Abstract
Purpose
Although most patients with estrogen receptor α (ER)-positive breast cancer initially respond to endocrine therapy, many ultimately develop resistance to antiestrogens. However, mechanisms of antiestrogen resistance and biomarkers predictive of such resistance are underdeveloped.
Experimental Design
We adapted four ER+ human breast cancer cell lines to grow in an estrogen-depleted medium. A gene signature of estrogen independence was developed by comparing expression profiles of long-term estrogen-deprived (LTED) cells to their parental counterparts. We evaluated the ability of the LTED signature to predict tumor response to neoadjuvant therapy with an aromatase inhibitor, and disease outcome following adjuvant tamoxifen. We utilized Gene Set Analysis (GSA) of LTED cell gene expression profiles and a loss-of-function approach to identify pathways causally associated with resistance to endocrine therapy.
Results
The LTED gene expression signature was predictive of high tumor cell proliferation following neoadjuvant therapy with anastrozole and letrozole, each in different patient cohorts. This signature was also predictive of poor recurrence-free survival in two studies of patients treated with adjuvant tamoxifen. Bioinformatic interrogation of expression profiles in LTED cells revealed a signature of MYC activation. The MYC activation signature and high MYC protein levels were both predictive of poor outcome following tamoxifen therapy. Finally, knockdown of MYC inhibited LTED cell growth.
Conclusions
A gene expression signature derived from ER+ breast cancer cells with acquired hormone independence predicted tumor response to aromatase inhibitors and associated with clinical markers of resistance to tamoxifen. In some cases, activation of the MYC pathway was associated with this resistance.
Keywords: aromatase, tamoxifen, myc, LTED, breast
INTRODUCTION
Estrogen receptor α (ER) and/or progesterone receptor (PR) are expressed in approximately 70% of breast cancers, which typically indicates that breast cancer cells depend upon estrogens for growth. Preferred therapies for such patients target ER signaling either by antagonizing binding of estrogens to the ER (tamoxifen), downregulating the ER (fulvestrant), or blocking estrogen biosynthesis (aromatase inhibitors, AIs). Although endocrine therapies have changed the natural history of hormone-dependent breast cancer, many tumors exhibit de novo or acquired resistance (1). While clinical data link ErbB2/HER2 overexpression with resistance to endocrine therapy (2-4), only <10% of hormone receptor-positive breast cancers overexpress HER2 (2), suggesting that mechanisms of escape from endocrine therapy remain undefined for the majority of patients.
Several studies have generated gene expression signatures using patient datasets with disease outcome to predict resistance to endocrine therapy. While this approach may yield predictive information, it does not provide a model in which to evaluate causal contributions of gene expression changes to an antiestrogen-resistant phenotype. In order to model breast cancers that are resistant to estrogen deprivation, we generated long-term estrogen-deprived (LTED) derivatives of four ER+ human breast cancer cell lines by maintaining them in conditions that mimic the low estrogen levels in patients treated with AIs [≤3 pM plasma 17β-estradiol (5)]. We compared LTED cells to respective parental controls to evaluate global changes in gene expression and defined a signature of 99 genes commonly deregulated across LTED lines. This LTED signature was predictive of a lower tumor response to neoadjuvant therapy with an AI, and poor outcome following adjuvant tamoxifen therapy in patients with hormone receptor-positive breast cancer. Furthermore, we bioinformatically derived a gene expression signature of MYC transcription factor activation in LTED cells that was predictive of poor outcome following tamoxifen therapy. High tumor MYC protein level was also predictive of shorter time-to-recurrence following adjuvant tamoxifen. Finally, siRNA-mediated depletion of MYC reduced the hormone-independent growth of LTED cells, suggesting that activated MYC could be a useful target for the treatment of antiestrogen-resistant breast cancer.
MATERIALS AND METHODS
Cell lines
MCF-7, ZR75-1, MDA-361, and HCC-1428 cells (ATCC) were maintained in IMEM/10% FBS (Gibco). Long-term estrogen-deprived (LTED) cell lines were generated by culturing cells under hormone-depleted conditions [phenol red-free IMEM/10% dextran-charcoal-treated FBS (DCC-FBS, Hyclone; contains <0.0367 pM 17β-estradiol) as described previously (6).
Gene expression microarray analysis of LTED cells
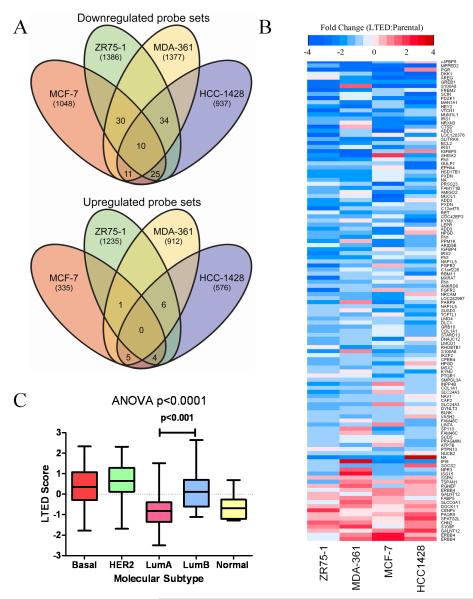
Raw data are available on the Gene Expression Omnibus (GEO; GSE19639). RNA harvested from cells incubated for 24 h in 10% DCC-FBS was analyzed using Affymetrix HG_U133_Plus_2 microarrays as described previously [(6); detailed in Supplemental Methods]. We compared the lists of probe sets altered across each LTED/parental comparison (≥1.5-fold, FDR-adjusted p≤0.05, Fig. 1A). Probe sets commonly deregulated in ≥3 LTED lines were included in the LTED gene signature.
Fig. 1.
A gene expression signature derived from cell lines with acquired estrogen independence is correlated with Luminal B breast cancer. A) Microarray probe sets down- or up-regulated ≥1.5-fold (p≤0.05) in LTED cells and parental controls were compared using Venn diagrams. Numbers of probe sets are indicated. B) Heatmap depicting the mean fold-change of each LTED line compared to respective parental controls for the 118 probe sets commonly deregulated in ≥3 LTED lines. C) LTED scores were calculated for 230 breast tumors (cohort POPOVICI230); tumors were subtyped according to the PAM50 classifier. LTED scores for each subtype were compared by ANOVA, followed by post-hoc t-test between subtypes.
Correlation of LTED signature genes with response to anastrozole
This patient cohort (SMITH69) was described previously (7). Core biopsies were collected prior to treatment (detailed in Supplemental Methods). Patients with hormone receptor-positive disease were treated with anastrozole. A repeat core biopsy was taken 14 days later. Ki67 immunohistochemical scoring was performed on 4-μm formalin-fixed, paraffin-embedded sections from biopsies. This study was approved by the Institutional Review Boards and Research and Ethics Committees of the recruiting centers.
RNA harvested from pretreatment and post-two-week-treatment frozen tumor samples was analyzed using Illumina HumanWG-6 v2 Expression BeadChips (detailed in Supplemental Methods). To examine the association between the 99-gene LTED signature and Ki67 score among patient tumors, 104 available probe sets mapping to 79/99 genes were extracted from the tumor dataset. Microarray data were filtered to extract the most variable probe set for each gene (in R software using package: genefilter). Next, all resulting signal intensities (log2-transformed) were multiplied by (−1) for genes upregulated in LTED cells, and (+1) for genes downregulated in LTED cells (Tables S1-S2). The sum value across all genes was then calculated for each patient tumor, generating an unscaled LTED ‘score’ that recapitulates the degree of similarity to the LTED cells. Thus, a high tumor LTED score corresponds to a signature highly concordant with resistance to estrogen deprivation. LTED scores were standardized to Z-scores by subtracting the mean score for the cohort, and then dividing by the standard deviation of the scores for the cohort (Table S3). Thus, the Z-scores for any given cohort have a mean of 0 with a standard deviation of 1. Z-scores were evaluated in a linear model in comparison to post-two-week-treatment Ki67 scores Gene expression and Ki67 data were available for 69 patients.
Correlation of LTED signature genes with response to letrozole
This patient dataset (MILLER48) was described previously (8). This study was approved by the local ethics committee (LREC; 2001/8/80 and 2001/8/81). Postmenopausal women with ER+ breast cancer were treated with neoadjuvant letrozole. Tumor gene expression data (.CEL files) were downloaded from GEO (GSE5462). Ki67 IHC scoring was performed as above. Gene expression and Ki67 data were available for 48 patients. Principal components analysis revealed a batch effect in the microarray data. LTED scores were calculated as above (using 78/99 available genes) and evaluated in a linear model (including the batch term) compared to post-treatment Ki67 values.
Correlation of LTED signature genes with response to tamoxifen
Tumor gene expression data (.CEL files) and patient characteristics from three cohorts (LOI164, LOI181, MDACC298) of patients with ER+ breast cancer treated with adjuvant tamoxifen for five years (9, 10) were downloaded from GEO. Patients from sites GUYT (n=87, GSE6532) and GUYT2 (n=77, GSE9195) comprised LOI164, which were analyzed on Affymetrix U133+2 microarrays (9). Patients from sites KIT and OXFT (n=181, GSE6532) comprised LOI181, which were analyzed on Affymetrix U133A and U133B microarrays. MDACC298 analysis was performed on Affymetrix U133A microarrays (GSE17705). Accession numbers of the patients which comprised LOI181 and LOI164 are annotated in Table S10, and analyses are described in Supplemental Methods. LTED scores were calculated as above (using 99/99, 97/99, and 78/99 available genes in LOI164, LOI181, and MDACC298, respectively). Patients were dichotomized based on median LTED score, and Kaplan-Meier survival curves were compared by log-rank test. The association of LTED score with relapse-free survival (RFS) was analyzed using Cox proportional hazards (PH) regression.
Molecular subtyping of clinical microarray datasets using the PAM50 classifier was performed using R package: genefu (11). Gene expression data from 230 breast cancers of mixed subtypes [GEO GSE20194 (12), POPOVICI230 cohort] were used to determine the association of LTED score with molecular subtype using robust scaling. Molecular subtyping was similarly performed on the tamoxifen-treated cohorts described above using no scaling, as all tumors were ER+.
Gene Set Analysis (GSA) is a variation on the Gene Set Enrichment Analysis algorithm and is described elsewhere (13, 14). Curated (c2) gene sets were downloaded from Molecular Signatures Database (14). GSA was performed using the two-class paired algorithm comparing each LTED cell line to its respective parental control in R (package: GSA; 1,000 permutations). Positively regulated gene sets (i.e., upregulated in LTED cells) were noted. Two gene sets specific to MYC activation were identified, which were combined to form a union set (231 genes), and filtered to identify genes scoring >1.0 by GSA; this yielded 46 genes. To test associations with clinical parameters, the expression values (log2-transformed) were summed across all 46 genes for each tumor, and the resulting MYC score was standardized to a Z-score within each cohort as above.
Reverse-phase protein array (RPPA) analysis
Forty-seven early-stage hormone receptor-positive primary breast tumor samples (MDACC47) were obtained from the Breast Tissue Frozen Tumor Bank at M.D. Anderson Cancer Center. These were from de-identified patients that received adjuvant tamoxifen therapy for five years. Specimens were collected under an IRB-approved protocol. Protein was extracted from tumors and analyzed by RPPA using a MYC antibody (Cell Signaling) as described previously [(6, 15, 16); detailed in Supplemental Methods).
siRNA targeting of MYC
Cells transfected with siRNA targeting MYC or non-silencing control (detailed in Supplemental Methods) were reseeded for cell proliferation and immunoblotting assays in 10% DCC-FBS. In proliferation assays, cells were trypsinized and counted after 6 days using a Coulter counter. Significant differences were determined by two-sided t-test (p<0.05). Protein lysates collected at two days post-transfection were analyzed by immunoblotting using antibodies against c-Myc (Santa Cruz) and actin (Sigma).
RESULTS AND DISCUSSION
Derivation of a gene expression signature of estrogen independence
We first compared gene expression profiles from four parental and their respective LTED cell lines. Venn diagrams were generated depicting microarray probe sets commonly altered across matching LTED/parental comparisons (Fig. 1A). We found 110 and 16 probe sets that were commonly down- or up-regulated across ≥3 LTED lines (≥1.5-fold, FDR-adjusted p≤0.05), respectively. These probe sets mapped to 86 down-regulated and 13 up-regulated genes, which we defined as the 99-gene LTED signature of estrogen independence (Table S4). A heatmap depicting the levels of expression of these 99 genes (118 probe sets) in LTED cells relative to respective parental controls is shown in Fig. 1B. Eight probe sets which mapped to multiple genes or were not annotated were excluded from further analysis.
Breast cancer is comprised of several molecular subtypes with different clinical outcomes (17). ER+ breast cancer can generally be segregated into luminal A and luminal B subtypes, with luminal B exhibiting reduced sensitivity to endocrine therapy and a worse prognosis compared to luminal A tumors (18). We hypothesized that the gene expression changes reflected in the LTED signature are associated with luminal B breast cancer. We performed molecular subtyping of 230 unselected breast tumors [POPOVICI230 cohort (12)] using the 50-gene PAM50 classifier (11). PAM50 subtypes displayed patterns consistent with clinical parameters (ER, PR, HER2). Using the 99 LTED signature genes (Fig. 1A), we generated an LTED score for each patient tumor (detailed in Methods). The tumor LTED score, which according to our analysis should reflect the degree of resistance to estrogen deprivation, significantly varied among subtypes (ANOVA p<0.001, Fig. 1C). A post-hoc analysis demonstrated that LTED score was significantly higher in luminal B than in luminal A tumors (t-test p<0.001), suggesting that the LTED signature associates with clinically relevant molecular subtypes of breast cancer.
LTED signature predicts clinical response to an aromatase inhibitor
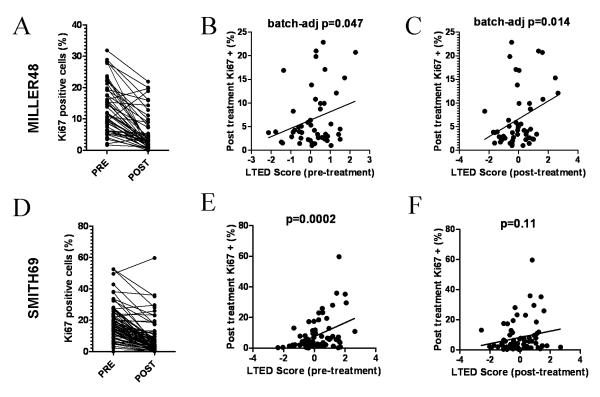
We next examined whether the expression profiles in LTED cells correlate with response to neoadjuvant letrozole. In a microarray dataset of primary tumor samples from 48 patients with ER+ breast cancer [MILLER48 cohort (7)], we calculated pretreatment and post-treatment (2 weeks) LTED scores for each tumor. To our knowledge, long-term follow-up in patients treated with an AI with tumor microarray data is currently unavailable. However, higher post-treatment Ki67 scores, a marker of high tumor cell proliferation (19), in primary cancers from patients treated with an AI for 2 weeks have been shown to predict poor long-term disease outcome (1). Thus, we performed Ki67 IHC on tumor biopsies obtained after 2 weeks of therapy with letrozole (Fig. 2A). Tumors with high pretreatment and post-treatment LTED scores showed significantly higher post-treatment (2 weeks) Ki67 scores (Fig. 2B-C; pretreatment LTED score r=0.27, batch-adjusted p=0.047; post-treatment LTED score r=0.32, batch-adjusted p=0.014). Therefore, the genes up- or down-regulated in LTED cells are expressed at higher or lower levels, respectively, in tumors where cell proliferation was not suppressed by letrozole. Notably, only 4/99 genes were classified as ‘cell growth/proliferation’ genes by Gene Ontology analysis (20), suggesting that the ability of the LTED signature to predict early response to the AI (as determined by Ki67) is not only based on quantification of cell proliferation genes. Therefore, the LTED score likely reflects the underlying biology of a tumor cell population refractory to estrogen deprivation rather than the proliferative state of the cancer. Since our previous analysis suggested that the LTED score is higher in luminal B than luminal A tumors (Fig. 1C), these results further corroborate our contention that the LTED score is capturing expression patterns indicative of reduced proliferative dependency on the ER pathway (i.e., luminal B tumors).
Fig. 2.
The LTED score predicts early tumor response to aromatase inhibitors. A) Forty-eight patients with ER+ breast cancer were treated with neoadjuvant letrozole for 2 weeks (cohort MILLER48). Ki67 scoring was performed on pre- and post-treatment (2 weeks) tumor biopsies. B-C) The LTED score for patient tumors from (A) was calculated from pretreatment (B) and post-treatment (C) biopsies. LTED scores were compared to the Ki67 scores at 2 weeks. Data were analyzed by linear regression with adjustment for tumor microarray batch effect. D) Sixty-nine patients with ER+ breast cancer were treated with neoadjuvant anastrozole for 14 days (cohort SMITH69). Ki67 scoring was performed on pre- and post-treatment tumor biopsies. E-F) Pretreatment (E) and post-treatment (F) LTED scores were calculated for tumors from (D) and compared to the 2-week Ki67 scores as in (B-C).
To confirm these results, we assessed whether the LTED signature can predict response to anastrozole in another cohort of 69 patients with ER+ breast cancer [SMITH69 cohort (7)]. Ki67 scoring was performed on tumor biopsies obtained after 14 days of therapy with the AI (Fig. 2D). Comparison of the pretreatment LTED scores with Ki67 scores (at 2 weeks) revealed that tumors with high LTED scores showed significantly higher Ki67 scores (Fig. 2E, r=0.43, p=0.0002). However, post-treatment LTED scores demonstrated a weaker correlation with Ki67 (Fig. 2F, r=0.19, p=0.11), suggesting that the pre-therapy LTED score may be a stronger predictor of response to estrogen deprivation.
LTED signature predicts disease outcome following adjuvant tamoxifen
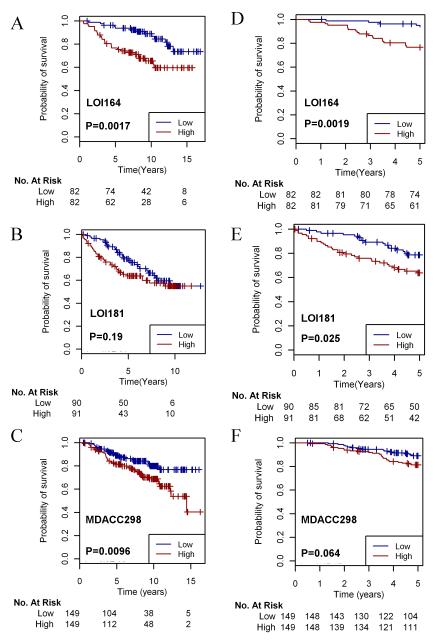
To determine whether the LTED signature is predictive of long-term outcome following adjuvant endocrine therapy, we assessed the correlation between pretreatment tumor LTED scores and relapse-free survival (RFS) in two cohorts of patients with ER+ breast cancer treated with adjuvant tamoxifen for five years [n=164 in cohort LOI164 , n=181 in cohort LOI181, with median follow-ups of 8.94 and 5.11 years, respectively (9)].
According to the PAM50 classifier, LOI164 was comprised of 77 (47%) luminal A, 74 (45%) luminal B, and 13 (8%) HER2-positive tumors. LOI181 was comprised of 156 (86%) luminal A, 21 (12%) luminal B, 3 (2%) HER2-positive, and 2 (1%) normal-like tumors. In univariate Cox PH models, the LTED score was associated with RFS in LOI164 (p=0.0065) and showed a trend in LOI181 (p=0.085). The LTED gene signature was a better predictor of RFS than 95.9% and 92.6% of 10,000 randomly generated 99-gene and 97-gene signatures by univariate analyses in the two datasets, respectively. In multivariate Cox PH models adjusting for age, tumor grade, tumor size, lymph node status, and molecular subtype in patients for which covariate data were available, the LTED score was independently predictive of RFS in LOI164 (p=0.0018, n=128, Table S5) but not in LOI181 (p=0.175, n=149, Table S6). Notably, the LTED score was independently predictive of RFS in the presence of molecular subtype in LOI164. We underscore that molecular subtype is linked with clinical outcome (18) but is routinely excluded from the majority of multivariate models predicting outcome in breast cancer. In order to visualize differences in RFS between patients with a high vs. low LTED score, we dichotomized patients based on median LTED score and generated Kaplan-Meier survival curves. Analysis of LOI164 showed that patients bearing tumors with high LTED scores (those demonstrating high concordance with the signature of LTED cells) had significantly worse outcome following adjuvant tamoxifen (Fig. 3A; log-rank test p<0.01). Again, a similar stratification of LOI181 did not produce statistically different RFS curves (Fig. 3B; p=0.19).
Fig. 3.
The LTED score predicts long-term disease outcome following adjuvant tamoxifen therapy. A-C) Patients in LOI164 (A), LOI181 (B), and MDACC298 (C) were dichotomized based on median LTED score, and Kaplan-Meier survival curves were compared using log-rank test. D-F) Patients in LOI164 (D), LOI181 (E), and MDACC298 (F) were censored at the five-year time point and analyzed as in (A-C).
Based on prior results, we speculate that cohort LOI181 contains a confounding factor which we were unable to identify that may affect the predictive power of the LTED signature in this cohort. A comparison of LOI181 with a large dataset of breast cancer patients with long-term follow-up further suggested this possibility. For example, among >2,500 ER+ tumors, Cheang et al. found a distribution of 59% luminal A, 33% luminal B, and 9% HER2-positive by molecular subtyping. In their study, the five-year RFS rate for 887 patients who received adjuvant tamoxifen was 83% for luminal A vs. 72% for luminal B (21). In contrast, LOI181 showed an abnormally high luminal A representation of 86% associated with an uncommonly poor five-year RFS rate of 55%. Despite the poor outcome in LOI181, this cohort showed a rate of node-positivity similar to LOI164 (47% vs. 57%, respectively)
In light of this discrepancy, we tested the predictive ability of the LTED score in a third cohort (MDACC298) comprised of 298 ER+ patients treated with adjuvant tamoxifen for 5 years [median follow-up of 8.17 years (10)]. According to the PAM50 classifier, MDACC298 was comprised of 168 (56%) luminal A, 91 (31%) luminal B, 9 (3%) HER2-positive, 26 (8%) normal-like tumors, and 4 (1.3%) basal-like tumors. Thus, MDACC298 appeared to have luminal A/B distributions similar to LOI164. We found that the LTED score was again strongly predictive of RFS in a univariate Cox PH model (p=0.004). Limited clinical data were available for MDACC298 (lymph node status in all patients). The LTED score was found to retain a significant correlation with RFS in the presence of node status (p=0.0004). LTED score was also predictive of RFS by Kaplan-Meier analysis (log-rank p=0.0096; Fig. 3C).
Since a fraction of tumors recurred in each of the three cohorts after the five-year course of tamoxifen, it is plausible that post-treatment recurrences were not due to tamoxifen resistance (i.e., tamoxifen may have been required to suppress cancer growth). Therefore, we further analyzed the correlation between LTED score and recurrence during the five years of treatment to better assess intrinsic drug resistance (24, 48, and 42 patients recurred within five years in LOI164, LOI181, and MDACC298, respectively). In this setting, a high LTED score was significantly predictive of early recurrence in all three cohorts (Fig. 3D-F and Tables S5-S6; univariate Cox PH model p=0.0019, p=0.025, and p=0.0055, respectively). Therefore, patients bearing primary tumors with gene expression profiles reflective of estrogen independence are more likely to recur both during and following five years of therapy.
Gene Set Analysis of LTED models reveals an activated MYC signature predictive of treatment outcome
The LTED signature appeared to contain both prognostic (long-term outcome following adjuvant therapy with tamoxifen) and predictive elements (recurrence during adjuvant tamoxifen and tumor cell proliferation – i.e., Ki67 – after short-term therapy with an AI). Although each of these elements is potentially useful to inform clinical outcome, neither can be directly used to determine therapeutically targetable mechanisms causally associated with resistance to endocrine therapy. The current clinical standard is to add chemotherapy to the treatment regimen when disease progresses and/or antiestrogen therapies are exhausted. However, rational therapies targeted against molecules or pathways causal to such resistance would be preferable. Thus, we next assessed whether the LTED gene expression pattern could be used to identify distinct molecular alterations or pathway activation signatures that inform novel drug targets.
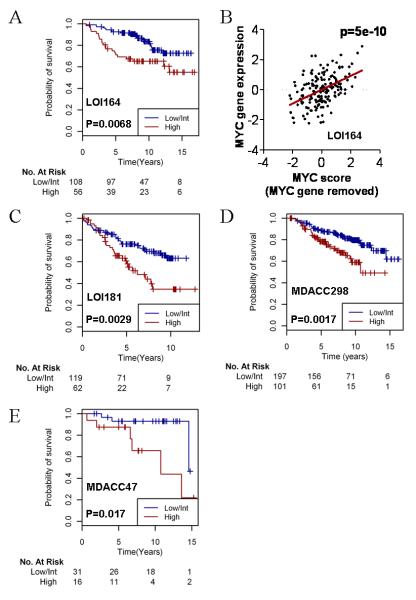
For this purpose, we utilized Gene Set Analysis (GSA). A paired design was invoked where each LTED cell line was compared to its parental control using the GSA algorithm. This analysis may be more informative for identifying coordinately regulated and biologically related gene sets. Among the gene sets enriched in LTED lines (Table S7), we found two signatures of MYC activation [COLLER_MYC_TARGETS_UP and MYC_ONCOGENIC_SIGNATURE (22, 23)]. The MYC oncogene is amplified in 15% of breast cancers (24). Further, breast cancer cells may overexpress MYC as a result of post-translational mechanisms (25). Given that prior reports have identified a large overlap in estrogen-responsive and MYC-responsive genes (26, 27), we hypothesized that MYC-coordinated gene expression promotes resistance to endocrine therapy by overriding the inhibitory effects of estrogen deprivation on genomic ER signaling. We created a union set of the genes activated by MYC from the two identified gene sets above, which yielded 231 genes (Table S8). We then determined which of these 231 genes were specifically upregulated in LTED cells versus the parental lines, and contributed most strongly to the correlation with the activated MYC signatures [gene score (s) >1]. We identified 46 such genes (Table S9), only one of which overlapped with the 99-gene LTED signature, suggesting that the 46-gene activated-MYC signature provides distinct biological information. The log2 signal intensities for these 46 genes were summed for each patient in cohort LOI164 to create an activated-MYC score. A high MYC score was associated with poor RFS (univariate Cox PH model p=0.004). When the data were stratified into ‘high’, ‘intermediate’, and ‘low’ MYC tertiles, it was apparent that the highest tertile represented a distinct fraction of the patients with a poor time-to-recurrence (not shown). Thus, we divided the patients into ‘high’ and ‘low/intermediate’ MYC score groups, and compared RFS curves by log-rank test (Fig. 4A; p=0.0068). After removing the contribution of the MYC gene itself, the MYC score was linearly associated with MYC mRNA levels (Fig. 4B; p=5×10−10). Interestingly, MYC mRNA levels alone were not significantly predictive of RFS (not shown). This was confirmed in the LOI181 and MDACC298 datasets (below), suggesting that a signature of MYC activation, which better reflects MYC transcriptional output, may be more informative than MYC gene expression alone.
Fig. 4.
An activated-MYC signature derived from LTED cells predicts poor outcome following adjuvant tamoxifen therapy. A) The MYC score was applied to cohort LOI164 and stratified by tertile. Kaplan-Meier survival curves were generated and the high tertile was compared to the combined intermediate and low tertiles by the log-rank test. B) MYC score (excluding MYC itself) was compared to MYC mRNA levels in patients from (A) by least squares regression. C-D) Cohorts LOI181 (C) and MDACC298 (D) were analyzed as in (A). E) MYC protein expression was quantified in 47 ER+ early-stage breast tumors (cohort MDACC47). Data were analyzed as in (A).
We also tested the ability of the activated-MYC signature to predict outcome in the LOI181 and MDACC298 cohorts. Although MYC score was not linearly associated with RFS in LOI181 or MDACC298 in univariate Cox PH models, Kaplan-Meier analyses comparing high vs. intermediate/low tertiles revealed an association with outcome (Fig. 4C-D; log-rank p=0.0029 and p=0.0017, respectively). Thus, an alternative explanation for the lack of significance in the Cox PH model could be that MYC-mediated endocrine resistance may behave as a binary ‘on/off’ characteristic, rather than a continuous variable where the degree of MYC activation is linearly related to the degree of antiestrogen resistance. To test if MYC expression is predictive of response to endocrine therapy in a large multi-study dataset, we utilized the Kaplan-Meier Plotter tool (28). In a population of 399 patients with ER+ breast cancer, MYC expression was associated with RFS in patients treated with endocrine therapy (Fig. S1, log-rank p=0.00069). In contrast, no difference in RFS curves was noted when the analysis was restricted to 609 patients not treated with adjuvant therapy. This analysis strongly suggests that MYC expression is predictive of response to endocrine therapy, and not simply a prognostic indicator. Correlations have been found between MYC and HER2 gene co-amplification and poor prognosis in breast cancer (29, 30), but other studies reported only rare co-amplification (31, 32). Similarly, we found a correlation between MYC score and HER2 mRNA levels (from microarray data) in the MDACC298 dataset (p=0.01, r=0.14) but not in LOI164 or LOI181. Therefore, the association between MYC and HER2 overexpression requires further study.
Next, we used RPPA to quantify MYC protein levels in 47 patients with early-stage ER+ breast cancer treated with adjuvant tamoxifen for five years (cohort MDACC47, median follow-up of 7.75 years). MYC levels were significantly associated with RFS following adjuvant tamoxifen (Cox PH model p=0.019). Stratification of the patients by high vs. low/intermediate MYC expression demonstrated a similar association (Fig. 4E; log-rank p=0.017).
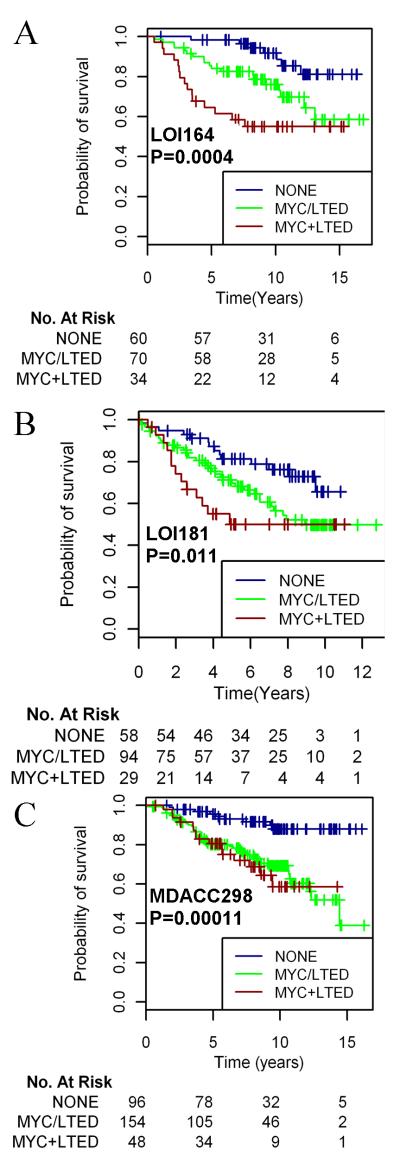
Finally, since the two gene signatures (‘LTED’ and ‘MYC activation’) derived from the LTED cells did not significantly overlap, we hypothesized that each signature may provide unique biological information. Thus, we asked whether stratification according to both factors (high LTED score and/or high MYC score)would serve as a better predictor of outcome. Indeed, patients with a high score for both signatures (MYC+LTED) demonstrated the shortest RFS in all three cohorts, while patients with low scores for both signatures (NONE) had the best outcome (Fig. 5). Patients with either a high MYC score or a high LTED score (MYC/LTED) had intermediate RFS rates.
Fig. 5.
Coexpression of patterns of MYC activation and estrogen independence predicts poor outcome following adjuvant tamoxifen. LOI164 (A), LOI181 (B), and MDACC298 (C) patients were stratified by MYC score (tertiles) and LTED score (median) as in Figs. 3-4. Patients demonstrating a high score in neither (NONE), either (MYC/LTED), or both scores (MYC+LTED) were grouped, and Kaplan-Meier survival curves were compared by log-rank test.
siRNA-mediated depletion of MYC inhibits estrogen-independent growth
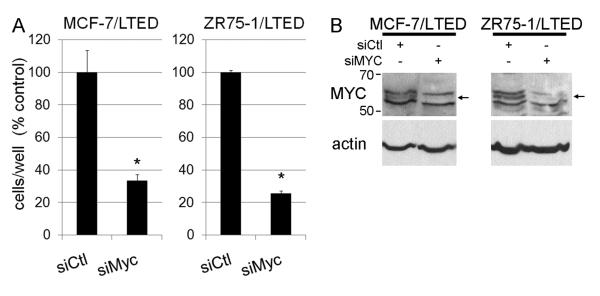
Since both MYC protein overexpression and a signature of MYC activation were predictive of antiestrogen resistance, we hypothesized that MYC inhibition should suppress estrogen-independent breast cancer cell growth. Thus, we utilized siRNA-mediated depletion of MYC in LTED cells to determine whether MYC contributes to hormone-independent growth. MYC knock-down suppressed the growth of MCF-7/LTED and ZR75-1/LTED cells (Fig. 6A). MYC knock-down was confirmed by immunoblotting in Fig. 6B. Similarly, MYC overexpression has been shown to confer resistance to tamoxifen in vitro (33), and siRNA-mediated downregulation of MYC impairs estrogen-induced growth (26). We were unable to effectively knock-down MYC expression in HCC-1428/LTED cells; MDA-361/LTED cells express very low MYC levels (not shown). These data suggest that MYC signaling may substitute for estrogen-induced ER signaling in a subset of breast cancers which are resistant to estrogen deprivation.
Fig. 6.
MYC is required for hormone-independent cell proliferation. Cells transfected with siRNA targeting MYC or non-silencing control (siCtl) were treated with 10% DCC-FBS, and adherent cells were counted after 6 days. Data are presented as % siCtl, mean of triplicates ± SD. *p<0.01 by t-test compared to siCtl. B) MYC and actin immunoblots of lysates from the same batches of cells used in (A).
In summary, data generated from these cellular models of ER+ breast cancer have revealed biomarkers that upon further development may prospectively identify patients at risk of early recurrence during or following endocrine therapy. These findings also support the combined use of antiestrogens with MYC-targeted therapies to abrogate or delay the emergence of endocrine resistance in patients with hormone receptor-positive breast cancer.
Supplementary Material
Statement of Translational Relevance.
Breast cancers which express estrogen receptor α (ER) and/or progesterone receptor typically exhibit response to endocrine therapies (i.e., tamoxifen, aromatase inhibitors). However, a significant fraction of such cancers exhibit or develop therapeutic resistance, and metastatic recurrences are uniformly incurable. For the majority of antiestrogen-resistant breast cancers, mechanisms and predictive biomarkers of resistance await discovery. We generated a gene expression signature of estrogen independence using ER+ breast cancer cell lines which acquired hormone-independent growth. In patients, this signature was predictive of 1) high tumor cell proliferation following neoadjuvant therapy with an aromatase inhibitor, and 2) shorter time-to-recurrence following adjuvant tamoxifen therapy. Additionally, we identified a signature of MYC transcription factor activation associated with estrogen-independent growth. A MYC activation signature and high MYC protein levels were predictive of shorter time-to-recurrence following adjuvant tamoxifen, thus offering MYC as a therapeutic target in breast cancers resistant to endocrine therapy.
Acknowledgments
Funding This work was supported by the National Institutes of Health F32CA121900 (T.W.M.), Breast Cancer Specialized Program of Research Excellence (SPORE) P50CA98131, Vanderbilt-Ingram Cancer Center Support Grant P30CA68485, M.D. Anderson Cancer Center Support Grant P30CA16672; a grant from the Breast Cancer Research Foundation (C.L.A.); ACS Clinical Research Professorship Grant CRP-07-234 (C.L.A.); the Lee Jeans Translational Breast Cancer Research Program (C.L.A.); Stand Up To Cancer grant “Targeting PI3K in Women’s Cancers” (C.L.A. & G.B.M.); Breakthrough Breast Cancer and Royal Marsden NIHR Biomedical Research Centre (M.D., Z.G., & A.D.).
Footnotes
Competing interests: M.D. has received research grant funds and honoraria for consulting and advisory board work from AstraZeneca and Novartis Pharmaceuticals Corp.
References
- 1.Forbes JF, Cuzick J, Buzdar A, Howell A, Tobias JS, Baum M. Effect of anastrozole and tamoxifen as adjuvant treatment for early-stage breast cancer: 100-month analysis of the ATAC trial. Lancet Oncol. 2008;9:45–53. doi: 10.1016/S1470-2045(07)70385-6. [DOI] [PubMed] [Google Scholar]
- 2.Ellis MJ, Tao Y, Young O, et al. Estrogen-independent proliferation is present in estrogen-receptor HER2-positive primary breast cancer after neoadjuvant letrozole. J Clin Oncol. 2006;24:3019–25. doi: 10.1200/JCO.2005.04.3034. [DOI] [PubMed] [Google Scholar]
- 3.Arpino G, Green SJ, Allred DC, et al. HER-2 amplification, HER-1 expression, and tamoxifen response in estrogen receptor-positive metastatic breast cancer: a southwest oncology group study. Clin Cancer Res. 2004;10:5670–6. doi: 10.1158/1078-0432.CCR-04-0110. [DOI] [PubMed] [Google Scholar]
- 4.De Laurentiis M, Arpino G, Massarelli E, et al. A meta-analysis on the interaction between HER-2 expression and response to endocrine treatment in advanced breast cancer. Clin Cancer Res. 2005;11:4741–8. doi: 10.1158/1078-0432.CCR-04-2569. [DOI] [PubMed] [Google Scholar]
- 5.Dixon JM, Renshaw L, Young O, et al. Letrozole suppresses plasma estradiol and estrone sulphate more completely than anastrozole in postmenopausal women with breast cancer. J Clin Oncol. 2008;26:1671–6. doi: 10.1200/JCO.2007.13.9279. [DOI] [PubMed] [Google Scholar]
- 6.Miller TW, Hennessy BT, Gonzalez-Angulo AM, et al. Hyperactivation of phosphatidylinositol-3 kinase promotes escape from hormone dependence in estrogen receptor-positive human breast cancer. J Clin Invest. 2010 doi: 10.1172/JCI41680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith IE, Walsh G, Skene A, et al. A phase II placebo-controlled trial of neoadjuvant anastrozole alone or with gefitinib in early breast cancer. J Clin Oncol. 2007;25:3816–22. doi: 10.1200/JCO.2006.09.6578. [DOI] [PubMed] [Google Scholar]
- 8.Miller WR, Larionov A, Renshaw L, et al. Gene expression profiles differentiating between breast cancers clinically responsive or resistant to letrozole. J Clin Oncol. 2009;27:1382–7. doi: 10.1200/JCO.2008.16.8849. [DOI] [PubMed] [Google Scholar]
- 9.Loi S, Haibe-Kains B, Desmedt C, et al. Predicting prognosis using molecular profiling in estrogen receptor-positive breast cancer treated with tamoxifen. BMC Genomics. 2008;9:239. doi: 10.1186/1471-2164-9-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Symmans WF, Hatzis C, Sotiriou C, et al. Genomic index of sensitivity to endocrine therapy for breast cancer. J Clin Oncol. 2010;28:4111–9. doi: 10.1200/JCO.2010.28.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parker JS, Mullins M, Cheang MC, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–7. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Popovici V, Chen W, Gallas BG, et al. Effect of training-sample size and classification difficulty on the accuracy of genomic predictors. Breast Cancer Res. 12:R5. doi: 10.1186/bcr2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Efron B, Tibshirani R. On testing the significance of sets of genes. Annals of Applied Statistics. 2007;1:107–29. [Google Scholar]
- 14.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hennessy B, Lu Y, Gonzalez-Angulo AM, Myhre S, Carey M, Ju Z, Coombes K, Liu W, Meric-Bernstam F, Bedrosian I, Yu Q, Yu S, Siwak D, McGahren M, Li J, Sahin A, Overgaard J, Alsner J, Neve RM, Kuo W-L, Gray JW, Borresen-Dale A-L, Mills GB. A technical assessment of the utility of reverse phase protein arrays for the study of the functional proteome in non-microdissected human breast cancers. Clinical Proteomics. 2010;6:129–51. doi: 10.1007/s12014-010-9055-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tibes R, Qiu Y, Lu Y, et al. Reverse phase protein array: validation of a novel proteomic technology and utility for analysis of primary leukemia specimens and hematopoietic stem cells. Mol Cancer Ther. 2006;5:2512–21. doi: 10.1158/1535-7163.MCT-06-0334. [DOI] [PubMed] [Google Scholar]
- 17.Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 18.Kennecke H, Yerushalmi R, Woods R, et al. Metastatic behavior of breast cancer subtypes. J Clin Oncol. 2010;28:3271–7. doi: 10.1200/JCO.2009.25.9820. [DOI] [PubMed] [Google Scholar]
- 19.Gerdes J, Lemke H, Baisch H, Wacker HH, Schwab U, Stein H. Cell cycle analysis of a cell proliferation-associated human nuclear antigen defined by the monoclonal antibody Ki-67. J Immunol. 1984;133:1710–5. [PubMed] [Google Scholar]
- 20.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheang MC, Chia SK, Voduc D, et al. Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J Natl Cancer Inst. 2009;101:736–50. doi: 10.1093/jnci/djp082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bild AH, Yao G, Chang JT, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–7. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 23.Coller HA, Grandori C, Tamayo P, et al. Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc Natl Acad Sci U S A. 2000;97:3260–5. doi: 10.1073/pnas.97.7.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deming SL, Nass SJ, Dickson RB, Trock BJ. C-myc amplification in breast cancer: a meta-analysis of its occurrence and prognostic relevance. Br J Cancer. 2000;83:1688–95. doi: 10.1054/bjoc.2000.1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer. 2008;8:83–93. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- 26.Musgrove EA, Sergio CM, Loi S, et al. Identification of functional networks of estrogen- and c-Myc-responsive genes and their relationship to response to tamoxifen therapy in breast cancer. PLoS One. 2008;3:e2987. doi: 10.1371/journal.pone.0002987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng AS, Jin VX, Fan M, et al. Combinatorial analysis of transcription factor partners reveals recruitment of c-MYC to estrogen receptor-alpha responsive promoters. Mol Cell. 2006;21:393–404. doi: 10.1016/j.molcel.2005.12.016. [DOI] [PubMed] [Google Scholar]
- 28.Gyorffy B, Lanczky A, Eklund AC, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2009 doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 29.Cuny M, Kramar A, Courjal F, et al. Relating genotype and phenotype in breast cancer: an analysis of the prognostic significance of amplification at eight different genes or loci and of p53 mutations. Cancer Res. 2000;60:1077–83. [PubMed] [Google Scholar]
- 30.Seshadri R, Matthews C, Dobrovic A, Horsfall DJ. The significance of oncogene amplification in primary breast cancer. Int J Cancer. 1989;43:270–2. doi: 10.1002/ijc.2910430218. [DOI] [PubMed] [Google Scholar]
- 31.Rodriguez-Pinilla SM, Jones RL, Lambros MB, et al. MYC amplification in breast cancer: a chromogenic in situ hybridisation study. J Clin Pathol. 2007;60:1017–23. doi: 10.1136/jcp.2006.043869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berns EM, Klijn JG, van Putten WL, van Staveren IL, Portengen H, Foekens JA. c-myc amplification is a better prognostic factor than HER2/neu amplification in primary breast cancer. Cancer Res. 1992;52:1107–13. [PubMed] [Google Scholar]
- 33.Prall OW, Rogan EM, Musgrove EA, Watts CK, Sutherland RL. c-Myc or cyclin D1 mimics estrogen effects on cyclin E-Cdk2 activation and cell cycle reentry. Mol Cell Biol. 1998;18:4499–508. doi: 10.1128/mcb.18.8.4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.