The 3,4-dihydroxyphenylalanine (dopa)-containing proteins of mussels and sandcastle worms provide attractive design paradigms for engineering synthetic polymers as wet adhesives and coatings.[1] Despite this, a generally accepted explanation of how dopa interacts with most surfaces is not available. Indeed, given the facile oxidation of dopa, even the question as to whether dopa (Figure 1a) is the only interacting functionality on all surfaces remains uncertain. The effect of uncontrollable Dopa redox on the dependability of catechol as an anchoring functionality for polymers is a recognized problem and best illustrated in coatings engineered for superparamagnetic nanoparticles used in magnetic resonance imaging.[2] A one-electron oxidation of dopa produces a semiquinone (Figure 1a) whereas a two-electron oxidation results in dopa o-quinone or dopaquinone (Figure 1b); both have wide ranging reaction chemistries. Semiquinones are present in the mussel adhesive but their contribution to adhesion is unknown.[3] Dopaquinone is far inferior to dopa at mediating adhesion to titania and mica surfaces,[4,5] but on amine-functionalized surfaces it provides up to 200-fold more strength than dopa itself.[4] In addition, under some solution conditions, tautomers of dopaquinone, namely dopaquinone methide (Figure 1c) and α,β-dehydrodopa (Δ-dopa) (Figure 1d) are more stable than the o-quinone parent. Quinone tautomers were previously detected in oxidized dopa peptides and proteins[6] and could be associated with structural changes in oxidized adhesive mussel foot proteins such as mfp-3.
Figure 1.
Reaction products for the stepwise 1- and 2-electron oxidation of dopa (a) to dopa semiquinone (a′), dopaquinone (b) and the quinone tautomers, dopa quinonemethide (c) and α, β–dehydrodopa (d). Dihedral torsional angles are shown around a dopaquinone side-chain (e); the essentially planar peptide bonds flank the side-chain on both sides. Migration of the double bond to between the α–β carbons in Δ–dopa abolishes optical activity and places NH, C=O, α-C and β-C into the same plane (f).
A better understanding of interfacial dopa chemistry is a crucial prerequisite to engineering the functionality for effective polymer adhesion. In the present study, we deposited thin films of mfp-3 on mica and tested whether the adhesion and film thickness (hard wall) detected by the surface forces apparatus (SFA) is influenced by the presence or absence of dopaquinone tautomers in the protein. Dopaquinones were formed in mfp-3 by autoxidation (raising the pH) or by periodate-mediated oxidation. Protein dopaquinone tautomerization is intriguing because of its effect on the α-carbons in the peptide backbone and the suggestion that the backbone flexibility of adhesive proteins may be adjusted by the selection of different resonance forms of dopa following oxidation. This runs against the conventional wisdom that oxidation of dopa-containing proteins serves exclusively as a cross-linking strategy.[7,8]
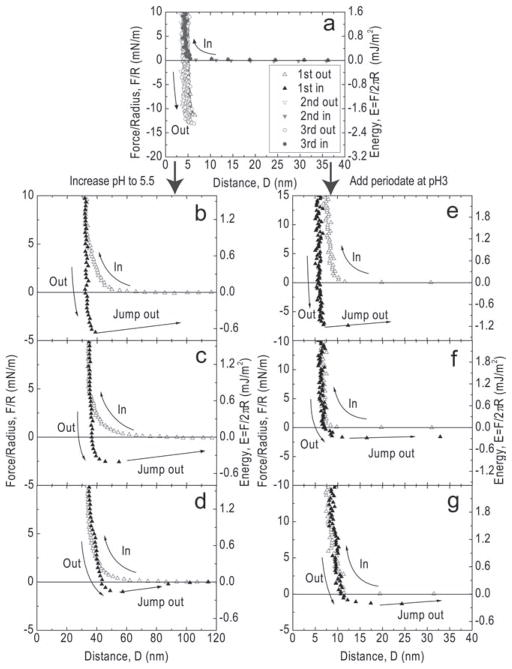
A strong adhesion force of ~12 mN/m equivalent to an adhesion energy of −2 mJ/m2 was reproducibly measured at pH 3 during separation of two mfp-3 films after 1 min compression times (Figure 2a). The effect of contact time is elaborated in the supporting results (Figure S1). Strong adhesion is also consistent with the abrupt jumping apart of two surfaces as manifested by the rapid movement of the fringes of equal chromatic order (FECO). Following introduction of pH 5.5 buffer into the gap solution, the surfaces were then brought into brief contact again without changing the contact position. An adhesion force of 4.5 mN/m was measured at pH 5.5, decreasing more than 60% from the mfp-3 mica adhesion at pH 3. During a second approach at pH 5.5 followed by separation, the measured adhesion force dropped to only 2.5 mN/m, and the adhesion associated with a third approach-separation cycle measured only ~1 mN/m. After this, adhesion exhibited no further change. The pH-dependent decrease in adhesion suggests a pH-dependent chemical or structural change in mfp-3. The most likely chemistry involved is dopa oxidation. Dopa is known to be unstable at elevated pH, undergoing facile oxidation to dopaquinone.[4,5,9] As dopaquinones accumulate in mfp-3, adhesion is diminished with each successive approach and separation cycle and/or time lapsed.
Figure 2.
Mfp-3 adhesion and dopa oxidation. a) The adhesion force of mfp-3 at pH3 during three successive approach and separation cycles. b) In the first approach-separation cycle at pH 5.5, an adhesion force of 4.5 mN/m was measured. c) The adhesion decreased to about 2.5 mN/m during the second cycle and d) to about 1 mN/m during the third. Oxidizing dopa in mfp-3 with increasing [periodate] follows similar trends in e) first periodate addition of 200 pmoles, f) second periodate 200 pmoles and g) third periodate 200 pmoles.
To explore the hypothesis that dopa oxidation is responsible a loss in adhesion to mica, a controlled dopa oxidation experiment was done by adding periodate at pH 3 where dopa undergoes negligible auto-oxidation (Figure 2e–f). Periodate, a strong 2-electron oxidant, stoichiometrically oxidizes catechols including dopa to o-quinones.[6,8] After achieving a dopa-mica adhesion at pH 3 of more than 12 mN/m, 200 pmoles of periodate (20 μL) were injected into the gap between the mfp-3 films. Adhesion dropped to 7.5 mN/m in the first approach-separation cycle following injection and further to 1.5 mN/m during the second force cycle. No further decrease in adhesion was observed during subsequent approach-separation cycles. The periodate results support the notion that oxidation of dopa to dopaquinone decreases mfp-3 adhesion to mica at pH 5.5 and 7. The obvious parallels between increasing the pH of the gap solution and periodate addition at pH 3 suggest that significant dopa oxidation happens even at pH 5.5 in the SFA. In bulk solution at pH 5.5, dopa is ordinarily stable for hours, not minutes. In contrast, at seawater pH ~8 and saturating levels of dissolved oxygen, the oxidation of dopa to quinone is nearly spontaneous.[10]
The SFA data reveal a second important trend: the hard wall, an indicator of repulsive interactions and an approximation of the hydrodynamic diameter of mfp-3, changed dramatically with pH. The film thickness of mfp-3 on mica at pH 3 was just ~3 nm (Figure 3a). After increasing the pH of the gap solution to 7.5 followed by a short equilibration time, repulsion was first noticeable at a distance of 40 nm during the approach, with an increase in the hard wall to ~20 nm (Figure 3a). The hard wall increase suggests that the mfp-3 film expands at higher pH. We know of only two chemical pathways relevant to quinone formation in mfps in this pH range: 1) protein cross-linking, and 2) the tautomerization of the dopaquinone to Δ-dopa. Cross-linking is not a plausible scenario for swelling because dilute mfp-1 in solution undergoes a significant diminution in hydrodynamic radius upon oxidation[11] and cross-linked mfp-1 films exhibit an irreversible decrease in film thickness as measured by surface plasmon resonance and the quartz crystal microbalance.[12] In contrast, the hard wall increase measured here was largely reversible, thus is more consistent with tautomerization than cross-linking. The tautomerization involves migration of the double bond from the quinone nucleus via the quinone methide to the alpha carbon in the backbone (Figure 1c–d). At pH 7, the Δ-dopa tautomer is favored, whereas at pH 3 the dopaquinone prevails.[6] The reversible reduction in the hard wall from 20 nm to 10 nm by decreasing the pH of the gap solution from 7.5 to 3 is consistent with the reversibility of quinone tautomers but does not prove their involvement.
Figure 3.
The effect of pH on mfp-3 structure. (a) After increasing the pH of the gap solution from 3 to 7.5, the film thickness measured by the SFA increased from 3 nm to about 20 nm. The hard wall of the protein layer was shifted back to 10 nm when the pH of the gap solution was brought back to 3, but was not accompanied by a recovery of substantial adhesion. (b) Sequential UV-vis spectra of mfp-3 (1 mg/ml) following oxidation by excess periodate in acetate buffer at pH 5.5. curve 1 (—): untreated; curve 2 (–·–): after adding periodate within a mixing time of 30 s; curve 3 (…): 30 min; curve 4 (—): 100 min. The maxima at 320 ~ 330 nm (D-dopa) and 390 ~ 400 nm (dopaquinone) increase and decrease, respectively, with time. c) CD spectra of mfp3 (0.3 mg/ml) in phosphate buffered saline buffer at pH 5.5 before and after adding excess periodate.
Tautomerization of dopaquinone to Δ-dopa needs a Lewis base to extract the acidic proton on the α-C (Figure 1e–f).[6] In our experiments, acetate and phosphate buffers, certain mfp-3 side-chains, and the mica surface[13] itself could act as Lewis bases to catalyze the rearrangement. UV–vis spectra were performed to test whether the dopaquinone to Δ-dopa tautomerization occurs in bulk mfp-3 solution upon dopa oxidation. Due to the precipitation of mfp-3 at near-neutral pH, UV–vis readings were only reliable up to pH 5.5. Dopa oxidation was enhanced at pH 5.5 (acetate buffer) by adding excess periodate. After periodate addition, a typical quinone spectrum (λmax ≈ 390–400 nm) developed within the mixing time (Figure 3b). As the oxidation proceeded, the quinone peak decayed, and an increase in absorbance at about 320–330 nm occurred; this is the characteristic maximum for Δ-dopa and is consistent with dopaquinone tautomerization to Δ-dopa in a low molecular weight peptide.[6] The UV–vis spectra of several model compounds of dopa oxidation e.g. 3-(3,4-dihydroxyphenyl)-2-propenoic acid (caffeic acid), 3-(2,3-dihydroxyphenyl)propanoic acid (hydrocaffeic acid) and its as well as its corresponding quinone support the structural designations (Figure S2). (Cyclic voltammetry was also performed but provided no diagnostic insights into the chemistry Figure S3). Caffeic acid with its α, β-unsaturated side-chain permits a clear qualitative differentiation from hydrocaffeic acid with its saturated side chain. Note that hydrocaffeic acid bears a remarkable similarity to the spectrum of mfp-3 before oxidation (Figure 3 b), whereas hydrocaffeic o-quinone and caffeic acid resemble the post-oxidation spectra thus providing support for the structural transformation of dopa groups in mfp-3. However, in contrast to the SFA experiments, periodate addition to the bulk solution reaction system was necessary to observe detectable oxidation. The greater susceptibility of mfp-3 to oxidation in the SFA compared with bulk solution is intriguing and worthy of greater scrutiny. Perhaps the confinement of mfp-3 between mica surfaces during an SFA experiment enhances its oxidation kinetics;[14] it is worth recalling in this regard that mica is an excellent Lewis base.[13] Another possibility is mechanochemical, i.e., a mechanical deformation of mfp-3 enhances dopa oxidation. For example, during wood milling lignins are degraded to benzoquinones,[15] and free radicals are generated from catechols during stirring.[16] It should be noted, however, there is no evidence for a mechanochemically driven oxidation in the three successive approach-separation cycles of Figure 2a.
Considering the double bond between the α-C and β-C atoms, the conformational flexibility of the protein backbone as well as the Δ-dopa side chain is likely to be more restricted. Without periodate addition, circular dichroism of mfp-3 exhibits an ellipticity suggestive of random coil structure. After adding periodate to mfp-3, the ellipticity at 199 nm decreased by more than 60%, indicating a loss of protein conformational flexibility and reduced optical activity (Figure 3c).
In the absence of folding, reduced flexibility of the protein backbone should increase the hydrodynamic size of the protein molecule. The increase in the hydrodynamic dimensions of mfp-3 was confirmed by dynamic light scattering (DLS). From DLS analysis, mfp-3 has a hydrodynamic radius of about 1–2 nm or a diameter of ~2–3 nm at pH 3. This diameter approximates the film thickness of the mfp-3 layer (~3–4 nm) measured by SFA experiment at the same pH (Figure 3a). At pH 5.5, however, the hydrodynamic radius remains at 1–2 nm–much less than the value deduced from SFA measurements (thickness 30–40 nm) and, as already mentioned, is attributed to confinement effects in the SFA. Upon raising the pH to 7.5 or adding periodate to pH 5.5 buffer, the hydrodynamic radius of mfp3 increased abruptly to 10–25 nm. Dialysis of a solution of mfp3 at pH 7 against acetic acid buffer at pH 3 decreased the hydrodynamic radius of mfp3 to 4–6 nm. Thus, the DLS-determined increase in hydrodynamic radius following dopa oxidation also qualitatively agrees with observations from SFA experiments. Although oxidation of mfp-3 in bulk solution is not necessarily a good approximation of mfp-3 reactivity on mica surfaces during SFA experiments, it still provides a reasonable basis for assessing the role of dopa oxidation and quinone tautomers in relation to the adhesion loss and hard wall increase with pH.
Freshly cleaved mica surfaces are known to consist of chemically inert polysiloxanes, and there is no evidence for a specific coordination complex as that proposed between dopa and titania surfaces.[17] The moderate adhesion of mfp-3 to mica is probably due to hydrogen bonding between the bidentate OH groups of the catechol and the O atoms in the mica crystal. The fit should be a snug one as the distance between adjacent O atoms in the mica crystal (~0.28 nm) matches the distance between OH groups in dopa, ~0.29 nm.[5] As predicted by Bell theory and by analogy to the A-T pairs in DNA, the well oriented bidentate hydrogen bonding (E = ~−28 kT) of dopa to mica should have a binding lifetime (τ = τoe−E/kT) that is 106 times as long as the monodentate form (E = ~−14 kT).[18]
At pH 3, maximal adhesion energy measured between an mfp-3 coated mica surface and bare mica after a 60 min contact time is Ead = −2.5 mJ/m2 (Supporting Information). Typically, the surface area of a mica sheet glued to the SFA cylindrical support is about 1 cm2. Assuming every molecule in the 20 μL of mfp-3 solution (20 μg/mL) is adsorbed to mica would deposit about 5.7 × 10−11 mole mfp-3s on each surface with a corresponding dopa density of about 5.7 × 10−6 mole/m2. Although we do not know the actual adsorption efficiency of dopa to each mica surface, a 50% adsorption to each surface (the maximum possible amount of dopa in direct adhesive contact with two mica surfaces) would give a lower boundary for the dopa-mica binding of 0.87 kJ/mol. This number is an order of magnitude smaller than the hydrogen bond energy, typically 10–40 kJ/mol. Assuming a lower coverage of 10%, however, would give a higher binding, i.e. 4.4 kJ/mol, and probably more realistic value.
Dopa redox plays a dual role in wet adhesion and cohesion within mussel plaques. Our SFA results indicate that dopa oxidizes to dopaquinone at even moderately acidic pH, causing a significant drop in adhesion. Once dopaquinone forms, it quickly undergoes tautomerization to Δ-dopa, which abolishes side-chain rotation and optical activity in the alpha carbon (Figure 1f). Tautomerization has also been observed in some Dopa-PEG constructs.[19] Because dopaquinone tautomerization to Δ-dopa in dopa-containing proteins and peptides imposes severe local configurational constraints, it would be expected to increase the hydrodynamic radius of a single protein chain. The double bond between the α and β carbons (Figure 1f) puts them in the same plane as the α-amine and carbonyl groups of the backbone with 120° angles around the sp2 α-carbon and a loss of optical activity.[20] Given the ten fairly evenly distributed dopa residues in the mfp-3 sequence,[21] converting all of these to Δ-dopa would be expected to decrease entropy in the chain and perhaps impose a new secondary conformation. The nature of this conformation in mfp-3 is unknown at this time but reasonable conjecture is possible. Mathur et al.[21] prepared a synthetic octapeptide with alternating Δ-Phe residues that exhibited a distinct preference for a 310 helix. This resembles an α-helix in being internally H-bonded but with fewer amino acids (i to i+3) between each donor-acceptor pair.[22]
It is not currently known whether Δ-dopa formation in mfp-3 on surfaces is adaptive or adventitious. The former would require a certain degree of local redox and pH control. Mussels could retard oxidative losses in the plaque by co-secreting a thiol containing protein antioxidant with mfp-3 and mfp-5 at relatively low pH regimes during plaque deposition.[23] A spatially tuned control of Δ-dopa formation would enable an extraordinary versatility in how the proteins are packed as they approach a solid surface. Indeed, the obvious porosity gradient in the plaque suggests such versatility.[24]
Experimental Section
Materials
Mfp-3 was purified according to a procedure previously described.[21] Purified mfp-3 performs consistently by SFA after storage for 100 days at pH 3 and −50 °C (figure s4). Before the experiment, 20 μL of mfp-3 solution (20 μg/mL) was injected onto one or both mica surfaces. After allowing 30 min absorption followed by rinsing with buffer, the two surfaces were then transferred into a SFA chamber with a droplet of buffer in between of the surfaces for measurement. The volume of the droplet solution is about 50 μL. Buffer preparation: 0.1 M acetic acid (EMD Chemicals, Gibbstown, NJ) and 0.25 M potassium nitrate (Aldrich, St. Louis, MO) (pH 3); 0.1 M sodium acetate (EM Science, Gibbstown, NJ) and 0.25 M potassium nitrate titrated by acetic acid to pH 5.5; 0.016 M potassium phosphate monobasic (Mallinckrodt, Hazelwood, MO) and 0.084 M potassium phosphate dibasic (EMD Chemicals, Gibbstown, NJ) (PH 7.5). Milli-Q water (Millipore, Bedford, MA) was used for all glassware cleaning and solution preparation.
Surface forces apparatus SFA
The adhesion between mfp-3 and mica was measured using a SFA in a configuration reported previously.[25] Details of the SFA technique have been described elsewhere.[26,27] The normal force measured by a SFA is typically normalized by the radius of the disk R. In a typical force-distance graph produced by SFA measurement, the positive value corresponds to the repulsive force, and the negative value represents the adhesive forces. Applying Derjaguin approximation F(D) = 2πRE(D), the normalized force F/R measured between two cylindrical surfaces can be easily converted to the energy E between two flat surfaces. In this paper, the adhesion energy is defined as the adhesion or “pull off” force measured divided by 2πR.
UV–vis spectrophotometry
UV–vis spectra were obtained using a Nanodrop 1000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE). Mfp3 (20 μL, 1 mg/mL), caffeic acid, hydrocaffeic acid were made in acetate buffer at pH 5.5, respectively, (0.1 M sodium acetate titrated with acetic acid). Excess sodium periodate was used to oxidize hydrocaffeic acid and dopa in mfp3.
Circular dichroism spectroscopy
Circular dichroism (CD) measurements were performed on an OLIS RSM circular dichroism spectropolarimeter. The cell was maintained at 25 °C using a Quantum temperature controller. Far-UV (250–190 nm) scans were performed in a micro cell (with a path-length of 0.05 cm) that required only 300 μL of solution. The computer-averaged trace of 10 scans was employed in all calculations. Signal due to solvent was subtracted. 0.3 mg/mL of mfp3 was measured in PBS buffer at pH 5.5 before and after adding 10 μL of 3.2 mg/mL NaIO4. The data were normally plotted as mean-residue-weight ellipticity ([θ]; units.degrees.cm2 dmol−1) versus wavelength in nm, calculated via the following equation:
where θobs is the observed ellipticity in degrees, m.r.w. is the mean residue weight, c is the concentration in g/mL and d is the path length in cm.
Dynamic light scattering
Dynamic light scattering (DLS) was used to evaluate the hydrodynamic dimensions of the protein samples. The hydrodynamic radius of mfp3 at pH 3 (acetic acid), 5.5 (sodium acetate), 7.5 (phosphate) and pH 5.5 (phosphate) was determined after adding excess amount of oxidant NaIO4 at 20 °C using a Dynapro DLS temperature controlled micro-sampler with a 824.7 nm laser diode (Protein Solutions, Charlottesville, VA). DLS scattering counts were recorded every 10 seconds (10 acquisitions/sample) and scattering intensity data were processed using Dynamics Dynapro Control software v.6.3.40.
Supplementary Material
Acknowledgments
This research was supported by the National Institutes of Health (R01 DE018468) and the MRSEC Program of the National Science Foundation under award No. DMR 05-20415. J. Yu and W. Wei contributed equally. This article is part of the Special Issue on Materials Research at the University of California, Santa Barbara.
Footnotes
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Jing Yu, Department of Chemical Engineering, University of California, Santa Barbara, CA 93106, USA.
Dr. Wei Wei, Materials Research Laboratory, University of California, Santa Barbara, CA 93106, USA
Eric Danner, Department of Molecular, Cell & Developmental Biology, University of California, Santa Barbara, CA 93106, USA.
Prof. Jacob N. Israelachvili, Department of Chemical Engineering, University of California, Santa Barbara, CA 93106, USA
Prof. J. Herbert Waite, Email: waite@lifesci.ucsb.edu, Department of Molecular, Cell & Developmental Biology, University of California, Santa Barbara, CA 93106, USA.
References
- 1.Dalsin JL, Messersmith BP. Mater Today. 2005;8:38. [Google Scholar]
- 2.Amstad E, Gillich T, Bilecka I, Textor M, Reimhult E. Nano Lett. 2009;9:4042. doi: 10.1021/nl902212q. [DOI] [PubMed] [Google Scholar]
- 3.Sever MJ, Weisser JT, Monahan J, Srinivasan S, Wilker JJ. Angew Chem Int Ed. 2004;43:448. doi: 10.1002/anie.200352759. [DOI] [PubMed] [Google Scholar]
- 4.Lee H, Scherer NF, Messersmith PB. Proc Nat Acad Sci USA. 2006;103:12999. doi: 10.1073/pnas.0605552103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson TJ, Yu J, Estrada AY, Waite JH, Israelachvili JN. Adv Funct Mater. 2010;20:4196. doi: 10.1002/adfm.201000932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rzepecki LM, Waite JH. Arch Biochem Biophys. 1991;285:27. doi: 10.1016/0003-9861(91)90324-c. [DOI] [PubMed] [Google Scholar]
- 7.Waite JH. Comp Biochem Physiol. 1990;97B:19. doi: 10.1016/0305-0491(90)90172-p. [DOI] [PubMed] [Google Scholar]
- 8.Liu B, Burdine L, Kodadek T. J Am Chem Soc. 2006;129:12348. doi: 10.1021/ja072904r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu M, Deming TJ. Macromolecules. 1998;31:4739. doi: 10.1021/ma980268z. [DOI] [PubMed] [Google Scholar]
- 10.Proudfoot GM, Ritchie IM. Austral J Chem. 1983;36:885. [Google Scholar]
- 11.Haemers S, Van Der Leeden MC, Koper GJM, Frens G. Langmuir. 2002;18:4903. [Google Scholar]
- 12.Höök F, Kasemo B, Nylander T, Fant C, Sott K, Elwing H. Anal Chem. 2001;73:5796. doi: 10.1021/ac0106501. [DOI] [PubMed] [Google Scholar]
- 13.Giese RF, van Oss CJ. J Dispersion Sci Technol. 1998;19:775. [Google Scholar]
- 14.Santiso EE, Kostov MK, George AM, Nardelli MB, Gubbins KE. Appl Surf Sci. 2007;253:5570. [Google Scholar]
- 15.Wu Z, Sumimoto M, Tanaka H. J Wood Chem Technol. 1995;15:27. [Google Scholar]
- 16.Tipikin DS, Lebedev YS, Rieker A. Chem Phys Lett. 1997;272:399. [Google Scholar]
- 17.Li SC, Chu LN, Gong XQ, Diebold U. Science. 2010;328:882. doi: 10.1126/science.1188328. [DOI] [PubMed] [Google Scholar]
- 18.Israelachvili JN. Intermolecular and Surface Forces. 3. Elsevier; London: 2010. [Google Scholar]
- 19.Lee BP, Dalsin JL, Messersmith PB. Biomacromolecules. 2002;3:1038. doi: 10.1021/bm025546n. [DOI] [PubMed] [Google Scholar]
- 20.Pieroni D, Fissi A, Pratesi C, Temussi PA, Ciardelli F. Biopolymers. 1993;33:1. doi: 10.1002/bip.360330102. [DOI] [PubMed] [Google Scholar]
- 21.Zhao H, Robertson NB, Jewhurst SA, Waite JH. J Biol Chem. 2006;281:1090. doi: 10.1074/jbc.M510792200. [DOI] [PubMed] [Google Scholar]
- 22.Mathur P, Ramakumar S, Chauhan VS. Biopolymers. 2004;76:150. doi: 10.1002/bip.10571. [DOI] [PubMed] [Google Scholar]
- 23.Zhao H, Waite JH. J Biol Chem. 2006;281:26150. doi: 10.1074/jbc.M604357200. [DOI] [PubMed] [Google Scholar]
- 24.Tamarin A, Lewis P, Askey J. J Morphol. 1976;149:199. doi: 10.1002/jmor.1051490205. [DOI] [PubMed] [Google Scholar]
- 25.Lin Q, Gourdon D, Sun CJ, Holten-Andersen N, Anderson TJ, Waite JH, Israelachvili JN. Proc Nat Acad Sci USA. 2007;104:3782. doi: 10.1073/pnas.0607852104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Israelachvili JN, Adams GE. J Chem Soc Faraday Trans I. 1978;74:975. [Google Scholar]
- 27.Israelachvili JN, Min Y, Akbulut M, Alig A, Carver G, Greene W, Kristiansen K, Meyer E, Pesika N, Rosenberg K, Zeng H. Rep Prog Phys. 2010;73:036601. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.