effects of the CAG250 repeat, such that now 61 ± 5% of the animals could climb at day 1, compared with 38 ± 7% in the absence of added Hsp70 (Fig. 5B). The effect of Hsp70 persisted with time such that by 14 days, whereas normally only 12 ± 7% of CAG250 flies could climb, with co-expressed Hsp70, now 27 ± 5% of CAG250 flies could climb. We confirmed that Hsp70 had no effect on the level of expression of the CAG250 repeat (Fig. 5C); rather, added Hsp70 mitigated the toxic effects of the CAG250 RNA. These data indicate that Hsp70 is a modulator of both pathogenic polyQ protein-based toxicity and toxicity conferred by a non-coding expanded CAG-repeat RNA. Identification of Hsp70 as a modifier of the CAG-repeat RNA-based toxicity raised the possibility that other modifiers of the pathogenic polyQ protein-based toxicity may also affect CAG RNA-based toxicity.
Figure 5.
Hsp70 rescues climbing defects of CAG250 flies. (A) Expressing GFP or Hsp70 alone does not cause climbing defects. In contrast, expressing CAG250 causes climbing defects in fly, and this defect progress with age. Genotypes of flies: da-GAL4 in trans to (GFP) UAS-GFP, (Hsp70) UAS-Hsp70 and (GFP+CAG250) is UAS-GFP UAS-DsRed-CAG250. (B) Co-expression of Hsp70 with CAG250 suppresses CAG250 toxicity and rescues climbing deficits. The climbing ability of CAG250 flies was examined at days 1, 7 and 14. CAG250/Hsp70 flies exhibit a significant improvement in climbing ability compared with age-matched CAG250 flies (mean ± SD, n= 3, Student's t-test, *P < 0.05). Genotypes of flies: da-GAL4 in trans to (CAG250+GFP) UAS-DsRed-CAG250 UAS-GFP and (CAG250+Hsp70) is UAS-DsRed-CAG250; UAS-Hsp70. (C) Levels of CAG-repeat RNA were examined by qRT-PCR and were not significantly altered when co-expressed with Hsp70 (3-day flies, Student's t-test, P > 0.05). Genotypes of flies from left to right: da-GAL4 in trans to UAS-GFP UAS-DsRed-CAG250 and UAS-DsRed-CAG250; UAS-Hsp70.
Additional modifiers of CAG-repeat RNA toxicity identified from a targeted modification screen
A previous genetic screen had identified a number of modifiers of Ataxin-3 protein-associated neurodegeneration (21). We therefore performed a targeted modification screen of these enhancer–promoter (EP)-element insertion lines for ability to modulate the CAG-repeat RNA toxicity. To assess effects, we examined several indicators of RNA-based toxicity when a CAG250 repeat is expressed in the animal: wing posture and motility deficits. Normally, flies hold their wings flat to the back, rarely showing an abnormal upward wing posture (Fig. 6A). However, 70% of CAG250 flies displayed an abnormal wing posture, holding one or both wings up (Fig. 6A and B). This abnormality may be due to muscle- and/or neural-based effects. Previous findings also show that flies display defects in climbing ability when expressing the toxic CAG-repeat RNA in neurons with elav-GAL4 (5) (Fig. 5A). We used these two indicators of the toxicity of the CAG250 RNA (held-up wings and loss of climbing ability) to assess whether additional previously defined modifiers of Ataxin-3 protein-based toxicity could affect RNA toxicity.
Figure 6.
Tpr2EB7-1A, dpldE546 and orb2B-8S suppress CAG-repeat RNA toxicity. (A) Abnormal wing posture (one or both wings held-up, lower panels) is observed in flies expressing CAG250 with a ubiquitous driver da-GAL4. (B) About 30% of animals expressing CAG250 have abnormal wing posture at day 1. Genotypes of flies from left to right: da-GAL4 in trans to UAS-GFP and UAS-GFP UAS-DsRed-CAG250. (C) Co-expression of Tpr2EB7-1A, dpldE546 and orb2B-8S, with CAG250 does not alter the CAG250 relative expression level examined by qRT-PCR (mean ± SD, n= 3, Student's t-test, P > 0.05). (D, F and H) Upregulation of Tpr2EB7-1A, dpldE546 and orb2B-8S significantly rescued abnormal wing posture of CAG250 flies at day 1. (E, G and I) Co-expression of Tpr2EB7-1A, dpldE546 and orb2B-8S significantly rescue climbing defects of CAG250 flies at day 1 (mean ± SD, n= 3 independent experiments, Student's t-test, *P < 0.05). Genotypes of flies: da-GAL4 in trans to (CAG250+GFP) UAS-DsRed-CAG250 UAS-GFP, (CAG250+Tpr2EB7-1A) UAS-DsRed-CAG250 Tpr2EB7-1A, (CAG250+dpldE546) UAS-DsRed-CAG250 dpldE546 and (CAG250+orb2B-8S) UAS-DsRed-CAG250; orb2B-8S.
Twenty-five different modifiers were previously isolated in a genome-wide screen for suppressors or enhancers of pathogenic polyQ Ataxin-3 protein toxicity (21). Of these, three mitigated both the wing posture and locomotor defects of CAG250 flies: the co-chaperone Tpr2EB7-1A, the lin-41 homologue dpldEP546 and RNA-binding protein orb2B8-S (Fig. 6D–I). While only 26 ± 4% of CAG250 flies had normal wing posture, now 66 ± 10% of flies with upregulated Tpr2 showed normal wing position (Fig. 6D). Tpr2EB7-1A also functionally rescued the climbing defects of CAG250 flies (58 ± 8% of Tpr2EB7-1A flies could climb, whereas only 36 ± 5% of CAG250 flies could climb in a similar manner in parallel control experiments) (Fig. 6E). Upregulation of dpld with the dpldEP546 allele rescued both wing posture and locomotor dysfunction, such that 58 ± 9% of CAG250 flies with upregulated dpldEP546 had normal wing posture, and climbing was restored such that 62 ± 9% of animals could climb (Fig. 6F and G). Upregulation of orb2 with the orb2B8-S allele resulted in 80 ± 14% of CAG250 flies now showing normal wing posture and 62 ± 8% of animals climbing normally (Fig. 6H and I). By qRT-PCR, we confirmed that these modifiers did not affect the expression levels of the CAG-repeat RNA (Fig. 6C). Taken together, these data indicated that these modifiers, originally identified as genes that when upregulated can mitigate Ataxin-3 pathogenic protein-based toxicity, also rescue CAG-repeat RNA-based toxicity.
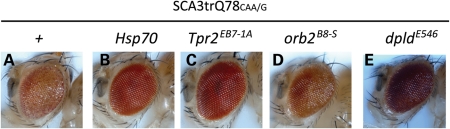
The genetic screen for modifiers of Ataxin-3 protein toxicity was performed with a transgene bearing a pure CAG-repeat RNA. As these four modifiers could modulate both the toxic effect of this transgene and the toxic effect of a non-coding CAG-repeat RNA, we were interested in whether the mitigation of the Ataxin-3 toxicity was due to a selective effect on the RNA versus an effect on both the protein and the RNA. To address this, we determined whether interrupting the RNA sequence of the Ataxin-3 pathogenic protein affected the interaction with these modulators. If these modifiers were no longer effective, then this would suggest that these modifiers were directed solely to the CAG-repeat RNA. We therefore co-expressed Hsp70, Tpr2EB7-1A, dpldEP546 and orb2B8-S, with an Ataxin-3-expressing transgene in which the polyQ domain is encoded by a CAA/G-interrupted repeat sequence. The SCA3trQ78CAA/G transgene encodes a toxic polyQ protein identical in sequence to the CAG-repeat encoding transgene SCA3trQ78CAG, differing only in whether the RNA is a pure CAG or interrupted by CAA codons. Expression of SCA3tr Q78CAA/G in the eye by gmr-GAL4 gave a disrupted eye effect (Fig. 7A), and co-expression of these modifiers mitigated the toxicity (Fig. 7B–E). These data indicate that these modifiers can modulate both Ataxin-3 pathogenic protein-based toxicity and CAG-repeat RNA-based toxicity, highlighting the overlap between RNA-based and polyQ protein-based toxicity mechanisms.
Figure 7.
Hsp70, Tpr2EB7-1A, dpldE546 and orb2B-8S suppress both CAG-repeat RNA-based toxicity and polyQ protein toxicity. (A) Eyes of flies expressing SCA3trQ78CAA/G, the toxic polyQ protein expressed from a transgene with an interrupted CAA/G polyQ repeat region, have degenerate eyes with reduced pigmentation. (B–E) Upregulation of Hsp70, Tpr2EB7-1A, dpldE546 and orb2B-8S ameliorates the degenerative effect of SCA3trQ78CAA/G. Genotypes of flies (A–E): gmr-GAL4 SCA3trQ78CAA/G in trans to +, UAS-Hsp70, Tpr2EB7-1A, dpldE546 and orb2B-8S.
DISCUSSION
PolyQ diseases have been considered toxic polyQ-based disorders, although recent data suggest that the expanded CAG trinucleotide repeat RNA encoding the polyQ domain may contribute to the disease (5). Given that mechanisms of RNA toxicity remain unclear, we provided insights into the nature of the CAG-repeat RNA toxicity by performing a microarray analysis to define the global transcriptional profile of the fly brain upon expression of toxic trinucleotide repeat RNA. These studies revealed that 160 genes were altered, including the Hsp70 genes. A targeted genetic screen then defined Hsp70, as well as a number of other genes, as modifiers of both the polyQ protein and toxic CAG-repeat RNA. These data indicate an overlap between mechanisms of the protein-based and RNA-based toxicities that may contribute to polyQ disease.
Microarray analysis identifies altered Hsp70: a modifier of CAG-repeat RNA-based toxicity
Our microarray analysis identified Hsp70 as upregulated in CAG100 flies. To confirm these findings, we performed immunostaining analysis for the Hsc70/Hsp70 protein in brains of animals expressing the toxic RNA. This revealed age-dependent accumulations. These data indicate that upregulated Hsp70 induced by the toxic CAG repeat occurs at both transcriptional and translational levels. Given that Hsc70/Hsp70 accumulations are induced by expressing CAG100, and Hsp70 is a powerful suppressor of several human neurodegenerative diseases, we then performed functional analysis. These findings showed that upregulation of Hsp70 rescues the locomotor dysfunction caused by the toxic CAG repeat. Similarly, in a Drosophila model of fragile X syndrome, which is also an RNA-mediated neurodegenerative situation, upregulation of Hsc70 modifies the CGG-repeat RNA-induced effects (22). Taken together, these data suggest that Hsc70/Hsp70 chaperones can modify toxicity conferred by both pathogenic RNA and proteins. Hsp70 is thought to prevent aggregation or to assist in the refolding of misfolded protein. Possible mechanisms of suppression by Hsp70 in RNA-mediated toxicity might involve a misfolded protein response triggered by deleterious consequence of the pathogenic repeat RNA.
Other ways in which Hsp70 may suppress include modulation of death pathways (23). Notably, several genes associated with the regulation of apoptosis were in the list of genes affected in the CAG100 flies. Upregulation of cytochrome c transcript is a general early apoptotic response, and studies show that Hsp70 suppresses apoptosis by acting downstream of cytochrome c release in mammalian cells (24,25). Intriguingly, cytochrome c distal (Cyt-c-d), one of the Drosophila cytochrome c genes, was upregulated pre-symptomatically in CAG100 flies in the microarray analysis (Cyt-c-d, 2.0-fold change). Moreover, upregulation of Hsp70 suppresses the symptomatic effects of toxicity of CAG250 flies. Potentially, Hsp70 may suppress RNA-based toxicity through modulation of apoptosis processes as an additional mechanism.
The molecular chaperone Hsp70 responds to several neurodegenerative disease situations in animal models. In the Drosophila model of SCA3, Hsp70 is localized to the nuclear accumulations, whereas normally there is a low level of Hsp70 in the cytoplasm (4). Upregulation of Hsp70 suppresses the toxicity caused by the pathogenic Ataxin-3 protein. In mouse models of spinal bulbar muscular atrophy (SBMA), due to polyQ expansions in the androgen receptor gene, Hsp70 levels are increased in the muscle, and upregulation of Hsp70 ameliorates pathogenicity of the SBMA model (26). In the Drosophila model of fragile X-associated tremor/ataxia syndrome, Hsp70-containing inclusions are induced by expressing long CGG repeats in the 5′-UTR of an enhanced green fluorescent protein (EGFP) reporter gene. Upregulation of Hsc70 modifies CGG-repeat-induced degeneration (22). RnrL, which suppresses non-coding CTG112 RNA-induced neurodegeneration in a Drosophila model for SCA8, was also identified in our present study (RnrL, 7.4-fold change) (27). These data suggest that candidate genetic modifiers can be identified through microarray analysis, defining altered genes in the disease-associated condition.
A recent study shows that modifier of mdg4 [mod(mdg4)] transcript levels is altered in Drosophila heads upon expression of CAG-, CUG- and AUUCU-repeat RNA and that mod(mdg4) is an enhancer of CAG- and CUG-repeat toxicity (28). We did not find that mod(mdg4) met our criteria in generating gene lists; however, we used only brain tissue and not heads with eye tissue included. Transcriptional profiles may differ in distinct cell types in response to toxic elements, and brain-enriched RNA may reveal distinct or more subtle changes in gene expression that are selective to neurons.
Candidate targets for further investigation of RNA-based toxicity mechanisms
Several functional classes of genes were differentially expressed in CAG100 flies (Fig. 3). These classes give insights into molecular pathways that respond to the toxic CAG-repeat RNA. For example, microglial activation and innate immune system activation are recognized as a disease feature in Alzheimer's (AD), PD, HD and ALS. Anti-inflammatory treatments may reduce susceptibility to AD and PD (29). Proteins involved in the innate immune response could be used as molecular biomarkers to identify pre-symptomatic stage of AD (30). Beyond changes in the cytochrome P450 superfamily, there were three upregulated transcripts associated with innate immune response, including attacin, Immune-induced molecule 2 and metchnikowin. These gene lists not only provide candidate targets for further genetic analysis, but might also provide insights into potential CAG-repeat RNA pathogenesis and the common characteristic of neurodegenerative diseases. Furthermore, a set of genes was selectively affected in CAA/G105 and not in CAG100 or CAG0 (Supplementary Material, Table S1c). These transcripts may be relevant to situations in which the polyQ repeat sequence in the disease gene is interrupted, as in ATXN2 polyQ expansions that present with parkinsonism or ALS rather than SCA2. Future studies may reveal insights with regard to potential pathways modulated.
Expanded CAG-repeat RNA and pathogenic polyQ protein trigger common pathways
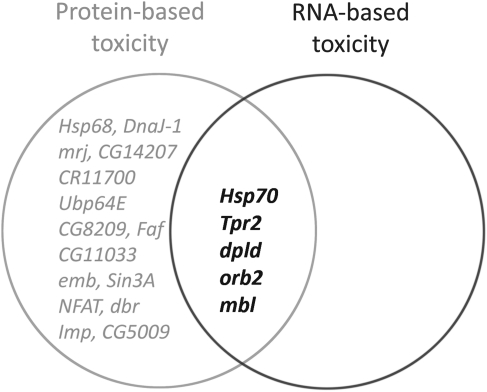
Beyond Hsp70, three genetic modifiers identified in the present study, Tpr2EB7-1A, dpldEP546 and orb2B8-S, also suppress both CAG-repeat RNA-based toxicity and polyQ protein-based toxicity. Previous studies show that these modifiers do not alter the Hsp70 expression level, thus function in a distinct manner (21). dTPR2 is homologous to the human tetratricopeptide repeat protein 2. Tpr2 is a co-chaperone containing two chaperone-binding TPR domains and a DnaJ homologous J domain (31,32). In previous studies, Hsp70 and five other modifiers of Ataxin-3 protein-associated neurodegeneration were categorized in the chaperone class (21). Surprisingly, two of these six also suppressed CAG-repeat RNA toxicity (Hsp70 and Tpr2EB7-1A) (Fig. 8). This raises a role for chaperones and ubiquitin-associated pathway components in CAG-repeat toxicity, although likely specific genes involved reflect specific processes primarily detected.
Figure 8.
Modifiers of Ataxin-3 protein-associated neurodegeneration affect both polyQ protein toxicity and CAG-repeat RNA-based toxicity. A previous genetic screen identified a number of modifiers of Ataxin-3 protein-associated neurodegeneration. We therefore performed a targeted screen of these modulators for ability to affect the CAG-repeat RNA toxicity. Hsp70 and another 18 genes were identified as suppressors of Ataxin-3 protein-based neurodegeneration. Among these 19 modulators, Hsp70, Tpr2EB7-1A, dpldE546 and orb2B8-S were found in the present study to also affect the CAG-repeat RNA-based toxicity. Previous studies also demonstrated that mbl affects the toxicity of both the Ataxin-3 protein and non-coding CAG-repeat RNA. These genes that can modulate both polyQ protein and CAG-repeat RNA-based toxicities highlight an overlap between RNA-based and polyQ protein based-toxicity mechanisms.
dpldEP546 was previously identified as a strong suppressor of Ataxin-3 toxicity and could suppress tau toxicity as well as a cell-death gene (hid). Intriguingly, data indicate that the mouse homologue of dpld (lin-41) is localized to P-granules, which are sites of RNA processing and degradation (33). This overlap between dpld as a modifier of polyQ protein toxicity and the CAG-RNA-based toxicity, coupled with the implication of P-granule involvement, supports the idea of RNA pathways going awry in both situations.
Drosophila orb2 is also known as Drosophila cytoplasmic polyadenylation element-binding protein (CPEB). CPEB belongs to a family of RNA-binding proteins which have two RRMs and a Zn-finger domain (34). CPEBs regulate translation of target mRNAs (35) and function in many processes, including synaptic plasticity, learning and memory (36–38). Recent studies show that Drosophila Orb2 binds directly to transcripts associated with long-term memory, and Orb2 targets are associated with neuronal growth, synapse formation and protein turnover (39). Taken together, these studies indicate that Orb2 is broadly involved in various neuronal functions. Moreover, Orb2 is co-localized in dFMR1-containing (fragile X mental retardation 1) foci in cultured Drosophila primary neurons (40). Fragile X mental retardation protein has been implicated in the translational regulation of target mRNAs in neurons and is the protein affected in fragile X syndrome (41–43). The pathogenic role of neuronal RNA granules in neurological disease suggests the possibility that CAG-repeat RNA might interfere with the normal function of Orb2 in neuronal RNA granules to cause toxicity. Homologues of mbl, which modifies Ataxin-3 protein toxicity, CAG-repeat RNA toxicity and CUG-repeat RNA toxicity, co-localize with RNA stress granules in cell culture (44,45). Thus, mbl may be broadly involved in other aspects of RNA metabolism that contribute to neurodegenerative disease. Future investigation to examine effects of the CAG-repeat RNA, and neural granules and P-bodies, together with studies of Orb2 will reveal further insight.
Intriguing recent data raise the possibility that expanded repeat RNAs may support protein translation in a manner not dependent on an AUG, as with classical protein-coding genes. That is, the hairpin repeat structure of triplet-repeat RNAs of a variety of sequences can lead to protein translation of the repeat region in all three reading frames. This so-called repeat-associated non-ATG translation (RAN translation) is reported to occur with SCA8 and DM1 CAG expansion transcripts (46). In the fly, previous studies failed to detect a protein generated by the non-coding CAG-repeat RNA (5); however, these studies were performed on young animals. Similarly, in a mouse model of CAG-repeat RNA toxicity, a polyQ-containing protein was not detected upon expression of CAG repeats in the 3′-UTR of the EGFP gene (47). RAN, however, may be selective to specific cell subtypes. Our finding of accumulation of Hsc70/Hsp70 with age in CAG-repeat expressing flies, suggestive of late-onset accumulation of misfolded protein, may be indicative of such mechanisms, as is the overlap in modifiers of the protein-based toxicity and RNA-based toxicity. Our studies provide a foundation to address the extent to which this mechanism, as well as others, contribution to the toxicity conferred by the expanded repeat CAG-RNA, and its integration with protein-based toxicity mechanisms.
MATERIALS AND METHODS
Drosophila stocks and crosses
General fly lines were from Bloomington Drosophila Stock Center and maintained at 25°C on standard medium, unless otherwise indicated. Transgenic lines UAS-DsRed-CAG0, UAS-DsRed-CAG100, UAS-DsRed-CAG250, UAS-DsRed-CAA/G105 and genetic modifier EP insertion lines have been described (5,21). Control CAG0 flies express a transgene encoding a DsRed reporter protein with no CAG repeat in the 3′-UTR. CAG100 and CAG250 flies express transgenes encoding the DsRed reporter with repeats of approximately 100 and 250 CAGs in the 3′-UTR, as described previously (5). CAA/G105 flies, a non-toxic trinucleotide control, express a transgene encoding DsRed with a repeat sequence of CAA and CAG of 105 triplet codons in the 3′-UTR, as described (5).
RNA isolation
Four independent replicates of flies expressing CAG0, CAG100 or CAA/G105 by elav-GAL4 were raised at 25°C with 12 h/12 h light–dark cycle (5). Three-day female flies from four independent crosses were collected and brains dissected. Fly brain tissue (about 20 brains per sample, dissected from head capsule, eyes, lamina and outer medulla removed) was isolated in cold phosphate-buffered saline and stored in Trizol Reagent (Invitrogen, Carlsbad, CA) at −80°C. Total brain RNA was extracted and purified using Trizol Reagent (Invitrogen) and the RNeasy Mini system (Qiagen, Valencia, CA) and treated with RNase-free DNase I (Qiagen). cDNA amplification (Ovation Pico WTA systems, NuGEN, San Carlos, CA), labelling and hybridization (Encore Biotin Module, NuGEN) were carried out by the microarray core facility at University of Pennsylvania.
Microarray analysis
Affymetrix Drosophila Genome 2.0 microarrays (Affymetrix) were used for the microarray analysis. Normalization of probe signal intensity levels across arrays was performed using the GC robust multichip average algorithm implemented in Partek Genomic suite 6.2 beta. Principle component analysis and Pearson's correlation coefficient were used to reveal outliner samples. Three-way analysis of variance (ANOVA) confirmed that the source of variation was not due to different batches of RNA. To generate lists of genes differentially expressed in different conditions, CAG100 and CAG250 were compared with CAG0 at the corresponding age (step-up P < 0.05). We used one-way ANOVA implemented in Partek Genomic suite 6.2 beta to generate a list of differentially expressed genes under each condition. Unannotated probe sets were discarded. When multiple probe sets were assigned to the same gene, the probe set with the smallest step-up P-value was included. Gene lists were restricted to genes that have at least a 2-fold change and step-up P-value less than 0.05. GEO accession number for the data is GSE31875.
Quantitative RT-PCR
qRT-PCR was performed on 3-day adult fly brain RNA. Brain dissections and total brain RNA were prepared and extracted, as described earlier. cDNA was synthesized using the High Capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA, USA). qRT-PCR was performed using SYBR Green master mix (Applied Biosystems). All RNA samples were analysed in triplicate or quadruplicate using a 7500 Fast Real-Time PCR System (Applied Biosystems). Data were analysed using the relative quantification method. The endogenous control was ribosomal protein 49 (Rp49). Primers identified using Primer Express 2.0 (Applied Biosystems) were synthesized by Integrated DNA Technologies, Inc. Primer sets are as follows:
rp49 forward: 5′-CAACATCGGTTACGGATCGA-3′, rp49 reverse: 5′-AATCCGGTGGGCAGCAT-3′; Roe1 forward: 5′-CCTAGTTTCTAAGAGATCTGTGACTG-3′, Roe1 reverse: 5′-GCCACGCGTATGGTACAAAT-3′; or63a forward: 5′-TGGAACAAGCCACATCTGAG-3′, or63a reverse: 5′-TGTTAACCTCGGTTGCAAAGT-3′; NFAT forward: 5′-ACACAATCCACGCCCAAG-3′, NFAT reverse: 5′-GAGCCACTCGCATTGGAC-3′; MTK forward: 5′-CCACCGAGCTAAGATGCAA-3′, MTK reverse: 5′-TCTGCCAGCACTGATGTAGC-3′; CTCF forward: 5′-GCTACACGTTCCGTGGTCA-3′, CTCF reverse: 5′-CGCTTTCGGTGTCGATTT-3′; CG9935 forward: 5′-GCAGACACTGGAACAATCACA-3′, CG9935 reverse: 5′-GGACCTGTCAGACCTTTCCA-3′; CG6656 forward: 5′-CGACCGATTGCAAAACATAA-3′, CG6656 reverse: 5′-CAACCCAGAACGACACACAC-3′; CG30104 forward: 5′-CATGGATGTGGAGTTCAACG-3′, CG30104 reverse: 5′-GATGCCCTGTGCCTTCAG-3′; CG18343 forward: 5′-ACGCAATACTTTTCGGCTTC-3′, CG18343 reverse: 5′-AGGGCGATAACCACTTTGAG-3′; CG12428 forward: 5′-CGCAAGTTCAACGACATTTTC-3′, CG12428 reverse: 5′-TTCATAGCTTGGATTCCTTG-3′; CG11893 forward: 5′-CTGGAATGGACAACTTTTTGAA-3′, CG11893 reverse: 5′-TTCTCAAAGTGTGGCTTGTACTTT-3′; 312 forward: 5′-TGGATCACTATTACGGCCTCA-3′, 312 reverse: 5′-GTCGTCCACATTCTCCTTGC-3′; DsRed forward: 5′-CCTCCTCCGAGAACGTCATC-3′, DsRed reverse: 5′-CCCTCCATGCGCACCTT-3′.
Histochemistry and image quantification
Cryosections were performed as described (48). For immunostaining, the primary antibody was anti-Hsc70/Hsp70 (SPA-822, 1:200, Stressgen-Enzo Life Sciences, Farmingdale, NY). The secondary antibody was anti-mouse conjugated to Alexa Fluor 488 (1:100, Molecular Probes, Eugene, OR). The number of Hsc70/Hsp70-positive particles was quantitated using the particle analysis method implemented in Image J (NIH). The thresholds were adjusted based on background signals in the central brain region. Images from three to four animals per genotype were analysed.
Climbing assay
Flies expressing the repeat gene by da-GAL4 were raised at 20°C to prevent lethality at pre-adult stages and shifted to 29°C after 1 day for a stronger effect and for time considerations. About 8–20 female flies were anaesthetized by CO2 and placed in a plastic vial. After a 1 h recovery period, flies were banged to the bottom of the vial, and then the number of flies that climbed higher than a 2 cm threshold in 10 s was scored. Three trials were performed at 5 min intervals for each cohort of animals. At least, 100 flies per genotype were examined in each repeat.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
FUNDING
This work received funding from the NIH (5R01NS043578, 5P01AG009215), the Ellison Medical Foundation, and the MDA. N.M.B. is an investigator of the Howard Hughes Medical Institute. Funding to pay the Open Access publication charges for this article was provided by the Howard Hughes Medical Institute.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs John Tobias and Shilpa Rao for help with microarray data analysis and interpretation and L.Y. Hao, Y. Fang and L. McGurk for critical comments.
REFERENCES
- 1.Li L.B., Bonini N.M. Roles of trinucleotide-repeat RNA in neurological disease and degeneration. Trends Neurosci. 2010;33:292–298. doi: 10.1016/j.tins.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ranum L.P., Cooper T.A. RNA-mediated neuromuscular disorders. Annu. Rev. Neurosci. 2006;29:259–277. doi: 10.1146/annurev.neuro.29.051605.113014. [DOI] [PubMed] [Google Scholar]
- 3.Gatchel J.R., Zoghbi H.Y. Diseases of unstable repeat expansion: mechanisms and common principles. Nat. Rev. Genet. 2005;6:743–755. doi: 10.1038/nrg1691. [DOI] [PubMed] [Google Scholar]
- 4.Warrick J.M., Chan H.Y., Gray-Board G.L., Chai Y., Paulson H.L., Bonini N.M. Suppression of polyglutamine-mediated neurodegeneration in Drosophila by the molecular chaperone HSP70. Nat. Genet. 1999;23:425–428. doi: 10.1038/70532. [DOI] [PubMed] [Google Scholar]
- 5.Li L.B., Yu Z., Teng X., Bonini N.M. RNA toxicity is a component of ataxin-3 degeneration in Drosophila. Nature. 2008;453:1107–1111. doi: 10.1038/nature06909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sobczak K., de Mezer M., Michlewski G., Krol J., Krzyzosiak W.J. RNA structure of trinucleotide repeats associated with human neurological diseases. Nucleic Acids Res. 2003;31:5469–5482. doi: 10.1093/nar/gkg766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Michlewski G., Krzyzosiak W.J. Molecular architecture of CAG repeats in human disease related transcripts. J. Mol. Biol. 2004;340:665–679. doi: 10.1016/j.jmb.2004.05.021. [DOI] [PubMed] [Google Scholar]
- 8.Sobczak K., Krzyzosiak W.J. CAG repeats containing CAA interruptions form branched hairpin structures in spinocerebellar ataxia type 2 transcripts. J. Biol. Chem. 2005;280:3898–3910. doi: 10.1074/jbc.M409984200. [DOI] [PubMed] [Google Scholar]
- 9.Corrado L., Mazzini L., Oggioni G.D., Luciano B., Godi M., Brusco A., D'Alfonso S. ATXN-2 CAG repeat expansions are interrupted in ALS patients. Hum. Genet. 2011 doi: 10.1007/s00439-011-1000-2. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 10.Yu Z., Zhu Y., Chen-Plotkin A.S., Clay-Falcone D., McCluskey L., Elman L., Kalb R.G., Trojanowski J.Q., Lee V.M., Van Deerlin V.M., et al. PolyQ repeat expansions in ATXN2 associated with ALS are CAA interrupted repeats. PLoS One. 2011;6:e17951. doi: 10.1371/journal.pone.0017951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim J.M., Hong S., Kim G.P., Choi Y.J., Kim Y.K., Park S.S., Kim S.E., Jeon B.S. Importance of low-range CAG expansion and CAA interruption in SCA2 parkinsonism. Arch. Neurol. 2007;64:1510–1518. doi: 10.1001/archneur.64.10.1510. [DOI] [PubMed] [Google Scholar]
- 12.Charles P., Camuzat A., Benammar N., Sellal F., Destee A., Bonnet A.M., Lesage S., Le Ber I., Stevanin G., Durr A., et al. Are interrupted SCA2 CAG repeat expansions responsible for parkinsonism? Neurology. 2007;69:1970–1975. doi: 10.1212/01.wnl.0000269323.21969.db. [DOI] [PubMed] [Google Scholar]
- 13.Miller J.W., Urbinati C.R., Teng-Umnuay P., Stenberg M.G., Byrne B.J., Thornton C.A., Swanson M.S. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 2000;19:4439–4448. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Machuca-Tzili L., Thorpe H., Robinson T.E., Sewry C., Brook J.D. Flies deficient in muscleblind protein model features of myotonic dystrophy with altered splice forms of Z-band associated transcripts. Hum. Genet. 2006;120:487–499. doi: 10.1007/s00439-006-0228-8. [DOI] [PubMed] [Google Scholar]
- 15.Reinke R., Krantz D.E., Yen D., Zipursky S.L. Chaoptin, a cell surface glycoprotein required for Drosophila photoreceptor cell morphogenesis, contains a repeat motif found in yeast and human. Cell. 1988;52:291–301. doi: 10.1016/0092-8674(88)90518-1. [DOI] [PubMed] [Google Scholar]
- 16.Luo L.Q., Martin-Morris L.E., White K. Identification, secretion, and neural expression of APPL, a Drosophila protein similar to human amyloid protein precursor. J. Neurosci. 1990;10:3849–3861. doi: 10.1523/JNEUROSCI.10-12-03849.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 18.Dennis G., Jr, Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 19.Hofmann T., Chubanov V., Chen X., Dietz A.S., Gudermann T., Montell C. Drosophila TRPM channel is essential for the control of extracellular magnesium levels. PLoS One. 2010;5:e10519. doi: 10.1371/journal.pone.0010519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scherzer C.R., Jensen R.V., Gullans S.R., Feany M.B. Gene expression changes presage neurodegeneration in a Drosophila model of Parkinson’s disease. Hum. Mol. Genet. 2003;12:2457–2466. doi: 10.1093/hmg/ddg265. [DOI] [PubMed] [Google Scholar]
- 21.Bilen J., Bonini N.M. Genome-wide screen for modifiers of ataxin-3 neurodegeneration in Drosophila. PLoS Genet. 2007;3:1950–1964. doi: 10.1371/journal.pgen.0030177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jin P., Zarnescu D.C., Zhang F., Pearson C.E., Lucchesi J.C., Moses K., Warren S.T. RNA-mediated neurodegeneration caused by the fragile X premutation rCGG repeats in Drosophila. Neuron. 2003;39:739–747. doi: 10.1016/s0896-6273(03)00533-6. [DOI] [PubMed] [Google Scholar]
- 23.Mosser D.D., Caron A.W., Bourget L., Meriin A.B., Sherman M.Y., Morimoto R.I., Massie B. The chaperone function of hsp70 is required for protection against stress-induced apoptosis. Mol. Cell. Biol. 2000;20:7146–7159. doi: 10.1128/mcb.20.19.7146-7159.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li C.Y., Lee J.S., Ko Y.G., Kim J.I., Seo J.S. Heat shock protein 70 inhibits apoptosis downstream of cytochrome c release and upstream of caspase-3 activation. J. Biol. Chem. 2000;275:25665–25671. doi: 10.1074/jbc.M906383199. [DOI] [PubMed] [Google Scholar]
- 25.Chandra D., Liu J.W., Tang D.G. Early mitochondrial activation and cytochrome c up-regulation during apoptosis. J. Biol. Chem. 2002;277:50842–50854. doi: 10.1074/jbc.M207622200. [DOI] [PubMed] [Google Scholar]
- 26.Adachi H., Katsuno M., Minamiyama M., Sang C., Pagoulatos G., Angelidis C., Kusakabe M., Yoshiki A., Kobayashi Y., Doyu M., et al. Heat shock protein 70 chaperone overexpression ameliorates phenotypes of the spinal and bulbar muscular atrophy transgenic mouse model by reducing nuclear-localized mutant androgen receptor protein. J. Neurosci. 2003;23:2203–2211. doi: 10.1523/JNEUROSCI.23-06-02203.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mutsuddi M., Marshall C.M., Benzow K.A., Koob M.D., Rebay I. The spinocerebellar ataxia 8 noncoding RNA causes neurodegeneration and associates with staufen in Drosophila. Curr. Biol. 2004;14:302–308. doi: 10.1016/j.cub.2004.01.034. [DOI] [PubMed] [Google Scholar]
- 28.van Eyk C.L., O'Keefe L.V., Lawlor K.T., Samaraweera S.E., McLeod C.J., Price G.R., Venter D.J., Richards R.I. Perturbation of the Akt/Gsk3-{beta} signalling pathway is common to Drosophila expressing expanded untranslated CAG, CUG and AUUCU repeat RNAs. Hum. Mol. Genet. 2011;20:2783–2794. doi: 10.1093/hmg/ddr177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwab C., McGeer P.L. Inflammatory aspects of Alzheimer disease and other neurodegenerative disorders. J. Alzheimers Dis. 2008;13:359–369. doi: 10.3233/jad-2008-13402. [DOI] [PubMed] [Google Scholar]
- 30.Ray S., Britschgi M., Herbert C., Takeda-Uchimura Y., Boxer A., Blennow K., Friedman L.F., Galasko D.R., Jutel M., Karydas A., et al. Classification and prediction of clinical Alzheimer's diagnosis based on plasma signaling proteins. Nat. Med. 2007;13:1359–1362. doi: 10.1038/nm1653. [DOI] [PubMed] [Google Scholar]
- 31.Brychzy A., Rein T., Winklhofer K.F., Hartl F.U., Young J.C., Obermann W.M. Cofactor Tpr2 combines two TPR domains and a J domain to regulate the Hsp70/Hsp90 chaperone system. EMBO J. 2003;22:3613–3623. doi: 10.1093/emboj/cdg362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kazemi-Esfarjani P., Benzer S. Genetic suppression of polyglutamine toxicity in Drosophila. Science. 2000;287:1837–1840. doi: 10.1126/science.287.5459.1837. [DOI] [PubMed] [Google Scholar]
- 33.Rybak A., Fuchs H., Hadian K., Smirnova L., Wulczyn E.A., Michel G., Nitsch R., Krappmann D., Wulczyn F.G. The let-7 target gene mouse lin-41 is a stem cell specific E3 ubiquitin ligase for the miRNA pathway protein Ago2. Nat. Cell Biol. 2009;11:1411–1420. doi: 10.1038/ncb1987. [DOI] [PubMed] [Google Scholar]
- 34.Hake L.E., Mendez R., Richter J.D. Specificity of RNA binding by CPEB: requirement for RNA recognition motifs and a novel zinc finger. Mol. Cell. Biol. 1998;18:685–693. doi: 10.1128/mcb.18.2.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richter J.D. CPEB: a life in translation. Trends Biochem. Sci. 2007;32:279–285. doi: 10.1016/j.tibs.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 36.Keleman K., Kruttner S., Alenius M., Dickson B.J. Function of the Drosophila CPEB protein Orb2 in long-term courtship memory. Nat. Neurosci. 2007;10:1587–1593. doi: 10.1038/nn1996. [DOI] [PubMed] [Google Scholar]
- 37.Kang H., Schuman E.M. A requirement for local protein synthesis in neurotrophin-induced hippocampal synaptic plasticity. Science. 1996;273:1402–1406. doi: 10.1126/science.273.5280.1402. [DOI] [PubMed] [Google Scholar]
- 38.Wu L., Wells D., Tay J., Mendis D., Abbott M.A., Barnitt A., Quinlan E., Heynen A., Fallon J.R., Richter J.D. CPEB-mediated cytoplasmic polyadenylation and the regulation of experience-dependent translation of alpha-CaMKII mRNA at synapses. Neuron. 1998;21:1129–1139. doi: 10.1016/s0896-6273(00)80630-3. [DOI] [PubMed] [Google Scholar]
- 39.Mastushita-Sakai T., White-Grindley E., Samuelson J., Seidel C., Si K. Drosophila Orb2 targets genes involved in neuronal growth, synapse formation, and protein turnover. Proc. Natl Acad. Sci. USA. 2010;107:11987–11992. doi: 10.1073/pnas.1004433107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cziko A.M., McCann C.T., Howlett I.C., Barbee S.A., Duncan R.P., Luedemann R., Zarnescu D., Zinsmaier K.E., Parker R.R., Ramaswami M. Genetic modifiers of dFMR1 encode RNA granule components in Drosophila. Genetics. 2009;182:1051–1060. doi: 10.1534/genetics.109.103234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Didiot M.C., Subramanian M., Flatter E., Mandel J.L., Moine H. Cells lacking the fragile X mental retardation protein (FMRP) have normal RISC activity but exhibit altered stress granule assembly. Mol. Biol. Cell. 2009;20:428–437. doi: 10.1091/mbc.E08-07-0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barbee S.A., Estes P.S., Cziko A.M., Hillebrand J., Luedeman R.A., Coller J.M., Johnson N., Howlett I.C., Geng C., Ueda R., et al. Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron. 2006;52:997–1009. doi: 10.1016/j.neuron.2006.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Merenstein S.A., Sobesky W.E., Taylor A.K., Riddle J.E., Tran H.X., Hagerman R.J. Molecular-clinical correlations in males with an expanded FMR1 mutation. Am. J. Med. Genet. 1996;64:388–394. doi: 10.1002/(SICI)1096-8628(19960809)64:2<388::AID-AJMG31>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 44.de Haro M., Al-Ramahi I., De Gouyon B., Ukani L., Rosa A., Faustino N.A., Ashizawa T., Cooper T.A., Botas J. MBNL1 and CUGBP1 modify expanded CUG-induced toxicity in a Drosophila model of myotonic dystrophy type 1. Hum. Mol. Genet. 2006;15:2138–2145. doi: 10.1093/hmg/ddl137. [DOI] [PubMed] [Google Scholar]
- 45.Onishi H., Kino Y., Morita T., Futai E., Sasagawa N., Ishiura S. MBNL1 associates with YB-1 in cytoplasmic stress granules. J. Neurosci. Res. 2008;86:1994–2002. doi: 10.1002/jnr.21655. [DOI] [PubMed] [Google Scholar]
- 46.Zu T., Gibbens B., Doty N.S., Gomes-Pereira M., Huguet A., Stone M.D., Margolis J., Peterson M., Markowski T.W., Ingram M.A., et al. Non-ATG-initiated translation directed by microsatellite expansions. Proc. Natl Acad. Sci. USA. 2011;108:260–265. doi: 10.1073/pnas.1013343108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hsu R.J., Hsiao K.M., Lin M.J., Li C.Y., Wang L.C., Chen L.K., Pan H. Long tract of untranslated CAG repeats is deleterious in transgenic mice. PLoS One. 2011;6:e16417. doi: 10.1371/journal.pone.0016417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chan H.Y., Warrick J.M., Andriola I., Merry D., Bonini N.M. Genetic modulation of polyglutamine toxicity by protein conjugation pathways in Drosophila. Hum. Mol. Genet. 2002;11:2895–2904. doi: 10.1093/hmg/11.23.2895. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.