Abstract
Analysis of phylogenetically conserved secondary structure has been important in the development of models for the secondary structure of structural RNAs. In this paper, we apply this type of analysis to several families of informational RNAs to evaluate its usefulness in developing secondary structure models for mRNAs and mRNA precursors. We observed many conserved helices in all mRNA groups analyzed. Three criteria were used to identify potential helices which were not conserved solely because of coding sequence constraints, and may therefore be important for the structure and function of the RNA. These results suggest that this approach will be useful in deriving secondary structure models for informational RNAs when used in conjunction with other complementary techniques, and in designing experiments to determine the functional significance of conserved base pairing interactions.
Full text
PDF









Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bannon G. A., Bowen J. K., Yao M. C., Gorovsky M. A. Tetrahymena H4 genes: structure, evolution and organization in macro- and micronuclei. Nucleic Acids Res. 1984 Feb 24;12(4):1961–1975. doi: 10.1093/nar/12.4.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer A. L., Miller O. L., Jr, McKnight S. L. Ribonucleoprotein structure in nascent hnRNA is nonrandom and sequence-dependent. Cell. 1980 May;20(1):75–84. doi: 10.1016/0092-8674(80)90236-6. [DOI] [PubMed] [Google Scholar]
- Brimacombe R., Maly P., Zwieb C. The structure of ribosomal RNA and its organization relative to ribosomal protein. Prog Nucleic Acid Res Mol Biol. 1983;28:1–48. doi: 10.1016/s0079-6603(08)60081-1. [DOI] [PubMed] [Google Scholar]
- Davies R. W., Waring R. B., Ray J. A., Brown T. A., Scazzocchio C. Making ends meet: a model for RNA splicing in fungal mitochondria. Nature. 1982 Dec 23;300(5894):719–724. doi: 10.1038/300719a0. [DOI] [PubMed] [Google Scholar]
- Fitch W. M. Calculating the expected frequencies of potential secondary structure in nucleic acids as a function of stem length, loop size, base composition and nearest-neighbor frequencies. Nucleic Acids Res. 1983 Jul 11;11(13):4655–4663. doi: 10.1093/nar/11.13.4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox G. E., Woese C. R. 5S RNA secondary structure. Nature. 1975 Aug 7;256(5517):505–507. doi: 10.1038/256505a0. [DOI] [PubMed] [Google Scholar]
- Grantham R., Gautier C., Gouy M. Codon frequencies in 119 individual genes confirm consistent choices of degenerate bases according to genome type. Nucleic Acids Res. 1980 May 10;8(9):1893–1912. doi: 10.1093/nar/8.9.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groudine M., Casimir C. Post-transcriptional regulation of the chicken thymidine kinase gene. Nucleic Acids Res. 1984 Feb 10;12(3):1427–1446. doi: 10.1093/nar/12.3.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintz N., Sive H. L., Roeder R. G. Regulation of human histone gene expression: kinetics of accumulation and changes in the rate of synthesis and in the half-lives of individual histone mRNAs during the HeLa cell cycle. Mol Cell Biol. 1983 Apr;3(4):539–550. doi: 10.1128/mcb.3.4.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz S., Horowitz A., Nilsen T. W., Munns T. W., Rottman F. M. Mapping of N6-methyladenosine residues in bovine prolactin mRNA. Proc Natl Acad Sci U S A. 1984 Sep;81(18):5667–5671. doi: 10.1073/pnas.81.18.5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury G., Gruss P., Dhar R., Lai C. J. Processing and expression of early SV40 mRNA: a role for RNA conformation in splicing. Cell. 1979 Sep;18(1):85–92. doi: 10.1016/0092-8674(79)90356-8. [DOI] [PubMed] [Google Scholar]
- Kost T. A., Theodorakis N., Hughes S. H. The nucleotide sequence of the chick cytoplasmic beta-actin gene. Nucleic Acids Res. 1983 Dec 10;11(23):8287–8301. doi: 10.1093/nar/11.23.8287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebhaber S. A., Begley K. A. Structural and evolutionary analysis of the two chimpanzee alpha-globin mRNAs. Nucleic Acids Res. 1983 Dec 20;11(24):8915–8929. doi: 10.1093/nar/11.24.8915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDevitt M. A., Imperiale M. J., Ali H., Nevins J. R. Requirement of a downstream sequence for generation of a poly(A) addition site. Cell. 1984 Jul;37(3):993–999. doi: 10.1016/0092-8674(84)90433-1. [DOI] [PubMed] [Google Scholar]
- Naora H., Deacon N. J., Buckle D. Further studies on similarity of terminal nucleotide sequences of eukaryote mRNA-a possible mechanism of RNA splicing. J Theor Biol. 1980 Jul 7;85(1):83–97. doi: 10.1016/0022-5193(80)90282-9. [DOI] [PubMed] [Google Scholar]
- Noller H. F. Structure of ribosomal RNA. Annu Rev Biochem. 1984;53:119–162. doi: 10.1146/annurev.bi.53.070184.001003. [DOI] [PubMed] [Google Scholar]
- Noller H. F., Woese C. R. Secondary structure of 16S ribosomal RNA. Science. 1981 Apr 24;212(4493):403–411. doi: 10.1126/science.6163215. [DOI] [PubMed] [Google Scholar]
- Nussinov R. Doublet frequencies in evolutionary distinct groups. Nucleic Acids Res. 1984 Feb 10;12(3):1749–1763. doi: 10.1093/nar/12.3.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
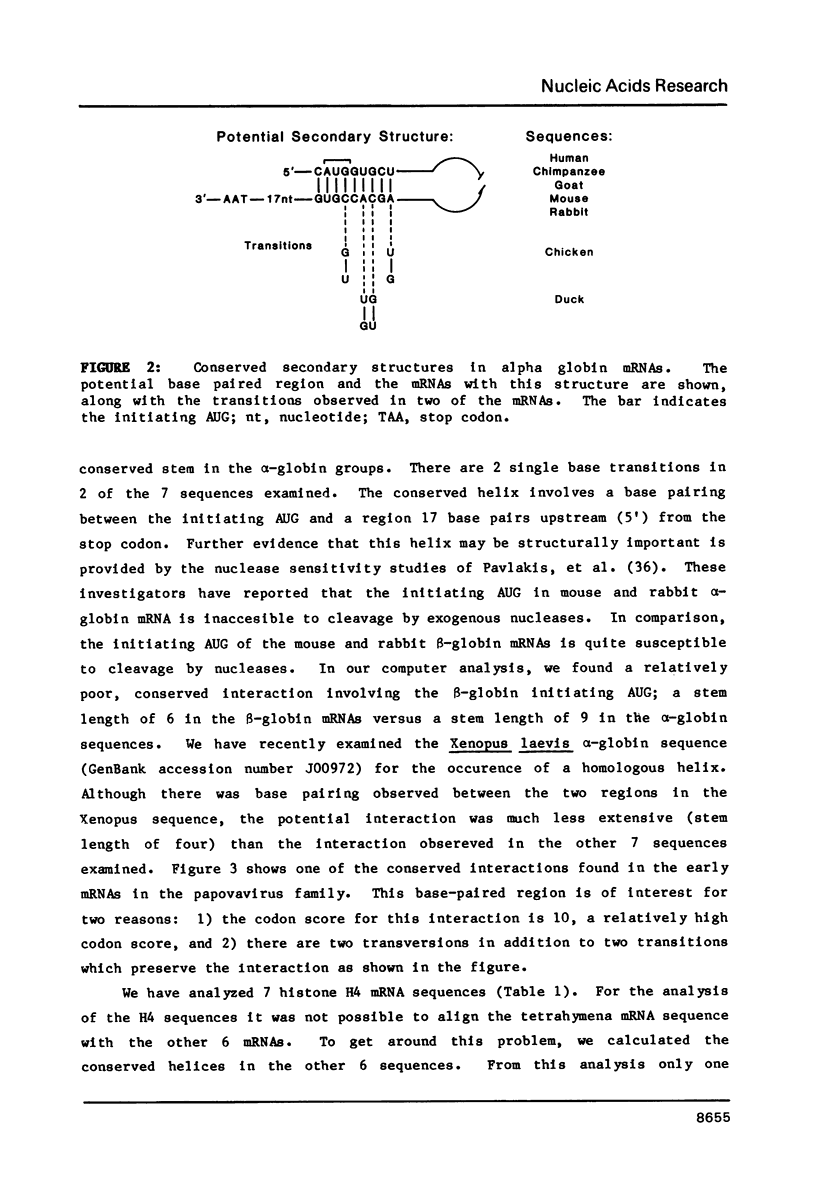
- Pavlakis G. N., Lockard R. E., Vamvakopoulos N., Rieser L., RajBhandary U. L., Vournakis J. N. Secondary structure of mouse and rabbit alpha- and beta-globin mRNAs: differential accessibility of alpha and beta initiator AUG codons towards nucleases. Cell. 1980 Jan;19(1):91–102. doi: 10.1016/0092-8674(80)90391-8. [DOI] [PubMed] [Google Scholar]
- Ponte P., Ng S. Y., Engel J., Gunning P., Kedes L. Evolutionary conservation in the untranslated regions of actin mRNAs: DNA sequence of a human beta-actin cDNA. Nucleic Acids Res. 1984 Feb 10;12(3):1687–1696. doi: 10.1093/nar/12.3.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray B. K., Brendler T. G., Adya S., Daniels-McQueen S., Miller J. K., Hershey J. W., Grifo J. A., Merrick W. C., Thach R. E. Role of mRNA competition in regulating translation: further characterization of mRNA discriminatory initiation factors. Proc Natl Acad Sci U S A. 1983 Feb;80(3):663–667. doi: 10.1073/pnas.80.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadofsky M., Alwine J. C. Sequences on the 3' side of hexanucleotide AAUAAA affect efficiency of cleavage at the polyadenylation site. Mol Cell Biol. 1984 Aug;4(8):1460–1468. doi: 10.1128/mcb.4.8.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schon E. A., Cleary M. L., Haynes J. R., Lingrel J. B. Structure and evolution of goat gamma-, beta C- and beta A-globin genes: three developmentally regulated genes contain inserted elements. Cell. 1981 Dec;27(2 Pt 1):359–369. doi: 10.1016/0092-8674(81)90419-0. [DOI] [PubMed] [Google Scholar]
- Smith M. M., Andrésson O. S. DNA sequences of yeast H3 and H4 histone genes from two non-allelic gene sets encode identical H3 and H4 proteins. J Mol Biol. 1983 Sep 25;169(3):663–690. doi: 10.1016/s0022-2836(83)80164-8. [DOI] [PubMed] [Google Scholar]
- Smith T. F., Waterman M. S., Sadler J. R. Statistical characterization of nucleic acid sequence functional domains. Nucleic Acids Res. 1983 Apr 11;11(7):2205–2220. doi: 10.1093/nar/11.7.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. An interactive graphics program for comparing and aligning nucleic acid and amino acid sequences. Nucleic Acids Res. 1982 May 11;10(9):2951–2961. doi: 10.1093/nar/10.9.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata T., Sasaki K., Iwabuchi M. The structural organization and DNA sequence of a wheat histone H4 gene. Nucleic Acids Res. 1983 Sep 10;11(17):5865–5875. doi: 10.1093/nar/11.17.5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell B. C., Tolstoshev P., Crystal R. G. Secondary structures for splice junctions in eukaryotic and viral messenger RNA precursors. Nucleic Acids Res. 1980 Aug 25;8(16):3659–3672. doi: 10.1093/nar/8.16.3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn J. C., Sperbeck S. J., Ramsey W. J., Lawrence C. B. A universal model for the secondary structure of 5.8S ribosomal RNA molecules, their contact sites with 28S ribosomal RNAs, and their prokaryotic equivalent. Nucleic Acids Res. 1984 Oct 11;12(19):7479–7502. doi: 10.1093/nar/12.19.7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Magrum L. J., Gupta R., Siegel R. B., Stahl D. A., Kop J., Crawford N., Brosius J., Gutell R., Hogan J. J. Secondary structure model for bacterial 16S ribosomal RNA: phylogenetic, enzymatic and chemical evidence. Nucleic Acids Res. 1980 May 24;8(10):2275–2293. doi: 10.1093/nar/8.10.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R., Roeder R. G., Heintz N. The primary structure and expression of four cloned human histone genes. Nucleic Acids Res. 1983 Nov 11;11(21):7409–7425. doi: 10.1093/nar/11.21.7409. [DOI] [PMC free article] [PubMed] [Google Scholar]



