Abstract
The 14-3-3 proteins were the first phosphoserine/phosphothreonine-binding proteins to be discovered, a finding that provided the foundation for their prominent role in cell signaling. 14-3-3 family members interact with a wide spectrum of proteins including transcription factors, biosynthetic enzymes, cytoskeletal proteins, signaling molecules, apoptosis factors, and tumor suppressors. The interaction with 14-3-3 can have a profound effect on a target protein, altering its localization, stability, conformation, phosphorylation state, activity, and/or molecular interactions. Thus, by modulating the function of a diverse array of binding partners, 14-3-3 proteins have become key regulatory components in many vital cellular processes – processes that are crucial for normal growth and development and that often become dysregulated in human cancer. This review will examine the recent advances that further elucidate the role of 14-3-3 proteins in normal growth and cancer signaling with a particular emphasis on the signaling pathways that impact cell proliferation, cell migration, and epithelial-to-mesenchymal transition.
Keywords: 14-3-3, cell proliferation, cell migration, actin cytoskeleton, EMT
1. Introduction
The phosphorylation and dephosphorylation of proteins is a key mechanism mediating cellular signal transduction. The 14-3-3 proteins are specific phosphoserine/phosphothreonine (pS/T) binding proteins and they have been tightly integrated into the phospho-regulatory signaling pathways that control many biological processes, including cell cycle regulation, protein trafficking, metabolic regulation, cell proliferation, and apoptosis (reviewed in [1]). There are seven mammalian 14-3-3 family members (β, γ, ε, σ, ζ, τ, η), all of which adopt a similar horseshoe-like structure capable of binding pS/T residues in a sequence specific context (RSXpS/TXP - mode 1 and RXXXpSXP - mode 2). It is important to note, however, that not all phosphorylation-dependent sites conform to these motifs and not all interactions are phosphorylation-dependent.
The 14-3-3 proteins exist as dimers in cells, forming homo and heterodimers. Each molecule in the dimer contains an independent ligand-binding channel and, as a result, a dimer can bind two pS/T sites simultaneously, found either on a single target or on separate binding partners [2]. Binding of a 14-3-3 dimer can alter the conformation of a target protein, mediate or prevent protein interactions, mask intrinsic localization motifs, and/or block the accessibility of a target protein to modifying enzymes such as kinases, phosphatases, or proteases. In this way, 14-3-3 binding interactions can regulate the function of a wide spectrum of proteins with diverse biological activities.
In this review, we will focus on the new developments that further elucidate the role of 14-3-3 in signaling pathways and cellular processes relevant to normal cell growth and tumorigenesis. In particular, we will give an update on the function and regulation of 14-3-3 binding interactions in pathways that control cell proliferation. We will also discuss recent reports that expand the function of 14-3-3 in cell migration and epithelial-to-mesenchymal transition (EMT), two processes contributing to cancer progression. Finally, we will examine recent studies evaluating the clinical significance of 14-3-3 proteins to human cancer.
2. 14-3-3 and cell proliferation: an update
2.1. Function and regulation of 14-3-3 binding interactions in RTK/Ras signaling
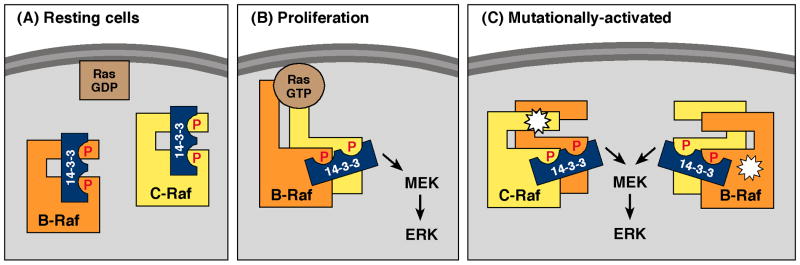
Unregulated cell proliferation is an early step in tumorigenesis, and the first indication that the 14-3-3 proteins would play a prominent role in cell proliferation came with the discovery in 1994 that they interacted with various proteins that had oncogenic potential, including members of the Raf kinase family [3]. Normal cell proliferation is often initiated by cell surface receptor tyrosine kinases (RTKs), and the Raf proteins are key signaling intermediates in RTK pathways, functioning as direct effectors of the Ras GTPase to promote signaling through the ERK casade. The role that 14-3-3 binding plays in Raf regulation is complex, due largely to the fact that all Raf family members contain at least two phosphorylation-dependent 14-3-3 binding sites and binding at different sites can have varying effects (reviewed in [4]; Fig. 1). All Raf proteins contain a 14-3-3 binding site in the N-terminal serine/threonine rich region and a site following the C-terminal catalytic domain. In the absence of signaling events, inactive Raf monomers are retained in the cytosol by the binding of a 14-3-3 dimer to both the Raf N- and C-terminal sites. Binding of 14-3-3 to the N-terminal site plays a critical role in inhibiting Raf signaling as mutations that prevent this interaction lead to constitutive membrane localization and Raf activation. However, during the Raf activation process, 14-3-3 serves a positive regulatory role and is required for the dimerization of the Raf kinases that is needed for catalytic activation. In this case, 14-3-3 binding to the N-terminal site is disrupted by dephosphorylation, and in the context of Ras-dependent clustering at the membrane, the 14-3-3 dimer can contact two Raf proteins simultaneously through their C-terminal sites, thus facilitating Raf dimerization [5-7].
Fig. 1.
The function of 14-3-3 in Raf kinase regulation. (A) In resting cells, a 14-3-3 dimer binds to phosphorylation sites found in the Raf N- and C-terminal regions, maintaining Raf in an inactive state in the cytosol. (B) In response to proliferative signals, 14-3-3 binding to the Raf C-terminal sites facilitates Raf dimerization in a Ras-dependent manner. (C) The N-terminal, negative regulatory 14-3-3 binding motif of C-Raf is frequently mutated in Noonan and LEOPARD syndromes. All mutationally-activated Raf proteins dimerize constitutively in a manner dependent on the Raf C-terminal 14-3-3 binding site.
Not only are the Raf kinases critical components of normal cellular proliferation, they can promote aberrant cell proliferation when activated by mutation. Somatic mutations in the B-Raf kinase are often observed in human cancer [8], and genetic mutations in either B-Raf or C-Raf can cause certain developmental disorders, including Noonan, LEOPARD, and cardio-facio-cutaneous syndromes [9]. Mutationally-activated Raf proteins dimerize constitutively in a manner that does not require the interaction with Ras, but is still dependent on 14-3-3 binding to the C-terminal site [5, 7](Fig. 1). Although Raf dimerization has little effect on the most prevalent V600E B-Raf mutant that has high enzymatic activity, it is required for the full proliferative potential of Raf mutants with moderate to low enzymatic activity [7]. Further demonstrating the importance of 14-3-3 in Raf regulation, residues critical for 14-3-3 binding to the N-terminal, negative regulatory site constitute a mutational “hot spot” for Noonan and LEOPARD syndrome patients with genomic mutations in C-Raf and represent one of the few somatic muations in C-Raf that has been implicated in human cancer [9, 10].
Another critical effector of RTK/Ras signaling is the PI3K/PDK/AKT cascade, and a prime reason for why the 14-3-3 proteins have been so tightly integrated into RTK/Ras signaling is the fact that AKT is a key kinase that generates 14-3-3 binding sites on a diverse array of target proteins [11]. Research in the last five years has continued to identify new AKT substrates that are regulated by 14-3-3 binding, two of which are particularly relevant to cancer signaling, Skp-2 and PACS-2. Skp-2 is a component of the SCF ubiquitin ligase complex that targets critical cell-cycle regulators, including the CDK inhibitor p27. In 2009, two reports identified AKT as a kinase that phosphorylates Skp-2 on S72 [12, 13]. AKT-dependent phosphorylation of S72 was shown to regulate the SCF-Skp-2 complex through multiple mechanisms, with binding of 14-3-3 to pS72 serving to relocalize Skp-2 from the nucleus to the cytoplasm. Interestingly, the work of Lin et al. [12] suggests a specific function for cytosolic Skp-2 in the positive regulation of cell migration, through a mechanism that is yet to be determined.
PACS-2 is a multifunctional sorting molecule that acts as both a secretory pathway trafficking protein and as a pro-apoptotic protein that mediates the translocation of Bid to the mitochondria in response to apoptotic inducers such as TRAIL [14]. In work conducted by Aslan and coworkers [15], AKT was found to phosphorylate PACS-2 on S437 and binding of 14-3-3 to this site repressed PACS-2's pro-apoptotic activity and was required for PACS-2 to mediate trafficking of membrane cargo. This study also reported that TRAIL-mediated apoptosis is dependent on PACS-2 expression and that activation of the TRAIL receptor induces the dephosphorylation of pS437, releasing 14-3-3 and switching PACS-2 to its pro-apoptotic function. Because binding of TRAIL to the apoptotic DR4 and DR5 receptors selectively kills cancerous and virus-infected cells in vivo without harming healthy cells, there has been much interest in the therapeutic potential of TRAIL. Interestingly, PACS-2 expression is lost in ∼50% of colorectal cancers [16], and Aslan et al. suggest that loss of PACS-2 expression may serve as a biomarker for TRAIL-resistant cancer.
Activation of the PI3K/PDK/AKT cascade is also required for the 14-3-3-dependent regulation of the Gab2 docking protein [17]. Gab2 binds select signal transducers involved in cell proliferation and cell migration, and its overexpression in several human malignancies is associated with increased metastatic potential [18]. Gab2 is recruited to specific receptor complexes via an interaction with the Grb2 adaptor, and studies by Brummer and coworkers [17] have found that 14-3-3 binding attenuates Gab2 signaling by promoting disassembly of the Gab2/Grb2 complex. 14-3-3 binding was mediated by pS210 and pT391 of Gab2, and phosphorylation of these sites was blocked when signaling through the PI3K/PDK/AKT cascade was inhibited. Interestingly, this negative feedback mechanism appears to be specific for Gab2 in that the 14-3-3 binding motifs are not conserved in the closely related Gab1 and Gab3 docking proteins.
Recent research has also identified a new mechanism that regulates AKT-dependent 14-3-3 binding interactions. This mechanism involves the methylation of arginine residues in the AKT consensus phosphorylation motif (RxRxxS). As reported by the laboratory of Dr. Akiyoshi Fukamizu, arginine methylation can disrupt AKT-mediated phosphorylation of the FOXO transcription factors and the Bad apoptosis regulator, thus preventing 14-3-3 binding to important functional sites [19, 20]. Whether this regulatory mechanism impacts other AKT substrates and the extent to which crosstalk between arginine methylation and phosphorylation occurs in vivo awaits further investigation.
2.2 New regulators of 14-3-3 binding interactions in the Hippo Pathway
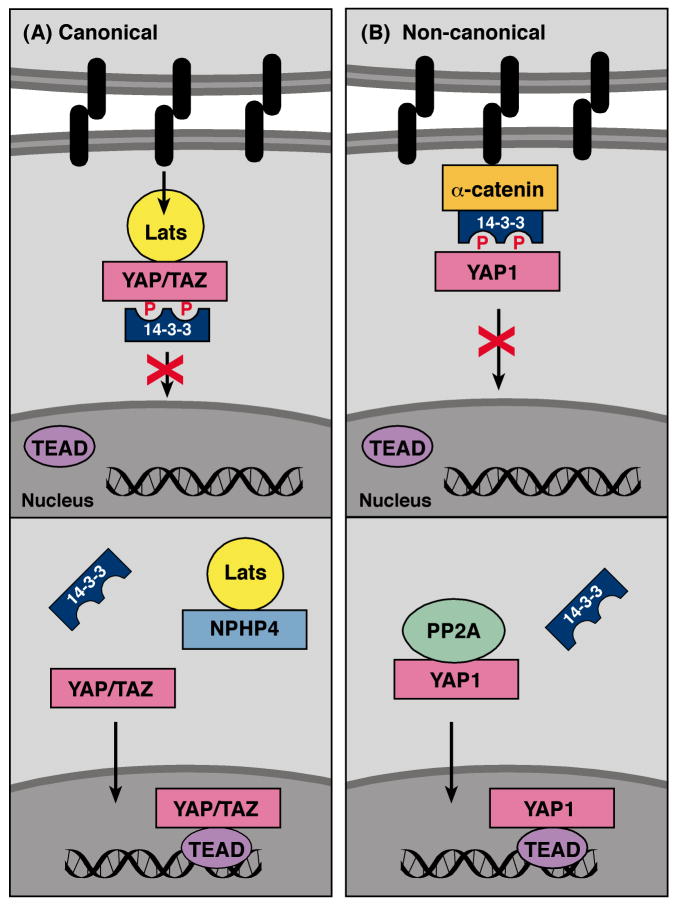
Another pathway to emerge as a critical regulator of cell proliferation is the Hippo pathway. This pathway was discovered in Drosophila and contributes to organ size control and tumor suppression by restricting proliferation and promoting apoptosis (for a review see [21]). During canonical Hippo signaling, activation of a kinase cascade results in the phosphorylation of the transcriptional coactivators YAP and TAZ on sites that bind 14-3-3 (Fig. 2A). 14-3-3 binding inactivates the pro-proliferative function of YAP/TAZ by sequestering them in the cytosol, thus preventing their interaction with the TEAD transcription factors. In 2011, additional proteins involved in regulating the binding of 14-3-3 to these transcriptional coactivators were reported. In particular, the work by Schlegelmilch and coworkers [22] identified a new mechanism for modulating 14-3-3 binding to YAP1 that is distinct from the canonical phosphorylation cascade (Fig. 2B). During studies investigating epidermal growth, activation of YAP1 was found to promote the proliferation of epidermal stem and progenitor cells in a manner dependent on the TEAD transcription factors. As expected, regulation of YAP1 localization and activity was critical for maintaining normal skin homeostasis, but surprisingly did not require the canonical kinase components of the Hippo pathway. To identify alternative upstream regulators, mass spectrometry analysis was performed on YAP1 protein complexes isolated from high-density skin culture cells predicted to contain high levels of inactive YAP1. This analysis revealed that α-catenin was a primary binding partner of inactive YAP. Interestingly, the interaction between α-catenin and YAP1 was not direct but required binding of 14-3-3 to the YAP1 pS127 site. Further examination revealed that the phosphorylation state of the YAP pS127 site was reduced in α-catenin depleted cells and that YAP1 selectively interacted with the catalytic subunit of PP2A in cells depleted of α-catenin, but not in those containing α-catenin. Schlegelmilch and colleagues go on to show that binding of α-catenin to the 14-3-3/YAP1 complex protects the YAP1 pS127 site from dephosphorylation mediated by PP2A, thus stabilizing the YAP1/14-3-3 interaction. In this mode of regulation, α-catenin, a critical and unique component of density-dependent cell-cell junctions, acts as a cell density sensor to inactivate YAP1 and prevent its entry into the nucleus.
Fig. 2.
14-3-3 negatively regulates Hippo pathway signaling. (A) In the canonical Hippo pathway, Lats generates a phosphorylation-dependent 14-3-3 binding site on YAP/TAZ under conditions of high cell density. The binding of 14-3-3 sequesters YAP/TAZ in the cytosol. NPHP4 can complete with YAP/TAZ for Lats binding, thus blocking Lats-mediated phosphorylation of YAP/TAZ and 14-3-3 binding. (B) In non-canonical Hippo signaling, α-catenin forms a complex with 14-3-3-bound YAP under high cell density, thus stabilizing the 14-3-3/YAP interaction. Under low cell density conditions, PP2A dephosphorylates the 14-3-3 docking site on YAP, resulting in its nuclear import.
In a second report, nephrophthisis protein 4 (NPHP4), a known cilia-associated protein, was identified as a negative regulator of 14-3-3 binding interactions in the canonical Hippo pathway [23] (Fig. 2A). Mutations in NPHP4, as well as the ten other nephrophthisis proteins (NPHPs), are associated with a progressive tubulo-interstitial kidney disorder known as nephrophthisis (NPH) [24]. Because mice null for the Hippo transcriptional coactivator TAZ develop a degenerative cystic kidney disease similar to NHP [25] and given that small kidney size is the most striking clinical features of patients with NPH [24], Habbig and coworkers [23] conducted studies to determine whether any of the NPHPs might regulate Hippo signaling. Through coimmunoprecipitation experiments, NPHP4 was found to specifically interact with Lats1, a kinase that generates 14-3-3 binding sites on TAZ/YAP in the canonical Hippo pathway. Focusing specifically on the TAZ coactivator, binding of NPHP4 to Lats1 was found to prevent Lats1 from interacting with and phosphorylating TAZ on the 14-3-3 docking site. As a result, TAZ accumulated in the nucleus. These findings indicate a pro-proliferative function for NPHP4, serving as an inhibitor of the canonical Hippo pathway by inducing TAZ/YAP activation. In terms of NPH, Habbig and colleagues speculate that loss of function mutations in NPHP4 may lead to insufficient TAZ/YAP-dependent proliferative signaling and contribute to the small kidney phenotype observed in NPH patients. Further studies will be required to determine if this is indeed the case and to further evaluate the importance of NPHP4 to Hippo signaling in vivo.
3. 14-3-3 and cell migration: role in regulating the actin cytoskeleton
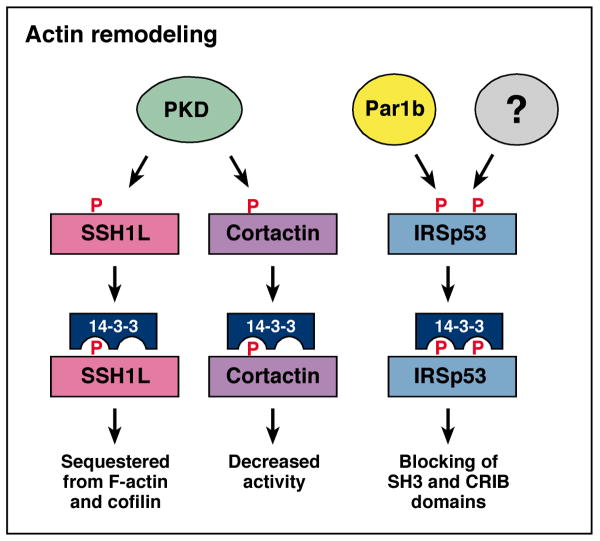
In cancer progression, a key property of metastatic tumor cells is the ability to migrate, and the first step in cell migration involves remodeling of the actin cytoskeletion. The dynamic remodeling processes occurring at the leading edge of migrating cells are controlled by a complex temporal and spatial interplay between Rho family GTPases, protein kinases, and protein phosphatases. In the last few years, a growing number of proteins involved in actin remodeling have been identified as 14-3-3 binding partners (Fig. 3), and protein kinase D has emerged as a kinase that generates 14-3-3 binding sites on several of these targets. One in this group is the cofilin regulator, slingshot 1-like (SSH1L). Cofilin plays an important role in actin remodeling by binding filamentous actin (F-actin) and severing actin filaments to generate free barbed ends [26]. Phosphorylation of cofilin on S3 disrupts its ability to bind and sever F-actin, whereas dephosphorylation of pS3 by the slingshot phosphatases activates cofilin [27]. Notably, the phosphatase activity of SSH1L is itself activated by F-actin binding, and the ability of SSH1L to interact with F-actin is inhibited by the association of SSH1L with 14-3-3 [28].
Fig. 3.
14-3-3 interacts with proteins involved in actin remodeling. PKD phosphorylates SSH1L and cortactin, whereas Par1b and unknown kinase(s) phosphorylate IRSp53 to generate 14-3-3 binding motifs. 14-3-3 binding inhibits the function of these actin regulatory proteins.
In 2009, two groups reported the identification of PKD as an F-actin-associated kinase that regulates the SSH1L/14-3-3 interaction [29, 30]. PKD was found to colocalize with SSH1L in F-actin-rich membrane structures, and in response to RhoA activation, phosphorylate SSH1L on S978 in its predicted F-actin binding region. Docking of 14-3-3 to pS978 relocalized SSH1L to the cytosol, thus sequestering SSH1L from both its activator F-actin and substrate cofilin. PKD-depletion and overexpression studies were conducted by both groups to demonstrate that PKD was the relevant pS978 kinase and that changes in SSH1L phosphorylation and localization had a direct effect on the phosphorylation state of cofilin. Further demonstrating the functional significance of these findings, overexpression of constitutively active PKD was found to block the migration of breast tumor cells, whereas depletion of PKD or overexpression of kinase-inactive PKD enhanced tumor cell migration.
The actin regulatory protein, cortactin, is another recently identified PKD substrate that is regulated by 14-3-3 binding [31]. Cortactin is an F-actin-binding protein enriched in dynamic actin structures such as membrane ruffles, lamellipodia of migrating cells, and invadipodia of invasive cancer cells [32]. Cortactin interacts with the Arp2/3 complex and, in conjunction with nucleation promoting factors such as WAVE2, functions to enhance actin polymerization rates [32]. PKD-mediated phosphorylation of cortactin on S298 generates a 14-3-3 docking motif, and as with SSH1L, this event appears to negatively regulate cortactin function. More specifically, Eiseler and coworkers [31] found that a phosphorylation-deficient cortactin mutant (S298A-Cortactin) had enhanced biological activity when examined in assays evaluating Wave2-Arp-driven actin polymerization, lamellipodia extension, and directed tumor cell migration.
14-3-3 has also been identified as a regulator of IRSp53, another molecule that plays a role in actin regulation. IRSp53 contains multiple protein interaction domains and acts as a signaling platform to connect the Cdc42 and Rac GTPases to downstream actin regulatory proteins [33]. Previous proteomic studies had isolated IRSp53 as a 14-3-3 binding partner [34], and recently, two reports have found that 14-3-3 binding modulates IRSp53 localization and function by controlling critical protein interactions [35, 36]. In the context of filopodium dynamics, Robens and colleagues [36] found that 14-3-3 docked to pT340 and pT360 located between two domains critical for IRSp53 function, the CRIB and SH3 domains. The kinase(s) that generates these sites was not determined; however, docking of 14-3-3 was found to block the accessibility of these domains and terminate IRSp53 signaling. In a second study, Cohen and coworkers identified IRSp53 as a substrate of the Par1b kinase and found that IRSp53 was critical for Par1b function in regulating basal actin organization and lumen polarity in epithelial cells. Par1b directly and indirectly induced the phosphorylation of IRSp53 on sites flanking the SH3 domain that bind 14-3-3 (S366 and S453/4/5). Although these binding sites are different from those reported in the preceding study, recruitment of 14-3-3 was again found to mask the IRSp53 SH3 domain and to disrupt docking of effectors that regulate actin polymerization. Notably, for all the proteins discussed, 14-3-3 binding acts to negatively regulate their function in actin remodeling.
4. 14-3-3 and EMT: implications for cancer progression
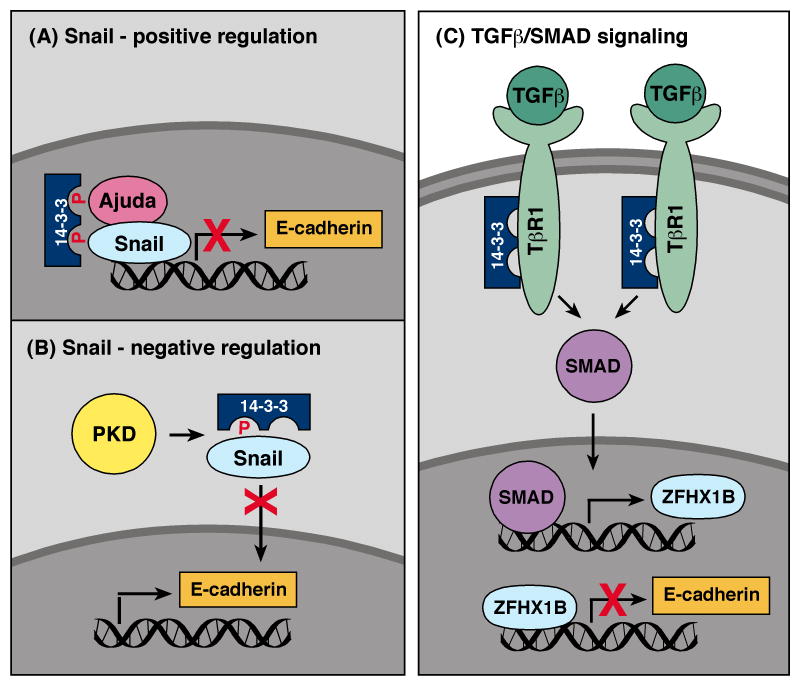
The transition of cells from an epithelial-to-mesenchymal phenotype (known as EMT) normally occurs during embryo development, and it is another biological process that contributes to cancer progression [37]. During EMT, epithelial cells acquire properties of mesenchymal cells, which include changes in morphology, loss of cell polarity, the expression of mesenchymal markers, and the ability to invade and migrate. As a result, a fundamental feature of EMT is the downregulation of E-cadherin and other cell adhesion molecules that inhibit cell motility. The Snail transcriptional repressor functions as a master regulator of EMT events and directly regulates genes affecting cell adhesion, motility, and polarity [38]. In 2010, two groups identified 14-3-3 as a new regulator of Snail [39, 40]. By sequence scanning, Hou and coworkers [40] observed two putative phosphorylation-dependent 14-3-3 binding motifs in Snail (S11 and T177), and in coexpression assays, Snail was shown to bind select 14-3-3 family members (γ, ε, η, and τ). While the phosphorylation state of S11 or T177 was not examined in this study, mutation of T177 disrupted 14-3-3 binding, whereas mutation of S11 had little effect. Further analysis revealed that Snail mutants unable to bind 14-3-3 at T177 were impaired in their ability to repress E-cadherin transcription and to induce EMT and cell migration when overexpressed in MCF10A cells. Hou et al. also found that the Snail co-repressor Ajuba interacted with 14-3-3 proteins and that both Ajuba and 14-3-3 proteins could be detected at the endogenous E-cadherin promoter in a Snail-dependent manner. These results implicate 14-3-3 as a positive regulator of Snail and suggest that 14-3-3 binding to the C-terminal T177 site may either stabilize the Snail repression complex and/or direct Snail complexes to chromatin (Fig. 4A).
Fig. 4.
Involvement of 14-3-3 in EMT. (A) 14-3-3 binding to the TGFβ receptor I (TβR1) prevents TβR1 ubiquitination and degradation, thus increasing TGF/SMAD signaling. Elevated TGF/SMAD signaling upregulates the transcriptional repressor ZFHX1B, which in turn downregulates E-cadherin expression. (B) Binding of 14-3-3 to the EMT regulator Snail at pT177 positively regulates Snail/Ajuda repressor complexes, thus downregulating E-cadherin expression. (C) PKD-mediated phosphorylation of S11 negatively regulates Snail function by sequestering Snail in the cytosol through 14-3-3 binding.
In a second study, Du and coworkers [39] identified Snail as a substrate of PKD, with PKD phosphorylating Snail on S11 in the N-terminal repression domain. To determine the relevance of S11 phosphorylation, Du et al. examined the intracellular localization of WT and S11A Snail when the proteins were overexpressed with various 14-3-3 family members and activated or kinase-inactive PKD. Results from these experiments suggest that S11 phosphorylation and binding of 14-3-3σ to this site sequesters Snail in the cytosol, thus inhibiting Snail's function as a transciptional repressor (Fig. 4B). Although this role for 14-3-3 binding appears to contradict the findings of the preceding study, it is possible that binding of 14-3-3 to specific Snail sites may elicit different effects. Taken together, these reports provide the framework for future studies that carefully analyze the phophorylation state of endogenous Snail to determine which sites are phosphorylated/dephosphorylated under conditions that induce EMT.
A role for 14-3-3 as a regulator of EMT has also been revealed in studies investigating the function of ErbB2 in breast cancer progression. Both clinical and experimental data indicate that increased expression of ErbB2 contributes to breast tumorigenesis; however, its overexpression alone is insufficient to promote invasive, metastatic disease [41]. In 2009, work from Dr. Dihua Yu's laboratory [42] identified 14-3-3ζ as a “second hit” that when overexpressed with ErbB2, increased the risk that a non-invasive ductal carcinoma in situ (DCIS) will progress to invasive breast cancer (IBC). In experiments examining the cellular processes altered by ErbB2 and 14-3-3ζ overexpression, Lu et al. found that overexpression of ErbB2 increased cell migration in a manner involving activation of the Src tyrosine kinase, whereas overexpression of 14-3-3ζ reduced cell-cell adhesion by promoting EMT. Collectively, overexpression of both proteins increased the invasive properties of normal mammary epithelial cells, as evidenced by loss of basement membrane integrity and movement of cells from acini into the culture matrix. Further investigation revealed that 14-3-3ζ exerted its effects through a pathway involving TGFβ/SMADs and the transcriptional repressor ZFHX1B, which like the Snail repressor downregulates E-cadherin and other cell adhesion molecules (Fig. 4C). More specifically, 14-3-3ζ was found to bind the TGFβ receptor I (TβRI) and inhibit its ubquitination and degredation by the proteosomal pathway, thus increasing receptor levels and elevating TGFβ/SMAD signaling. In turn, increased TGFβ/SMAD pathway activation resulted in the upregulation of transcriptional repressor ZFHX1B. Analysis of patient samples further indicated that 14-3-3ζ overexpression correlated with increased TβRI levels and reduced expression of E-cadherin in both DCIS and IBC. Importantly, patients whose breast tumors overexpressed both ErbB2 and 14-3-3ζ were found to have higher rates of metastatic recurrence and death than those whose tumors overexpressed only ErbB2 or 14-3-3ζ [43]. More recently, work from this group has also found that 14-3-3 overexpression in breast tumors correlates with increased levels of activated AKT. Their findings suggest that in addition to its effects on EMT, 14-3-3ζ may impact AKT activation and tumor cell survival by binding the p85 subunit of PI3K and increasing the membrane localization and signaling activity of PI3K [44].
5. 14-3-3 proteins: clinical significance to human cancer
As the understanding of 14-3-3 function grows and the analysis of patient tumors increases, the clinical significance of 14-3-3 proteins to human cancer has begun to emerge. To date, the two family members that have been most implicated in cancer processes are 14-3-3σ and 14-3-3ζ. 14-3-3σ is unique among the 14-3-3 proteins in that it is expressed primarily in epithelial cells and forms homodimers almost exclusively [45]. For many years, 14-3-3σ has been considered a tumor suppressor due to its positive role in regulating p53 and in mediating a G2/M checkpoint following DNA damage. In addition, the down-regulation of 14-3-3σ has been observed in numerous cancers of epithelial origin, including lung carcinomas and most primary breast cancers (Reviewed in[46]). Recently, Dr. William Muller's lab has reported that in the context of breast tumors overexpressing ErbB2, 14-3-3σ plays a critical role in maintaining cell polarity and that its loss may contribute to disease progression [47]. However, despite these findings, other studies have reported that 14-3-3σ overexpression correlates with and may predict poor prognosis of colorectal, prostate, and pancreatic cancers, and that its expression may contribute to resistance to DNA-damaging drugs in some tumor types [46]. Thus, the role of 14-3-3σ as a “general” tumor suppressor in human cancer has been brought into question, and more research is needed to investigate its function in specific cancer types.
In contrast to 14-3-3σ, 14-3-3ζ is a ubiquitously expressed 14-3-3 family member that has been implicated to have oncogenic potential through its interaction with target proteins involved in cancer initiation (Rafs, p85PI3K, Bad, FOXO transcription factors) as well as cancer progression (Snail, TβRI). Elevated expression of 14-3-3ζ has been observed in a variety of tumor types, and studies now indicate that 14-3-3ζ over-expression may be a marker for disease recurrence and poor survival in patients with breast cancer, head-and-neck/oral squamous cell carcinomas, and non-small cell lung carcimomas [48]. Elevated levels of 14-3-3ζ have also been linked to drug resistance to certain anticancer therapies, including tamoxifen treatment. Tamoxifen has been found to upregulate the expression of 14-3-3ζ in estrogen receptor (ER)-positive breast cancer lines, and high expression of 14-3-3 in primary breast tumors correlates with a poor clinical outcome [49]. Recent work by Bergamaschi and Katzenllenbourgen [50] has revealed that tamoxifen, but not other selective ER modulators, downregulates the microRNA miR451 that specifically targets 14-3-3ζ and as a result, increases 14-3-3ζ expression. In this report, the levels of 14-3-3ζ and miR-451 were found to inversely correlate in tamoxifen-resistant breast cancer cells and increased expression of miR-451 could re-sensitize cells to tamoxifen. Down-regulation of 14-3-3ζ by siRNA treatment has also been shown to increase the sensitivity of head and neck cancers, lung carcinomas, and diffuse large B cell lymphomas to apoptosis-inducing agents [51-53]. Taken together, these findings indicate the need for developing clinical tests that assay the expression of 14-3-3 family members in multiple tumor types and suggest that monitoring 14-3-3 levels during cancer treatment may be of importance.
6. Concluding comments
Research in the last few years has revealed exciting advances that further elucidate the vast role of 14-3-3 proteins in cell signaling. These studies have identified new 14-3-3 binding partners and led to the discovery of a new mechanism for modulating 14-3-3 binding interactions that involves arginine methylation. In addition, PKD has emerged as another protein kinase that serves an important function in generating 14-3-3 binding sites on critical targets, including proteins involved in regulating cell migration and EMT. In the future, it is likely that more binding partners will be uncovered and that the involvement of 14-3-3 in vital cellular processes will expand. Moreover, as the analysis of patient tumors continues, it is likely that additional information will be learned regarding the involvement of various 14-3-3 proteins in cancer progression and whether their presence will guide treatment options. Also on the horizon is the pursuit of small molecule inhibitors that target either global or specific 14-3-3 interactions. Such reagents will undoubtedly further our understanding of 14-3-3 function and may prove useful in the design of new therapeutic treatments.
Highlights.
Function and regulation of 14-3-3 binding interactions in RTK/Ras signaling
New regulators of 14-3-3 binding interactions in the Hippo pathway
14-3-3 and cell migration: role in regulating the actin cytoskeleton
14-3-3 and EMT: implications for cancer progression
14-3-3 proteins: clinical significance in human cancer
Acknowledgments
Our work is supported by federal funds from the National Cancer Institute.
Abbreviations
- RTK
receptor tyrosine kinase
- EMT
epithelial-to-mesenchymal transition
- pS/T
phosphoserine/phosphothreonine
- PP2A
protein phosphatase 2A
- SCF
Skp1/Cul-1/F-box
- CDK
cyclin-dependent kinase
- SSH1L
slingshot 1-like
- F-actin
filamentous actin
- NPH
nephrophthisis
- NPHP
nephrophthisis protein
- TβRI
TGFβ receptor I
- DCIS
ductal carcinoma in situ
- IBC
invasive breast cancer
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Aitken A. 14-3-3 proteins: a historic overview. Semin Cancer Biol. 2006;16:162–72. doi: 10.1016/j.semcancer.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 2.Gardino AK, Smerdon SJ, Yaffe MB. Structural determinants of 14-3-3 binding specificities and regulation of subcellular localization of 14-3-3-ligand complexes: a comparison of the X-ray crystal structures of all human 14-3-3 isoforms. Semin Cancer Biol. 2006;16:173–82. doi: 10.1016/j.semcancer.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 3.Morrison D. 14-3-3: modulators of signaling proteins? Science. 1994;266:56–7. doi: 10.1126/science.7939645. [DOI] [PubMed] [Google Scholar]
- 4.Wellbrock C, Karasarides M, Marais R. The RAF proteins take centre stage. Nat Rev Mol Cell Biol. 2004;5:875–85. doi: 10.1038/nrm1498. [DOI] [PubMed] [Google Scholar]
- 5.Garnett MJ, Rana S, Paterson H, Barford D, Marais R. Wild-type and mutant B-RAF activate C-RAF through distinct mechanisms involving heterodimerization. Mol Cell. 2005;20:963–9. doi: 10.1016/j.molcel.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 6.Rushworth LK, Hindley AD, O'Neill E, Kolch W. Regulation and role of Raf-1/B-Raf heterodimerization. Mol Cell Biol. 2006;26:2262–72. doi: 10.1128/MCB.26.6.2262-2272.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ritt DA, Monson DM, Specht SI, Morrison DK. Impact of feedback phosphorylation and Raf heterodimerization on normal and mutant B-Raf signaling. Mol Cell Biol. 2010;30:806–19. doi: 10.1128/MCB.00569-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dhomen N, Marais R. New insight into BRAF mutations in cancer. Curr Opin Genet Dev. 2007;17:31–9. doi: 10.1016/j.gde.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 9.Tartaglia M, Gelb BD. Disorders of dysregulated signal traffic through the RAS-MAPK pathway: phenotypic spectrum and molecular mechanisms. Ann N Y Acad Sci. 2010;1214:99–121. doi: 10.1111/j.1749-6632.2010.05790.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kobayashi T, Aoki Y, Niihori T, Cave H, Verloes A, Okamoto N, et al. Molecular and clinical analysis of RAF1 in Noonan syndrome and related disorders: dephosphorylation of serine 259 as the essential mechanism for mutant activation. Hum Mutat. 2010;31:284–94. doi: 10.1002/humu.21187. [DOI] [PubMed] [Google Scholar]
- 11.Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–74. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin HK, Wang G, Chen Z, Teruya-Feldstein J, Liu Y, Chan CH, et al. Phosphorylation-dependent regulation of cytosolic localization and oncogenic function of Skp2 by Akt/PKB. Nat Cell Biol. 2009;11:420–32. doi: 10.1038/ncb1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao D, Inuzuka H, Tseng A, Chin RY, Toker A, Wei W. Phosphorylation by Akt1 promotes cytoplasmic localization of Skp2 and impairs APCCdh1-mediated Skp2 destruction. Nat Cell Biol. 2009;11:397–408. doi: 10.1038/ncb1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simmen T, Aslan JE, Blagoveshchenskaya AD, Thomas L, Wan L, Xiang Y, et al. PACS-2 controls endoplasmic reticulum-mitochondria communication and Bid-mediated apoptosis. EMBO J. 2005;24:717–29. doi: 10.1038/sj.emboj.7600559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aslan JE, You H, Williamson DM, Endig J, Youker RT, Thomas L, et al. Akt and 14-3-3 control a PACS-2 homeostatic switch that integrates membrane traffic with TRAIL-induced apoptosis. Mol Cell. 2009;34:497–509. doi: 10.1016/j.molcel.2009.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kruyt FA. TRAIL and cancer therapy. Cancer Lett. 2008;263:14–25. doi: 10.1016/j.canlet.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 17.Brummer T, Larance M, Herrera Abreu MT, Lyons RJ, Timpson P, Emmerich CH, et al. Phosphorylation-dependent binding of 14-3-3 terminates signalling by the Gab2 docking protein. EMBO J. 2008;27:2305–16. doi: 10.1038/emboj.2008.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wohrle FU, Daly RJ, Brummer T. Function, regulation and pathological roles of the Gab/DOS docking proteins. Cell Commun Signal. 2009;7:22. doi: 10.1186/1478-811X-7-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sakamaki J, Daitoku H, Ueno K, Hagiwara A, Yamagata K, Fukamizu A. Arginine methylation of BCL-2 antagonist of cell death (BAD) counteracts its phosphorylation and inactivation by Akt. Proc Natl Acad Sci U S A. 2011;108:6085–90. doi: 10.1073/pnas.1015328108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamagata K, Daitoku H, Takahashi Y, Namiki K, Hisatake K, Kako K, et al. Arginine methylation of FOXO transcription factors inhibits their phosphorylation by Akt. Mol Cell. 2008;32:221–31. doi: 10.1016/j.molcel.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 21.Chan SW, Lim CJ, Chen L, Chong YF, Huang C, Song H, et al. The Hippo pathway in biological control and cancer development. J Cell Physiol. 2011;226:928–39. doi: 10.1002/jcp.22435. [DOI] [PubMed] [Google Scholar]
- 22.Schlegelmilch K, Mohseni M, Kirak O, Pruszak J, Rodriguez JR, Zhou D, et al. Yap1 acts downstream of alpha-catenin to control epidermal proliferation. Cell. 2011;144:782–95. doi: 10.1016/j.cell.2011.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Habbig S, Bartram MP, Muller RU, Schwarz R, Andriopoulos N, Chen S, et al. NPHP4, a cilia-associated protein, negatively regulates the Hippo pathway. J Cell Biol. 2011;193:633–42. doi: 10.1083/jcb.201009069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hildebrandt F, Attanasio M, Otto E. Nephronophthisis: disease mechanisms of a ciliopathy. J Am Soc Nephrol. 2009;20:23–35. doi: 10.1681/ASN.2008050456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hossain Z, Ali SM, Ko HL, Xu J, Ng CP, Guo K, et al. Glomerulocystic kidney disease in mice with a targeted inactivation of Wwtr1. Proc Natl Acad Sci U S A. 2007;104:1631–6. doi: 10.1073/pnas.0605266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andrianantoandro E, Pollard TD. Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol Cell. 2006;24:13–23. doi: 10.1016/j.molcel.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 27.Huang TY, DerMardirossian C, Bokoch GM. Cofilin phosphatases and regulation of actin dynamics. Curr Opin Cell Biol. 2006;18:26–31. doi: 10.1016/j.ceb.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 28.Nagata-Ohashi K, Ohta Y, Goto K, Chiba S, Mori R, Nishita M, et al. A pathway of neuregulin-induced activation of cofilin-phosphatase Slingshot and cofilin in lamellipodia. J Cell Biol. 2004;165:465–71. doi: 10.1083/jcb.200401136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eiseler T, Doppler H, Yan IK, Kitatani K, Mizuno K, Storz P. Protein kinase D1 regulates cofilin-mediated F-actin reorganization and cell motility through slingshot. Nat Cell Biol. 2009;11:545–56. doi: 10.1038/ncb1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peterburs P, Heering J, Link G, Pfizenmaier K, Olayioye MA, Hausser A. Protein kinase D regulates cell migration by direct phosphorylation of the cofilin phosphatase slingshot 1 like. Cancer Res. 2009;69:5634–8. doi: 10.1158/0008-5472.CAN-09-0718. [DOI] [PubMed] [Google Scholar]
- 31.Eiseler T, Hausser A, De Kimpe L, Van Lint J, Pfizenmaier K. Protein kinase D controls actin polymerization and cell motility through phosphorylation of cortactin. J Biol Chem. 2010;285:18672–83. doi: 10.1074/jbc.M109.093880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weaver AM, Young ME, Lee WL, Cooper JA. Integration of signals to the Arp2/3 complex. Curr Opin Cell Biol. 2003;15:23–30. doi: 10.1016/s0955-0674(02)00015-7. [DOI] [PubMed] [Google Scholar]
- 33.Scita G, Confalonieri S, Lappalainen P, Suetsugu S. IRSp53: crossing the road of membrane and actin dynamics in the formation of membrane protrusions. Trends Cell Biol. 2008;18:52–60. doi: 10.1016/j.tcb.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 34.Mackie S, Aitken A. Novel brain 14-3-3 interacting proteins involved in neurodegenerative disease. FEBS J. 2005;272:4202–10. doi: 10.1111/j.1742-4658.2005.04832.x. [DOI] [PubMed] [Google Scholar]
- 35.Cohen D, Fernandez D, Lazaro-Dieguez F, Musch A. The serine/threonine kinase Par1b regulates epithelial lumen polarity via IRSp53-mediated cell-ECM signaling. J Cell Biol. 2011;192:525–40. doi: 10.1083/jcb.201007002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robens JM, Yeow-Fong L, Ng E, Hall C, Manser E. Regulation of IRSp53-dependent filopodial dynamics by antagonism between 14-3-3 binding and SH3-mediated localization. Mol Cell Biol. 2010;30:829–44. doi: 10.1128/MCB.01574-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nieto MA. The Ins and Outs of the Epithelial to Mesenchymal Transition in Health and Disease. Annu Rev Cell Dev Biol. 2010 doi: 10.1146/annurev-cellbio-092910-154036. [DOI] [PubMed] [Google Scholar]
- 38.Barrallo-Gimeno A, Nieto MA. The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development. 2005;132:3151–61. doi: 10.1242/dev.01907. [DOI] [PubMed] [Google Scholar]
- 39.Du C, Zhang C, Hassan S, Biswas MH, Balaji KC. Protein kinase D1 suppresses epithelial-to-mesenchymal transition through phosphorylation of snail. Cancer Res. 2010;70:7810–9. doi: 10.1158/0008-5472.CAN-09-4481. [DOI] [PubMed] [Google Scholar]
- 40.Hou Z, Peng H, White DE, Wang P, Lieberman PM, Halazonetis T, et al. 14-3-3 binding sites in the snail protein are essential for snail-mediated transcriptional repression and epithelial-mesenchymal differentiation. Cancer Res. 2010;70:4385–93. doi: 10.1158/0008-5472.CAN-10-0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nofech-Mozes S, Spayne J, Rakovitch E, Hanna W. Prognostic and predictive molecular markers in DCIS: a review. Adv Anat Pathol. 2005;12:256–64. doi: 10.1097/01.pap.0000184177.65919.5e. [DOI] [PubMed] [Google Scholar]
- 42.Lu J, Guo H, Treekitkarnmongkol W, Li P, Zhang J, Shi B, et al. 14-3-3zeta Cooperates with ErbB2 to promote ductal carcinoma in situ progression to invasive breast cancer by inducing epithelial-mesenchymal transition. Cancer Cell. 2009;16:195–207. doi: 10.1016/j.ccr.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neal CL, Yao J, Yang W, Zhou X, Nguyen NT, Lu J, et al. 14-3-3zeta overexpression defines high risk for breast cancer recurrence and promotes cancer cell survival. Cancer Res. 2009;69:3425–32. doi: 10.1158/0008-5472.CAN-08-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neal CL, Xu J, Li P, Mori S, Yang J, Neal NN, et al. Overexpression of 14-3-3zeta in cancer cells activates PI3K via binding the p85 regulatory subunit. Oncogene. 2011 doi: 10.1038/onc.2011.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilker EW, Grant RA, Artim SC, Yaffe MB. A structural basis for 14-3-3sigma functional specificity. J Biol Chem. 2005;280:18891–8. doi: 10.1074/jbc.M500982200. [DOI] [PubMed] [Google Scholar]
- 46.Li Z, Liu JY, Zhang JT. 14-3-3sigma, the double-edged sword of human cancers. Am J Transl Res. 2009;1:326–40. [PMC free article] [PubMed] [Google Scholar]
- 47.Ling C, Zuo D, Xue B, Muthuswamy S, Muller WJ. A novel role for 14-3-3sigma in regulating epithelial cell polarity. Genes Dev. 2010;24:947–56. doi: 10.1101/gad.1896810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Neal CL, Yu D. 14-3-3zeta as a prognostic marker and therapeutic target for cancer. Expert Opin Ther Targets. 2010;14:1343–54. doi: 10.1517/14728222.2010.531011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frasor J, Chang EC, Komm B, Lin CY, Vega VB, Liu ET, et al. Gene expression preferentially regulated by tamoxifen in breast cancer cells and correlations with clinical outcome. Cancer Res. 2006;66:7334–40. doi: 10.1158/0008-5472.CAN-05-4269. [DOI] [PubMed] [Google Scholar]
- 50.Bergamaschi A, Katzenellenbogen BS. Tamoxifen downregulation of miR-451 increases 14-3-3zeta and promotes breast cancer cell survival and endocrine resistance. Oncogene. 2011 doi: 10.1038/onc.2011.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fan T, Li R, Todd NW, Qiu Q, Fang HB, Wang H, et al. Up-regulation of 14-3-3zeta in lung cancer and its implication as prognostic and therapeutic target. Cancer Res. 2007;67:7901–6. doi: 10.1158/0008-5472.CAN-07-0090. [DOI] [PubMed] [Google Scholar]
- 52.Maxwell SA, Li Z, Jaye D, Ballard S, Ferrell J, Fu H. 14-3-3zeta mediates resistance of diffuse large B cell lymphoma to an anthracycline-based chemotherapeutic regimen. J Biol Chem. 2009;284:22379–89. doi: 10.1074/jbc.M109.022418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Matta A, DeSouza LV, Ralhan R, Siu KW. Small interfering RNA targeting 14-3-3zeta increases efficacy of chemotherapeutic agents in head and neck cancer cells. Mol Cancer Ther. 2010;9:2676–88. doi: 10.1158/1535-7163.MCT-10-0312. [DOI] [PubMed] [Google Scholar]