Abstract
Mutations of the Interleukin-1-receptor accessory protein like 1 (IL1RAPL1) gene are associated with cognitive impairment ranging from non-syndromic X-linked mental retardation to autism. IL1RAPL1 belongs to a novel family of IL1/Toll receptors, which is localized at excitatory synapses and interacts with PSD-95. We previously showed that IL1RAPL1 regulates the synaptic localization of PSD-95 by controlling c-Jun N-terminal kinase activity and PSD-95 phosphorylation. Here, we show that the IgG-like extracellular domains of IL1RAPL1 induce excitatory pre-synapse formation by interacting with protein tyrosine phosphatase delta (PTPδ). We also found that IL1RAPL1 TIR domains interact with RhoGAP2, which is localized at the excitatory post-synaptic density. More interestingly, the IL1RAPL1/PTPδ complex recruits RhoGAP2 at excitatory synapses to induce dendritic spine formation. We also found that the IL1RAPL1 paralog, IL1RAPL2, interacts with PTPδ and induces excitatory synapse and dendritic spine formation. The interaction of the IL1RAPL1 family of proteins with PTPδ and RhoGAP2 reveals a pathophysiological mechanism of cognitive impairment associated with a novel type of trans-synaptic signaling that regulates excitatory synapse and dendritic spine formation.
INTRODUCTION
IL1RAPL1 belongs to the IL1/Toll receptor family and shares 52% homology with the IL-1 receptor accessory protein (IL-1RacP). Similar to other members of the IL-1 receptor family, it is characterized by three extracellular Ig-like domains, a transmembrane domain and an intracellular TIR domain. However, unlike the family members, 150 additional amino acids are located at the C-terminus. The homology with IL-RacP is evenly distributed throughout the protein with the exception of the last 150 amino acids, which are present only in IL1RAPL1 and its paralog, IL1RAPL2. The first identified mutation in the IL1RAPL1 gene, which was described by Carrie et al. (1), is associated with a non-syndromic form of mental retardation (MR). Similar to some other genes involved in cognitive impairment (2–4), IL1RAPL1 mutations are associated with a spectrum of cognitive impairments ranging from MR to autism (5–11).
It has been previously demonstrated that the intracellular C-terminal domain of IL1RAPL1 interacts with NCS-1 (12) and that this interaction mediates the regulatory effect of IL1RAPL1 over-expression on N-type voltage-gated calcium channel activity in PC12 cells (13). More recently, we found that the C-terminal tail of IL1RAPL1 interacts with PSD-95 and regulates PSD-95 localization to synapses by stimulating c-Jun N-terminal kinase (JNK) phosphorylation at Ser-295 (14).
In this study, we found that both IL1RAPL1 and IL1RAPL2 can induce excitatory pre-synapse differentiation and dendritic spine formation. Interestingly, although the extracellular domain is sufficient for inducing pre-synaptic differentiation, both extracellular and intracellular TIR domains are required for dendritic spine formation. Using affinity chromatography, we identified protein tyrosine phosphatase delta (PTPδ) as a binding partner of IL1RAPL1 through its extracellular domain. This interaction was confirmed biochemically and with an HEK cell trans-clustering assay using both IL1RAPL1 and IL1RAPL2. Using yeast two-hybrid screening, we found that the IL1RAPL1 intracellular TIR domain interacts with RhoGAP2, which is localized at the excitatory post-synaptic density. The interaction of IL1RAPL1 with RhoGAP2 is required to induce dendritic spine formation. Interestingly, we found that blocking the IL1RAPL1/PTPδ interaction abolished RhoGAP2 recruitment at excitatory synapses, suggesting that IL1RAPL1 is involved in a novel trans-synaptic signaling pathway that regulates excitatory synapse and dendritic spine formation.
RESULTS
IL1RAPL1 induces the formation of functional excitatory synapses
We have previously demonstrated that IL1RAPL1 over-expression in neurons increases the excitatory synapse number (measured as an increase in pre-synaptic contacts and dendritic spine number) and that this ability does not depend on the interaction between IL1RAPL1 and PSD-95 (14).
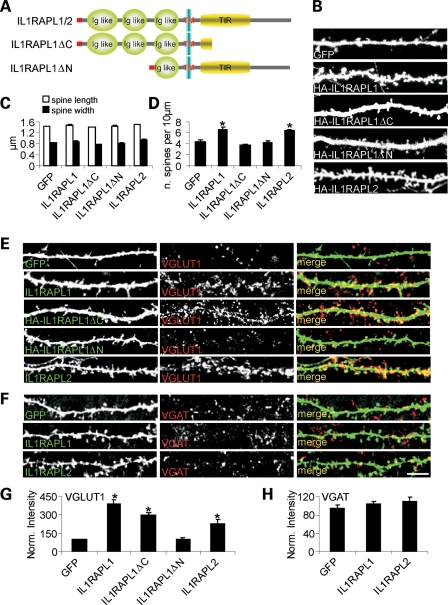
To better understand which domains of IL1RAPL1 are involved in this function, we over-expressed full-length IL1RAPL1 or two different mutants in neurons at days in vitro 9 (DIV 9). The two mutants investigated were HA-IL1RAPL1▵C, which mimics the Y459X mutation that was described in a boy affected by MR (1) and lacks part of the TIR domain and the entire C-terminal tail (Fig. 1A), and HA-IL1RAPL1▵N, which has a deletion of the two external N-terminal Ig-like domains on the extracellular surface (Fig. 1A). Seven days after transfection, neurons were analyzed for the dendritic spine number and shape, and the pre-synaptic excitatory and inhibitory contacts were stained using VGLUT1 and VGAT antibodies, respectively.
Figure 1.
IL1RAPL1 and IL1RAPL2 promote dendritic spine and excitatory synapse formation. (A) A schematic representation of IL1RAPL1/2, IL1RAPL1▵C and IL1RAPL1▵N. (B) Hippocampal neurons were transfected at DIV 9 with GFP alone or with GFP and HA-IL1RAPL1, HA-IL1RAPL1▵C, HA-IL1RAPL1▵N or HA-IL1RAPL2 constructs as indicated and stained at DIV 16 for GFP and HA (only the GFP channel is shown) (scale bars = 10 μm). (C and D) Quantification of spine density (number of spines per 10 μm of dendrite length), head width (μm) and length (μm) in neurons transfected as described in (A) (16 neurons examined for each construct). We then calculated mean and SEM between the values we obtained for the each neurons transfected with the same cDNA. Histograms represent the mean ± SEM (*P < 0.01). (E and F) Hippocampal neurons were transfected at DIV 9 with GFP, HA-IL1RAPL1, HA-IL1RAPL1▵C, HA-IL1RAPL1▵N or HA-IL1RAPL2 and stained at DIV 16 with GFP or HA and VGLUT1 (E) or VGAT (F) antibodies. Each row of images shows double-labeling for GFP or HA (green, left panel) and VGLUT1 or VGAT (red, middle panel); the merged images are shown in color in right panel (scale bar = 10 μm). (G and H) Quantification of VGLUT1 (G) and VGAT (H) cluster intensity in neurons over-expressing the IL1RAPL1 and two constructs (at least nine neurons were analyzed for each construct). Bar graphs show the mean ± SEM of the dendritic VGLUT1 and VGAT intensity normalized to the GFP-transfected neurons (*P < 0.01).
As shown in Figure 1B and quantified in the graph in Figure 1C and D, both the extracellular and the intracellular domains of IL1RAPL1 were necessary to increase the dendritic spine number (Fig. 1D, mean ± SEM dendritic spine number per 10 μm in neurons expressing GFP: 4.4 ± 0.2; HA-IL1RAPL1: 6.6 ± 0.3; HA-IL1RAPL1▵C: 3.6 ± 0.3; HA-IL1RAPL1▵N: 4.1 ± 0.4; *P < 0.05). However, the full-length and mutant IL1RAPL1 constructs did not modify the dendritic spine shape (Fig. 1C). Only the extracellular domain of IL1RAPL1 was required to increase the excitatory (VGLUT1-positive, Fig. 1E and G), but not the inhibitory (VGAT-positive, Fig. 1F and H), pre-synaptic staining (Fig. 1G, mean ± SEM normalized fluorescent intensity of VGLUT1 staining in neurons expressing GFP: 92.9 ± 6.3; HA-IL1RAPL1: 388.1 ± 33.9; HA-IL1RAPL1▵C: 298.2 ± 21.9; HA-IL1RAPL1▵N: 102.1 ± 10.5; *P < 0.01).
We also studied the role of the IL1RAPL1 gene paralog, IL1RAPL2, in neurons to understand whether this protein shares properties with IL1RAPL1. Similar to IL1RAPL1, IL1RAPL2 over-expression increased the dendritic spine number without affecting morphology (Fig. 1B–D, number of spines per 10 μm in neurons expressing IL1RAPL2 compared with GFP: 6.3 ± 0.3 versus 4.4 ± 0.2, *P < 0.05) and promoted excitatory (Fig. 1E and G, VGLUT1 staining in neurons expressing HA-IL1RAPL2 compared with neurons expressing GFP: 227.2 ± 31.2 versus 92.9 ± 6.3; *P < 0.01), but not inhibitory, pre-synaptic contact formation in connecting neurons (Fig. 1F and H).
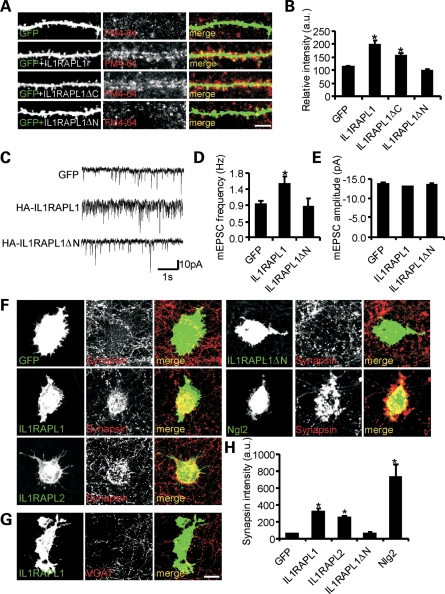
To visualize whether these extra synapses actively release neurotransmitters, we performed an FM4–64 labeling experiment. Neurons were transfected at DIV 9 with full-length IL1RAPL1 or mutants (IL1RAPL1▵C and IL1RAPL1▵N) and GFP to visualize transfected neurons, using immunofluorescence. Seven days after transfection, pre-synaptic vesicle recycling was stimulated with elevated K+ (90 mm KCl for 1 min), and the cells were loaded with FM4–64, washed in low-KCl buffer containing tetrodotoxin (TTX) to remove excess FM4–64 and then fixed. Over-expression of IL1RAPL1 and IL1RAPL1▵C increased the number of active pre-synaptic compartments in contacting neurons (Fig. 2A and B, mean ± SEM FM4–64 relative intensity in neurons expressing GFP: 113.0 ± 3.9; HA-IL1RAPL1: 198.2.1 ± 17.4; HA-IL1RAPL1▵C: 156.3 ± 11.0; HA-IL1RAPL1▵N: 98.3.1 ± 4.2; *P < 0.01).
Figure 2.
IL1RAPL1 and IL1RAPL2 promote functional excitatory synapse formation. (A) FM1–64 staining (at DIV 16) of functional pre-synaptic terminals on hippocampal neurons transfected at DIV 9 with GFP alone or with GFP and HA-IL1RAPL1, HA-IL1RAPL1▵C or HA-IL1RAPL1▵N (scale bar = 10 μm). (B) Quantification of puncta density of FM1–64 staining of transfected cells (at least nine neurons analyzed for each construct). Histograms represent the mean ± SEM FM1–64 staining intensity normalized to GFP-transfected neurons (*P < 0.01). (C) Typical recording of mEPSCs of neurons transfected with GFP alone or GFP plus HA-IL1RAPL1 or HA-IL1RAPL1▵N. (D and E) Bar graphs representing the mean mEPSC frequency ± SEM (D) and mean mEPSC amplitude ± SEM (E) (at least 16 neurons were recorded for each construct; *P < 0.01). (F and G) IL1RAPL1 and IL1RAPL2 are able to induce synapsin I clustering in contacting axons of co-cultured neurons. HEK293T cells expressing GFP alone or GFP plus IL1RAPL1, IL1RAPL2, IL1RAPL1▵N or Ngl2 were co-cultured with hippocampal neurons and stained for synapsin I (F), or VGAT (G) (scale bar = 20 μm). (H) Quantification of the mean ± SEM intensity of synapsin I clusters induced by IL1RAPL1, IL1RAPL2, IL1RAPL1▵N or Ngl2. Integrated fluorescence intensity of synapsin I was normalized to the cell area (at least 20 cells were analyzed; *P < 0.01).
Finally, in IL1RAPL1 over-expressing neurons, but not in neurons over-expressing IL1RAPL1▵N, an increase in the number of pre-synaptic contacts was associated with an increase of the miniature excitatory post-synaptic current (mEPSC) frequency (Fig. 2C and D, mean ± SEM mEPSC frequency of neurons expressing GFP: 0.9 ± 0.1 Hz; HA-IL1RAPL1: 1.5 ± 0.2 Hz, P < 0.5; HA-IL1RAPL1▵N: 0.85 ± 0.25 Hz; *P < 0.05), whereas no modification of the mEPSC amplitude was observed (Fig. 2C and E; mEPSC amplitude of neurons expressing GFP: 13.6 ± 0.3 pA; HA-IL1RAPL1: 12.8 ± 0.3 pA; P = 0.07; HA-IL1RAPL1▵N: 13.3 ± 0.5 Hz). These data suggest that IL1RAPL1 may act as a synaptogenic protein and that the extracellular domain is required to induce pre-synaptic formation, although both the extracellular and intracellular domains are required for inducing dendritic spine formation.
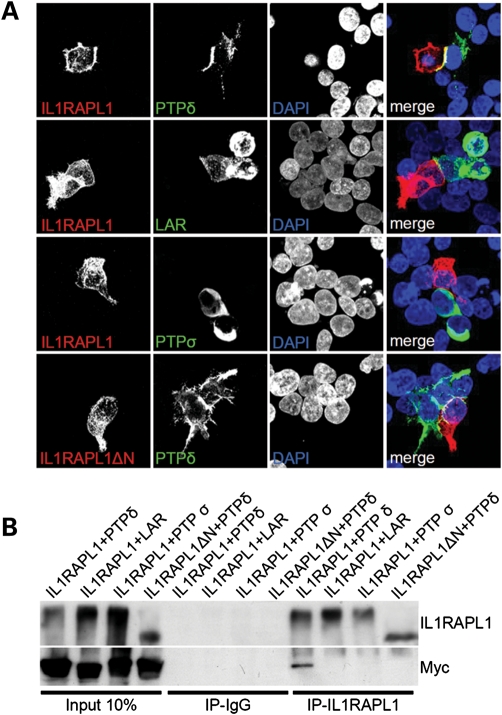
Figure 3.
The extracellular domain of IL1RAPL1 interacts with that of PTPδ. (A) Two sets of HEK293FT cells were transfected with IL1RAPL1 or IL1RAPL1▵N and with the Myc-tagged constructs of LAR-ecto-pDis (LAR), PTPδ-ecto-pDis (PTPδ) and PTPσ-ecto-pDis (PTPσ) for 16 h using the calcium phosphate precipitation method. Cells expressing the proper construct were plated together on a 16 mm cover slip and grown for 24 h before fixation and staining with IL1RAPL1 or Myc (35). (B) Cells were transfected as described in (A), and the lysate was mixed 1:1 and immunoprecipitated with antibodies against IL1RAPLA1. The eluent was analyzed by western blot using IL1RAPL1 and Myc antibodies.
The extracellular domain of IL1RAPL1 is necessary to induce pre-synaptic clustering
Our data suggest that both IL1RAPL1 and IL1RAPL2 may function as synaptic cell adhesion proteins. To corroborate this hypothesis, we performed a neuron-fibroblast co-culture assay (15).
COS-7 cells transfected with GFP (negative control), neuroligin-2 (positive control), HA-IL1RAPL1 (either full-length or lacking the extracellular domain, IL1RAPL1▵N) or HA-IL1RAPL2 were co-cultured with hippocampal neurons at DIV 8. After 3 days of co-culture, we fixed the cells, and the pre-synapses were visualized using an antibody against synapsin I.
COS-7 cells expressing wild-type (wt) IL1RAPL1, IL1RAPL2 and neuroligin-2, but not IL1RAPL1▵N, induced pre-synaptic differentiation in contacting axons, showing significantly higher levels of synapsin I intensity compared with the COS-7 cells expressing GFP (Fig. 2F and H, mean ± SEM synapsin intensity staining measured over COS-7 cells transfected with GFP: 55.9 ± 4.0; IL1RAPL1: 321.2 ± 32.3; IL1RAPL2: 253.9 ± 19.6; IL1RAPL1▵N: 62.5 ± 10.0; neuroligin-2: 731.5 ± 154.3; *P < 0.05). However, IL1RAPL1-expressing COS-7 cells were not able to recruit VGAT-positive pre-synaptic terminals (Fig. 2G).
These data demonstrate that IL1RAPL1 and IL1RAPL2 may act as synaptogenic molecules for excitatory synapses that interact with an unknown pre-synaptic partner.
IL1RAPL family proteins specifically interact with PTPδ
To identify the receptor for IL1RAPL1, we used the IL1RAPL1 extracellular domain coupled to Fc (Fc-IL1RAPL1-N) or control Fc proteins and protein A beads for column purification of synaptosomal membrane proteins from P18 rat brains that bound Fc-IL1RAPL1 (Supplementary Material, Fig. S1). IL1RAPL1-associated proteins were then analyzed by MudPit tandem mass spectrometry (16). We found less than 200 proteins that co-precipitated with Fc-IL1RAPL1-N (Supplementary Material, Fig. S1). Among these, some proteins were common with Fc alone, and other proteins were cytosolic molecules or enzymes. Among the few transmembrane pre-synaptic proteins, PTPδ was precipitated. PTPδ, together with LAR and PTPσ, is a member of the LAR family of proteins. These are synaptic transmembrane proteins that bind to netrin-G ligand-3 (NGL-3) and induce pre- and post-synaptic differentiation in neurons.
To confirm the interaction, we initially examined whether membrane-associated PTPδ could regulate the accumulation of IL1RAPL1 at contact sites. HEK293FT cells were transfected with PTPδ-ecto-pDisplay or HA-IL1RAPL1. One day after transfection, the cells were co-cultured, and the subcellular distribution of the two proteins was observed. Interestingly, HA-IL1RAPL1, but not HA-IL1RAPL1▵N, showed enhanced accumulation at sites of contact with PTPδ-transfected cells (Fig. 3A).
To determine whether IL1RAPL1 specifically bound PTPδ and not other PTP family members, we performed a similar set of experiments with LAR and PTPσ; neither protein showed enhanced accumulation at sites of contact with IL1RAPL1-transfected cells (Fig. 3A). Similarly, IL1RAPL2 specifically interacted with PTPδ but not with the other members of the PTP family (Supplementary Material, Fig. S2).
We also tested the interaction between IL1RAPL1 and other transmembrane synaptic partners, including neurexins. Again, we did not find other interactors among the analyzed proteins (Supplementary Material, Fig. S3).
The specific interaction between IL1RAPL1 and PTPδ was also confirmed by immunoprecipitation. For this experiment, HEK293FT cells were transfected with full-length IL1RAPL1 or IL1RAPL1▵N. Another set of cells was transfected with PTPδ, LAR or PTPσ. The lysates obtained from the transfected cells were mixed 1:1 and immunoprecipitated using antibodies specific for the intracellular domain of IL1RAPL1 (14). As shown in Figure 3B, IL1RAPL1 but not IL1RAPL1▵N immunoprecipitated only PTPδ.
These results suggest that post-synaptic IL1RAPL1 enhances the maturation of pre-synaptic terminals by its trans-synaptic interaction with membrane-bound PTPδ.
The intracellular domain of IL1RAPL1 interacts with RhoGAP2
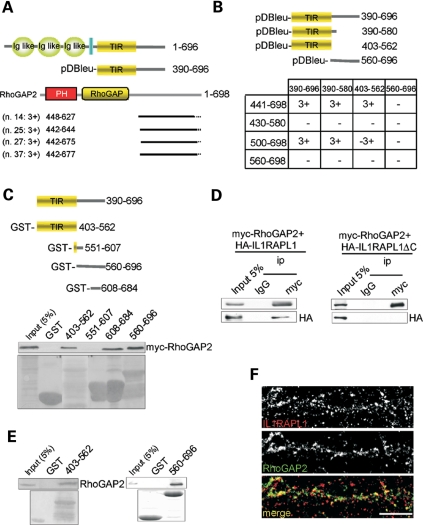
As shown in Figure 1, the IL1RAPL1 intracellular domain is required to induce dendritic spine formation, and this activity is not dependent on the interaction with PSD-95 (14). Therefore, we sought to identify proteins that bind to IL1RAPL1 in the intracellular domain that might be required for dendritic spine formation. We used the yeast two-hybrid system with the intracellular C-terminal tail of IL1RAPL1 (amino acids 390–696; Fig. 4A) as bait to screen a human fetal brain cDNA library. Four prey cDNA clones were isolated (clones 14, 25, 27 and 37; Fig. 4A), all of which encoded RhoGAP2, a novel RhoGTPase-activating protein II.
Figure 4.
The intracellular domain of IL1RAPL1 interacts with RhoGAP2. (A) Schematic representation of IL1RAPL1 and the C-terminal domain of IL1RAPL1, which was used as bait for the yeast two-hybrid screening. Also shown are the results of the yeast two-hybrid screening with a schematic representation of the four positive clones for RhoGAP2. The two-hybrid interaction was quantified based on the activation of the three reporter genes (HIS3, LacZ and URA 3) (3+: activation of all three reporter genes; 2+: activation of HIS3 and LacZ reporters; 1+: activation of HIS3). The small numbers refer to amino acid residues. (B) Mapping of the IL1RAPL1/RhoGAP2 interaction using the two-hybrid assay. We used the full-length C-terminus of IL1RAPL1 (amino acids 390–696), the TIR domain (amino acids 403–562) and two other fragments from the C-terminus of IL1RAPL1 (amino acids 390–580 and 560–696) as bait and four fragments of the C-terminus of RhoGAP2 (amino acids 441–698, 430–580, 500–698, 560–698) as prey. The full-length IL1RAPL1 C-terminus and the TIR domain constructs interacted with the RhoGAP2 C-terminal tail (3+). (C–E) GST pull-down experiments using IL1RAPL1 and RhoGAP2. Diagram of the four fragments of the C-terminal tail of IL1RAPL1 used in the GST pull-down assay (top). In COS7 cells transfected with mycRhoGAP2, the TIR domain of IL1RAPL1 (amino acids 403–562), as well as the amino acids 608–684 and amino acids 560–696 fragments, pulled down full-length RhoGAP2 (mycRhoGAP2). The purified GST-amino acids 403–562 and GST-amino acids 560–696 fragments bound to RhoGAP2 in lysates from the rat brain. (D). Interaction between IL1RAPL1 and RhoGAP2 was confirmed by co-immunoprecipitation experiments using COS7 cells. Myc-tagged wt RhoGAP2 co-immunoprecipitated with HA-IL1RAPL1, whereas the mutated form of IL1RAPL1 (HA-IL1RAPL1 ΔC) did not. (F) Representative images of the staining and co-localization of endogenous IL1RAPL1 and RhoGAP2 in hippocampal neurons (scale bar = 10 μm).
The sites of interaction between IL1RAPL1 and RhoGAP2 were further studied using a yeast two-hybrid system using the full-length IL1RAPL1 C-terminus containing the TIR domain (amino acids 390–696), the TIR domain plus 18 amino acids of the distal region of the C-terminus (amino acids 390–580), the TIR domain alone (amino acids 403–562) or the IL1RAPL1 C-terminus lacking the TIR domain (amino acids 560–696) as bait.
For prey, the following constructs were tested (Fig. 4B): the RhoGAP full-length C-terminus (amino acids 441–698) and three C-terminal fragments (amino acids 430–580, 500–698 and 560–698) (Fig. 4B).
Only two RhoGAP2 constructs [full-length RhoGAP2 C-terminus and a C-terminal fragment (amino acids 500–698)] interacted with wt IL1RAPL1 (with an intact C-terminal tail) and with the TIR domain (amino acids 390–580, 403–562). None of the RhoGAP2 constructs interacted with the IL1RAPL1 construct missing the TIR domain (amino acids 560–696) (Fig. 4B). These data show that IL1RAPL1 and RhoGAP2 interact and that the TIR domain of IL1RAPL1 is necessary for interaction in the two-hybrid system.
We further investigated the interaction between IL1RAPL1 and RhoGAP2 in transfected COS-7 cells and neuronal protein extracts using pull-down and co-immunoprecipitation experiments. First, we performed a GST pull-down assay using COS7 cells. GST was fused to the IL1RAPL1 TIR domain (amino acids 403–562) or the IL1RAPL1 C-terminal fragments (amino acids 551–607, 608–684 or 560–696) and tested for binding to full-length myc-RhoGAP2. As expected, the TIR domain of IL1RAPL1 precipitated full-length myc-RhoGAP2. In addition, the 560–696 and 608–684 fragments precipitated myc-RhoGAP2. We then generated an antibody that specifically recognizes RhoGAPII (see Experimental Procedures, Supplementary Material, Fig. S4A for details) and showed that the TIR domain and the last 132 amino acids of the IL1RAPL1 C-terminus interact with RhoGAP2 present in rat brain lysates (Fig. 4E).
These data indicate that the IL1RAPL1–RhoGAP2 interaction is complex and involves different regions of the C-terminal end of IL1RAPL1 (Fig. 4C and E).
Next, we found that HA-IL1RAPL1 was specifically co-immunoprecipitated with myc-RhoGAP2 in transfected COS-7 cells, but HA-IL1RAPL1 with the C-terminus deleted (HA-IL1RAPL1▵C) was not (Fig. 4D). Finally, immunofluorescence labeling of hippocampal neurons showed that endogenous IL1RAPL1 and RhoGAP2 co-localized to dendritic spines (Fig. 4F). These findings demonstrate the association of IL1RAPL1 with RhoGAP2.
RhoGAP2 is localized to excitatory synapses
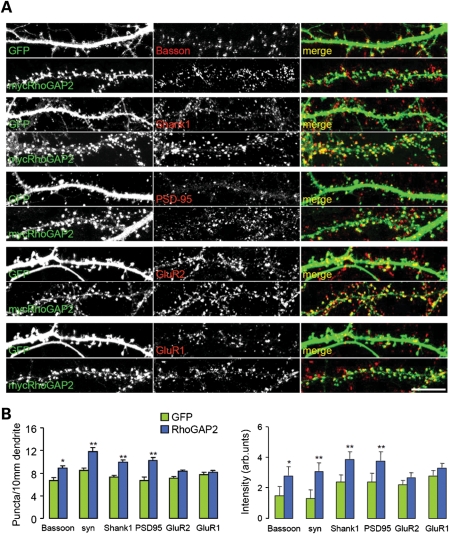
We then asked whether RhoGAP2 is localized to the post-synaptic site of the excitatory synapses, as suggested by its interaction with IL1RAPL1. We found that RhoGAP2 is expressed in the cortex, the cerebellum and the hippocampus (Supplementary Material, Fig. S4A) and in the cortex and hippocampus in mice during development (Supplementary Material, Fig. S4B). Using subcellular fractionation, we also showed that RhoGAP2 is present in the synaptosomal fraction (Supplementary Material, Fig. S2B). The synaptic subcellular localization of RhoGAP2 was further studied by multiple-immunolabeling experiments in cultured hippocampal neurons, using anti-GluR1, anti-GluR2, anti-PSD-95, anti-Shank, anti-Bassoon, anti-synaptophysin and anti-V-GAT antibodies. Endogenous RhoGAP2 was mostly co-localized with PSD-95 and Shank1 proteins (percent of IL1RAPL1 co-localized with GluR2/3: 77.3% ± 2.3; with GluR1: 74.4% ± 3.6; with PSD-95: 71.2% ± 1.9; with Shank1: 66.8% ± 4.2; with Bassoon: 52.3% ± 2.8; and with synaptophysin 56: 3% ± 3.9) (Supplementary Material, Fig. S2C and D), which are markers of excitatory post-synapses. Moreover, only 5% ± 1.2 of the endogenous RhoGAP2 co-localized with V-GAT (Supplementary Material, Fig. S4C and D). These results suggest that RhoGAP2 is enriched in the post-synaptic compartment.
We also examined the effect of over-expressing RhoGAP2 in mature neurons on the staining of endogenous pre- and post-synaptic markers. In neurons over-expressing mycRhoGAP2, there was a significant increase in the number of puncta containing synaptophysin (relative to GFP-transfected neurons, 11.86 ± 0.64 versus 8.5 ± 0.44; P< 0.01), Bassoon (relative to control, 8.9 ± 0.39 versus 6.42 ± 0.51; P < 0.05), Shank1 (relative to GFP transfected neurons, 9.9 ± 0.45 versus 7.3 ± 0.34; P < 0.01) and PSD-95 (relative to control; 10.3 ± 0.63 versus 6.7 ± 0.45; P < 0.01) but not GluR1 (relative to GFP transfected neurons, 8.16 ± 0.33 versus 7.7 ± 0.41) or GluR2 (relative to GFP-transfected neurons, 8.4 ± 0.21 versus 7.1 ± 0.33) (Fig. 5A and B).
Figure 5.
RhoGAP2 increases the number of synapses and the accumulation of endogenous synaptic proteins. (A) Hippocampal neurons at DIV 14 were transfected with mycRhoGAP2 or with the GFP control and were stained at DIV 22. Each row of images shows a double-labeling of mycRhoGAP2 or GFP (green, left panel) and Bassoon, synaptophysin, PSD-95, Shank1, GluR1 or GluR2 (red, middle panel); the merged images are shown in color in the right panel. Individual channels are shown in gray scale (scale bars = 10 μm). (B) Quantification of puncta density and synaptic staining intensity of Bassoon, synaptophysin, PSD-95, Shank1, GluR1 and GluR2 after over-expression of mycRhoGAP2 (>7 neurons analyzed for each protein; 40–70 synapses scored per neuron). Quantification of the mean percent of co-localization (6 SEM) of endogenous RhoGAP2 clusters with endogenous GluR2/3, GluR1, PSD-95, Shank1, Bassoon, synaptophysin and V-GAT.
We also found that spines of mycRhoGAP2-over-expressing cells showed increased staining intensity for synaptophysin (relative to control, 3.1 ± 0.55 versus 1.3 ± 0.21; P < 0.01), Bassoon (relative to control, 2.8 ± 0.58 versus 1.5 ± 0.6; P < 0.05), Shank1 (relative to control, 3.9 ± 0.45 versus 2.4 ± 0.7; P < 0.01) and PSD-95 (relative to control, 3.8 ± 0.55 versus 2.4 ± 0.63; P < 0.01), but not GluR1 (relative to control, 3.3 ± 0.27 versus 2.8 ± 0.31) or GluR2 (relative to control, 2.7 ± 0.27 versus 2.2 ± 0.45) (Fig. 5A and B).
These data demonstrate that RhoGAP2 is a novel synaptic RhoGAP that interacts with IL1RAPL1 and might contribute to excitatory synapse formation.
RhoGAP2 is recruited to synapses by the IL1RAPL1/PTPδ interaction
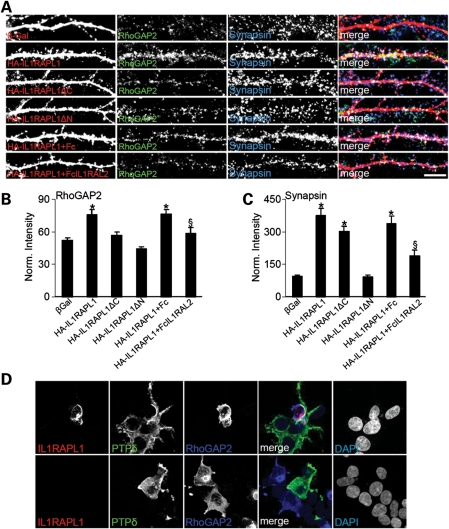
We then determined whether the interaction between IL1RAPL1 and PTPδ is required for RhoGAP2 recruitment and function in excitatory synapses. We over-expressed wt IL1RAPL1 and mutants and stained for endogenous RhoGAP2 expression. We found that the over-expression of IL1RAPL1 significantly increased both endogenous RhoGAP2 and synapsin staining compared with β-Gal-over-expressing neurons (Fig. 6A–C). The over-expression of both IL1RAPL1▵C (unable to bind to RhoGAP2) and IL1RAPL1▵N (unable to bind to PTPδ) did not increase the staining of endogenous RhoGAP2 (Fig. 6A and B), whereas only IL1RAPL1▵C increased the synapsin staining (Fig. 6A and C) (Fig. 6B, mean ± SEM normalized intensity staining of endogenous RhoGAP2 in neurons over-expressing β-Gal: 51.9 ± 2.7; IL1RAPL1: 76.0 ± 4.3; IL1RAPL1▵C: 57.0 ± 2.9; IL1RAPL1▵N: 44.6 ± 1.9; *P< 0.05. Fig. 6C, mean ± SEM normalized staining intensity of endogenous synapsin in neurons over-expressing β-Gal: 93.6 ± 6.3; IL1RAPL1: 376.5 ± 31.7; IL1RAPL1▵C: 301.5.0 ± 25.0; IL1RAPL1▵N: 91.5 ± 10.5; *P < 0.05). These data suggest that IL1RAPL1 binds and recruits RhoGAP2 to synapses via its intracellular domain, and binding of PTPδ at the extracellular domain is also required. To confirm these findings, neurons over-expressing IL1RAPL1 were incubated with purified Fc or Fc-IL1RAPL2 for 6 days, which should compete with the binding between IL1RAPL1 and PTPδ. We found that Fc-IL1RAPL2 was able to significantly reduce both RhoGAP2- and synapsin-induced recruitment by IL1RAPL1 (Fig. 6A–C) (Fig. 6B, mean ± SEM normalized intensity staining of endogenous RhoGAP2 in neurons over-expressing IL1RAPL1 and incubated with Fc: 76.6.9 ± 3.9; incubated with Fc-IL1RAPL2: 58.5.0 ± 5.5; §P < 0.05. Fig. 6C, mean ± SEM normalized intensity staining of endogenous synapsin in neurons over-expressing IL1RAPL1 and incubated with Fc: 337.2 ± 6.3; incubated with Fc-IL1RAPL2: 189.2 ± 25.5; §P < 0.05). We also measured the level of co-localization of endogenous RhoGAP2 with synapsin and found that the over-expression of IL1RAPL1▵C does not reduce significantly the level of RhoGAP2 that co-localizes with synapsin (data not shown), suggesting that IL1RAPL1 might not be necessary for RhoGAP2 localization at synapses.
Figure 6.
The IL1RAPL1/PTPδ complex recruits RhoGAP2 to synapses. (A) Hippocampal neurons were transfected with β-Gal, HA-IL1RAPL1, HA-IL1RAPL1▵C or HA-IL1RAPL1▵N at DIV 9, fixed at DIV 16 and triple-stained for β-Gal, IL1RAPL1, RhoGAP2 and synapsin. Two sets of neurons transfected with IL1RAPL1 were treated with Fc or FcIL1RAPL2, as indicated in the panels. Each row of images shows triple-labeling for HA (red, left panel), RhoGAP2 (green, middle left panel) and synapsin (blue, middle right panel); merged images are shown in color in the right panel (scale bar = 10 μm). (B and C) Quantification of RhoGAP2 (B) and synapsin (D) cluster intensity in neurons over-expressing IL1RAPL1 constructs (at least nine neurons were analyzed for each construct). Bar graphs show the mean ± SEM of dendritic RhoGAP2 and synapsin intensity normalized to GFP-transfected neurons (*P < 0.01). Two sets of HEK293FT cells were transfected with IL1RAPL1 with or without RhoGAP2 and with the Myc-tagged construct PTPδ-ecto-pDis (PTPδ) for 16 h using the calcium phosphate precipitation method. Cells expressing the proper constructs were plated together on a 16 mm cover slip and grown for 24 h before fixation and staining for IL1RAPL1, Myc and RhoGAP2.
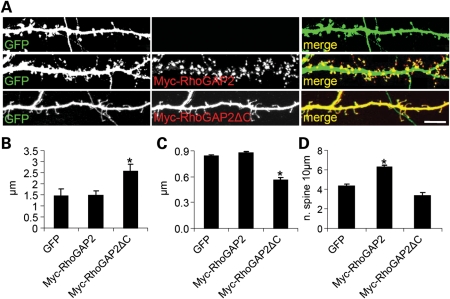
Thus, our data suggest that the interaction between IL1RAPL1 and PTPδ induces pre-synaptic maturation, and the interaction with RhoGAP2 increases post-synaptic cluster and dendritic spine formation. To further support this finding, we over-expressed wt RhoGAP2 or RhoGAP2▵C (unable to bind IL1RAPL1; we inserted a stop codon at amino acid 500) and GFP in hippocampal neurons and determined the dendritic spine number and morphology. We found that the over-expression of RhoGAP2 increased the dendritic spine number without changing the shape, whereas the over-expression of RhoGAP2▵C significantly altered the shape of dendritic spines, which became longer and thinner compared with neurons over-expressing GFP (Fig. 7A) (Fig. 7B, mean ± SEM dendritic width, µm, in neurons expressing GFP: 0.8 ± 0.01; RhoGAP2: 0.9 ± 0.02; RhoGAP2▵C: 0.5 ± 0.03; *P < 0.05. Fig. 7B, mean ± SEM dendritic length, µm, in neurons expressing GFP: 1.4 ± 0.3; RhoGAP2: 1.5 ± 0.2; RhoGAP2▵C: 2.5 ± 0.3; *P < 0.05. Fig. 7C, mean ± SEM dendritic spines number in neurons expressing GFP: 4.3 ± 0.2; RhoGAP2: 6.2 ± 0.3; RhoGAP2▵C: 3.4 ± 0.3; *P < 0.05).
Figure 7.
Dendritic spine number and morphology of hippocampal neurons are regulated by RhoGAP2. (A) Hippocampal neurons at DIV 9 were transfected with GFP alone or with GFP and Myc-RhoGAP2 or GFP and Myc-RhoGAP2▵C, as indicated on the panels. After 1 week, neurons were fixed and stained for GFP and Myc. (B–D) Quantification of the mean (±SEM) length (B), width (C) and number (per 10 μm) of dendritic spines. Over 14 transfected neurons from four independent experiments were measured for each transfection (*P < 0.01, Student's t-test) (scale bar = 10 µm).
DISCUSSION
In this study, we showed that the MR-related protein IL1RAPL1 is a synaptic adhesion protein that binds trans-synaptically to PTPδ through its extracellular domain and to RhoGAP2 through its intracellular domain.
Cell adhesion proteins are thought to be key regulators of synaptogenesis. In particular, these proteins are involved in both the initial phase of synaptogenesis, during cell-type-specific target recognition, and the synapse maturation phase, during which the protein components of the pre-synaptic release machinery and the post-synaptic signaling apparatus are recruited to the nascent synaptic contact (17). These proteins are known to induce synaptic specialization in neurons when over-expressed in fibroblasts during co-culture experiments.
We previously showed that IL1RAPL1 is a transmembrane protein located at post-synaptic densities of excitatory synapses (14) that can induce pre-synaptic differentiation when over-expressed in neuronal cultures. Here, we found that the IL1RAPL1 extracellular domain interacts with PTPδ, a pre-synaptic protein involved in synapse formation.
This interaction was found using non-biased Fc-IL1RAPL1 affinity chromatography and mass-spectrometry analysis using brain extract and then confirmed by HEK cell trans-clustering assays and immunoprecipitation by trans-interaction.
PTPδ is a member of the LAR phosphor-tyrosine phosphatase family, which also includes LAR and PTPσ (18). All three members of the family are specifically involved in synapse formation by binding to NGL-3 through the FNIII domain (15,19). It has also been found that the Ig domain of PTPσ binds to the neurotrophin receptor TrkC, suggesting that multiple proteins can contribute to the pre-synaptic differentiation of the LAR-PTP families (20). In this study, we found that PTPδ can specifically trans-synaptically bind to a second and a third partner, IL1RAPL1 and its paralog IL1RAPL2.
It was previously demonstrated that PTPδ is the only LAR family member that is not able to induce post-synaptic differentiation, such as PSD-95 accumulation, when added to neuronal cultures (19). However, our data showed that the ability of IL1RAPL1 to induce dendritic spine formation requires both the extracellular and the intracellular domains.
Using yeast two-hybrid screening, we identified RhoGAP2 as a new intracellular partner of IL1RAPL1, and our data suggest that the IL1RAPL1 and RhoGAP2 interaction is required for the ability of IL1RAPL1 to induce dendritic spine formation.
Indeed, we previously showed that IL1RAPL1 binds to PSD-95; however, this interaction is not required for the IL1RAPL1-mediated increase in the number of dendritic spines (14). In this study, we showed that the intracellular domain also binds to RhoGAP2, a GTPase-activating protein previously shown to inhibit Rac1 activity (21). However, because the deletion of most of the N- or C-terminal region of IL1RAPL1 used in our experiments not only disrupts binding of IL1RAPL1 to PTPδ and RhoGAP2, but also binding to any other possible known and unknown interacting proteins, we cannot totally exclude that other IL1RAPL1 partners are important for its activity on excitatory synapse formation.
Our biochemical and morphological data suggest that RhoGAP2 is localized to excitatory synapses, and its C-terminal tail interacts with the TIR domain of IL1RAPL1, although pull-down experiments suggest a possible additional interaction at the C-terminal tail.
RhoGAP proteins increase the intrinsic GTPase activity to inactivate the RhoGTPase switch and guanine-nucleotide dissociation inhibitors (22). Rho proteins have been implicated in different aspects of neuronal morphogenesis, including dendritic arbor development and spine morphogenesis (23). In particular, Rac1 positively regulates and RhoA negatively regulates dendritic spine morphogenesis (24).
Surprisingly, we found that the over-expression of RhoGAP2 induces both excitatory synapse and dendritic spine formation. However, the over-expression of a RhoGAP2 mutant that does not bind to IL1RAPL1 causes a drastic change in spine shapes which are converted to filopodia-like shape. Thus, our data suggest that the RhoGAP2 activity on dendritic spines is regulated by the interaction with IL1RAPL1/PTPδ complex.
Although the function of the IL1RAPL1–RhoGAP2 interaction remains to be defined, we found that RhoGAP2 is recruited to synapses by the PTPδ–IL1RAPL1 interaction, as suggested by the finding that endogenous RhoGAP2 staining is increased at synapses when IL1RAPL1 is over-expressed or when the interaction between IL1RAPL1 and PTPδ is inhibited by Fc-IL1RAPL2.
In conclusion, we show for the first time that IL1RAPL1 and its paralog ILRAPL2 are synaptic adhesion molecules that contribute to synapse formation, and alteration of this pathway might contribute to the development of ID in patients with IL1RAPL1 mutations.
MATERIALS AND METHODS
Antibodies
The following antibodies were used: rabbit anti-IL1RAPL1 (K10) (14), rabbit anti-RhoGAP2 (QQ15) raised against the peptide CysGHRRASSGDRLKDTGSVQRLSTYD (amino acids 659–677), rabbit anti-VGlut1 (Synaptic System), mouse anti-VGAT (NeuroMab), mouse anti-synapsin (Sigma), mouse anti-Bassoon (Synaptic System), guinea pig anti-Shank1 (gift from E. Kim, KAIST), mouse anti-PSD-95 K28/43 (NeuroMab), mouse anti-GluR2 (Chemicon, International S.C.), rabbit anti-GluR1C-term (Chemicon International), mouse anti-synaptophysin (Sigma), rabbit anti-HA-tag (Santa Cruz Biotechnology), mouse anti-HA-tag (Roche Applied Science), rabbit anti-Myc-tag (Sigma) and mouse anti-β-galactosidase (Promega, Madison, WI, USA).
cDNA constructs
Full-length or deletion HA-IL1RAPL1 constructs have been previously described (14). IL1RAPL2 cDNA was first obtained by Kristi Palmer (Amgen) and then sub-cloned into the GW1–2b-GluR2 Signal peptide-HA vector. For the IL1RAPL1▵C construct, we inserted a stop codon at amino acid 459, and for the IL1RAPL1▵N construct, we deleted the first 242 amino acids.
For the chromatography assay, the IL1RAPL1 extracellular domain (amino acids 1–353) was cloned into the NheI/KpnI sites of the pEGFP-N1 vector in which EGFP was replaced with a human Fc domain (19). Myc- and HA-tagged LAR-ecto-pDis, PTPδ-ecto-pDis and PTPσ-ecto-pDis have been previously described (19). Neuroligin2-HA has been previously described (25). Neurexin1β-CFP, Neurexin1α-CFP, Neurexin2α-CFP, Neurexin3α-CFP, Myc-Netrin-G1, Myc-Netrin-G2, Myc-DASM-1, SALM1-GFP, SALM2-GFP, SALM3-GFP, SALM4-GFP, NGL-3-GFP and NCAM140-GFP constructs have been previously described (26,27). RhoGAP2 cDNA was bought from Invitrogen and then sub-cloned into the GW12b-myc plasmid. To develop RhoGAP2▵C, we inserted a stop codon at amino acid 500.
Cell culture, transfection, staining and quantification of primary rat hippocampal neurons
Low-density hippocampal neuronal cultures were prepared from E18–E19 rat hippocampi as previously described with minor modifications (28,29) and were grown in 12-well Petri dishes (Iwaky Sterilin, Caerphilly, UK). Neurons were transfected using the calcium phosphate precipitation method on DIV 9, and experiments were performed 7 days after transfection.
African green monkey kidney (COS-7) or HEK293T cells at 50–70% confluency (24 h after plating in six-well plates; Iwaky Sterilin, Caerphilly, UK) were transfected using Lipofectamine® 2000 Transfection Reagent (Invitrogen, San Diego, CA, USA) with cDNA expression constructs (1–2 μg DNA/well) for 2–3 h in 5% CO2 at 37°C. Cells were washed twice with PBS, fed with DMEM containing 10% FBS and 1% penicillin/streptomycin and grown for 24–48 h before lysis for co-immunoprecipitation or pull-down assays.
Hippocampal neurons were fixed in 4% paraformaldehyde (PFA)–4% sucrose or 100% methanol at −20° for 10 min. Primary (1:50–1:400) and secondary (1:200) antibodies were applied in GDB buffer [30 mm phosphate buffer (pH 7.4) containing 0.2% gelatin, 0.5% Triton X-100 and 0.8 m NaCl].
Confocal images were obtained using a Zeiss 510 confocal microscope (Carl Zeiss; a gift from F. Monzino) with a 63× objective (numerical aperture 1.4) with sequential acquisition settings of 1024 × 1024 pixels. Each image is a z-series projection of about 7–15 images that were each averaged two to four times and taken at 0.4–0.7 µm depth intervals. Morphometric analysis and quantification of dendritic spines, synaptic protein staining and FM4–64 staining were performed using the MetaMorph software (Molecular Devices, Downingtown, PA, USA) by investigators who were blind to the type of transfection and experimental manipulation. Labeled, transfected neurons were chosen randomly for quantification from six cover slips from six independent experiments for each construct.
The dendritic spine number and dimensions and the synapse number were measured as described previously (29,30) with minor modifications. For each neuron, we measured the number of spines/synapses present in all the dendrites along their entire length, thus we calculated the number of spines/synapses present in the entire neuron. Then, we calculated mean and SEM for the neurons transfected with the same cDNA.
Fluorescence images and morphometric measurements were made as described (14). FM1–43 staining was performed by incubating neurons for 1 min in 6 μm FM1–43 (Molecular Probes) in a high-potassium buffer followed by two washes in Tyrode solution in the presence of 1 μm TTX (Tocris) as previously described (31).
Electrophysiological recording of cultured hippocampal neurons
Whole-cell patch-clamp recordings were taken from GFP-, IL1RAPL1- or IL1RAPL1▵N-transfected rat hippocampal neurons as described above. Patch electrodes, fabricated from thick borosilicate glass, were pulled and fire-polished to a final resistance of 3–4 MΩ and filled with the standard internal solution [(in mm) 100 CsMES, 20 CsCl, 2 MgCl2, 5 ethylene glycol tetraacetic acid, 10 HEPES, 4 ATP and 15 phosphocreatine (pH 7.4)]. EPSCs were investigated in cultured neurons by superfusing the whole-cell clamped neuron with a Tyrode solution containing (in mm): 150 NaCl, 2 CaCl2, 1 MgCl2, 4 KCl, 10 glucose and 10 HEPES (pH 7.4). Neurons were voltage-clamped at –70 mV, and TTX (0.3 µm) was added to block spontaneous action potential propagation during the recording of mEPSCs. Each experiment was performed at room temperature (22–24°C). Statistical analysis was performed using a two-tailed t-test.
Mixed-culture assay
Mixed co-culture assays were performed as described previously (32,33). Briefly, cultured hippocampal neurons at DIV 8 were co-cultured with COS-7 cells expressing GFP, HA-IL1RAPL1 (full-length or deletion mutants), HA-IL1RAPL2 or HA-neuroligin2 in the presence of 0.5 µm cytosine arabinoside to inhibit COS-7 cell proliferation. Three days later, cells were fixed and immunostained. Quantification of synapsin staining in the transfected COS-7 cells was performed using the MetaMorph software (Molecular Devices).
Affinity chromatography
Affinity chromatography for mass spectrometric identification of IL1RAPL1-binding proteins was performed as described previously (34). Briefly, HEK293FT cells were transfected with the IL1RAPL1-ecto-Fc expression constructs and grown in Opti-MEM (Invitrogen) for 10 days. Every 3 days, the IL1RAPL1-ecto-Fc protein was purified from conditioned 293FT media using Protein A sepharose.
Fifteen P18 rat brains were homogenized in homogenization buffer (0.32 m sucrose, 4 mm HEPES, pH 7.5, and protease inhibitors) using a Dounce homogenizer. Homogenates were centrifuged at 1000g for 15 min at 4°C. Supernatants were centrifuged again at 1000g for 15 min and then the resulting supernatants were centrifuged for at 10 000g for 20 min. The P2 pellet containing crude synaptosomes was resuspended in homogenization buffer and centrifuged at 10 000g for 20 min, yielding the P2′ pellet containing washed crude synaptosomes. The P2′ pellet was extracted in 20 mm Tris (pH 8.0), 0.1 mm CaCl2 and 1% Triton X-100 for 30 min at 4°C to enrich for pre-synaptic proteins (35). The extracts were centrifuged at 10 000g for 30 min, and the supernatants were diluted 1:1 with extraction buffer. Protein A beads bound to 50 µg human Fc control protein or 50 µg IL1RAPL1 ecto-Fc protein were added and rotated overnight at 4°C. Bound proteins were eluted from the beads by incubation with 0.2 m glycine (pH 2.2) and TCA-precipitated overnight.
The precipitate was boiled for 5 min and separated on Invitrogen Nupage gels. The nanoflow LC−MS/MS analysis was performed using a Q-STAR mass spectrometer (PE-Sciex, Canada) equipped with a nanoelectrospray ion source (Proxeon Biosystems, Odense, Denmark).
HEK293FT cell adhesion proteins
HEK293FT cells at 70–80% confluency (24 h after plating in 12-well plates) were transfected using the calcium phosphate precipitation method with the cDNA expression constructs (1–2 µg DNA per well) for 16 h, grown for an additional 3 h, collected and resuspended in DMEM (Lonza). To examine protein interaction, cells expressing the proper construct were plated together on a 16 mm cover slip and grown for 24 h before fixation with 4% PA supplemented with 4% sucrose (36).
Yeast two-hybrid screening and cDNA constructs
For the two-hybrid experiments, a fragment corresponding to the C-terminus of IL1RAPL1 (amino acids 390–696) was cloned into the pDBLeu vector in frame with the GAL4-binding domain and used as bait to screen a human fetal brain cDNA library (ProQuest Pre-made cDNA Libraries), which had been cloned into the pPC86 vector. Positive colonies grew on plates containing 10 mm 3-AT without tryptophan, leucine or histamine and expressed all three reporter genes: HIS3, LacZ and URA3. cDNA plasmids from positive clones were recovered using Escherichia coli DH5α plated on Amp and sequenced.
For further two-hybrid experiments, four DNA fragments corresponding to the IL1RAPL1 C-terminus—amino acids 390–580, 390–580, 403–562 and 560–696—were sub-cloned into pDBLeu and used as bait; the DNA fragments corresponding to amino acids 441–698, 430–580, 500–698 and 560–698 of RhoGAP2 were sub-cloned into the pPC86 plasmid and used as prey. RhoGAP2 with the myc tag was sub-cloned into the GW1-CMV expression vector (British Biotechnology, UK). Truncated HA-ILRAPL1 and HA-IL1RAPL1▵C were developed using PCR amplification and the appropriate oligonucleotides; these were also sub-cloned into the GW1-CMV expression vector with a HA-tag at the N-terminus as previously described (14).
The GST-Il-RAPL1 fragments corresponding to amino acids 403–562, 551–607, 608–684 and 560–696 were cloned into the pGEX4T-3 vector.
Co-immunoprecipitation and GST pull-down assays
Transfected COS7 cells were lysed with buffer containing 50 mm Tris–HCl, 150 mm NaCl, 1 mm EDTA, 1% NP40, 0.5% deoxycholate, 0.05% SDS and protease inhibitors (lysis buffer). Lysate samples (100 µg protein) were incubated overnight at 4°C with mouse anti-myc-tag antibodies (Santa Cruz Biotechnology, CA, USA) at 5 µg/ml in buffer A (200 mm NaCl, 10 mm EDTA, 10 mm Na2HPO4, 0.5% NP-40, 0.1% SDS, 10 mm NaF and Ser/Thr- and Tyr-phosphatase inhibitor cocktails).
Protein A agarose beads (Santa Cruz Biotechnology) washed in buffer A were added, and the incubation continued for 2 h. The beads were pelleted by centrifugation, washed five times with buffer A, resuspended in sample buffer for SDS–PAGE and boiled for 5 min. The beads were again pelleted by centrifugation, and the supernatants were applied to 7.5% SDS–PAGE. Protein bands were transferred to nitrocellulose membranes (Amersham) at 80 V for 120 min at 4°C. Primary antibodies were applied overnight in blocking buffer [20 mm Tris (pH 7.4), 150 mm NaCl, 0.1% Tween20 and 3% dried non-fat milk]. Secondary antibodies (HRP-conjugated anti-mouse or anti-rabbit) (Amersham) were used at a 1:2000 dilution. The signal was detected using an ECL detection system.
GST fusion proteins were prepared in E. coli BL21 and purified by standard procedures. African green monkey kidney COS-7 cells or rat brain homogenates were lysed in lysis buffer [50 mm Tris–HCl (pH 7.4), 150 mm NaCl, 1 mm EDTA, 1% NP40 and 0.5% deoxycholate]. Lysates were then incubated with 30 mg of GST fusion protein immobilized on GST 4B beads (GE Healthcare) for 3 h at 4°C, washed extensively five times in the lysis buffer and resuspended in 25 ml of 3× SDS sample buffer. GST alone was used as a control. Samples were separated by SDS–PAGE followed by western blot analysis. For GST pull-down experiments, hippocampal neuron lysates and rat brain extracts were incubated with 30 µg GST fusion protein immobilized on GST 4B beads (GE Healthcare) for 3 h at 4°C, washed five times in lysis buffer, and resuspended in 25 µl SDS sample buffer. GST alone served as a control. Samples were separated by SDS–PAGE followed by western blot analysis using the appropriated antibodies.
The following antibodies and dilutions were used: rabbit anti-Myc-tag (1:200; Upstate Cell Signaling Solutions) and rabbit anti-RhoGAP2 (1:500; in-house).
SUPPLEMENTARY MATERIAL
FUNDING
C.S. and M.P. were supported by Regione Lombardia (project number SAL-50-16983 TERDISMENTAL). J.K. was supported by Human Frontier Science Program Organization (grant number LT00021/2008-L). M.P. was supported by Telethon, Italy (grant number S01014TELU) and Fondazione Cariplo (project number 2008-2318). C.S. was supported by Telethon, Italy (grant number GGP09196), Fondazione CARIPLO (project number 2009.264), RSTL-CNR, Italian Institute of Technology, Seed Grant and Ministry of Health in the frame of ERA-NET NEURON. Funding to pay the Open Access publication charges for this article was provided by Telethon, Italy.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Alice Zanchi for technical work and Eunjoon Kim (Kaist) and A.M. Craig for the generous gift of cDNA expressing LAR-ecto-pDis, PTPδ-ecto-pDis, PTPσ-ecto-pDis, Neurexin1β-CFP, Neurexin1α-CFP, Neurexin2α-CFP, Neurexin3α-CFP, Myc-Netrin-G1, Myc-Netrin-G2, Myc-DASM-1, SALM1-GFP, SALM2-GFP, SALM3-GFP, SALM4-GFP, NGL-3-GFP and NCAM140-GFP.
Conflict of Interest statement: None declared.
REFERENCES
- 1.Carrie A., Jun L., Bienvenu T., Vinet M.C., McDonell N., Couvert P., Zemni R., Cardona A., Van Buggenhout G., Frints S., et al. A new member of the IL-1 receptor family highly expressed in hippocampus and involved in X-linked mental retardation. Nat. Genet. 1999;23:25–31. doi: 10.1038/12623. [DOI] [PubMed] [Google Scholar]
- 2.Laumonnier F., Shoubridge C., Antar C., Nguyen L.S., Van Esch H., Kleefstra T., Briault S., Fryns J.P., Hamel B., Chelly J., et al. Mutations of the UPF3B gene, which encodes a protein widely expressed in neurons, are associated with nonspecific mental retardation with or without autism. Mol. Psychiatry. 2010;15:767–776. doi: 10.1038/mp.2009.14. [DOI] [PubMed] [Google Scholar]
- 3.Bourgeron T. A synaptic trek to autism. Curr. Opin. Neurobiol. 2009;19:231–234. doi: 10.1016/j.conb.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 4.Kumar R.A., Christian S.L. Genetics of autism spectrum disorders. Curr. Neurol. Neurosci. Rep. 2009;9:188–197. doi: 10.1007/s11910-009-0029-2. [DOI] [PubMed] [Google Scholar]
- 5.Behnecke A., Hinderhofer K., Bartsch O., Nümann A., Ipach M.L., Damatova N., Haaf T., Dufke A., Riess O., Moog U. Intragenic deletions of IL1RAPL1: report of two cases and review of the literature. Am. J. Med. Genet. A. 2011;155A:372–379. doi: 10.1002/ajmg.a.33656. [DOI] [PubMed] [Google Scholar]
- 6.Bhat S.S., Ladd S., Grass F., Spence J.E., Brasington C.K., Simensen R.J., Schwartz C.E., Dupont B.R., Stevenson R.E., Srivastava A.K. Disruption of the IL1RAPL1 gene associated with a pericentromeric inversion of the X chromosome in a patient with mental retardation and autism. Clin. Genet. 2008;73:94–96. doi: 10.1111/j.1399-0004.2007.00920.x. [DOI] [PubMed] [Google Scholar]
- 7.Franek K.J., Butler J., Johnson J., Simensen R., Friez M.J., Bartel F., Moss T., DuPont B., Berry K., Bauman M., et al. Deletion of the immunoglobulin domain of IL1RAPL1 results in nonsyndromic X-linked intellectual disability associated with behavioral problems and mild dysmorphism. Am. J. Med. Genet. A. 2011;155A:1109–1114. doi: 10.1002/ajmg.a.33833. [DOI] [PubMed] [Google Scholar]
- 8.Gao X., Xi G., Niu Y., Zhang S., Fu R., Zheng Z., Zhang K., Lv S., He H., Xue M., et al. A study on the correlation between IL1RAPL1 and human cognitive ability. Neurosci. Lett. 2008;438:163–167. doi: 10.1016/j.neulet.2008.03.084. [DOI] [PubMed] [Google Scholar]
- 9.Piton A., Michaud J.L., Peng H., Aradhya S., Gauthier J., Mottron L., Champagne N., Lafrenière R.G., Hamdan F.F., Joober R., et al. Mutations in the calcium-related gene IL1RAPL1 are associated with autism. Hum. Mol. Genet. 2008;17:3965–3974. doi: 10.1093/hmg/ddn300. [DOI] [PubMed] [Google Scholar]
- 10.Tabolacci E., Pomponi M.G., Pietrobono R., Terracciano A., Chiurazzi P., Neri G. A truncating mutation in the IL1RAPL1 gene is responsible for X-linked mental retardation in the MRX21 family. Am. J. Med. Genet. A. 2006;140:482–487. doi: 10.1002/ajmg.a.31107. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Z., Yagi M., Okizuka Y., Awano H., Takeshima Y., Matsuo M. Insertion of the IL1RAPL1 gene into the duplication junction of the dystrophin gene. J. Hum. Genet. 2009;54:466–473. doi: 10.1038/jhg.2009.63. [DOI] [PubMed] [Google Scholar]
- 12.Bahi N., Friocourt G., Carrie A., Graham M.E., Weiss J.L., Chafey P., Fauchereau F., Burgoyne R.D., Chelly J. IL1 receptor accessory protein like, a protein involved in X-linked mental retardation, interacts with Neuronal Calcium Sensor-1 and regulates exocytosis. Hum. Mol. Genet. 2003;12:1415–1425. doi: 10.1093/hmg/ddg147. [DOI] [PubMed] [Google Scholar]
- 13.Gambino F., Pavlowsky A., Béglé A., Dupont J.L., Bahi N., Courjaret R., Gardette R., Hadjkacem H., Skala H., Poulain B., et al. IL1-receptor accessory protein-like 1 (IL1RAPL1), a protein involved in cognitive functions, regulates N-type Ca2+-channel and neurite elongation. Proc. Natl Acad. Sci. USA. 2007;104:9063–9068. doi: 10.1073/pnas.0701133104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pavlowsky A., Gianfelice A., Pallotto M., Zanchi A., Vara H., Khelfaoui M., Valnegri P., Rezai X., Bassani S., Brambilla D., et al. A postsynaptic signaling pathway that may account for the cognitive defect due to IL1RAPL1 mutation. Curr. Biol. 2010;20:103–115. doi: 10.1016/j.cub.2009.12.030. [DOI] [PubMed] [Google Scholar]
- 15.Woo J., Kwon S.K., Choi S., Kim S., Lee J.R., Dunah A.W., Sheng M., Kim E. Trans-synaptic adhesion between NGL-3 and LAR regulates the formation of excitatory synapses. Nat. Neurosci. 2009;12:428–437. doi: 10.1038/nn.2279. [DOI] [PubMed] [Google Scholar]
- 16.Washburn M.P., Wolters D., Yates J.R. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 17.Yamagata M., Sanes J.R., Weiner J.A. Synaptic adhesion molecules. Curr. Opin. Cell Biol. 2003;15:621–632. doi: 10.1016/s0955-0674(03)00107-8. [DOI] [PubMed] [Google Scholar]
- 18.Johnson K.G., Van Vactor D. Receptor protein tyrosine phosphatases in nervous system development. Physiol. Rev. 2003;83:1–24. doi: 10.1152/physrev.00016.2002. [DOI] [PubMed] [Google Scholar]
- 19.Kwon S.K., Woo J., Kim S.Y., Kim H., Kim E. Trans-synaptic adhesions between netrin-G ligand-3 (NGL-3) and receptor tyrosine phosphatases LAR, protein-tyrosine phosphatase delta (PTPdelta), and PTPsigma via specific domains regulate excitatory synapse formation. J. Biol. Chem. 2010;285:13966–13978. doi: 10.1074/jbc.M109.061127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takahashi H., Arstikaitis P., Prasad T., Bartlett T.E., Wang Y.T., Murphy T.H., Craig A.M. Postsynaptic TrkC and presynaptic PTPσ function as a bidirectional excitatory synaptic organizing complex. Neuron. 2011;69:287–303. doi: 10.1016/j.neuron.2010.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanz-Moreno V., Gadea G., Ahn J., Paterson H., Marra P., Pinner S., Sahai E., Marshall C.J. Rac activation and inactivation control plasticity of tumor cell movement. Cell. 2008;135:510–523. doi: 10.1016/j.cell.2008.09.043. [DOI] [PubMed] [Google Scholar]
- 22.Jaffe A.B., Hall A. Rho GTPases: biochemistry and biology. Annu. Rev. Cell Dev. Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- 23.Govek E.E., Newey S.E., Van Aelst L. The role of the Rho GTPases in neuronal development. Genes Dev. 2005;19:1–49. doi: 10.1101/gad.1256405. [DOI] [PubMed] [Google Scholar]
- 24.Govek E.E., Newey S.E., Akerman C.J., Cross J.R., Van der Veken L., Van Aelst L. The X-linked mental retardation protein oligophrenin-1 is required for dendritic spine morphogenesis. Nat. Neurosci. 2004;7:364–372. doi: 10.1038/nn1210. [DOI] [PubMed] [Google Scholar]
- 25.Levinson J.N., Chéry N., Huang K., Wong T.P., Gerrow K., Kang R., Prange O., Wang Y.T., El-Husseini A. Neuroligins mediate excitatory and inhibitory synapse formation: involvement of PSD-95 and neurexin-1beta in neuroligin-induced synaptic specificity. J. Biol. Chem. 2005;280:17312–17319. doi: 10.1074/jbc.M413812200. [DOI] [PubMed] [Google Scholar]
- 26.Graf E.R., Zhang X., Jin S.X., Linhoff M.W., Craig A.M. Neurexins induce differentiation of GABA and glutamate postsynaptic specializations via neuroligins. Cell. 2004;119:1013–1026. doi: 10.1016/j.cell.2004.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mah W., Ko J., Nam J., Han K., Chung W.S., Kim E. Selected SALM (synaptic adhesion-like molecule) family proteins regulate synapse formation. J. Neurosci. 2010;30:5559–5568. doi: 10.1523/JNEUROSCI.4839-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sala C., Piech V., Wilson N.R., Passafaro M., Liu G., Sheng M. Regulation of dendritic spine morphology and synaptic function by Shank and Homer. Neuron. 2001;31:115–130. doi: 10.1016/s0896-6273(01)00339-7. [DOI] [PubMed] [Google Scholar]
- 29.Verpelli C., Piccoli G., Zanchi A., Gardoni F., Huang K., Brambilla D., Di Luca M., Battaglioli E., Sala C. Synaptic activity controls dendritic spine morphology by modulating eEF2-dependent BDNF synthesis. J. Neurosci. 2010;30:5830–5842. doi: 10.1523/JNEUROSCI.0119-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Piccoli G., Verpelli C., Tonna N., Romorini S., Alessio M., Nairn A.C., Bachi A., Sala C. Proteomic analysis of activity-dependent synaptic plasticity in hippocampal neurons. J. Proteome Res. 2007;6:3203–3215. doi: 10.1021/pr0701308. [DOI] [PubMed] [Google Scholar]
- 31.Hering H., Lin C.C., Sheng M. Lipid rafts in the maintenance of synapses, dendritic spines, and surface AMPA receptor stability. J. Neurosci. 2003;23:3262–3271. doi: 10.1523/JNEUROSCI.23-08-03262.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim S., Burette A., Chung H.S., Kwon S.K., Woo J., Lee H.W., Kim K., Kim H., Weinberg R.J., Kim E. NGL family PSD-95-interacting adhesion molecules regulate excitatory synapse formation. Nat. Neurosci. 2006;9:1294–1301. doi: 10.1038/nn1763. [DOI] [PubMed] [Google Scholar]
- 33.Biederer T., Scheiffele P. Mixed-culture assays for analyzing neuronal synapse formation. Nat. Protoc. 2007;2:670–676. doi: 10.1038/nprot.2007.92. [DOI] [PubMed] [Google Scholar]
- 34.de Wit J., Sylwestrak E., O'Sullivan M.L., Otto S., Tiglio K., Savas J.N., Yates J.R., Comoletti D., Taylor P., Ghosh A. LRRTM2 interacts with Neurexin1 and regulates excitatory synapse formation. Neuron. 2009;64:799–806. doi: 10.1016/j.neuron.2009.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Phillips G.R., Huang J.K., Wang Y., Tanaka H., Shapiro L., Zhang W., Shan W.S., Arndt K., Frank M., Gordon R.E., et al. The presynaptic particle web: ultrastructure, composition, dissolution, and reconstitution. Neuron. 2001;32:63–77. doi: 10.1016/s0896-6273(01)00450-0. [DOI] [PubMed] [Google Scholar]
- 36.Krivosheya D., Tapia L., Levinson J.N., Huang K., Kang Y., Hines R., Ting A.K., Craig A.M., Mei L., Bamji S.X., et al. ErbB4-neuregulin signaling modulates synapse development and dendritic arborization through distinct mechanisms. J. Biol. Chem. 2008;283:32944–32956. doi: 10.1074/jbc.M800073200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.