ABSTRACT
In nature, the complex composition and structure of the plant cell wall pose a barrier to enzymatic degradation. Nevertheless, some anaerobic bacteria have evolved for this purpose an intriguing, highly efficient multienzyme complex, the cellulosome, which contains numerous cellulases and hemicellulases. The rod-like cellulose component of the plant cell wall is embedded in a colloidal blend of hemicelluloses, a major component of which is xylan. In order to enhance enzymatic degradation of the xylan component of a natural complex substrate (wheat straw) and to study the synergistic action among different xylanases, we have employed a variation of the designer cellulosome approach by fabricating a tetravalent complex that includes the three endoxylanases of Thermobifida fusca (Xyn10A, Xyn10B, and Xyn11A) and an Xyl43A β-xylosidase from the same bacterium. Here, we describe the conversion of Xyn10A and Xyl43A to the cellulosomal mode. The incorporation of the Xyl43A enzyme together with the three endoxylanases into a common designer cellulosome served to enhance the level of reducing sugars produced during wheat straw degradation. The enhanced synergistic action of the four xylanases reflected their immediate juxtaposition in the complex, and these tetravalent xylanolytic designer cellulosomes succeeded in degrading significant (~25%) levels of the total xylan component of the wheat straw substrate. The results suggest that the incorporation of xylanases into cellulosome complexes is advantageous for efficient decomposition of recalcitrant cellulosic substrates—a distinction previously reserved for cellulose-degrading enzymes.
IMPORTANCE
Xylanases are important enzymes for our society, due to their variety of industrial applications. Together with cellulases and other glycoside hydrolases, xylanases may also provide cost-effective conversion of plant-derived cellulosic biomass into soluble sugars en route to biofuels as an alternative to fossil fuels. Xylanases are commonly found in multienzyme cellulosome complexes, produced by anaerobic bacteria, which are considered to be among the most efficient systems for degradation of cellulosic biomass. Using a designer cellulosome approach, we have incorporated the entire xylanolytic system of the bacterium Thermobifida fusca into defined artificial cellulosome complexes. The combined action of these designer cellulosomes versus that of the wild-type free xylanase system was then compared. Our data demonstrated that xylanolytic designer cellulosomes displayed enhanced synergistic activities on a natural recalcitrant wheat straw substrate and could thus serve in the development of advanced systems for improved degradation of lignocellulosic material.
Introduction
Xylanases catalyze the breakdown of xylan, the second most abundant polymer on Earth after cellulose (1), into xylooligosaccharides and xylose. These enzymes can contribute in combination with cellulases to the efficient conversion of cellulosic biomass to soluble sugars en route to biofuels (2–6). Improvement of xylanolytic activity has considerable potential for a broad variety of applications: e.g., biobleaching of kraft pulps in the paper industries to avoid the use of chlorine as a bleaching agent, for food and animal feed, or for the production of oligosaccharides from isolated xylans, which are then used as functional food additives or alternative sweeteners with certain beneficial properties (7–10).
In past studies, we initiated the conversion of the simple cellulolytic free enzyme system of the aerobic bacterium Thermobifida fusca (both cellulases and xylanases) to a cellulosomal system using designer cellulosome technology, in order to enhance the combined synergistic activities of the enzymes towards synthetic substrates (cellulose and xylan) and a natural complex cellulosic substrate (wheat straw) (11–16). Designer cellulosomes serve as a platform for promoting synergistic action among enzyme components (17). This concept is based on the very high affinity (18, 19) and specific interaction (20–22) between cohesin and dockerin modules from the same species. Cohesins from different species are recombined into a single protein component, termed “chimeric scaffoldin,” which serves to incorporate enzyme hybrids bearing matching dockerins.
Previous research on cellulosomes and designer cellulosomes has shown that cellulosomal cellulases act together in a heightened synergistic manner in the degradation of recalcitrant cellulosic substrates. The observed enhancement in synergy has been linked to both enzyme proximity and/or common targeting of the enzymes to appropriate sites on the substrate (12–17, 23–28). Moreover, it has been demonstrated that the inclusion of xylanases together with cellulases in designer cellulosomes also causes enhanced synergy on a natural cellulosic wheat straw substrate (15, 16). Indeed, xylanases are defined components in native cellulosomes as well as noncellulosomal complexes (29–33), although it is less evident why complexation of xylanases would be necessary for degradation of the presumably less recalcitrant polysaccharide.
In a previous publication (15), we described the conversion of T. fusca Xyn10B and Xyn11A endoxylanases into the cellulosomal mode and their integration into designer cellulosomes. In the present article, we report the conversions of two additional T. fusca enzymes, endoxylanase Xyn10A and β-xylosidase Xyl43A, into the cellulosomal mode by grafting divergent dockerins onto the enzymes via recombinant means. The latter enzymes were combined together with previously described dockerin-containing forms of Xyn10B and Xyn11A into a tetravalent cellulosome complex via an appropriate chimeric scaffoldin, and the resultant complex was analyzed for its synergistic capacity to degrade wheat straw.
RESULTS
Thermobifida fusca xylanases.
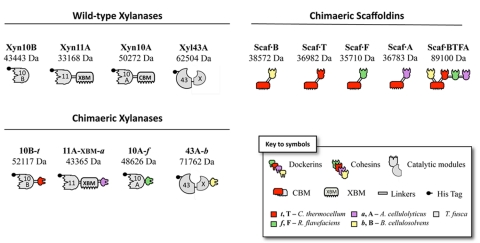
The schematic modular content of the wild-type enzymes used in this study is shown in Fig. 1. Four different wild-type T. fusca xylanases were used: Xyn10B, Xyn11A, Xyn10A, and Xyl43A. Xyn10B and Xyn11A were used as designer cellulosome components in previous communications (15, 16). Xyn10A and the Xyl43A enzyme were developed specifically in the present study for incorporation into designer cellulosomes. In most cases, native cellulosomal enzymes contain a dockerin at their C termini, which was thus the default position in this work. However, if a native noncellulosomal enzyme bears an N-terminal carbohydrate-binding module (CBM), it was thus replaced by an N-terminal dockerin.
FIG 1 .
Schematic representation of the recombinant proteins used in this study. The source of the representative module (see key) is indicated as follows: light gray, T. fusca; green, R. flavefaciens; red, C. thermocellum; lavender, A. cellulolyticus; and yellow, B. cellulosolvens. In the shorthand notation for the engineered enzymes, the numbers 10, 11, and 43 refer to the corresponding GH family (GH10, GH11, and GH43) of the catalytic module; uppercase letters (B, F, T, and A) indicate the source of the cohesin module, and lowercase italic letters (b, f, t, and a) indicate the source of the dockerin module: B. cellulosolvens, R. flavefaciens, C. thermocellum, and A. cellulolyticus, respectively.
The wild-type Xyn10B and Xyn11A xylanases have been characterized previously (34, 35). The wild-type Xyn10B enzyme lacks a CBM. In contrast, Xyn11A contains a family 2 CBM (CBM2), which exhibits binding specificity for both cellulose and xylan (34). For the purposes of this study, this CBM is designated “XBM” (xylan-binding module), due to its capacity to bind to xylan and its previously determined effect on enzyme activity (15). The β-xylosidase Xyl43A carries an X module at its C terminus with no apparent binding function but which is critical to enzyme activity (S. Moraïs et al., unpublished data).
Wild-type Xyn10A contains a family 2 CBM, and its biochemical properties have not been characterized previously. Xyn10A was thus examined for xylan degradation on three different xylan substrates: birchwood xylan, beechwood xylan, and oat spelt xylan (Table 1). The latter was the substrate most efficiently degraded by this enzyme. The overall structural characteristics of xylan from beechwood and birchwood xylan are similar (more than 90% β-1,4-linked xylose residues), whereas oat spelt xylan is a kind of arabinoxylan, the main chain of which is largely branched with arabinose residues (36, 37). These structural differences between xylan substrates may account for the different specific activities observed for Xyn10A (38). The endoxylanase Xyn10A exhibited comparable xylanolytic activities on the different substrates compared with those of the family 10B xylanase, both of which appear to be selective for substituted xylans (arabinoxylans). The family 11A xylanase, however, was generally more effective than the family 10 enzymes in xylan degradation and appeared to degrade both the less-branched and heavily branched model xylan substrates in an equivalent manner (Table 1) (15).
TABLE 1 .
Specific activities of individual recombinant enzymes on various xylans
| Substrate | Sp act (kat/mol of enzyme ± SD) |
|||||
|---|---|---|---|---|---|---|
| Xyn10A | 10A-f | Xyn10Ba | 10B-ta | Xyn11Aa | 11A-XBM-aa | |
| Birchwood xylan | 57 ± 10 | 77 ± 7 | 90 ± 5 | 136 ± 8 | 430 ± 2 | 425 ± 7 |
| Oat spelt xylan | 125 ± 5 | 138 ± 10 | 125 ± 2 | 152 ± 2 | 449 ± 3 | 433 ± 9 |
| Beechwood xylan | 15 ± 1 | 44 ± 5 | 40 ± 2 | 97 ± 7 | 440 ± 1 | 445 ± 6 |
Results from previous publication (15).
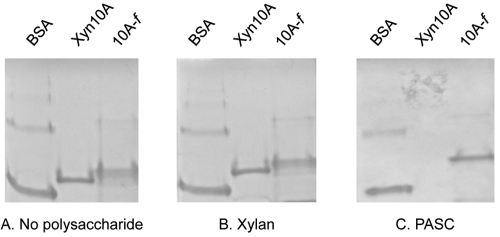
The binding properties of Xyn10A towards cellulose and insoluble xylan were examined herein (Fig. 2) by affinity electrophoresis. Xyn10A exhibited binding ability to cellulose but not to xylan. As expected, the bovine serum albumin (BSA) negative control failed to bind either xylan or cellulose. In a related species, Thermomonospora alba, comparable binding abilities were demonstrated for the family 2 CBM of a similar family 10 xylanase (39).
FIG 2 .
Affinity electrophoresis of family 10A xylanases. Affinity electrophoresis performed in the absence (A) or in the presence of 0.1% (wt/vol) oat spelt xylan (B) or 0.1% (wt/vol) PASC (C). Samples included BSA (lane 1), wild-type Xyn10A (lane 2), and 10A–f (lane 3).
Cooperation between wild-type enzymes.
Xylanases Xyn10B and Xyn11A were previously shown to display synergistic activities on wheat straw (15). For the purpose of this study, we examined each possible enzyme combination among the four T. fusca xylanases in the free mode; several enhanced activities were observed and are noted in Table 2, the strongest of which was observed for Xyn11A in combination with Xyl43A. The subsequent additions of the third xylanase, Xyn10A, and β-xylosidase Xyl43A to the latter enzymes (Xyn10B and Xyn11A) served to increase the final level of reducing sugars; it was therefore of interest to combine the four xylanases (three endoxylanases and the β-xylosidase) and to compare their activities as free enzymes or in a designer cellulosome format. For this purpose, dockerin modules of different specificities were attached to each xylanase, and their combined performance was evaluated in subsequent experiments.
TABLE 2 .
Enhancement of reducing sugar production by mixtures of T. fusca wild-type xylanases after 18 h of wheat straw degradation
| Enzyme(s) | Total concn of reducing sugars (µM) |
Enhancementa |
|---|---|---|
| Xyn10B | 48 ± 5 | |
| Xyn11A | 121 ± 3 | |
| Xyn10A | 90 ± 6 | |
| Xyl43A | 15 ± 2 | |
| Xyn10B + Xyn11A | 241 ± 4 | 1.4 ± 0.08 |
| Xyn11A + Xyn10A | 173 ± 2 | 0.8 ± 0.03 |
| Xyn10B + Xyn10A | 156 ± 5 | 1.1 ± 0.05 |
| Xyn10A + Xyl43A | 142 ± 1 | 1.4 ± 0.09 |
| Xyn10B + Xyl43A | 58 ± 1 | 0.9 ± 0.09 |
| Xyn11A + Xyl43A | 201 ± 5 | 1.5 ± 0.02 |
| Xyn10B + Xyn11A + Xyn10A | 276 ± 2 | 1.1 ± 0.05 |
| Xyn10B + Xyn11A + Xyl43A | 258 ± 4 | 1.4 ± 0.05 |
| Xyn11A + Xyn10A + Xyl43A | 233 ± 3 | 1 ± 0.03 |
| Xyn10B + Xyn10A + Xyl43A | 178 ± 4 | 1.2 ± 0.07 |
| Xyn10B + Xyn11A + Xyn10A + Xyl43A | 317 ± 3 | 1.2 ± 0.06 |
Enhancement of reducing sugar production = activity of the mixture/sum of individual activities.
Construction and expression of designer cellulosome components.
The recombinant dockerin-appended proteins designed for use in this study are shown schematically in Fig. 1. In order to convert T. fusca enzymes into the cellulosomal mode, a dockerin module of divergent specificity was fused to each catalytic module of the relevant enzyme.
Dockerin-bearing Xyn10B and Xyn11A have been the subject of previous studies (15), in which dockerins of different specificities were added at the C termini of the native xylanases, thereby generating the chimeric dockerin-containing enzymes, herein designated “10B-t” and “11A-XBM-a.” In order to integrate Xyn10B into an enzymatic complex, a Clostridium thermocellum dockerin (40) was fused at its C terminus, resulting in chimera 10B-t. Interestingly, the incorporation of the resultant 10B-t into a chimeric CBM-containing scaffoldin imparts a cellulose-binding component by default to this enzyme, which is inherently lacking in the wild-type enzyme, due to its lack of a CBM. In chimera 11A-XBM-a, a dockerin from Acetivibrio cellulolyticus was appended to the C terminus of the original Xyn11A, whereby the original catalytic module and the essential xylan-binding CBM (XBM) were both retained.
In the present work, the Xyn10A enzyme was converted to the cellulosomal mode by replacing its native CBM using a third type of divergent dockerin (chimera 10A-f). Chimera 10A-f is thus a recombinant xylanolytic enzyme comprising two fused modules: a catalytic module of the T. fusca family 10 xylanase A and a dockerin from Ruminococcus flavefaciens (41). In this case, the cellulose-binding function was deemed unnecessary to the action of this enzyme, since the binding ability will be recovered upon its incorporation into the chimeric CBM-containing scaffoldin. The examination of 10A-f binding properties by affinity electrophoresis towards cellulose and xylan was determined as described for the wild-type Xyn10A (Fig. 2). As predicted, the cellulose-binding capacity was essentially abolished in 10A-f, demonstrating that the C-terminal CBM of the protein is indeed responsible for this binding property. In addition, similarly to the wild-type enzyme, the chimera failed to bind to insoluble xylan.
The chimeric form of Xyl43A, 43-b, was designed to contain a divergent type II dockerin from Bacteroides cellulosolvens at its C terminus. The X-module is essential to β-xylosidase activity and was thus retained in the chimera. The wild-type enzyme exhibits binding activity for xylan but not cellulose, the latter of which will be acquired by the chimeric enzyme through incorporation into a chimeric CBM-containing scaffoldin (Moraïs et al., unpublished).
In order to incorporate the complement of dockerin-bearing xylanases into a defined functional designer cellulosome, an elaborate scaffoldin, containing a CBM and appropriate matching cohesins is required. The tetravalent chimeric scaffoldin Scaf·BTFA was described in an earlier work (16) and includes four different cohesin types together with the cellulose-binding module, as shown schematically in Fig. 1. From the N terminus to the C terminus, the cohesin modules of the chimeric scaffoldin include B from B. cellulosolvens, T from C. thermocellum, F from R. flavefaciens, and A from A. cellulolyticus, with a potent cellulose-binding C. thermocellum family 3a carbohydrate-binding module (CBM3a) positioned between B and T. The cohesin counterparts of Scaf·BTFA allow the position-specific incorporation of the above-described matching dockerin-containing xylanases, and its CBM will direct the complex to the complex cellulosic substrate.
All purified recombinant proteins showed a single major band on SDS-PAGE (not shown), and in each case, their mobility was consistent with their molecular mass.
Enzymatic properties of converted enzymes.
In our previous publication (15), we determined that specific activities observed for chimera 10B-t were higher than those of wild-type Xyn10B on all types of xylan substrates used for characterization of the wild-type enzymes (Table 1). In contrast, the chimeric version of the family 11 xylanase remained in the same range of activities as the wild-type enzyme. Addition of the dockerin modules in enzymes could reduce the enzyme affinity to its substrate (i.e., increase Km) and thus improve the rate of product release (enhance enzymatic activity), which is rate limiting in many glycoside hydrolases (42).
Chimera 10A-f was tested for xylan degradation on the same three xylan substrates. 10A-f displayed similar but somewhat heightened xylanolytic activities, compared to those of the wild-type enzyme, on birchwood and oat spelt xylans. The chimeric enzyme was observed to be much more active on beechwood xylan than the wild-type enzyme and thus had properties that resembled those of the other dockerin-containing family 10 chimera, 10B-t.
The dockerin-bearing β-xylosidase, 43A-b, was examined for its activity on the colorimetric substrate pNPX, and the apparent Km value was determined to be 0.23 mM, while the kcat was 4.83 s−1. Temperature and pH studies were comparable to those achieved for the wild-type enzyme (Moraïs et al., unpublished); pH optima ranged from 5.5 to 6 and temperature optima from 55 to 60°C (data not shown).
Cohesin-dockerin specificity.
A sensitive, affinity-based, enzyme-linked, microtiter plate assay was used to examine the specificity of the scaffoldin-borne cohesins for the chimeric dockerin-bearing enzymes (43). All of the scaffoldin-borne cohesins specifically bound their respective dockerin modules and failed to bind (or bound very poorly) nonmatching dockerin-bearing proteins (data not shown). The interaction between the cohesins and their matching dockerins was very similar to that of the individual monovalent scaffoldins, indicating that the binding capacities of the scaffoldins were reliable and selective. The observed cohesin-dockerin interactions for each scaffoldin were of similar intensity, thus suggesting stoichiometric ratios in each case.
Analysis of complex formation.
Designer cellulosome complexes were examined by nondenaturing PAGE, as detailed in previous publications (13, 16, 27, 28). Using this approach, complex formation is characterized by a single major band with minor altered mobility (band strengthened and usually shifted), thus indicating complete or near-complete complex formation. The position of a given band depends on the size, shape, and charge characteristics of the given protein. In the present case, stoichiometric mixtures of the enzymes together with the scaffoldin resulted in a complex characterized by a single major band with but minor altered mobility relative to the scaffoldin band in the absence of enzymes (data not shown). The observed minor change in mobility of large protein complexes limits the efficacy of the nondenaturing PAGE technique, particularly as the number of protein components increases in higher-order complexes. Moreover, as in previous studies of designer cellulosomes, some of the chimeric scaffoldin and enzyme preparations contained very minor residual contaminating bands, observed by denaturing and/or nondenaturing PAGE, which tended to obscure the analysis.
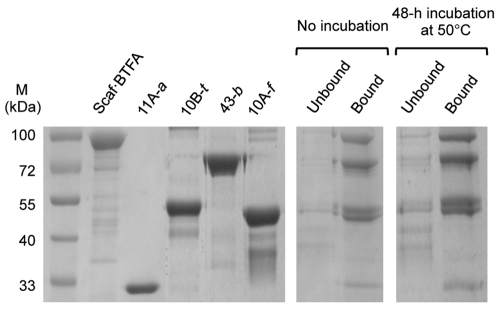
Therefore, as a complementary approach for complex formation, we employed the ability of the complex to bind microcrystalline cellulose by virtue of its resident CBM. For this purpose, the designer cellulosome preparations were subjected to interaction with cellulose, and the bound and unbound fractions were examined by SDS-PAGE. Dockerin-bearing enzymes that remained in the unbound fractions indicated that they failed to interact properly with the matching cohesins of the scaffoldin protein. As seen in Fig. 3, it appears that complex formation was near complete, as all of the designer cellulosome components were present in the bound fractions, and only negligible amounts of the protein bands were found in the unbound fraction. The experiment was repeated after incubating the complex 48 h at 50°C, and the results demonstrated the general stability of the designer cellulosome complexes under those conditions. In the absence of scaffoldin, the dockerin-bearing enzymes failed to bind to the cellulose matrix and remained in the unbound fraction (data not shown).
FIG 3 .
Cellulose-binding ability of the designer cellulosome complex. Samples included molecular mass markers (M) (lane 1), Scaf·BTFA (lane 2), 11A-a (lane 3), 10B-t (lane 4), 43-b (lane 5), 10A-f (lane 6), unbound and bound fractions immediately after complex formation (lanes 7 and 8, respectively), and unbound and bound fractions following 48-h incubation at 50°C after complex formation (lanes 9 and 10, respectively).
The same procedure was performed using wheat straw as a substrate to determine the binding properties of the designer cellulosome assemblies to the natural complex substrate. The results demonstrated that the potent binding capacity of the scaffoldin-borne CBM3a towards the cellulose component of the wheat straw was conserved (data not shown).
Enzymatic assays of designer cellulosomes on hatched wheat straw.
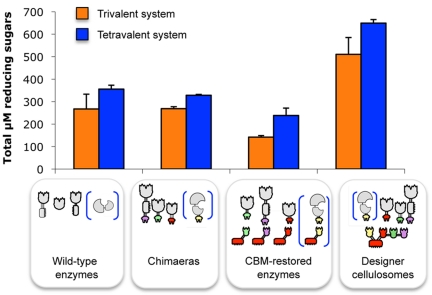
In order to examine the potential advantage of incorporating the Xyl43A β-xylosidase into a tetravalent chimeric scaffoldin containing the three characterized xylanases from T. fusca (Xyn10B, Xyn11A, and Xyn10A), several enzyme combinations with or without the β-xylosidase were assayed on hatched wheat straw (Fig. 4). Each combination contained equimolar mixtures of the desired enzymes for proper comparison. Free combinations of dockerin-bearing chimeras were compared to the wild-type xylanases, and no significant difference in straw degradation was observed, suggesting that, in this case, the addition of the dockerins had little or no effect on the activities of the catalytic module of each enzyme present in the reaction mixtures. Monovalent scaffoldin-bearing enzymes were assayed to evaluate the CBM targeting effect, and they appeared to be surprisingly less active in straw degradation relative to the free wild-type enzymes and free dockerin-containing xylanases. In this context, the attached CBMs could possibly prevent enzyme mobility on the substrate, thereby preventing access to the appropriate cell wall substrate component(s), compared to the enzymes in the free state. In this context, it has been demonstrated in a recent article (44) that adding a CBM to endoglucanases was not always beneficial for effective degradation of pretreated natural cellulosic substrates.
FIG 4 .
Comparative degradation of wheat straw by the various complexes and free enzyme systems after 24 h. The composition of the complexes and free enzyme systems is indicated in symbolic form. Orange bars represent the combination of the three xylanases (Xyn10B plus Xyn11A plus Xyn10A), and dark blue bars represent the combination of the four xylanases (Xyn10B plus Xyn11A plus Xyn10A plus Xyl43A). Enzymatic activity is defined as µM total reducing sugars. Each reaction was performed in triplicate, and standard deviations for straw hydrolysis are indicated.
Interestingly, not only did the trivalent designer cellulosome appear to degrade hatched wheat straw more efficiently than the free wild-type enzymes (about 2-fold enhancement), but the beneficial effect of Xyl43A was also demonstrated. Its incorporation in the enzyme mixtures served to increase significantly (and in all cases) the final level of reducing sugar production. Reaction yields after 24 h comprised about 5.2% for the tetravalent designer cellulosome, versus the corresponding yield of 2.9% for the wild-type enzymes (Fig. 4). Kinetic experiments were conducted with combinations of the wild-type enzymes versus those of designer cellulosomes (Fig. 5). The trivalent and tetravalent designer cellulosome activities were significantly higher than those of the corresponding combinations of free (wild-type) enzyme. Consequently, placing the enzymes in close proximity via the chimeric scaffoldin was advantageous and doubled the complex substrate degradation. Combinations in which Xyl43A was introduced also appeared to be significantly more effective (P < 0.01), suggesting its concerted action with the other xylanases.
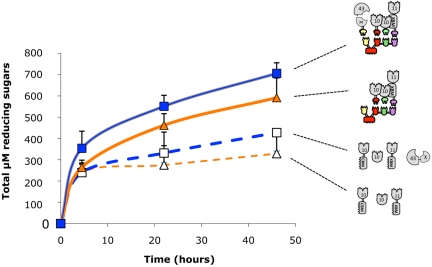
FIG 5 .
Kinetic studies of wheat straw hydrolysis by the various complexes and free enzyme systems. The curves shown represent degradation by the tetravalent designer cellulosome (■), degradation by the trivalent designer cellulosome (▲), degradation by the four wild-type xylanases (□), and degradation by three wild-type xylanases (∆). Enzymatic activity is defined as µM total reducing sugars. Triplicates of each reaction were carried out. Standard deviations are indicated.
Sugar identification and analysis.
Identity and concentration of saccharides were determined by high-performance anion-exchange chromatography (HPAEC), using known concentrations of standards. Relative amounts of products following enzymatic action of the different preparations on the wheat straw substrate were calculated via integration of the identified peaks. Combinations of free and scaffoldin-borne enzymes were allowed to interact with hatched wheat straw, and the degradation products were analyzed after 42 h of incubation. Different quantities of xylose and xylobiose were found as major final products in the samples (Table 3). As expected, xylobiose appeared only in samples in which Xyl43A was absent and was detected at higher concentrations in the free enzymes than in trivalent designer cellulosome. These results are consistent with those of a recent publication (45), in which it was demonstrated that smaller sugar products are obtained with designer cellulosomes than those using free enzyme preparations. Xylose was detected at increasing concentrations, upon addition of Xyl43A and incorporation of the dockerin-containing enzyme into the chimeric scaffoldin, consistent with the levels of reducing sugars described in Fig. 4 to 6.
TABLE 3 .
Soluble sugar production following digestion of hatched wheat straw by various enzyme combinations with 42 h of incubationa
| Enzyme combination | Production of: |
||||
|---|---|---|---|---|---|
| Arabinose | Xylose | Xylobiose | Xylotriose | Cellulose degradation products |
|
| Wild-type free enzymes | |||||
| Xyn10B + Xyn11A + Xyn10A | NDb | 5.1 ± 0.9 (34.1) | 9.9 ± 0.8 (35.0) | ND | ND |
| Xyn10B + Xyn11A + Xyn10A + Xyl43A | ND | 12.5 ± 1.4 (83.4) | ND | ND | ND |
| Trivalent designer cellulosome | |||||
| Scaffoldin (10B-t + 11A-XBM-a +10A-f) | ND | 14.0 ± 0.7 (93.3) | 6.2 ± 1.7 (21.9) | ND | ND |
| Scaffoldin (10B-t + 11A-XBM-a +10A-f +43-b) | ND | 26.6 ± 2.1 (177) | ND | ND | ND |
Values are given in mg/g substrate ± standard deviation (with µmol/g substrate included parenthetically). Data were obtained by high-performance liquid chromatography (HPLC) analysis. Absence of glucose was confirmed by using a glucose assay kit. Values for xylose were corroborated using a xylose assay kit. An unidentified peak, present only after enzymatic treatments, eluted at ~4 min (between the xylose and xylobiose peaks), possibly a modified monosaccharide, as previously described (2, 15, 16).
ND, not detected.
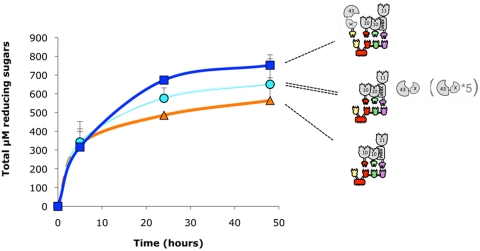
FIG 6 .
Kinetic studies of wheat straw hydrolysis by the various complexes. The curves shown represent degradation by the tetravalent designer cellulosome (■), degradation by the trivalent designer cellulosome (▲), degradation by the trivalent designer cellulosome and the free wild-type Xyl43A (○), and degradation by the trivalent designer cellulosome and the 5-fold-concentrated free wild-type Xyl43A (●). Enzymatic activity is defined as µM total reducing sugars. Triplicates of each reaction were carried out. Standard deviations are indicated.
Yield calculations.
In an earlier report (27), the washed wheat-straw substrate was found to contain 3.3 mmol of acid-extractable reducing sugar/g of dry matter, comprising approximately 2.3 mmol of glucose, 0.8 mmol of xylose, and 0.1 mmol of arabinose per g of dry matter. Based on these values, in the present work, the reaction yields for xylose after 42 h comprised about 25% and 20.7% for the tetravalent and trivalent designer cellulosome systems, respectively, versus 14.3% and 11.4% for the corresponding complement of wild-type enzymes.
Influence of Xyl43A incorporation into xylanolytic designer cellulosomes on hatched wheat straw degradation.
Wild-type free Xyl43A β-xylosidase was tested for its cooperative effect on the activity of the trivalent designer cellulosome (bearing 10B-t, 11A-XBM-a, and 10A-f), and the activity profile on straw was compared to that of the tetravalent designer cellulosome (bearing 10B-t, 11A-XBM-a, and 10A-f together with 43-b) (Fig. 6). Adding free wild-type Xyl43A to the trivalent designer cellulosome served to increase reducing sugar production, by driving xylan degradation to completion and releasing higher levels of xylose. Interestingly, adding the free Xyl43A β-xylosidase to the trivalent designer cellulosome was less efficient than including the enzyme in the tetravalent designer cellulosome complex, even when the concentration of free Xyl43A was elevated five times. This suggests that the dockerin-containing 43-b plays a fundamental enzymatic role as a component of the designer cellulosome.
DISCUSSION
Native cellulosomes comprise large multicomponent complexes, which are heterogeneous in content. The observed heterogeneity reflects the basic needs of the bacterial system as the plant cell wall polysaccharides are being degraded. The process is intricate and dynamic, during which the recalcitrant and complex nature of the cellulosic substrate is greatly transformed. Complementary types of enzymes are necessary to hydrolyze the intractable portions of the substrate, and new types of enzymes are required to deal with both the accumulating degradation products and the newly exposed polysaccharide components.
The designer cellulosome approach was established for two major reasons. First, in order to address the contribution to substrate degradation by specific cellulosomal components, a synthetic biology strategy would be advantageous, whereby the content of individual cellulosome assemblies can be precisely controlled. Second, this approach may offer an effective means for preparing efficient artificial systems for a variety of biotechnological and nanotechnological applications, notably for the conversion of cellulosic biomass into biofuels. The present work describes our continued efforts in the development of the designer cellulosome platform towards these goals.
In previous work, we developed the free enzyme system of Thermobidfida fusca for conversion into the cellulosomal mode. T. fusca possesses a concise set of only six cellulases and four xylanases. This very limited panel of enzymes allows the very attractive possibility of converting the entire enzymatic system into the cellulosomal mode, which eliminates the difficulties in selecting enzymes for inclusion into designer cellulosomes. In this context, more sophisticated enzymatic systems, like those of C. thermocellum, produce more than 70 dockerin-bearing proteins, and selection of preferred components would be more difficult. In the case of the highly cellulolytic T. fusca, nature has taken care of the selection process for us. T. fusca thus represents an excellent system for preparation of distinct artificial designer cellulosomes and for comparison of their performance with that of the free wild-type counterparts.
Initial research using the T. fusca system focused on the conversion of its free cellulase system to the cellulosomal mode (11–14). One advantage of using this system lies in the fact that the enzymes are not inherently cellulosomal, but can be easily converted to dockerin-containing enzymes which then fit into complementary scaffoldins. In subsequent investigations, we considered the combined action of cellulases and xylanases together in the same designer cellulosome complex (15, 16). In the present article, we converted the entire xylanolytic system of T. fusca into a defined designer cellulosome complex for comparison with the free xylanase system.
In the past, a variety of naturally occurring xylanase complexes have been described. In some bacteria, several types of xylanases appear to be organized in multifunctional supermolecular complexes. The presence of a complex, presumed to be analogous to the cellulosome paradigm but with predominant xylanase activity, was first reported in Butyrivibrio fibrisolvens (30, 31, 46). Cell-associated xylanase complexes were also observed in the anaerobic thermophilic bacterium Thermoanaerobacterium saccharolyticum grown on insoluble xylan (47). High xylanolytic activity was also reported in large multienzyme complexes (termed “xylanosomes”) derived from aerobic fungi (32). More recently, a multienzyme complex from Bacillus licheniformis SVD1 was examined, and seven xylanases were identified (together with two cellulases, two mannanases, and a pectinase) (48). The most extensively investigated “xylanosome” was found in Streptomyces olivaceoviridis. In this bacterium, the multienzyme complex was found to exhibit mainly xylanolytic activities and very low cellulolytic activity (29, 49). Endoxylanases in the complex were related to families 10 and 11 (50, 51). The family 10 xylanase holds a xylan-binding module, which was suggested to confer the ability to bind specifically to xylan (52). Intriguingly, scaffoldin subunits were not found in this complex, and the question of how the xylanases aggregate remained unanswered. Moreover, the xylanosome complex was not associated with the bacterial cell wall.
It is of interest to consider whether or not one should use the term “xylanosome” as a general description of xylanase-containing multienzyme complexes. Specifically, is the term used in the correct context, and can it be applied to such enzymes complexes? Indeed, with reference to naturally occurring cellulosomes, it should be noted that in addition to cellulases, they also contain hemicellulases and a variety of other plant cell wall-degrading enzymes, such as pectinases and carbohydrate esterases (53–55). We therefore contend that “cellulosome” should be generally employed as a generic term to describe enzyme complexes that exhibit the features of one or more scaffoldins that serve to incorporate dockerin-bearing enzymes via their cohesins, independent of the exact type(s) of enzyme components included in the complex. In this context, Clostridium papyrosolvens C7 was found to produce several types of multienzyme complexes, some of which were found to comprise xylanases exclusively (56). In this particular case, C. papyrosolvens produces defined cellulosomes, characterized by a cohesin-containing scaffoldin and dockerin-containing enzymatic subunits, and further division into cellulosomes, xylanosomes, etc., would be unwarranted.
The organization of enzymes into cellulosomes has been proposed in the past to provide a particularly attractive platform for enhancing the synergistic activity of complementary enzymes, notably the cellulosomal cellulases, on recalcitrant cellulosic substrates. On the molecular level, the mechanisms that could account for such a phenomenon would include proximity and substrate-targeting effects (23, 24, 27), as well as substrate channeling (57–59). The trivalent and tetravalent xylanase-containing designer cellulosomes produced in the present work exhibited enhanced levels of degradation of the natural recalcitrant wheat straw substrate compared to the combined action of the corresponding free wild-type enzymes. As demonstrated recently for the wheat straw substrate (11–13, 15, 25–27), enzyme proximity has also been shown in the present work to be the major source of improvement in substrate degradation, and little or no contribution could be attributed to the substrate-targeting effect. It is perhaps surprising that a designer cellulosome that contains only xylanases would exhibit such a strong proximity effect on the natural wheat straw substrate. Nevertheless, the strong binding of the designer cellulosome to the hatched wheat straw substrate via the scaffoldin-borne CBM3a indicates that the cellulose fibrils of the substrate are readily accessible, suggesting that the tethered xylanases would thus act in an enhanced synergistic manner to degrade the neighboring xylan components.
The Xyl43A β-xylosidase served to further enhance the activity of the three xylanases of T. fusca in both the free and cellulosomal modes, presumably by driving the enzymatic reaction more to completion and producing xylose as a major end product. The location of the Xyl43A enzyme as a bona fide component of the designer cellulosome was associated with further enhancement of reducing sugar production. It is currently difficult to assign an exact mechanistic explanation for this observation. Clearly, the effect is not directly associated with an overall xylosidase-mediated relief of enzyme inhibition, since excess free (wild-type) Xyl43A failed to approach the levels observed in the designer cellulosomes. An alternative effect may be related to substrate channeling between one or more of the other enzyme components of the designer cellulosome and the 43A-b β-xylosidase.
This work demonstrates the incorporation of a multiplicity of xylanases into a single designer cellulosome complex, without complementary cellulases, thereby substantiating the utility of this approach for enhancing xylanolytic activities on natural substrates. Indeed, even in the absence of chemical or thermal pretreatment of the hatched wheat straw substrate, encouraging xylose yields (up to 25%) were achieved. Xylanolytic designer cellulosomes may thus be appropriate for future environmentally friendly industrial applications, notably for the conversion of plant biomass to commodity products and biofuels.
MATERIALS AND METHODS
Cloning.
Plasmids encoding the wild-type enzymes (pXyn10B, pXyn11A, p10B-t, and p11-XBM-a) and scaffoldins (pScaf·B, pScaf·T, pScaf·F, pScaf·A, and pScaf·BTAF), were cloned as described earlier (15, 16, 34, 35, 60). Xyl43A (Tfu 1616; GenBank accession no. AAZ555651) was cloned from Thermobifida fusca YX genomic DNA. Primers were designed with the program Oligo Primer Analysis Software version 5.1 and ordered at the Weizmann Institute of Sciences facility (N-terminal primer, 5′ TCATGACATATGCACCATCACCATCACCATACTTCTCCCCAAGTCACGTCCT 3′; C-terminal primer, 5′ TGATTGCTCGAGTTAGGAGGGGGACTGAG GCCGGTA 3′) (NdeI and XhoI sites in boldface). The PCR product was inserted and ligated into linearized pET21a to form pXyl43A.
Xyn10A was amplified from T. fusca YX genomic DNA using the 5′ CCG CCGCTAGCATGCACCATCACCATCACCACACTGAAACCCGTCACCGTCCTTC 3′ and 5′ TAATTTGAGCTCTCATGAACAGGTGGCTCCGTTGAGGG 3′ primers (NheI and SacI sites in boldface) and ligated into linearized pET21a to form pXyn10A. To form p10A-f, the Xyn10A catalytic module was cloned using the wild-type forward primer described above and 5′ TCCCAAGAGCTCCGTCGAACTAGTGCCGTCACCGGGGCCGCCC 3′ (NheI and SacI sites in boldface) and ligated to the Ruminococcus flavefaciens dockerin from ScaA amplified using the 5′ CCATCGAGCTCACAACTCCTCAGCCCGGCAC 3′ and 5′ GGTGGTCTCGAGTTACTGAGGAAGTGTGATGA 3′ primers (SacI and XhoI sites in boldface). The Bacteroides cellulosolvens dockerin from Cel48 was amplified from the bacterial genomic DNA using the 5′ TTATTAACTAGTGGTCTCATCTATCCAAAAGGCACAG 3′ and 5′ TTATTAGAATTCTTA TTTTTGTTCTGCTGGGAACTCG 3′ primers (SpeI and EcoRI sites in boldface) and was added to the Xyl43A catalytic module C terminal (cloned using above wild-type forward primer and 5′ CTATGAACTAGTGGAGGGGGACTGAGGCCGGTA 3′ [SpeI site in boldface]) as a reverse primer to form p43-b.
PCRs were performed using ABGene Reddymix ×2 (Advanced Biotechnologies, Ltd., Epsom, United Kingdom), DNA samples were purified using a HiYield gel/PCR fragment extraction kit (Real Biotech Corporation, RBC, Banqiao City, Taiwan).
Protein expression and purification.
Wild-type and chimeric enzymes and recombinant scaffoldins (Xyn10B, Xyn11A, 10B-t, 11A-XBM-a, Scaf·B, Scaf·T, Scaf·F, Scaf·A, and Scaf·BTAF) were prepared as described earlier (34, 35, 61). The plasmids containing the genes coding for Xyn10A, 10A-f, Xyl43A, and 43-b were expressed in Escherichia coli BL21(DE3)/pLysS cells, and the relevant His-tagged proteins were purified on a Ni-nitrilotriacetic acid (NTA) column (Qiagen) according to the previously reported protocol (11). Scaffoldin plasmids were expressed in the same E. coli strain and purified on phosphoric acid-swollen cellulose (PASC) at 7.5 mg ml/ml and pH 7 by virtue of their resident CBM3a (60). SDS-PAGE (12% gels) was performed to examine the purity of the recombinant proteins. The concentration of each of the purified proteins was estimated by absorbance (280 nm), based on the known amino acid composition of the protein (http://www.expasy.org/tools/protparam.html). Typical yields of purified proteins for the preparations of the chimeric enzymes 10B-t, 11A-XBM-a, 10A-f, and 43A-b were 93.9, 4.3, 84.8, and 8.7 mg/liter, respectively. Wild-type enzyme preparations yielded 48.7, 5.4, 79.8, and 31.2 mg/liter of purified proteins for Xyn10B, Xyn11A, Xyn10A, and Xyl43A, respectively. Proteins were stored in 50% (vol/vol) glycerol at −20°C.
Affinity-based enzyme-linked immunosorbent assay.
Cohesin-dockerin specificity was determined by the matching fusion-protein procedure of Barak et al. (11, 43).
Cellulose-binding assay.
Equimolar mixtures of purified proteins (70 pmol each in 50 mM citrate buffer, pH 6.0, 12 mM CaCl2, 2 mM EDTA) were incubated for 2 h at 37°C and mixed with 20 mg of microcrystalline cellulose (FMC Biopolymer, Philadelphia, PA) in a final volume of 200 µl. The tubes were rotated gently at 4°C for 1 h and centrifuged at 14,000 rpm for 2 min. The supernatant fluids (containing unbound protein) were discarded, and the cellulose pellet was washed twice by resuspension in 100 µl of the same buffer (supplemented with 0.05% of Tween 20 to eliminate nonspecific binding) and centrifugation at 14,000 rpm for 2 min. The resultant pellet was resuspended in 60 µl of SDS-containing buffer and heated at 100°C for 10 min to dissociate any bound protein. Equimolar mixtures of enzymes without the scaffoldin were used as a negative control to ensure specificity of binding. The chimeric enzyme 11A-a (lacking the XBM [15]) was used in the latter control samples to avoid direct binding of the 11A-a enzyme to cellulose. Bound and unbound fractions and individual proteins were analyzed by SDS-PAGE (10% gels). A similar assay was also performed after an incubation period of 48 h at 50°C, in order to examine the stability of the different cohesin-dockerin interactions—notably those derived from mesophilic bacteria—under the conditions of the cellulose-degrading activity assay, as performed in this study.
Wheat straw binding assay.
The above-described procedure was applied to assay the binding abilities of the designer cellulosomes to the wheat straw substrate, by substituting 10 mg of blended hatched wheat straw in place of microcrystalline cellulose in the assay mixture.
Affinity electrophoresis-based carbohydrate-binding assay.
The procedure of Tomme et al. (62) was followed with minor modifications. Nondenaturing continuous PAGE (12.5% acrylamide gels) was performed in 1.5 M Tris-HCl buffer (pH 8.2). For ligand-containing gels, oat spelt xylan or PASC was added to the gel mixtures at a final concentration of 0.1% (wt/vol) prior to polymerization. Native gel without ligand was run simultaneously under the same conditions. Electrophoresis was carried out at room temperature and 100 V. Bovine serum albumin (BSA) (MP Biomedicals, LLC, United States) was used as a negative nonbinding control. Separated proteins were revealed by staining with Coomassie blue.
Nondenaturing PAGE.
Differential mobility on nondenaturing gels was used to examine the interaction between the chimeric scaffoldin and enzymes. Equimolar concentrations of each protein (4 to 8 µg) incorporated into the designer cellulosome complexes were mixed in a 30-µl reaction mixture, which contained 15 µl of Tris-buffered saline, pH 7.4 (TBS), supplemented with 10 mM CaCl2 and 0.05% Tween 20. The protein mixtures were incubated for a minimum of 1.5 h at 37°C to enable the binding interaction. Sample buffer (7.5 µl in the absence of SDS) was added to 15 µl of the reaction mixture, and the samples were loaded onto nondenaturating gels (4.3% stacking gel and 9% separating gel). A parallel SDS-PAGE gel (10%) was performed on the remaining 15-µl sample supplemented with 7.5 µl of sample buffer containing SDS.
Enzymatic activity.
The dinitrosalicylic acid (DNS) method (63, 64) was used to determine xylanase activity by measuring reducing sugars released from the xylan substrate. The composition of a typical assay mixture consisted of the enzyme (0 to 10 nM) added to 100 µl of 50 mM citrate buffer, pH 6.0, supplemented with 12 mM CaCl2 and 2 mM EDTA. Aliquots (100 µl) of the desired xylan substrate, either birchwood, beechwood, or oat spelt xylan (Sigma, Chemical Co., St. Louis, MO), were added to the reaction mixture at a 2% concentration (suspended in 50 mM citrate buffer pH 6.0), and the reaction tubes were incubated for 20 min at 50°C. The tubes were transferred to an ice-water bath and centrifuged, and 100 µl of the supernatant was added to 150 µl DNS reagent. The tubes were then boiled for 10 min, and absorbance (540 nm) was measured. Generation of reducing sugars was determined using xylose as a standard.
Before use, the hatched wheat straw substrate (0.2 to 0.8 mm; Valagro, Poitiers, France) was subjected to a blending protocol as described previously (15, 27, 65). The concentration of the hatched wheat straw was set at 3.5 g/liter, and the concentration of xylanases was set at 0.3 µM, in an assay mixture of similar composition as described above. Prior to assay, the dockerin-containing enzymes were incubated for 2 h at 37°C (in the absence of substrate) in the presence of equimolar concentrations of scaffoldin. All assays were performed in triplicate.
Sugar identification and analysis.
Sugar content was analyzed using a high-performance anion-exchange chromatography (HPAEC) system equipped with a PA1 column (Dionex, Sunnyvale, CA). Samples, comprising the supernatant fluids of the reaction mixtures obtained after centrifugation, were loaded onto the PA1 column, and NaOH (200 mM) served as the effluent. Arabinose, xylose, xylobiose, and xylotriose standards were loaded separately to determine elution time and peak areas as a function of the sugar concentration, in order to identify the different sugars present in each sample as well as their concentrations. Arabinose and xylose were observed in blank controls at low levels (samples consisted of double-distilled water instead of the enzymatic mixture); these values were deducted in all the samples.
Concentrations of xylose and glucose (or the absence thereof) were confirmed by a d-xylose assay kit (Megazyme, Wicklow, Ireland); and a glucose assay kit (GAGO20; Sigma), respectively, in both cases according to the manufacturer’s instructions.
ACKNOWLEDGMENTS
We are grateful for the technical assistance of Dan Goldman (Technion, Haifa, Israel) and Tali Scherf (Weizmann Institute of Sciences, Rehovot, Israel).
S.M. greatly appreciates a scholarship received from the Ministry of Immigrant Absorption, Jerusalem, Israel. This research was supported by grants from the United States-Israel Binational Science Foundation (BSF), Jerusalem, Israel, by the Alternative Energy Research Initiative Bioenergy Consortium, by the Brazilian friends of the Weizmann Institute of Science Alternative Energy Research Initiative (research grants from Charles Rothschild, Mario Fleck, and Roberto and Renata Ruhman), by the Technion-Niedersachsen Research Cooperation Program, a grant to E.A.B. and R.L. from the Israel Ministry of Science (IMOS), by grants (no. 966/09, 159/07, and 24/11 to E.A.B. and R.L., respectively) by the Israel Science Foundation and establishment of an Israeli Center of Research Excellence (I-CORE Center no. 152/11, E.A.B. and Y.S.) managed by the Israel Science Foundation. Y.S. holds the Erwin and Rosl Pollak Chair in Biotechnology at Technion, and E.A.B. is the incumbent of the Maynard I. and Elaine Wishner Chair of Bio-Organic Chemistry.
Footnotes
Citation Moraïs S, et al. 2011. Assembly of xylanases into designer cellulosomes promotes efficient hydrolysis of the xylan component of a natural recalcitrant cellulosic substrate. mBio 2(6):e00233-11. doi:10.1128/mBio.00233-11.
REFERENCES
- 1. Beg QK, Kapoor M, Mahajan L, Hoondal GS. 2001. Microbial xylanases and their industrial applications: a review. Appl. Microbiol. Biotechnol. 56:326–338 [DOI] [PubMed] [Google Scholar]
- 2. Selig MJ, Knoshaug EP, Adney WS, Himmel ME, Decker SR. 2008. Synergistic enhancement of cellobiohydrolase performance on pretreated corn stover by addition of xylanase and esterase activities. Bioresour. Technol. 99:4997–5005 [DOI] [PubMed] [Google Scholar]
- 3. Margeot A, Hahn-Hagerdal B, Edlund M, Slade R, Monot F. 2009. New improvements for lignocellulosic ethanol. Curr. Opin. Biotechnol. 20:372–380 [DOI] [PubMed] [Google Scholar]
- 4. Himmel ME, et al. 2007. Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science 315:804–807 . (Erratum, 316:982, 2008 [DOI] [PubMed] [Google Scholar]
- 5. Lynd LR, et al. 2008. How biotech can transform biofuels. Nat. Biotechnol. 26:169–172 [DOI] [PubMed] [Google Scholar]
- 6. Lynd LR, Weimer PJ, van Zyl WH, Pretorius IS. 2002. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 66:506–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mechaly A, et al. 2000. Overproduction and characterization of seleno-methionine xylanase T-6. J. Biotechnol. 78:83–86 [DOI] [PubMed] [Google Scholar]
- 8. Suurnakki A, Tenkanen M, Buchert J, Viikari L. 1997. Hemicellulases in the bleaching of chemical pulps. Adv. Biochem. Eng. Biotechnol. 57:261–287 [DOI] [PubMed] [Google Scholar]
- 9. Viikari L, Pauna M, Kantelinen A, Sundquist J, Linko M. 1986. Bleaching with enzymes. In Proceedings of the 3rd International Conference on Biotechnololgy in the Pulp and Paper Industry, p 67–69Swedish Forest Products Research Laboratory, Stockholm, Sweden [Google Scholar]
- 10. Woodward J. 1984. Xylanases: functions, properties and applications. Top. Enzyme Ferment. Biotechnol. 8:9–30 [Google Scholar]
- 11. Caspi J, et al. 2006. Thermobifida fusca family-6 cellulases as potential designer cellulosome components. Biocatal. Biotransform. 24:3–12 [Google Scholar]
- 12. Caspi J, et al. 2008. Conversion of noncellulosomal Thermobifida fusca free exoglucanases into cellulosomal components: comparative impact on cellulose-degrading activity. J. Biotechnol. 135:351–357 [DOI] [PubMed] [Google Scholar]
- 13. Caspi J, et al. 2009. Effect of linker length and dockerin position on conversion of a Thermobifida fusca endoglucanase to the cellulosomal mode. Appl. Environ. Microbiol. 75:7335–7342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Caspi J, et al. 2010. Thermobifida fusca exoglucanase Cel6B is incompatible with the cellulosomal mode in contrast to endoglucanase Cel6A. Syst. Synth. Biol. 4:193–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Moraïs S, et al. 2010. Contribution of a xylan-binding module to the degradation of a complex cellulosic substrate by designer cellulosomes. Appl. Environ. Microbiol. 76:3787–3796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Moraïs S, et al. 2010. Cellulase-xylanase synergy in designer cellulosomes for enhanced degradation of a complex cellulosic substrate. mBio 1:e00285–00310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bayer EA, Morag E, Lamed R. 1994. The cellulosome—a treasure-trove for biotechnology. Trends Biotechnol. 12:378–386 [DOI] [PubMed] [Google Scholar]
- 18. Pagès S, et al. 1999. Sequence analysis of scaffolding protein CipC and ORFXp, a new cohesin-containing protein in Clostridium cellulolyticum: comparison of various cohesin domains and subcellular localization of ORFXp. J. Bacteriol. 181:1801–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fierobe H-P, et al. 1999. Cellulosome from Clostridium cellulolyticum: molecular study of the dockerin/cohesin interaction. Biochemistry 38:12822–12832 [DOI] [PubMed] [Google Scholar]
- 20. Yaron S, Morag E, Bayer EA, Lamed R, Shoham Y. 1995. Expression, purification and subunit-binding properties of cohesins 2 and 3 of the Clostridium thermocellum cellulosome. FEBS Lett. 360:121–124 [DOI] [PubMed] [Google Scholar]
- 21. Pagès S, et al. 1997. Species-specificity of the cohesin-dockerin interaction between Clostridium thermocellum and Clostridium cellulolyticum: prediction of specificity determinants of the dockerin domain. Proteins 29:517–527 [PubMed] [Google Scholar]
- 22. Lytle B, Myers C, Kruus K, Wu JHD. 1996. Interactions of the CelS binding ligand with various receptor domains of the Clostridium thermocellum cellulosomal scaffolding protein, CipA. J. Bacteriol. 178:1200–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lamed R, Setter E, Bayer EA. 1983. Characterization of a cellulose-binding, cellulase-containing complex in Clostridium thermocellum. J. Bacteriol. 156:828–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lamed R, Setter E, Kenig R, Bayer EA. 1983. The cellulosome—a discrete cell surface organelle of Clostridium thermocellum which exhibits separate antigenic, cellulose-binding and various cellulolytic activities. Biotechnol. Bioeng. Symp. 13:163–181 [Google Scholar]
- 25. Fierobe H-P, et al. 2001. Design and production of active cellulosome chimeras. Selective incorporation of dockerin-containing enzymes into defined functional complexes. J. Biol. Chem. 276:21257–21261 [DOI] [PubMed] [Google Scholar]
- 26. Fierobe H-P, et al. 2002. Degradation of cellulose substrates by cellulosome chimeras. Substrate targeting versus proximity of enzyme components. J. Biol. Chem. 277:49621–49630 [DOI] [PubMed] [Google Scholar]
- 27. Fierobe H-P, et al. 2005. Action of designer cellulosomes on homogeneous versus complex substrates: controlled incorporation of three distinct enzymes into a defined trifunctional scaffoldin. J. Biol. Chem. 280:16325–16334 [DOI] [PubMed] [Google Scholar]
- 28. Vazana Y, Moraïs S, Barak Y, Lamed R, Bayer EA. 2010. Interplay between Clostridium thermocellum family 48 and family 9 cellulases in cellulosomal versus noncellulosomal states. Appl. Environ. Microbiol. 76:3236–3243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jiang ZQ, et al. 2004. A novel, ultra-large xylanolytic complex (xylanosome) secreted by Streptomyces olivaceoviridis. Biotechnol. Lett. 26:431–436 [DOI] [PubMed] [Google Scholar]
- 30. Lin LL, Thomson JA. 1991. An analysis of the extracellular xylanases and cellulases of Butyrivibrio fibrisolvens H17c. FEMS Microbiol. Lett. 68:197–203 [DOI] [PubMed] [Google Scholar]
- 31. Lin LL, Thomson JA. 1991. Cloning, sequencing and expression of a gene encoding a 73 kDa xylanase enzyme from the rumen anaerobe Butyrivibrio fibrisolvens H17c. Mol. Gen. Genet. 228:55–61 [DOI] [PubMed] [Google Scholar]
- 32. Ohtsuki T, et al. 2005. Production of large multienzyme complex by aerobic thermophilic fungus Chaetomium sp. nov. MS-017 grown on palm oil mill fibre. Lett. Appl. Microbiol. 40:111–116 [DOI] [PubMed] [Google Scholar]
- 33. Sunna A, Antranikian G. 1997. Xylanolytic enzymes from fungi and bacteria. Crit. Rev. Biotechnol. 17:39–67 [DOI] [PubMed] [Google Scholar]
- 34. Irwin D, Jung ED, Wilson DB. 1994. Characterization and sequence of a Thermomonospora fusca xylanase. Appl. Environ. Microbiol. 60:763–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kim JH, Irwin D, Wilson DB. 2004. Purification and characterization of Thermobifida fusca xylanase 10B. Can. J. Microbiol. 50:835–843 [DOI] [PubMed] [Google Scholar]
- 36. Gruppen H, et al. 1992. Characterisation by 1H NMR spectroscopy of enzymically derived oligosaccharides from alkali-extractable wheat-flour arabinoxylan. Carbohydr. Res. 233:45–64 [DOI] [PubMed] [Google Scholar]
- 37. Kormelink FMJ, Gruppen H, Viëtor RJ, Voragen AGJ. 1993. Mode of action of the xylan-degrading enzymes from Aspergillus awamori on alkali-extractable cereal arabinoxylans. Carbohydr. Res. 249:355–367 [DOI] [PubMed] [Google Scholar]
- 38. Guo B, Chen XL, Sun CY, Zhou BC, Zhang YZ. 2009. Gene cloning, expression and characterization of a new cold-active and salt-tolerant endo-b-1,4-xylanase from marine Glaciecola mesophila KMM 241. Appl. Microbiol. Biotechnol. 84:1107–1115 [DOI] [PubMed] [Google Scholar]
- 39. Blanco J, Coque JJ, Velasco J, Martín JF. 1997. Cloning, expression in Streptomyces lividans and biochemical characterization of a thermostable endo-β-1,4-xylanase of Thermomonospora alba ULJB1 with cellulose-binding ability. Appl. Microbiol. Biotechnol. 48:208–217 [DOI] [PubMed] [Google Scholar]
- 40. Wang WK, Kruus K, Wu JHD. 1993. Cloning and DNA sequence of the gene coding for Clostridium thermocellum cellulase SS (CelS), a major cellulosome component. J. Bacteriol. 175:1293–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ding S-Y, et al. 2001. Cellulosomal scaffoldin-like proteins from Ruminococcus flavefaciens. J. Bacteriol. 183:1945–1953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Asenjo JA. 1983. Maximizing the formation of glucose in the enzymatic hydrolysis of insoluble cellulose. Biotechnol. Bioeng. 25:3185–3190 [DOI] [PubMed] [Google Scholar]
- 43. Barak Y, et al. 2005. Matching fusion-protein systems for affinity analysis of two interacting families of proteins: the cohesin-dockerin interaction. J. Mol. Recognit. 18:491–501 [DOI] [PubMed] [Google Scholar]
- 44. Kim TW, et al. 2010. Binding modules alter the activity of chimeric cellulases: effects of biomass pretreatment and enzyme source. Biotechnol. Bioeng. 107:601–611 [DOI] [PubMed] [Google Scholar]
- 45. Molinier AL, et al. 2011. Synergy, structure and conformational flexibility of hybrid cellulosomes displaying various inter-cohesins linkers. J. Mol. Biol. 405:143–157 [DOI] [PubMed] [Google Scholar]
- 46. Mannarelli BM, Evans S, Lee D. 1990. Cloning, sequencing, and expression of a xylanase gene from the anaerobic ruminal bacterium Butyrivibrio fibrisolvens. J. Bacteriol. 172:4247–4254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zeikus J, Lee C, Lee Y, Saha B. 1991. Thermostable saccharidases: new sources, uses, and biodesigns. ACS Symp. Ser. 460:36-51 [Google Scholar]
- 48. Van Dyk J, Sakka M, Sakka K, Pletschke B. 2010. Identification of endoglucanases, xylanases, pectinases and mannanases in the multi-enzyme complex of Bacillus licheniformis SVD1. Enzyme Microb. Technol. 47:112–118 [Google Scholar]
- 49. Deng W, et al. 2005. Variation of xylanosomal subunit composition of Streptomyces olivaceoviridis by nitrogen sources. Biotechnol. Lett. 27:429–433 [DOI] [PubMed] [Google Scholar]
- 50. Fujimoto Z, et al. 2000. Crystal structure of Streptomyces olivaceoviridis E-86 β-xylanase containing xylan-binding domain. J. Mol. Biol. 300:575–585 [DOI] [PubMed] [Google Scholar]
- 51. Kaneko S, et al. 2000. Purification and characterization of a family G/11 b-xylanase from Streptomyces olivaceoviridis E-86. Biosci. Biotechnol. Biochem. 64:447–451 [DOI] [PubMed] [Google Scholar]
- 52. Jiang Z, et al. 2006. Subunit composition of a large xylanolytic complex (xylanosome) from Streptomyces olivaceoviridis E-86. J. Biotechnol. 126:304–312 [DOI] [PubMed] [Google Scholar]
- 53. Schwarz WH. 2001. The cellulosome and cellulose degradation by anaerobic bacteria. Appl. Microbiol. Biotechnol. 56:634–649 [DOI] [PubMed] [Google Scholar]
- 54. Tamaru Y, Doi RH. 2001. Pectate lyase A, an enzymatic subunit of the Clostridium cellulovorans cellulosome. Proc. Natl. Acad. Sci. U. S. A. 20:4125–4129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Raman B, et al. 2009. Impact of pretreated switchgrass and biomass carbohydrates on Clostridium thermocellum ATCC 27405 cellulosome composition: a quantitative proteomic analysis. PLoS One 4:e5271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pohlschröder M, Leschine SB, Canale-Parola E. 1994. Multicomplex cellulase-xylanase system of Clostridium papyrosolvens C7. J. Bacteriol. 176:70–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lamed R, Bayer EA. 1988. The cellulosome of Clostridium thermocellum. Adv. Appl. Microbiol. 33:1–46 [Google Scholar]
- 58. Mayer F, Coughlan MP, Mori Y, Ljungdahl LG. 1987. Macromolecular organization of the cellulolytic enzyme complex of Clostridium thermocellum as revealed by electron microscopy. Appl. Environ. Microbiol. 53:2785–2792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhang Y. 2011. Substrate channeling and enzyme complexes for biotechnological applications. Biotechnol. Adv. 29:715–725 [DOI] [PubMed] [Google Scholar]
- 60. Haimovitz R, et al. 2008. Cohesin-dockerin microarray: diverse specificities between two complementary families of interacting protein modules. Proteomics 8:968–979 [DOI] [PubMed] [Google Scholar]
- 61. Irwin DC, Zhang S, Wilson DB. 2000. Cloning, expression and characterization of a family 48 exocellulase, Cel48A, from Thermobifida fusca. Eur. J. Biochem. 267:4988–4997 [DOI] [PubMed] [Google Scholar]
- 62. Tomme P, Boraston A, Kormos JM, Warren RA, Kilburn DG. 2000. Affinity electrophoresis for the identification and characterization of soluble sugar binding by carbohydrate-binding modules. Enzyme Microb. Technol. 27:453–458 [DOI] [PubMed] [Google Scholar]
- 63. Ghose TK. 1987. Measurements of cellulase activity. Pure Appl. Chem. 59:257–268 [Google Scholar]
- 64. Miller GL. 1959. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Biochem. 31:426–428 [Google Scholar]
- 65. Tabka MG, Herpoël-Gimbert I, Monod F, Asther M, Sigoillot JC. 2006. Enzymatic saccharification of wheat straw for bioethanol production by a combined cellulase xylanase and feruloyl esterase treatment. Enzyme Microb. Technol. 39:897–902 [Google Scholar]