ABSTRACT
We previously reported widespread differential expression of long non-protein-coding RNAs (ncRNAs) in response to virus infection. Here, we expanded the study through small RNA transcriptome sequencing analysis of the host response to both severe acute respiratory syndrome coronavirus (SARS-CoV) and influenza virus infections across four founder mouse strains of the Collaborative Cross, a recombinant inbred mouse resource for mapping complex traits. We observed differential expression of over 200 small RNAs of diverse classes during infection. A majority of identified microRNAs (miRNAs) showed divergent changes in expression across mouse strains with respect to SARS-CoV and influenza virus infections and responded differently to a highly pathogenic reconstructed 1918 virus compared to a minimally pathogenic seasonal influenza virus isolate. Novel insights into miRNA expression changes, including the association with pathogenic outcomes and large differences between in vivo and in vitro experimental systems, were further elucidated by a survey of selected miRNAs across diverse virus infections. The small RNAs identified also included many non-miRNA small RNAs, such as small nucleolar RNAs (snoRNAs), in addition to nonannotated small RNAs. An integrative sequencing analysis of both small RNAs and long transcripts from the same samples showed that the results revealing differential expression of miRNAs during infection were largely due to transcriptional regulation and that the predicted miRNA-mRNA network could modulate global host responses to virus infection in a combinatorial fashion. These findings represent the first integrated sequencing analysis of the response of host small RNAs to virus infection and show that small RNAs are an integrated component of complex networks involved in regulating the host response to infection.
IMPORTANCE
Most studies examining the host transcriptional response to infection focus only on protein-coding genes. However, mammalian genomes transcribe many short and long non-protein-coding RNAs (ncRNAs). With the advent of deep-sequencing technologies, systematic transcriptome analysis of the host response, including analysis of ncRNAs of different sizes, is now possible. Using this approach, we recently discovered widespread differential expression of host long (>200 nucleotide [nt]) ncRNAs in response to virus infection. Here, the samples described in the previous report were again used, but we sequenced another fraction of the transcriptome to study very short (about 20 to 30 nt) ncRNAs. We demonstrated that virus infection also altered expression of many short ncRNAs of diverse classes. Putting the results of the two studies together, we show that small RNAs may also play an important role in regulating the host response to virus infection.
Introduction
Mammalian genomes transcribe many short and long non-protein-coding RNAs (ncRNAs), but whether these RNAs play a role in the host response to virus infection remains an enigma. It is known that some small RNAs, such as microRNAs (miRNAs) (1), are involved in virus-host interactions. For example, in vitro studies have shown that the liver-specific miR-122 is required for hepatitis C virus (HCV) RNA replication (2). Distinct expression profiles of cellular miRNAs enabled researchers to differentiate infection by the lethal 1918 pandemic influenza virus from nonlethal seasonal influenza virus A/Texas/36/91 infection (3). It was also reported that HIV-1 virus is able to suppress expression of the polycistronic miRNA cluster miR-17/92 to enable efficient viral replication (4). However, changes in expression of other small RNAs during virus infection have not been systematically studied.
Using next-generation deep-sequencing technology, we recently discovered widespread differential expression of host long ncRNAs in response to virus infection (5), but the experimental protocol used was not designed to capture small RNAs (6). In this study, we used deep-sequencing technology to perform a complementary small RNA transcriptome analysis of the same severe acute respiratory syndrome coronavirus (SARS-CoV)-infected lung samples collected from four mouse strains as previously reported. In addition, lung samples collected from the same four influenza virus-infected mouse strains were included in the new analysis. Our results show that many known miRNAs responded differently to the two virus infections and that many of them were also differentially regulated during lethal influenza virus infection, as shown in a previous study (3). We also discovered many non-miRNA small RNAs and unannotated small RNAs that were differentially expressed during infection. The integration of transcriptome sequencing analysis of long transcripts and small RNAs showed the intricate interactions of short and long RNAs during virus infection. The changes in miRNAs positively correlated with the changes in long transcripts cotranscribing from the same locus, indicating that the miRNAs were transcriptionally regulated during virus infection. We predicted that differentially expressed miRNAs could target the majority of differentially expressed long transcripts during virus infection.
RESULTS
Deep sequencing of small RNAs in lung samples from mice infected with SARS-CoV or influenza virus.
To systematically investigate the regulation of small RNAs during viral infection, we performed two batches of small RNA sequencing analysis using lung samples from virus-infected mice. To follow up our previous study on long ncRNAs, we first sequenced the small RNAs of the same lung samples collected from four mouse strains infected with SARS-CoV: 129S1/SvImJ (129/S1), WSB/EiJ (WSB), PWK/PhJ (PWK), and CAST/EiJ (CAST) (5). For comparisons, we performed small RNA sequencing analysis using another set of lung samples collected from additional mice of the same four mouse strains infected with influenza virus. These mouse strains were selected because of their differential range of susceptibility phenotypes as revealed following infection with SARS-CoV or influenza virus (5) and because of the opportunity they presented of pursuing downstream quantitative trait locus (QTL) mapping of regulation and function in the Collaborative Cross, a recombinant inbred mouse resource for mapping complex traits (7). Directional cDNA libraries were constructed using standard Illumina protocols for small RNA analysis, which targeted small RNAs of around 19 to 22 nucleotides (nt).
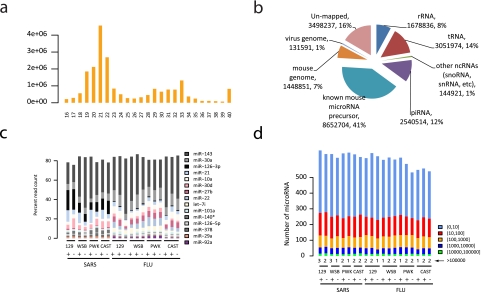
In total, we obtained ~369 million adaptor-trimmed reads ranging from 16 to 40 nt in length from 20 mouse lung samples (over 18 million reads per sample on average). As expected, the reads exhibited a large peak in abundance in the length range of 19 to 22 nt (Fig. 1a; see Fig. S1 in the supplemental material). For each sample, the majority (88% on average) of total short reads were mapped into different classes of abundant small RNAs. miRNAs composed the most abundant class, accounting for about 45% of total reads on average (Fig. 1b; see also Table S1 in the supplemental material). We observed a wide range of counts of reads (from 1 to an average of 2,577,006 for the most abundant miRNA in a given sample) mapped to individual miRNAs in both normal and virus-infected samples, indicating that techniques with dynamic ranges as large as 106 are required for comprehensive profiling of miRNA expression. On average, about 616 of 1,055 annotated mature mouse miRNAs were detected with at least one read in a given sample. The detected miRNAs showed very distinctive expression patterns in both normal and virus-infected mouse lung samples. In all samples, the top 20 most abundant miRNAs by read count accounted for about 90% of miRNA reads and the top 10 miRNAs for 80% of miRNA reads. The most abundant miRNAs and the overall abundance distribution of all detected miRNAs are shown in Fig. 1c and d.
FIG 1 .
Overview of deep-sequencing analysis of the small RNA transcriptome in lungs from virus-infected mice. (a) Length distribution of small RNA short reads in an arbitrarily chosen sample. Reads were adaptor trimmed. Reads < 16 nt were discarded, and reads > 40 nt were shortened to 40 nt by clipping from the 3 ′ end. (b) Global classification of small RNA transcriptional activity of the sample described in the panel a legend. Short reads were sequentially mapped to sequences of ribosomal RNAs (rRNA), transfer RNAs (tRNA), small cytoplasmic RNAs, small nuclear RNAs, small nucleolar RNAs (other ncRNAs), piwi-associated small RNAs (piRNA), microRNA precursor sequences, a mouse reference genome, and the SARS-CoV genome. (c) Relative abundances of the most abundant mature miRNAs in lung samples from virus-infected mice. The 10 most abundant mature miRNAs in at least 4 samples across all 20 samples are shown. (d) Distribution of detected mature miRNAs grouped by normalized read count (the number of the reads per million mapped reads) across all 20 samples. Numbers in parentheses represent ranges. SARS, SARS-CoV; FLU, influenza virus.
Differential expression of known host miRNAs during virus infection.
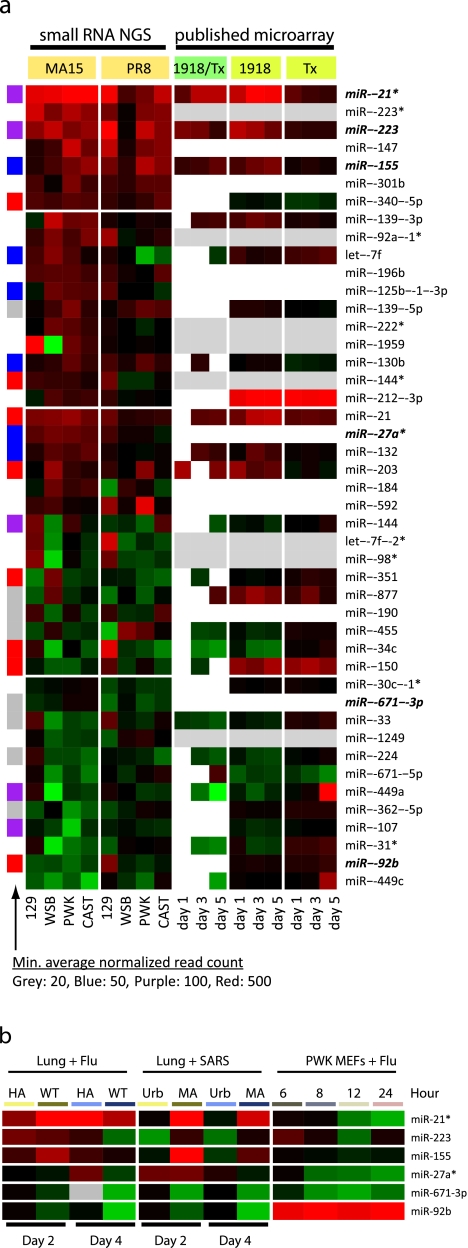
We first studied annotated miRNAs, as host miRNAs represented the most abundant class of small RNAs observed and the host miRNA response to SARS-CoV infection has not been reported previously. We identified 45 mature miRNAs that were differentially expressed during SARS-CoV or influenza virus infection. Thirty were consistently upregulated or consistently downregulated by more than 1.5-fold across three or more mouse strains during SARS-CoV infection, and 10 were consistently upregulated or consistently downregulated by more than 1.5-fold across three or more mouse strains during influenza virus infection (Fig. 2a). In addition, 24 of these miRNAs were consistently upregulated or consistently downregulated by more than 1.5-fold in two or more mouse strains during both SARS-CoV and influenza virus infection. Additional quantitative PCR (qPCR) analyses of replicate samples with a large subset of differentially expressed miRNAs showed good concordance between replicates and statistical congruence (see Fig. S2 in the supplemental material). Interestingly, there were 7 miRNAs that were upregulated in at least three mouse strains during both SARS-CoV and influenza virus infection, suggesting that a subset of miRNAs such as these commonly responded to different virus infections. However, 84% (38/45) of the differentially expressed miRNAs showed different expression profiles across four mouse strains in SARS-CoV and influenza virus infections (Fig. 2a), suggesting that expression of most miRNAs was likely both host and virus dependent. Although the physiological relevance of small changes in miRNA expression remains to be investigated, other studies have indicated that 1.5-fold changes in expression can have significant biological impact (8). In addition, because we profiled whole lung samples, some of the observed changes in expression could have been due to the infiltration of immune cells into the infected lung.
FIG 2 .
miRNAs differentially expressed during virus infection. (a) Overview of 45 miRNAs differentially expressed in mouse lung samples during SARS-CoV (MA15) or influenza virus (PR8) infection. Colors on the heat map indicate the log2 ratios of expression (representing normalized read counts) in virus-infected samples to expression in matched mock-infected samples. Red, upregulation; green, downregulation. The data in the eight columns under “small RNA NGS” represent log2 ratios of expression in virus-infected samples to expression in mock-infected samples as determined on the basis of small RNA transcriptome sequencing analysis. NGS, next-generation sequencing. The nine columns under “published microarray” represent log2 ratios determined on the basis of the microarray measurements of the corresponding miRNAs in lung samples from a previous study (3) in which mice were infected with the fully reconstructed 1918 pandemic (1918) or a nonlethal seasonal (Tx) influenza virus. The three columns under “1918/Tx” show the results of direct comparisons of miRNA expression in 1918-infected samples to expression in Tx-infected samples at 1, 3, and 5 days after infection; note that only changes of at least 1.5-fold are indicated. The log2 ratios of expression in infected samples to expression in mock-infected samples are shown separately under “1918” and “Tx,” where rows in grey indicate that the corresponding miRNAs were not probed by the miRNA microarray and rows in white indicate undetected miRNAs. The left sidebar shows the relative abundances of the corresponding miRNAs represented as the minimum average normalized read counts across all samples. (b) Patterns of changes in expression of selected miRNAs across a panel of virus infection samples measured by qPCR. Six miRNAs from panel a (indicated in boldface and italics) were selected; two, namely, miR-155 in macrophages (10) and miR-223 in neutrophils (11), are known to be related to immune cells. The other four miRNAs came from a small-scale screening performed using qPCR with a subset of randomly selected differentially expressed miRNAs in PWK/PhJ mouse embryonic fibroblasts (MEFs) infected with influenza virus (data not shown). Also, miR-27a* and miR-671-3p were not measured by previous microarray (see panel a). Colors on the heat map indicate the log2 ratios (the differences between medians of normalized Ct values of replicate samples) of expression in virus-infected samples to expression in matched mock-infected samples. Red, upregulation; green, downregulation. The “Lung + Flu” data contrast miRNA expression changes in lung samples from mice infected with highly pathogenic influenza virus (WT, VN1203) and with minimally pathogenic influenza virus (HA, VN1203 with a mutation in HA protein) at two time points: days 2 and 4 after infections. Similarly, the “Lung + SARS” data contrast miRNA expression changes in lung samples from mice infected with highly pathogenic SARS-CoV (MA, MA15) and minimally pathogenic SARS-CoV (Urb, Urbani). The “PWK MEFs + Flu” data represent temporal miRNA expression changes in samples from cultured PWK MEFs infected with the mouse-adapted A/PR/8/34 influenza virus across four time points after infection. A more detailed representation of individual replicate experiments is shown in Fig. S3. WT, wild type.
To investigate whether the differential expression patterns of the miRNAs were related to viral pathogenesis, we first compared these findings with those from our previous microarray study of host miRNAs in mouse lungs infected with the fully reconstructed 1918 pandemic influenza virus (r1918) (3). We found that 17 of the 45 differentially expressed mature miRNAs identified here were not on the arrays (8 miRNAs) or not detected (9 miRNAs) by the previous microarray technology (Fig. 2a), showing the benefits of deep-sequencing technology, in which the measurement of RNAs is independent of prior knowledge of transcript annotations and is achieved with a dynamic range of up to 106 (9). Of those 28 miRNAs profiled on the microarray in the previous study, 75% (21/28) showed at least a 1.5-fold difference between the response to the highly pathogenic r1918 virus infection and the response to minimally pathogenic A/Texas/36/91 virus infection for at least one of three time points studied. These findings suggest that many of these miRNAs may be commonly involved in viral pathogenesis.
Next, we surveyed the changes in expression of 6 differentially expressed miRNAs during virus infection across a set of diverse conditions (Fig. 2b and Materials and Methods). Even with only 6 miRNAs surveyed and relatively large variations among replicate samples, we were able to observe distinctive miRNA expression changes. These changes were detected under conditions of infection with different viruses and were seen with samples generated in vivo versus in vitro (see Fig. S3 in the supplemental material). First, we observed that 5 of 6 miRNAs had very different expression patterns in the highly pathogenic SARS-CoVMA15 versus the minimally pathogenic Urbani strain (Fig. 2b; Fig. S3). Different expression patterns were also seen with 3 of 6 miRNAs in comparisons of the highly pathogenic VN1203 influenza virus to the attenuated VN1203 hemagglutinin (HA) mutant strain. This argues that some of the identified miRNAs may be associated with pathogenic outcomes. Second, many factors may have contributed to the different expression patterns; however, it appears unlikely that the observed miRNA changes were due to pure immune cell infiltration, as 4 miRNAs showed strong evidence of differential expression in cultured fibroblasts during infection. Third, and not surprisingly, several miRNAs showed expression patterns that differed between the in vivo and in vitro models. The differences could be attributed to cell type specificity or to different environmental and/or growth conditions, as two miRNAs that did not show obvious differential expression patterns in cultured fibroblasts, miR-155 in macrophages (10) and miR-223 in neutrophils (11), are known to be related to different immune cell types. In summary, these results suggest that miRNAs are likely involved in viral pathogenesis and that the choice of experimental systems used for miRNA studies should be considered a critical and informing component in the study design.
Differential expression of non-miRNA small RNAs and novel small RNAs during virus infection.
As shown in Table S1 in the supplemental material, there were still many (about 17 million in total) “leftover” reads which did not map to annotated or predicted abundant small RNAs but were aligned to the mouse reference genome, suggesting that there might be some novel host small RNAs relevant to virus infection. Therefore, starting by mapping all short reads directly to the mouse reference genome, we carried out a genome-wide search of novel small RNAs (see Materials and Methods).
In total, of 4,473,273 start positions in the genome with at least one uniquely mapped read, we found that about 5% (233,236) gave at least 4 reads of the same length in a sample, resulting in 16,054 nonredundant candidate loci for putative small RNAs. About 1.7% (276/16,054) of the candidate loci (median length, 39 nt) were differentially expressed during SARS-CoV and/or influenza virus infection (see Table S2 and Fig. S4a in the supplemental material); 46 of those candidate loci overlapped with annotated miRNA precursors (miRBase version 16). We next used a conservative filtering strategy to remove 60 of 276 candidate loci from further analysis because of the possibility of misalignment of short reads originating from other highly expressed loci (see Materials and Methods). We considered the remaining 216 to represent putative small RNA loci, as we reasoned that their showing consistent responses to virus infection suggested that they were more likely to be biologically functional rather than to represent random noise. To validate the analytical approach used to identify these differentially expressed putative small RNA loci, we compared the expression ratios calculated for infection versus matched mock infection that we used to identify these putative small RNA loci to the corresponding expression ratios that we calculated for the overlapping miRNA precursors on the basis of the reads directly mapped to annotated precursor sequences. We obtained very good agreement (analysis of variance [ANOVA] F test, P = 9.4e-101; Pearson correlation coefficient, r = 0.85) between the two estimations. This result confirmed that the analytical approach used here performed well, since we performed an unbiased genome-wide search for all small RNAs without considering any known annotations instead of looking only at annotated miRNA sequences.
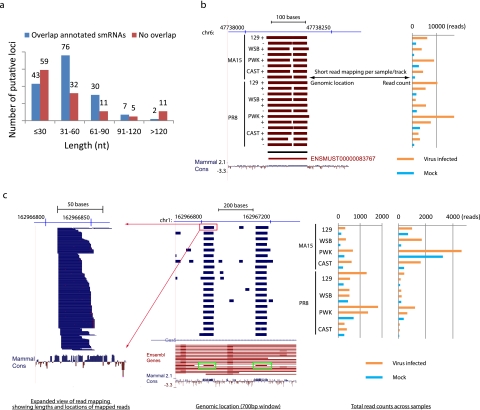
To investigate the genomic origins of the putative small RNA loci, we compared the loci to those included in a previously compiled nonredundant set of mouse genome annotations (5). We found that 63% (135/216) overlapped (sense or antisense) with annotated loci, including coding RNAs, ncRNAs, and those with unannotated genomic regions (Table 1), suggesting that the majority of small RNAs originated from genomic regions encoding long transcripts. Also, the result suggested that long transcripts from some of these regions have not been well annotated or that some regions formed independent transcriptional units exclusive to small RNAs, as 37% of putative small RNA loci did not overlap annotated long transcript-transcribing regions. To understand the classes of putative small RNAs produced from the loci, we then compared them to annotated or predicted abundant small RNAs. Interestingly, many putative small RNA loci overlapped non-miRNA small RNAs such as small nucleolar RNAs (snoRNAs) and piwi-associated small RNAs (piRNAs) (Table 1). snoRNAs guide the chemical modification of ribosomal RNAs and other ncRNAs and are essential for major biological processes, including protein translation and mRNA splicing (12). piRNAs represent a class of small RNAs that are 24 to 32 nt long and have been linked to transcriptional gene silencing in germ line cells (13). piRNAs have also been identified in various somatic tissues (14, 15). Also, we observed length distributions of small RNA loci overlapping annotated small RNA loci that were similar to those of small RNA loci with no overlap (Fig. 3a), indicating that our identified loci were indeed producing small RNAs. Examples of detailed read mapping of loci overlapped with annotated small RNAs are shown in Fig. 3b and c; similar results determined using read mapping of putative novel small RNA loci are shown in Fig. 4. We observed that 32 putative small RNA loci overlapped with annotated snoRNAs on the basis of the updated Ensembl annotation and that almost all (29 of 32) of the loci also overlapped with annotated coding or ncRNA loci, suggesting that, in addition to miRNAs, snoRNAs might represent another large class of small RNAs differentially expressed in response to virus infection and originating from long transcript-encoding loci.
TABLE 1 .
Genomic overlap of 216 differentially expressed putative small RNA loci with existing annotationsa
| Overlap status of putative loci |
Annotated locus category |
Total no. of putative small RNA loci |
No. of putative small RNA loci in indicated category of overlap with annotated small RNAs |
||||
|---|---|---|---|---|---|---|---|
| miRNA | snoRNA | piRNA | RNA repeat | Total (%) | |||
| Overlapped with annotative loci | Protein coding | 95 | 25 | 23 | 4 | 7 | 54 (57) |
| Antisense to protein coding |
22 | 3 | 0 | 2 | 2 | 5 (23) | |
| ncRNA | 19 | 2 | 6 | 3 | 6 | 14 (74) | |
| Antisense to ncRNA |
10 | 0 | 3 | 2 | 5 | 8 (80) | |
| Genomic region | 6 | 2 | 2 | 0 | 1 | 5 (83) | |
| Antisense to genomic region |
3 | 0 | 0 | 0 | 0 | 0 (0) | |
| Total sense | 118 | 29 | 29 | 7 | 14 | 71 (60) | |
| Total antisense | 35 | 3 | 3 | 4 | 7 | 13 (37) | |
| Total | 135 | 31 | 29 | 9 | 16 | 75 (56) | |
| No overlap | 81 | 15 | 3 | 16 | 37 | 53 (65) | |
Annotated loci data represent the set of nonredundant annotations of mouse protein-coding loci and ncRNAs compiled in reference (5), combined with the set of unannotated genomic regions identified in the same study. Annotated small RNAs are as follows: miRNA, miRNA precursors; snoRNA, small nucleolar RNAs; piRNA, piwi-associated small RNAs. RNA repeat data were downloaded from the UCSC genome browser (http://genome.ucsc.edu/). Numbers of putative small RNA loci represent the numbers of putative small RNA loci overlapped with different classes of annotated loci in terms of genomic coordinates; sense and antisense loci are counted separately. For example, 95 putative small RNA loci overlapped with annotated protein-coding loci on the sense strand (column 3, row 1). Total sense data represent the total numbers of putative small RNA loci that overlapped with annotated loci on the sense strand. Total antisense data represent the total numbers of putative small RNA loci overlapped with annotated loci on the antisense strand. The putative small RNA loci that overlapped with annotated loci were further classified by checking whether they also overlapped with different annotated small loci in terms of genomic coordinates. For example, of 95 small RNA loci that overlapped with annotated loci, 25 also overlapped with annotated miRNAs (column 4, row 1).
FIG 3 .
Length distributions of identified putative small RNA loci and two examples of non-miRNA small RNAs differentially expressed during SARS-CoV and influenza virus infection. (a) The length distribution of the identified putative small RNA loci. Data for “overlap annotated smRNAs” represent loci overlapping annotated or predicted small RNAs, including miRNAs, snoRNAs, piRNAs, and RNA repeats. “No overlap” data represent loci that do not overlap any small RNAs. (b) UCSC genome browser (http://genome.ucsc.edu) display of the locations of reads mapped to a 350-bp region of chromosome 6 (chr6). On the left, each track represents short reads collected from small RNA transcriptome sequencing analysis of a single mouse lung sample. Because there were too many reads to be displayed individually, total reads were condensed (dense mode) to display the genomic regions with mapped reads. The red color indicates that the reads were mapped to the negative strand. The black horizontal bar under the sample tracks shows the location of the predicted small RNA locus. The red horizontal bar represents a Y RNA, a small noncoding RNA component of the Ro ribonucleoprotein particle, as annotated by the use of Ensembl software. The raw total read counts of the corresponding small RNA locus across all 20 samples are represented on the right in the same order as shown on the left. Read counts indicate that expression of the small RNA was upregulated during virus infection. Mammal cons, UCSC track (http://genome.ucsc.edu) showing the placental mammal basewise conservation by PhyloP. (c) An overview similar to that shown in panel b for a 700-bp region of chromosome 1 covering two predicted small RNA loci. Each track represents a condensation of the short reads from a single sample (middle panel). The blue color indicates the reads were mapped to the positive strand. Green boxes show two predicted small RNA loci overlapped with two snoRNAs (the red horizontal bar inside each green box) annotated by the use of Ensembl software (version 61) and located in the introns of Gas5, a known long ncRNA located on the positive strand. Different isoforms of Gas5 are shown on the track labeled Ensembl Genes. An expanded view of the read mapping highlighted by the red box is displayed on the left. Distributions of the lengths and mapped locations of short reads from the single sample are shown. Raw read counts of two corresponding small RNA loci across all 20 samples are shown on the right (in the same order as shown in the middle panel). Both small RNA loci were upregulated during SARS-CoV and influenza virus infection.
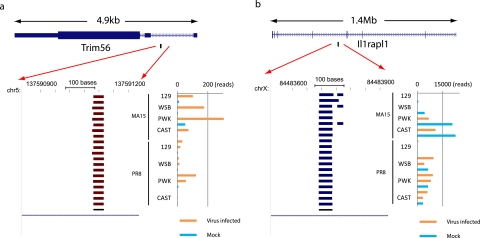
FIG 4 .
Examples of two nonannotated small RNAs. (a) An overview of a 400-bp region of chromosome 5 similar to that shown in Fig. 3b. The predicted small RNA is located in an intron of the Trim56 protein-coding gene (top). Both Trim56 and the predicted small RNA are on the negative strand. (b) An overview of a 400-bp region of chromosome X similar to that shown in panel a. The predicted small RNA is antisense to an intronic region of the Il1rapl1 protein-coding gene (top). Ll1rapl1 is on the negative strand, and the predicted small RNA is on the positive strand.
Coupling changes in host short and long RNA expression through parallel small RNA and whole-transcriptome sequencing.
To better understand the host response to virus infection at the system level, we performed an integrative analysis of small RNA transcriptome sequencing with our previously reported whole-transcriptome sequencing method using the same set of SARS-CoV-infected samples (5). To the best of our knowledge, studies have profiled host miRNA expression changes during virus infection (1, 3, 16, 17), but regulation of such miRNA expression changes has not been systematically investigated. miRNA biogenesis can be regulated at different steps (18), including at least three steps of RNA processing: (i) the transcription of miRNA primary transcripts (pri-miRNA) of several thousand base pairs, (ii) the processing of the long pri-miRNAs to miRNA precursors (70 to 100 nt), and (iii) the processing of miRNA precursors into mature miRNAs (about 20 to 22 nt). As we had previously generated whole-transcriptome data using the set of lung samples infected with SARS-CoV with which we quantified long transcripts (especially those >200 nt in length) (5), we reasoned that the long pri-miRNAs were also quantified.
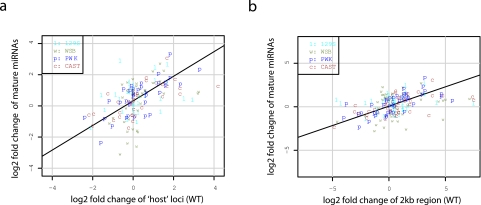
Though the transcript structures of pri-miRNAs have not been completely annotated, it is known that many mature miRNAs originate from annotated long (protein-coding or noncoding) transcripts. We reasoned that those annotated loci overlapping miRNA precursors would be a good proxy for pri-miRNAs, as it has been shown that many mammalian miRNAs are transcriptionally linked to expression of the genes from which they originate (19, 20). We then compared the changes in expression of identified mature miRNAs obtained during SARS-CoV infection to the corresponding changes in expression of overlapping loci obtained from the previous whole-transcriptome sequencing analysis (5). Interestingly, we found that the changes in expression of mature miRNAs and the overlapping loci significantly positively correlated (ANOVA P = 7e-11; Pearson correlation coefficient = 0.51) (Fig. 5a). Further, we took the 2-kb genomic regions surrounding each annotated miRNA precursor (1 kb upstream and 1 kb downstream, excluding the precursor region) as another approximation of pri-miRNAs and observed a similarly high positive correlation with changes in expression (ANOVA P = 1.3e-08; Pearson correlation coefficient = 0.42) (Fig. 5b). These results show for the first time that the transcriptional regulation of pri-miRNAs could be largely responsible for the differential regulation of mature miRNAs during virus infection.
FIG 5 .
Correlations of mature miRNA expression changes with the corresponding pri-miRNA estimates. (a) Scatterplot of the log2 fold changes of mature miRNA expression measured by small RNA transcriptome sequencing analysis (y axis) and the log2 fold changes in expression of the annotated loci overlapped with the corresponding miRNAs measured by whole-transcriptome sequencing analysis using the same samples (x axis). In total, 36 mature miRNA loci overlapped annotated loci and were included in the plot. (b) Data are presented as described for panel a, with the y axis data showing the log2 fold changes in expression of the 2-kb genomic regions surrounding the corresponding miRNA precursors measured by whole-transcriptome sequencing analysis of the same samples. In total, 42 mature miRNAs were included in the plot, as 3 of 45 genomic regions surrounding differentially expressed miRNAs were not subjected to differential expression analysis due to the lack of a sufficient number of reads for quantification in the whole-transcriptome sequencing data.
To investigate the potential functional impact of differential expression of mature miRNAs, we combined the data showing the mRNA expression changes measured by the whole-transcriptome sequencing analysis with that from the miRNA target predictions. We found that the majority (78%) of mRNA loci were predicted as targets of at least 1 of the 45 differentially expressed mature miRNAs identified above (see Fig. S5a in the supplemental material) and that the differentially expressed mRNA loci listed were significantly (P < 2.2e-16 [chi-square test]) enriched with predicted targets of those differentially expressed miRNAs. The ratio of miRNA targets versus nontargets for differentially expressed mRNA loci was ~3.5 compared to ~1.6 for those loci that were not differentially expressed during infection, representing an enrichment of about 2.2-fold. These results strongly suggest that collectively differentially expressed miRNAs modulate the global host responses to virus infection. Notably, about 80% of those differentially expressed targets were predicted targets of two or more identified mature miRNAs, indicating that a single target could be regulated in vivo by multiple miRNAs at the same time during virus infection. Importantly, 82% of the targets predicted to be regulated by multiple miRNAs were targeted by both upregulated and downregulated miRNAs at the same time, showing that additional studies are necessary to elucidate which specific miRNAs, if any, play a regulatory role in the changes of individual targets in vivo (Fig. S5b). Functional analysis of predicted miRNA targets also indicated that many important aspects of the host response (such as innate immunity and cytokine production) were likely modulated by miRNAs (see Table S3 in the supplemental material). Figure S6 shows a subnetwork of differentially expressed miRNAs and their predicted targets in several antiviral pathways.
DISCUSSION
Studies of the host small RNA response to virus infection have been focused on miRNAs (1, 3, 16, 17), but there is growing evidence that the mammalian transcriptome comprises a diversity of small RNAs in addition to miRNAs. For example, human and protozoan snoRNAs have been reported to be processed into miRNA-like RNAs (21, 22), and the members of a class of novel small RNAs have been reported to be derived from many snoRNAs (23). Abundant small RNAs are also derived from tRNAs in a Dicer-dependent manner (9), and human tRNA-derived small RNAs appear to be involved in the global control of small RNA silencing (24). To our knowledge, the present study was the first to use comprehensive deep-sequencing technology to characterize virus infection-induced changes in expression of miRNAs and other classes of small RNAs, including many nonannotated small RNAs. As the biological functions of these diverse types of small RNAs are largely unknown, virus infection models also offer a unique platform for studying the regulation and biology of these diverse small RNAs. For example, the influenza virus NS1 protein inhibits host pre-mRNA splicing through its interaction with snoRNAs (25–27). Also, it has been shown that SARS-CoV and other nidoviruses encode a protein with sequence similarity to XendoU, a eukaryotic endoribonuclease that is involved in cellular snoRNA processing (28, 29). Though the experimental protocol used here is not optimized for the capture of RNA products with 2′,3′-cyclic phosphates as specifically generated by XendoU (30), we did observe that the fold changes of identified small RNAs tended to be larger in SARS-CoV-infected samples than in influenza virus-infected samples (see Fig. S4b and c in the supplemental material), suggesting that further investigation of the impact of viral proteins on host small RNA processing could represent another approach for the study of virus-host interactions.
It is known that the characteristics of their secondary structures could be indicative of the types of putative small RNAs such as miRNAs and therefore of their general functional mechanisms. We therefore investigated whether the putative small RNAs identified here tended to have stable secondary RNA structures (see Materials and Methods). In total, 89 (41%) putative small RNA loci were predicted by secondary structure analysis; 20 of those loci did not overlap with those of the annotated small RNAs (see Table S4 in the supplemental material). Interestingly, those that overlapped with annotated small RNAs were significantly (chi-square test; P = 9.26e-06) more likely to be predicted by RNA secondary structure analysis (54% [69/128]) compared to those which did not overlap any annotated small RNAs (23% [20/88]), suggesting that the existing annotation of small RNAs might be biased toward those with stable local secondary structures. For example, out of 46 putative small RNA loci that overlapped with known miRNA precursors, 76% (one prediction only) to 87% (all four predictions combined) were predicted with secondary structures, indicating that our automated strategy for structure prediction performed well in terms of finding local RNA structures such as hairpins. For those putative loci that overlapped with other classes of small RNAs, however, the percentage predicted with secondary structures was much lower, ranging from 40.6% to 59% for snoRNAs to 24% for piRNAs. A closer look at those loci overlapping snoRNAs showed that 90% (9/10) of the loci that overlapped with H/ACA box family snoRNAs but only about 45% (10/22) of the loci that overlapped with C/D box family snoRNAs were predicted with secondary structures. Since snoRNAs are broadly classified into two families, corresponding to a C/D box with one short (~5 nt) hairpin structure and an H/ACA box with at least two large hairpin structures (31), these results suggest that investigations based solely on a typical RNA secondary structure-based approach might miss many small RNAs that do not form stable local RNA structures per se and that approaches utilizing transcriptome sequencing might be able to identify broader classes of small RNAs.
Compared to the current knowledge regarding noncanonical small RNAs, the functional mechanism of miRNAs is relatively better understood. However, the regulation of miRNA expression changes during virus infection has not been systematically investigated. Through integrative analysis of parallel sequencing of both small RNAs and long transcripts from the same samples, we were able to infer the regulatory relationships both upstream and downstream of miRNAs with respect to the differences in expression. Not only did we show that the differential expression of miRNAs was likely due to transcriptional regulation, but our data also indicate that those miRNAs may collectively play a role in modulating the global host response. Since it has been shown that mechanistically mammalian miRNAs mainly act to decrease target mRNA levels, thus leading to reduced protein output (32), these results argue that a better understanding of the small RNAs, including miRNAs, would be required for complete knowledge of the regulation of host responses to virus infections. As evidenced here and in other studies (33), multiple miRNAs can simultaneously regulate the same targets, arguing that experimental design and computational strategies of greater complexity, such as we proposed previously (17), are necessary to elucidate the combinatorial nature of miRNA-mRNA regulatory networks in vivo.
Our integrated sequencing analysis also offers several additional benefits, such as the investigation of the functions of some long ncRNAs. We previously reported that many long ncRNAs of unknown function respond to virus infection (5), and it is known that long ncRNAs can serve as precursors for small RNAs (34). In this study, we identified a number of small RNAs that overlapped with long ncRNAs (Table 1). For example, miR-155, one of the upregulated miRNAs identified here (Fig. 2a), is produced from an exon of the long ncRNA BIC, which was also observed to be upregulated by whole-transcriptome sequencing analysis (5). Both miR-155 and BIC were induced in primary murine macrophages stimulated by beta interferon (INF-β) (10). Loss-of-function studies showed that mice lacking a functional BIC/miR-155 gene exhibited a defective immune response in vivo and were deficient in cytokine production (35, 36). Also, Fig. 3 shows two small RNAs overlapping snoRNAs encoded in the introns of Gas5, another long ncRNA that was previously identified as an apoptosis regulator (37). It is unclear whether snoRNAs from Gas5 play any functional roles in response to virus infection, but it has been hypothesized that the functional effect of Gas5 on T-cell growth may be mediated through its intronic snoRNAs (38). These examples suggest that a detailed investigation of the connections between long ncRNAs and the coexpressed small RNAs might facilitate a better understanding of the regulation of host responses. Moreover, even though the study of viral small RNAs is beyond the scope of this report, it is worth noting that we also observed many small sequence reads from both viral genomes (see Table S1 in the supplemental material). Though isolation of small RNAs from SARS-CoV infections has not been reported, it was shown recently that influenza virus expresses many small RNAs (39, 40). In summary, our results convincingly show that, in the future, integrated sequencing analysis of different fractions of the complex transcriptome should facilitate a full understanding of virus-host interactions and viral pathogenesis.
MATERIALS AND METHODS
Mouse lines and virus infection.
The small RNA transcriptome deep-sequencing analysis was performed on lung samples from our previously published study (5). Briefly, we infected four of the eight founder mouse strains used in generating the Collaborative Cross, a recombinant inbred mouse resource for mapping complex traits (41). These strains included 129S1/SvImJ (129/S1), WSB/EiJ (WSB), PWK/PhJ (PWK), and CAST/EiJ (CAST) mice. Ten-week-old mice were intranasally infected with phosphate-buffered saline (PBS) alone or with 1 × 105 PFU of mouse-adapted severe acute respiratory syndrome coronavirus (SARS-CoV; rMA15), or 500 PFU of influenza A virus strain A/Pr/8/34 (H1N1; PR8).
To match the previous whole-transcriptome analysis, we performed small RNA transcriptome sequencing analysis on the same eight samples from mice with SARS-CoV infections, including one SARS-CoV rMA15-infected mouse and one matched mock-infected mouse from each of the four strains at 2 days postinfection (dpi). In addition, we sequenced the small RNA transcriptome for 12 samples obtained from influenza virus-infected mice, including two PR8-infected mice and one matched mock-infected mouse from each of the four strains at 2 dpi. The additional replicate samples from matched infections were used for evaluation by quantitative reverse transcription-PCR (qRT-PCR).
Additional virus infections for qPCR assay.
Twenty-week-old C57BL/6 mice were infected by intranasal instillation of influenza virus VN1203 (1 × 103 PFU), VN1203 HA avirulent (VN1203 with a mutation of the HA protein) (1 × 104 PFU), SARS-CoV rMA15 (1 × 105 PFU), or infectious-clone SARS (icSARS) Urbani (1 × 105 PFU) in 50 µl of PBS or were subjected to mock infection with PBS alone. In this experiment, VN1203 was treated as a highly pathogenic strain and VN1203 HA avirulent as a minimally pathogenic strain of influenza virus, as the 50% mouse lethal doses were 1 PFU for VN1203 and 104.3 PFU for VN1203 HA (unpublished data). Similarly, MA15 was treated as a highly pathogenic strain and Urbani as a minimally pathogenic strain of SARS-CoV based on a previous study (42). Lung tissues were removed on days 2 and 4 postinfection, and total RNA was harvested from an individual lung lobe. There were 3 mice for each time point and 3 animals for the time-matched mock-infected samples.
PWK/PhJ mouse embryonic fibroblasts (MEFs) were infected with A/PR/8/34 influenza virus at a multiplicity of infection (MOI) of 2.0. Allantoic fluid was used for mock treatments (n = 3 per set of conditions at each time point). Following infection, cells were washed once and incubated in complete medium at 37°C. At 6, 8, 12, and 24 h postinfection, cells were harvested in TRIzol and samples were processed using an miRNeasy Mini kit (Qiagen) according to the manufacturer’s instructions.
RNA preparation.
The individual lung lobes were homogenized using Trizol and a Magnalyser system (Roche) according to the manufacturer’s instructions. RNA was further purified using an miRNeasy Mini kit (Qiagen) according to the manufacturer’s instructions. RNA samples were spectroscopically verified for purity, and the quality of the intact RNA was assessed using an Agilent 2100 Bioanalyzer. The assay also confirmed that the RNA samples were free of genomic DNA contamination.
Library construction.
For SARS MA15-infected samples, small RNA libraries were prepared with a Small RNA v1.5 sample preparation kit following the manufacturer’s instructions (Illumina, San Diego, CA). Total RNA was ligated with a 3 ′ RNA adaptor (5′-/5rApp/ATCTCGTATGCCGTCTTCTGCTTG/3ddC/) specifically modified to target miRNAs and other small RNAs that have a 3 ′ hydroxyl group resulting from cleavage by Dicer and other RNA-processing enzymes and then with a 5 ′ RNA adaptor (5′-GUUCAGAGUUCUACAGUCCGACGAUC) at the 5 ′ end of RNA with a phosphate group. The 5 ′ adaptor also included the sequencing primer. RT-PCR amplification was then done using the adaptors as primers, selectively enriching the fragments that had adaptors on both ends. The resulting double-stranded DNA libraries were polyacrylamide gel electrophoresis (PAGE) (6% Novex Tris-borate-EDTA [TBE] PAGE; Invitrogen) purified and size selected to eliminate dimerized adaptors.
For influenza virus-infected samples, small RNA libraries were prepared using a TruSeq Small RNA sample preparation kit (Illumina, San Diego, CA) following the manufacturer’s instructions. This protocol is very similar to that of the version 1.5 sample preparation kit but with a change in the 3 ′ adapter (5 ′ TGGAATTCTCGGGTGCCAAGG) that also targets the 3 ′ hydroxyl resulting from enzymatic cleavage by Dicer.
Sequencing and read mapping.
All libraries were sequenced to 50 nt (SARS-CoV infection) or 54 nt (influenza virus infection) of read length on individual lanes on a Genome Analyzer II system following the protocols of the manufacturer (Illumina, San Diego, CA).
The adaptor sequences were first trimmed from small RNA reads. After adaptor trimming, reads < 16 nt in length were discarded, and reads > 40 nt were shortened to 40 nt by clipping from the 3 ′ end. Bowtie software (43) was used to align the trimmed reads to reference sequences, allowing up to two mismatches and with options selected as “-k 1 –m1 –best –strata,” which kept only uniquely mapped reads.
For the global classification, the trimmed reads were aligned sequentially to mouse ribosomal sequences (rRNA) from GenBank, tRNA sequences (tRNA) from the genome browser of the University of California, Santa Cruz (UCSC), (http://genome.ucsc.edu), small cytoplasmic RNA (scRNA), small nuclear RNA (snRNA), and small nucleolar RNAs (snoRNA) sequences from NCBI RefSeq annotation, piwi-associated small RNA (piRNA) sequences from the functional RNA database (44), microRNA (miRNA) precursor sequences from miRBase (http://www.mirbase.org, release 16), the mouse reference genome (mm9, July 2007, NCBI build 37, reference strain C57BL/6J), and SARS-CoV (GenBank accession no. DQ497008) or influenza virus (GenBank accession no. AF389115 to AF389122) genomic sequences. Also, unmapped reads after rRNA alignment were mapped directly to the same mouse reference genome to search for nonannotated small RNAs. For visualization, BAM files were generated using SAMtools (45) and displayed using the UCSC genome browser.
Differential expression analysis of small RNAs from annotated loci.
To quantify transcript expression, we estimated transcript abundance by counting the total number of reads mapped to each transcript. Read sequences that mapped to more than one location were excluded from expression quantification. To compare transcript expression data across different sets of conditions, the transcript abundances, i.e., the raw read counts, were scaled first as the number of reads per million (rpm) mapped for each sample. Next, we chose the geometric mean of all samples as a reference. The quantile-quantile (qq) plots of the distribution of differences between the reference and remaining samples in the numbers of log2-transformed revolutions per minute were compared. We determined a scaling factor for each sample as the median difference between the corresponding quantile values of the individual sample and the reference. An offset of 1 was added to all normalized values to facilitate comparisons involving one or more values of zero and to reduce the variability of the log ratios for low expression values.
For quantification of annotated miRNAs, we first removed reads mapped to other abundant small RNAs (rRNAs, tRNAs, snoRNAs, snRNAs, scRNAs, and piRNAs) and used the results to map the remaining reads directly to miRNA precursors. To accommodate the variability in the RNA sequences of individual mature miRNAs, i.e., the so-called isomiRs (46), we counted all reads mapped with start positions located within a 3-nt window for each annotated mature miRNA. Only reads of 19 to 25 nt were used for miRNA quantification.
For other annotated loci, we used the mapping results of the reads to search the mouse reference genome. A nonredundant set of annotations that included annotations of long (>200 nt) noncoding RNAs was compiled as previously described (5). In brief, we clustered the overlapping annotated transcripts into single loci. We found 10,986 nonoverlapping ncRNA loci (8,008 of which were larger than 200 nt) in addition to 21,565 protein-coding loci. Read sequences that mapped to multiple locations on the corresponding reference sequences were excluded from the counts during all differential expression analyses. The repeat information was downloaded as a RepeatMasker track using the UCSC genome browser.
Identification of non-miRNA and novel small RNAs by a genome-wide search.
We carried out a genome-wide search of novel small RNAs, starting by mapping all short reads directly to the mouse reference genome. We located start positions in the genome mapped with short reads from all 20 samples and counted the number of reads of the same length from the same sample for each start position mapped. We then identified the (top 5%) most abundant start positions by the read count of all start positions located as described above as candidate loci for putative small RNAs. We next merged overlapping candidate loci into single loci to create a set of nonredundant candidate loci for putative small RNAs. We counted the number of reads that were 16 to 36 nt in length that mapped to each of nonredundant candidate loci across all samples and estimated the changes in expression of these candidate loci during virus infection by the use of a method similar to that used for miRNA differential analysis. The differentially expressed candidate loci were predicted as putative small RNA loci.
To minimize the possibility that an identification of a non-miRNA small RNA was due to a partial misalignment of short reads that originated from very abundant RNAs sharing high sequence similarities, we located all genomic regions with high sequence similarity to each putative small RNA locus. These “homologous” genomic regions were identified as follows: (i) for putative small RNA loci that were relatively long (>40 nt), we used the standalone BLAT program with the default setting for DNA alignment to align the genomic sequence of the small RNA locus to the reference genome sequence (47); (ii) for small RNA locus that were relatively short (40 nt or less), we extracted all reads uniquely mapped to the small RNA locus as described above. We then used the Bowtie program as described above (43) but with criteria that were less stringent (i.e., with a change from 2 mismatches to 3 mismatches) and with adjustments of the program settings to report all valid alignments (Bowtie option “-all”) to realign the extracted reads to the reference genomes. For both alignment approaches, we treated the genomic region corresponding to each obtained alignment as a genomic region “homologous” to the small RNA locus. To remove redundancies, we merged overlapping homologous genomic regions into single homologous genomic regions. For each identified homologous region, we counted the number of uniquely mapped reads obtained across all samples as described above. We then compared the read counts of these homologous regions to that of the genomic region corresponding to the original small RNA locus. We kept a putative small locus for further analysis only when the number of uniquely mapped reads in the genomic region corresponding to the small RNA locus was at least twice as large (on average, across all samples) as that of the homologous region with the highest number of uniquely mapped reads.
We used RNAfold (48) and RNAz (49) to perform RNA secondary structure predictions for putative small RNA loci. Conserved RNA secondary structures (P > 0.5) were predicted using 30-way multiple alignments downloaded from the UCSC genome browser (http://genome.ucsc.edu) and RNAz (49). For predictions performed using RNAfold (48), we extracted three genomic windows of different sizes (100, 150, and 200 nt) surrounding each putative small RNA loci. When a locus was shorter than the desired genomic window, we expanded the locus by first adding neighboring candidate loci within the desired genomic window and then extending the sequence at both ends to achieve the desired width. To evaluate the statistical significance of the secondary structures predicted by RNAfold, we generated 500 randomly permutated sequences from the original sequence, with dinucleotide frequencies preserved using uShuffle (50), and used RNAfold to similarly fold all random sequences. We calculated the percentage of times that a random sequence exhibited a higher minimum free energy level than the original sequence as the P value of the predicted secondary structure for the locus. A prediction with P < 0.05 was considered significant.
Quantitative real-time PCR (qPCR).
Quantitative real-time PCR was used to validate expression of selected miRNAs on replicate samples. For each miRNA qRT-PCR assay, the total RNA input used was 20 ng per sample. qPCR (including reverse transcription- and miRNA-specific primer sets) was performed using a miRCURY LNA Universal RT microRNA PCR system (Exiqon, Woburn, MA). qPCR was performed with an ABI 7900 real-time PCR system and SYBR green chemistry. Each assay was run in triplicate with Power SYBR green PCR master mix (Applied Biosystems, Carlsbad, CA) for miRNA detections in a 10-µl total reaction volume.
SUPPLEMENTAL MATERIAL
Length distribution of small RNA short reads for all samples, presented as described for Fig. 1a. Reads were adaptor trimmed. Reads < 16 nt were discarded, and reads > 40 nt were shortened to 40 nt by clipping from the 3 ′ end. The second peak of 18 to 24 nt corresponds to mature miRNAs, and the second peak between 31 and 38 nt corresponds to the mixture of different types of abundant small RNAs such as tRNA plus some reads which were not mapped to the reference genomes. Download Figure S1, PDF file, 0.1 MB.
Comparison of ratios of expression resulting from infection versus mock infection for 12 differentially expressed miRNAs. (a) Scatter plot of the log2 infection/mock infection expression ratios for 12 differentially expressed miRNAs originally obtained by small RNA next-generation sequencing (NGS) to the infection/mock infection ratios obtained by qPCR performed on independently collected lung samples from mice infected with SARS-CoV (MA15) or influenza virus (PR8). The ratios were shown to be significantly correlated (ANOVA P = 1.2e-16; Pearson correlation coefficient = 0.72). (b) Heat map representation of individual infection/mock infection expression ratios as shown in panel a. Each column represents an independent sample collected from one infected mouse compared to a matched mock-infected sample, except for the qPCR assay of samples from MA15-infected CAST mice, for which the mock infection values represent the averages of the results obtained with two samples (one from a male mouse and another from a female mouse). Also, the data from CAST replicate 2 (R2) represent the results obtained with a sample from a male mouse. All samples were collected 2 days after infection. Red on the heat map indicates upregulation during infection, while green indicates downregulation during infection. Grey indicates that no PCR product was generated or that there was a >3-fold difference between the results obtained in the two qPCR replicate experiments. Raw threshold cycle (CT) values were normalized using the geometric means of the results obtained with two internal controls (5S RNA and RNU5G). Download Figure S2, PDF file, 0.1 MB.
Changes in expression of 6 selected miRNAs surveyed across a large number of conditions. The data are presented as described for Fig. 2b but represent measurements of individual replicate samples. Raw CT values were normalized using the geometric means of the results obtained with two internal controls (5S RNA and RNU5G). On the y axis, delta delta CT values represent the differences between the normalized CT values determined for infected samples and the median CT values (normalized) determined for matched mock-infected samples. Download Figure S3, PDF file, 0.1 MB.
(a) Overview of 216 putative small RNAs differentially expressed in mouse lung samples during SARS-CoV (MA15) and influenza virus (PR8) infection. The small RNAs were classified into three groups consisting of 46 small RNAs overlapped with annotated miRNAs (miR), 30 small RNAs overlapped with annotated snoRNAs (snoRNA; there were 2 additional small RNAs that overlapped with both miRNAs and snoRNAs that are not shown in this block but are included in the “miR” block described above), and 140 small RNAs that did not overlap with annotated miRNA and snoRNAs (other). Colors on the heat map indicate the log2 ratios of expression (normalized read counts) in virus-infected mouse lung samples to expression in matched mock-infected samples. Red, upregulation; green, downregulation. The left sidebar shows the relative abundances of the corresponding small RNAs represented as the minimum average normalized read counts across all samples. (b) Box plots showing the differences between the corresponding absolute log2 fold changes of expression in MA15-infected lung samples and PR8-infected samples in the same mouse strains across three different groups of small RNAs as described for panel a. Red, miR; blue, snoRNA; cyan, other. P values correspond to the results of paired t tests comparing the corresponding absolute log2 fold changes of expression in MA15-infected samples versus PR8-infected samples across three different groups of small RNAs in the same mouse strains. (c) Data are presented as described for panel b for 161 small RNAs with nonzero read counts in all 20 samples. Download Figure S4, PDF file, 0.3 MB.
Global regulation of host responses by differentially expressed mature miRNAs. (a) Heat map showing an overview of the concordance of changes in expression of mature miRNAs (columns) and their predicted targets (rows): red represents changes in expression of mature miRNA and its predicted targets in the same direction (positive correlation) during SARS-CoV infection, and green represents changes in the opposite direction (negative correlation). White indicates that the gene is not a predicted target of the corresponding miRNA. The block at the bottom shows the log2 ratios of changes in expression (red, upregulation; green, downregulation) of 29 differentially expressed miRNAs during SARS-CoV infection in four mouse strains that had at least one differentially expressed predicted target. Similarly, the block on the right shows the corresponding changes in expression of 1,014 predicted miRNA targets. Target prediction was based on the results obtained with TargetScan software (release 5.1). Since the TargetScan predictions were determined on the basis of RefSeq annotations, we compiled a nonredundant set of 20,166 loci by grouping all overlapping RefSeq transcripts and estimated the changes in expression based on whole-transcriptome sequencing analysis of the same samples, 1,305 of which consistently showed changes of at least 1.8-fold across at least 3 of 4 mouse strains and were considered differentially expressed. The contingency table on the bottom right shows the distribution of loci based on target prediction (Target) and differential expression (DE). (b) The distribution of the differentially expressed miRNA targets based on the numbers of positively correlated and negatively correlated miRNAs. For each mRNA locus, the differentially expressed miRNAs were separated into two groups: miRNAs that were predicted to target the loci and had changes in expression opposite those of the mRNA locus during SARS-CoV infection (“negatively correlated”) and miRNAs that were predicted to target the same mRNA locus and had changes in expression in the same direction (“positively correlated”). The number of miRNAs in each group was counted for each mRNA locus. All mRNA loci were then grouped based on the number of positively correlated (y axis) and negatively correlated (x axis) miRNAs, and the numbers of mRNA loci in each group were plotted accordingly. The size of each circle is proportional to the number of mRNA loci (labeled) within each group. Download Figure S5, PDF file, 0.8 MB.
A subnetwork of differentially expressed miRNAs and their predicted targets. Nodes represent miRNAs and predicted miRNA targets; arrows connect miRNAs to predicted targets. Nodes are colored based on changes in expression during SARS-CoV infection: red represents upregulation, green represents downregulation. Predicted mRNA targets of differentially expressed miRNAs were analyzed using IPA software (Ingenuity Systems, Inc., Redwood City, CA). For simplicity, only the miRNA-mRNA pairs with opposite changes in expression are included. The network represents the genes in the enriched Ingenuity canonical pathways (P < 0.05) that were previously reported to be involved in antiviral responses as follows: NF-κB signaling, interferon signaling, role of PKR in interferon induction and antiviral response, role of JAK1, JAK2, and TYK2 in interferon signaling, role of pattern recognition receptors in recognition of bacteria and viruses, activation of IRF by cytosolic pattern recognition receptors. Download Figure S6, PDF file, 0.1 MB.
Overview of small RNA sequencing data.
Identified small RNA loci and associated expression patterns and annotations.
Enriched canonical pathways. Pathway enrichment analyses were separately performed using three sets of genes: (i) the complete set of differentially expressed mRNA loci; (ii) the subset of differentially expressed mRNA loci that were predicted to be targets of differentially expressed miRNAs; (iii) the subset of differentially expressed mRNA loci that were predicted to be targets of differentially expressed miRNAs and that had changes in expression opposite those of the corresponding miRNAs. The analysis was done using IPA software (Ingenuity Systems, Inc., Redwood City, CA). Only canonical pathways with a P value < 0.05 are shown in the table.
Summary of RNA secondary structure prediction for putative small RNA loci in comparison to annotated small RNAs. +, RNA secondary structure predicted using RNAfold; one hundred nanot, a genomic window 100 nt in length was taken for loci shorter than 100 nt. RNAfold predictions were also repeated with genomic windows 150 nt (150 nt) and 200 nt (200 nt) in length for each putative small RNA loci. RNAz, conserved RNA secondary structure predicted using RNAz. Annotated small RNAs are same as those listed in Table 1.
ACKNOWLEDGMENTS
We thank Elizabeth Rosenzweig and Jean Chang (University of Washington, Seattle) for generating qPCR data. We thank Lynn Law and Janine Bryan (University of Washington, Seattle) for helpful suggestions.
This work was supported by the National Institutes of Health grant U54 AI081680 (PNWRCE) and by funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, contract no. HHSN272200800060C.
L.G., M.T.F., M.B.F., J.R.T., and C.L. performed viral infection experiments. M.J.T. and S.L. performed deep-sequencing experiments. X.P. analyzed the data. X.P., S.P., M.H., G.P.S., T.M.T., Y.K., R.S.B., and M.G.K. contributed to the concept, strategy, study design, and project management. X.P., L.G., M.T.F., M.J.T., M.J.K., J.R.T., M.B.F., G.P.S., C.L., R.S.B., and M.G.K. contributed to the writing of the manuscript.
Footnotes
Citation Peng X, et al. 2011. Integrative deep sequencing of the mouse lung transcriptome reveals differential expression of diverse classes of small RNAs in response to respiratory virus infection. mBio 2(6):e00198-11. doi:10.1128/mBio.00198-11.
REFERENCES
- 1. Gottwein E, Cullen BR. 2008. Viral and cellular microRNAs as determinants of viral pathogenesis and immunity. Cell Host Microbe 3:375–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. 2005. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science 309:1577–1581 [DOI] [PubMed] [Google Scholar]
- 3. Li Y, et al. 2010. MicroRNA expression and virulence in pandemic influenza virus-infected mice. J. Virol. 84:3023–3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Triboulet R, et al. 2007. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science 315:1579–1582 [DOI] [PubMed] [Google Scholar]
- 5. Peng X, et al. 2010. Unique signatures of long noncoding RNA expression in response to virus infection and altered innate immune signaling. mBio 1:e00206-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Armour CD, et al. 2009. Digital transcriptome profiling using selective hexamer priming for cDNA synthesis. Nat. Methods 6:647–649 [DOI] [PubMed] [Google Scholar]
- 7. Aylor DL, et al. 2011. Genetic analysis of complex traits in the emerging collaborative cross. Genome Res. 21:1213–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mestdagh P, et al. 2009. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 10:R64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cole C, et al. 2009. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA 15:2147–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. O’Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. 2007. MicroRNA-155 is induced during the macrophage inflammatory response. Proc. Natl. Acad. Sci. U. S. A. 104:1604–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Johnnidis JB, et al. 2008. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature 451:1125–1129 [DOI] [PubMed] [Google Scholar]
- 12. Matera AG, Terns RM, Terns MP. 2007. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 8:209–220 [DOI] [PubMed] [Google Scholar]
- 13. Klattenhoff C, Theurkauf W. 2008. Biogenesis and germline functions of piRNAs. Development 135:3–9 [DOI] [PubMed] [Google Scholar]
- 14. Ro S, et al. 2007. Cloning and expression profiling of testis-expressed piRNA-like RNAs. RNA 13:1693–1702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yan Z, et al. 2011. Widespread expression of piRNA-like molecules in somatic tissues. Nucleic Acids Res. 39:6596–6607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Houzet L, et al. 2008. MicroRNA profile changes in human immunodeficiency virus type 1 (HIV-1) seropositive individuals. Retrovirology 5:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Peng X, et al. 2009. Computational identification of hepatitis C virus associated microRNA-mRNA regulatory modules in human livers. BMC Genomics 10:373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim VN, Han J, Siomi MC. 2009. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 10:126–139 [DOI] [PubMed] [Google Scholar]
- 19. Baskerville S, Bartel DP. 2005. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA 11:241–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. 2004. Identification of mammalian microRNA host genes and transcription units. Genome Res. 14:1902–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ender C, et al. 2008. A human snoRNA with microRNA-like functions. Mol. Cell 32:519–528 [DOI] [PubMed] [Google Scholar]
- 22. Saraiya AA, Wang CC. 2008. snoRNA, a novel precursor of microRNA in Giardia lamblia. PLoS Pathog. 4:e1000224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Taft RJ, et al. 2009. Small RNAs derived from snoRNAs. RNA 15:1233–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Haussecker D, et al. 2010. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA 16:673–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lu Y, Qian XY, Krug RM. 1994. The influenza virus NS1 protein: a novel inhibitor of pre-mRNA splicing. Genes Dev. 8:1817–1828 [DOI] [PubMed] [Google Scholar]
- 26. Qiu Y, Nemeroff M, Krug RM. 1995. The influenza virus NS1 protein binds to a specific region in human U6 snRNA and inhibits U6-U2 and U6-U4 snRNA interactions during splicing. RNA 1:304–316 [PMC free article] [PubMed] [Google Scholar]
- 27. Wang W, Krug RM. 1998. U6atac snRNA, the highly divergent counterpart of U6 snRNA, is the specific target that mediates inhibition of AT-AC splicing by the influenza virus NS1 protein. RNA 4:55–64 [PMC free article] [PubMed] [Google Scholar]
- 28. Gioia U, et al. 2005. Functional characterization of XendoU, the endoribonuclease involved in small nucleolar RNA biosynthesis. J. Biol. Chem. 280:18996–19002 [DOI] [PubMed] [Google Scholar]
- 29. Renzi F, et al. 2006. The structure of the endoribonuclease XendoU: from small nucleolar RNA processing to severe acute respiratory syndrome coronavirus replication. Proc. Natl. Acad. Sci. U. S. A. 103:12365–12370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schutz K, Hesselberth JR, Fields S. 2010. Capture and sequence analysis of RNAs with terminal 2′,3′-cyclic phosphates. RNA 16:621–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bachellerie JP, Cavaillé J, Hüttenhofer A. 2002. The expanding snoRNA world. Biochimie 84:775–790 [DOI] [PubMed] [Google Scholar]
- 32. Guo H, Ingolia NT, Weissman JS, Bartel DP. 2010. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466:835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Krek A, et al. 2005. Combinatorial microRNA target predictions. Nat. Genet. 37:495–500 [DOI] [PubMed] [Google Scholar]
- 34. Wilusz JE, Sunwoo H, Spector DL. 2009. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 23:1494–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rodriguez A, et al. 2007. Requirement of bic/microRNA-155 for normal immune function. Science 316:608–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Thai TH, et al. 2007. Regulation of the germinal center response by microRNA-155. Science 316:604–608 [DOI] [PubMed] [Google Scholar]
- 37. Williams GT, et al. 2006. Isolation of genes controlling apoptosis through their effects on cell survival. Gene Ther. Mol. Biol. 10:255–262 [PMC free article] [PubMed] [Google Scholar]
- 38. Williams GT, Mourtada-Maarabouni M, Farzaneh F. 2011. A critical role for non-coding RNA GAS5 in growth arrest and rapamycin inhibition in human T-lymphocytes. Biochem. Soc. Trans. 39:482–486 [DOI] [PubMed] [Google Scholar]
- 39. Perez JT, et al. 2010. Influenza A virus-generated small RNAs regulate the switch from transcription to replication. Proc. Natl. Acad. Sci. U. S. A. 107:11525–11530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Umbach JL, Yen HL, Poon LL, Cullen BR. 2010. Influenza A virus expresses high levels of an unusual class of small viral leader RNAs in infected cells. mBio 1:e00204-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Churchill GA, et al. 2004. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat. Genet. 36:1133–1137 [DOI] [PubMed] [Google Scholar]
- 42. Roberts A, et al. 2007. A mouse-adapted SARS-coronavirus causes disease and mortality in BALB/c mice. PLoS Pathog. 3:e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mituyama T, et al. 2009. The Functional RNA Database 3.0: databases to support mining and annotation of functional RNAs. Nucleic Acids Res. 37:D89–D92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li H, et al. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Morin RD, et al. 2008. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 18:610–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kent WJ. 2002. BLAT—the BLAST-like alignment tool. Genome Res. 12:656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hofacker IL, et al. 1994. Fast folding and comparison of RNA secondary structures. Monatsh. Fur Chem. 125:167–188 [Google Scholar]
- 49. Washietl S, Hofacker IL, Stadler PF. 2005. Fast and reliable prediction of noncoding RNAs. Proc. Natl. Acad. Sci. U. S. A. 102:2454–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jiang M, Anderson J, Gillespie J, Mayne M. 2008. uShuffle: a useful tool for shuffling biological sequences while preserving the k-let counts. BMC Bioinform. 9:192 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Length distribution of small RNA short reads for all samples, presented as described for Fig. 1a. Reads were adaptor trimmed. Reads < 16 nt were discarded, and reads > 40 nt were shortened to 40 nt by clipping from the 3 ′ end. The second peak of 18 to 24 nt corresponds to mature miRNAs, and the second peak between 31 and 38 nt corresponds to the mixture of different types of abundant small RNAs such as tRNA plus some reads which were not mapped to the reference genomes. Download Figure S1, PDF file, 0.1 MB.
Comparison of ratios of expression resulting from infection versus mock infection for 12 differentially expressed miRNAs. (a) Scatter plot of the log2 infection/mock infection expression ratios for 12 differentially expressed miRNAs originally obtained by small RNA next-generation sequencing (NGS) to the infection/mock infection ratios obtained by qPCR performed on independently collected lung samples from mice infected with SARS-CoV (MA15) or influenza virus (PR8). The ratios were shown to be significantly correlated (ANOVA P = 1.2e-16; Pearson correlation coefficient = 0.72). (b) Heat map representation of individual infection/mock infection expression ratios as shown in panel a. Each column represents an independent sample collected from one infected mouse compared to a matched mock-infected sample, except for the qPCR assay of samples from MA15-infected CAST mice, for which the mock infection values represent the averages of the results obtained with two samples (one from a male mouse and another from a female mouse). Also, the data from CAST replicate 2 (R2) represent the results obtained with a sample from a male mouse. All samples were collected 2 days after infection. Red on the heat map indicates upregulation during infection, while green indicates downregulation during infection. Grey indicates that no PCR product was generated or that there was a >3-fold difference between the results obtained in the two qPCR replicate experiments. Raw threshold cycle (CT) values were normalized using the geometric means of the results obtained with two internal controls (5S RNA and RNU5G). Download Figure S2, PDF file, 0.1 MB.
Changes in expression of 6 selected miRNAs surveyed across a large number of conditions. The data are presented as described for Fig. 2b but represent measurements of individual replicate samples. Raw CT values were normalized using the geometric means of the results obtained with two internal controls (5S RNA and RNU5G). On the y axis, delta delta CT values represent the differences between the normalized CT values determined for infected samples and the median CT values (normalized) determined for matched mock-infected samples. Download Figure S3, PDF file, 0.1 MB.
(a) Overview of 216 putative small RNAs differentially expressed in mouse lung samples during SARS-CoV (MA15) and influenza virus (PR8) infection. The small RNAs were classified into three groups consisting of 46 small RNAs overlapped with annotated miRNAs (miR), 30 small RNAs overlapped with annotated snoRNAs (snoRNA; there were 2 additional small RNAs that overlapped with both miRNAs and snoRNAs that are not shown in this block but are included in the “miR” block described above), and 140 small RNAs that did not overlap with annotated miRNA and snoRNAs (other). Colors on the heat map indicate the log2 ratios of expression (normalized read counts) in virus-infected mouse lung samples to expression in matched mock-infected samples. Red, upregulation; green, downregulation. The left sidebar shows the relative abundances of the corresponding small RNAs represented as the minimum average normalized read counts across all samples. (b) Box plots showing the differences between the corresponding absolute log2 fold changes of expression in MA15-infected lung samples and PR8-infected samples in the same mouse strains across three different groups of small RNAs as described for panel a. Red, miR; blue, snoRNA; cyan, other. P values correspond to the results of paired t tests comparing the corresponding absolute log2 fold changes of expression in MA15-infected samples versus PR8-infected samples across three different groups of small RNAs in the same mouse strains. (c) Data are presented as described for panel b for 161 small RNAs with nonzero read counts in all 20 samples. Download Figure S4, PDF file, 0.3 MB.
Global regulation of host responses by differentially expressed mature miRNAs. (a) Heat map showing an overview of the concordance of changes in expression of mature miRNAs (columns) and their predicted targets (rows): red represents changes in expression of mature miRNA and its predicted targets in the same direction (positive correlation) during SARS-CoV infection, and green represents changes in the opposite direction (negative correlation). White indicates that the gene is not a predicted target of the corresponding miRNA. The block at the bottom shows the log2 ratios of changes in expression (red, upregulation; green, downregulation) of 29 differentially expressed miRNAs during SARS-CoV infection in four mouse strains that had at least one differentially expressed predicted target. Similarly, the block on the right shows the corresponding changes in expression of 1,014 predicted miRNA targets. Target prediction was based on the results obtained with TargetScan software (release 5.1). Since the TargetScan predictions were determined on the basis of RefSeq annotations, we compiled a nonredundant set of 20,166 loci by grouping all overlapping RefSeq transcripts and estimated the changes in expression based on whole-transcriptome sequencing analysis of the same samples, 1,305 of which consistently showed changes of at least 1.8-fold across at least 3 of 4 mouse strains and were considered differentially expressed. The contingency table on the bottom right shows the distribution of loci based on target prediction (Target) and differential expression (DE). (b) The distribution of the differentially expressed miRNA targets based on the numbers of positively correlated and negatively correlated miRNAs. For each mRNA locus, the differentially expressed miRNAs were separated into two groups: miRNAs that were predicted to target the loci and had changes in expression opposite those of the mRNA locus during SARS-CoV infection (“negatively correlated”) and miRNAs that were predicted to target the same mRNA locus and had changes in expression in the same direction (“positively correlated”). The number of miRNAs in each group was counted for each mRNA locus. All mRNA loci were then grouped based on the number of positively correlated (y axis) and negatively correlated (x axis) miRNAs, and the numbers of mRNA loci in each group were plotted accordingly. The size of each circle is proportional to the number of mRNA loci (labeled) within each group. Download Figure S5, PDF file, 0.8 MB.
A subnetwork of differentially expressed miRNAs and their predicted targets. Nodes represent miRNAs and predicted miRNA targets; arrows connect miRNAs to predicted targets. Nodes are colored based on changes in expression during SARS-CoV infection: red represents upregulation, green represents downregulation. Predicted mRNA targets of differentially expressed miRNAs were analyzed using IPA software (Ingenuity Systems, Inc., Redwood City, CA). For simplicity, only the miRNA-mRNA pairs with opposite changes in expression are included. The network represents the genes in the enriched Ingenuity canonical pathways (P < 0.05) that were previously reported to be involved in antiviral responses as follows: NF-κB signaling, interferon signaling, role of PKR in interferon induction and antiviral response, role of JAK1, JAK2, and TYK2 in interferon signaling, role of pattern recognition receptors in recognition of bacteria and viruses, activation of IRF by cytosolic pattern recognition receptors. Download Figure S6, PDF file, 0.1 MB.
Overview of small RNA sequencing data.
Identified small RNA loci and associated expression patterns and annotations.
Enriched canonical pathways. Pathway enrichment analyses were separately performed using three sets of genes: (i) the complete set of differentially expressed mRNA loci; (ii) the subset of differentially expressed mRNA loci that were predicted to be targets of differentially expressed miRNAs; (iii) the subset of differentially expressed mRNA loci that were predicted to be targets of differentially expressed miRNAs and that had changes in expression opposite those of the corresponding miRNAs. The analysis was done using IPA software (Ingenuity Systems, Inc., Redwood City, CA). Only canonical pathways with a P value < 0.05 are shown in the table.
Summary of RNA secondary structure prediction for putative small RNA loci in comparison to annotated small RNAs. +, RNA secondary structure predicted using RNAfold; one hundred nanot, a genomic window 100 nt in length was taken for loci shorter than 100 nt. RNAfold predictions were also repeated with genomic windows 150 nt (150 nt) and 200 nt (200 nt) in length for each putative small RNA loci. RNAz, conserved RNA secondary structure predicted using RNAz. Annotated small RNAs are same as those listed in Table 1.