Abstract
Glioblastomas (GBM), the most common primary brain tumors, infiltrate the brain, grow rapidly, and are refractory to current therapies. Signature genetic lesions in glioblastomas include mutation of the EGFR receptor tyrosine kinase and activating mutations in components of the PI-3 kinase (PI3K) pathway. Despite years of study, how these pathways specifically regulate glial pathogenesis is unclear. To address the genetic and cellular origins of this disease, a novel Drosophila GBM model has been developed in which glial progenitor cells give rise to proliferative and invasive neoplastic cells that create transplantable tumors in response to constitutive co-activation of the EGFR-Ras and PI3K pathways. Standing with a rich literature demonstrating the direct relevance of Drosophila to studies on human cancer, neurological disease, and neurodevelopment, this model represents a robust cell-type specific Drosophila neurological disease model in which malignant cells are created by mutations in genetic pathways thought to be driving forces in a homologous human disease. Using lineage analysis and cell-type specific markers, neoplastic glial cells were found to originate from committed glial progenitor cells, rather than from multipotent neuroblasts. Genetic analyses demonstrated that EGFR-Ras and PI3K induce fly glial neoplasia through activation of a combinatorial genetic network composed, in part, of other genetic pathways also commonly mutated in human glioblastomas. In the future, large scale forward genetic screens with this model may reveal new insights into the origins and treatments of human glioblastoma.
INTRODUCTION
Human gliomas, a group of both high-grade and low-grade neoplasms of glial cells and their precursors, are the most common and deadly primary tumors of the central nervous system (CNS). Glioblastoma (GBM) is the most frequent and malignant form of high-grade glioma. GBMs are resistant to current treatments, and are highly proliferative and diffusely invasive. This latter property renders GBMs largely incurable by surgical resection as the invasive cells form rapidly growing secondary tumors (Giese et al. 2003). One key to developing new, effective therapies against these tumors is to understand the fundamental genetic, cellular, and molecular logic underlying gliomagenesis.
Cellular and genetic origins of malignant glioma: Glia, EGFR, and PI-3 kinase
Normal glia have diverse and essential roles in the CNS (Lemke 2001). The mammalian brain contains multiple glial cell types, such as astrocytes, and glial progenitor cells, including oligodendrocyte precursor cells, many of which normally migrate and maintain a proliferative capacity throughout life (Colognato and ffrench-Constant 2004; Suh et al. 2009). Neural stem cells are self-renewing, proliferative cells that divide to yield progeny that exhibit multilineage differentiation. Current hypotheses favor that GBMs and other high-grade gliomas arise from neural stem cells since stem-cell-like cells can be isolated from surgical specimens that recapitulate tumors upon transplantation (Singh et al. 2004). Yet, transforming mutations need not occur in neural stem cells, as experimental models show that both astrocytic glia and oligodendrocyte precursor cells can give rise to GBM and high-grade gliomas in response to mutations associated with GBM (Bachoo et al. 2002; Marumoto et al. 2009; Uhrbom et al. 2002; Uhrbom et al. 2005). Moreover, the distinction between neural stem cells and glia is rather blurred given that stem cells in certain regions of the mammalian brain are actually astrocytic glia (Doetsch et al. 1999; Sanai et al. 2004; Suh et al. 2009). Thus, misregulation of gliomagenic pathways can confer unrestricted proliferative and self-renewal capacities to a range of glial cell types. Understanding how gliomagenic mutations transform these different cell types may reveal shared cellular characteristics required for tumorigenesis that could provide new therapeutic opportunities.
Gliomagenesis involves mutations in many genes, most of which normally regulate neural development (Furnari et al. 2007). The most frequent genetic lesions in GBM and high-grade gliomas include amplification and mutation of receptor tyrosine kinases, most often Epidermal Growth Factor Receptor tyrosine kinase (EGFR), which play crucial roles in the normal specification, survival, proliferation, and migration of neural stem cells as well as astroglial and oligodendroglial precursor cells (Aguirre et al. 2007; Burrows et al. 1997; Caric et al. 2001; Kornblum et al. 1998; McLendon et al. 2008; Parsons et al. 2008; Sun et al. 2005). GBMs with EGFR amplification often harbor mutant forms of EGFR, such as EGFRvIII, which display constitutive kinase activity and stimulate Ras signaling to drive proliferation, promote migration, and block apoptosis (Boockvar et al. 2003; Ekstrand et al. 1992; Huang et al. 1997; Nagane et al. 1996; Prigent et al. 1996; Sugawa et al. 1990). The importance of receptor tyrosine kinase (RTK) signaling in GBM is strengthened by the occurrence of activating mutations and amplifications of other RTKs, such as PDGFRalpha and FGFR (McLendon et al. 2008; Parsons et al. 2008). Other common mutations in GBM and high-grade glioma target the phosphatidyl-inositol-3 kinase (PI3K) signaling pathway, which normally stimulates cellular proliferation, growth, and migration in various neural cell types (reviewed in Endersby and Baker 2008). Mutations occur in PIK3CA, which encodes the p110α catalytic subunit of PI3K, and PIK3R1, which encodes the p85α adaptor subunit of PI3K (Broderick et al. 2004; McLendon et al. 2008; Samuels et al. 2004). Akt kinases, key effectors of PI3K, are often activated or amplified in GBMs (Haas-Kogan et al. 1998; McLendon et al. 2008). Loss of PTEN occurs through deletion, point mutations, and epigenetic silencing in approximately 70–80% of GBMs (Baeza et al. 2003; Li et al. 1997; McLendon et al. 2008; Parsons et al. 2008; Steck et al. 1997). Through its lipid phosphatase activity, PTEN antagonizes PI3K signaling, and, thus, the major effect of PTEN loss is constitutive activation of the PI3K-Akt pathway (reviewed in Sulis and Parsons 2003). In the mouse CNS, PTEN loss provokes proliferation and inappropriate migration in otherwise wild-type neural stem cells and astrocytes (Fraser et al. 2004; Groszer et al. 2001; Marino et al. 2002). Conversely, restoring PTEN expression in PTEN-deficient GBM cells suppresses growth and tumorigenicity (Furnari et al. 1997). Components of the Rb-E2F pathway, which regulates cell cycle entry and exit, are also frequently mutated in GBM. Loss of the cdk inhibitor p16ink4a/Arf or tumor suppressor Rb, which normally restricts proliferation of neural stem cells and glial progenitor cells, are the most common mutations (McLendon et al. 2008; Parsons et al. 2008).
Although mutations in EGFR, PI3K, and E2F/Rb pathway frequently occur in GBMs, activation of EGFR-Ras or PI3K alone is not sufficient to transform glia or neural stem cells. Rather, multiple mutations that co-activate EGFR-Ras and PI3K signaling, or activate EGFR-Ras and inactivate p16ink4a/Arf, are required to induce GBM or high-grade glioma (Bachoo et al. 2002; Holland et al. 2000; Holland et al. 1998; Marumoto et al. 2009). For example, in mouse models, constitutively active Ras causes GBM when combined with constitutively active Akt or PTEN loss in either neural stem cells or astrocytic glia, (Holland et al. 2000; Hu et al. 2005; Marumoto et al. 2009) demonstrating that co-activation and co-mutation of these pathways are required for transformation.
The full range of signaling events acting downstream of EGFR-Ras and PI3K to elicit neoplastic transformation and inappropriate migration remains to be determined. While studies have established that several core pathway effectors are utilized by EGFR-Ras and PI3K in development and in GBM (reviewed in Furnari et al. 2007), constitutive activation of EGFR-Ras and PI3K may deploy distinct outputs, not used in development, that allow particular cells to escape normal physiological cues that restrain proliferation and self-renewal. Combined pathway activation may produce quantitative or qualitative changes that either pathway alone cannot accomplish. For example, in the developing Drosophila eye, PI3K signaling upregulates the expression of EGFR pathway components, which alters EGFR-dependent transcriptional output (McNeill et al. 2008). Diffuse infiltrative behavior is characteristic of malignant primary brain tumors and is not often observed in brain metastases from other malignant tumors with EGFR and/or PI3K mutations, indicating that cell-type specific factors, rather than intrinsic activities of EGFR and/or PI3K signaling, dictates invasive behavior. Consistent with this, molecules known to modulate glioblastoma invasion also regulate neural migration and adhesion (reviewed in Hoelzinger et al. 2007). However, interactions between many of these molecules and EGFR-Ras or PI3K signaling have not yet been examined.
Though activation of EGFR-Ras and PI3K pathways can stimulate gliomagenesis, treatments with pharmacologic inhibitors of these pathways does not show significant efficacy in GBM patients (Cloughesy et al. 2008; Mellinghoff et al. 2005; Raizer et al. 2010). Numerous explanations have been proffered for these results. For example, epigenetic alterations in GBM cells caused by transformation may promote drug resistance; drugs may not reach active local concentrations in the brain due to sequestration by stromal cells; paracrine signals from the tumor microenvironment may promote resistance; and/or mutations in additional genes may confer resistance to inhibition of these pathways. Distinguishing among these and other possibilities remains a looming experimental challenge. Other experimental efforts are aimed at creating improved therapeutics to target these pathways (Perera et al. 2007). Yet, inhibition of EGFR-Ras and PI3K pathways may only be cytostatic, at best, because GBM cells may simply revert to a more normal glial cell fate rather than actively undergo apoptosis. Thus, more effective therapeutic strategies must be developed that not only inhibit these pathways, but that also actively stimulate the death of GBM cells.
To find new therapeutic targets, large-scale tumor genomics projects, such as the Cancer Genome Atlas (TCGA) project, are cataloging genetic mutations and gene expression patterns in hundreds of GBM tumor samples (Brennan et al. 2009; McLendon et al. 2008; Parsons et al. 2008; Verhaak et al. 2010). These projects have confirmed the frequency of EGFR and PI3K pathway mutations in GBM and have identified potentially hundreds of other genetic alterations associated with tumors, many of which have not been previously shown to have a causal role in tumorigenesis. These projects have also generated data consistent with the occurrence of two to five different molecular subtypes of glioblastoma as defined by patterns of gene expression (Brennan et al. 2009; Colman et al. 2010; Phillips et al. 2006; Verhaak et al. 2010). However, the mechanistic relationship between different gene expression patterns and different mutations observed in these tumors has not been thoroughly examined. Indeed, assessing the functional significance of the new genetic alterations and gene expression patterns identified by the TCGA glioblastoma project and other large-scale tumor genomics projects presents another enormous experimental challenge.
Mammalian glioblastoma models
To address these challenges, investigators employ a range of experimental systems. Established serum-cultured GBM cell lines have been extensively and successfully used to explore important aspects of gliomagenesis, such as oncogenic signal transduction, tumor genetics, and mutational progression. However, serum-cultured GBM cells lines do not retain the full repertoire of genetic and phenotypic characteristics observed in tumor cells in vivo (Lee et al. 2006b), such as EGFR amplification or invasive growth, limiting their usefulness. New culture techniques with neural-stem cell media, which contains the RTK ligands bFGF and EGF, have been used to propagate proliferative, tumorigenic cells derived from fresh surgical samples, underscoring the importance of EGFR signaling in these tumors (Laks et al. 2009; Singh et al. 2004). Tumorigenic GBM cells isolated and characterized in this way have been called glioblastoma stem cells (GSCs), or glioblastoma propagating cells. GSCs retain more genetic and phenotypic characteristics of tumors than serum-cultured GBM cell lines (Bao et al. 2006; Chen et al. 2010; Cheng et al. 2010; Lee et al. 2006b; Singh et al. 2004), but have some technical limitations as well. For example, bFGF- and EGF-cultured GSCs can only be established and/or maintained from a subset of GBM tumors (Chen et al. 2010; Laks et al. 2009). Moreover, much controversy still exists as to the methods for defining and characterizing GSCs and their relationship to the normal cells from which they originate. Cultured GSCs display some properties of normal cultured neural stem cells, such as expression of neural stem cell markers and multilineage differentiation. Yet, this should not be construed as evidence that GBMs arise solely from neural stem cells given that differentiated glia can dedifferentiate and acquire neural stem cell-like properties when cultured in neural stem cell media (Buffo et al. 2008). Rather, this supports the idea that GBM is propagated by transformed cells that, whether they were once neural stem cells or differentiated glia, acquire unregulated and inappropriate stem cell-like properties in response to gliomagenic mutations.
Cell culture GBM models are limited because they do not reproduce the complex environment inherent in the CNS, or in the tumors themselves. Vasculature, neurons, reactive astrocytes, and macrophage-like microglia within the CNS can be sources of signals that may promote proliferation and survival of tumorigenic GBM cells (reviewed in Pong and Gutmann 2010). Recent reports demonstrate genetic and cellular heterogeneity within individual tumors, even among cells with EGFR mutations and amplifications. For example, EGFRvIII-mutant GBM cells secrete cytokines that promote the survival and proliferation of tumor cells harboring amplification of the wild-type EGFR receptor, demonstrating that EGFR signaling plays both cell autonomous and nonautonomous roles within these tumors (Inda et al. 2010). To replicate the complex cellular environment in vivo, investigators turn to the mouse models. In xenograft models, tumorigenic cells from fresh surgical samples or cell cultures can be propagated intracranially in immunocompromised mice, and these tumors can show characteristics of the original tumors (Sarkaria et al. 2006; Singh et al. 2004). Other models, in which tumors are created in the mouse brain by transgenic or viral induction of neural stem cells or lineage-restricted glial cells, have provided valuable insights into the role of known mutations and pathways in gliomagenesis in vivo, as discussed above (reviewed in Fomchenko and Holland 2006; Hu and Holland 2005). However, transgenic and xenograft-based mouse models have not been widely used to screen for new genes or new therapeutic targets involved in GBM or to test the causal role of the many rarer mutations and combinations of mutations uncovered by the TCGA, largely because forward functional genetic assays are very laborious and expensive to do in these systems.
To address the many challenges in this field, we need new approaches that complement the strengths and limitations of cell culture and mouse models, and that efficiently integrate existing data and new technologies. To this end, we turn to Drosophila melanogaster, a highly versatile genetic model system in which cell-type specific gene function can be manipulated with single-cell precision in vivo in an intact complex nervous system.
Drosophila as a human disease model system
Drosophila melanogaster has proven to be a powerful model system for studying the genetics and biology of human diseases (reviewed in Bilder 2004; Brumby and Richardson 2005; Hirth 2010). Genomic sequencing shows that approximately 70% of all known human disease genes have functional orthologs in Drosophila (Reiter and Bier 2002). Among the most evolutionarily conserved genes are components of major signal transduction pathways that play essential roles in neural development and cancer, including EGFR/RTK-Ras, PI3K, Notch, Wnt, Jak-STAT, Hedgehog, and TGF-β. Many core components of these pathways were first discovered in genetic studies of fly neural development and tumorigenesis (Gao et al. 2000; Karim et al. 1996; Lee and Orr-Weaver 2003; Pan 2010; Simon 2000; Simon et al. 1991; Voas and Rebay 2004; Wilson and Chuang 2010). In fact, Drosophila was one of the first experimental systems to show that cancer is linked to heritable genetic mutations; one of the first recessive lethal mutations isolated in Morgan’s laboratory, lethal 7, caused transplantable, malignant tumors (Stark 1918; Stark 1919). In the 1960s and 70s, Drosophila geneticists identified several lethal mutants that develop malignant tumorous growths in several tissues, including the brain (Gateff 1978; Gateff and Schneiderman 1969). These tumors can be maintained by serial transplantation, and are characterized by continuous proliferation, impaired differentiation, and invasiveness (Akai et al. 1967; Gateff and Schneiderman 1969). This established some of earliest experimental evidence for the existence of tumor suppressor genes that unleash tumorigenesis when homozygous mutant (Comings 1973). In recent years, specific Drosophila models have been created for several human diseases, including cancers and neurological disorders, and these models have produced observations about human disease that have been validated in mammalian systems (Choi et al. 2010; McBride et al. 2005; Read et al. 2005; Steffan et al. 2001). Presently, most Drosophila disease models focus on genetic and phenotypic analysis of tissue-specific gene function in fly cell types with obvious human counterparts, such as stem cells in the gut and neurons in the brain. Given that many human diseases are governed by intrinsic properties of their host tissue or cell-of-origin, cell-type specific fly disease models are likely to yield insights relevant to their respective human diseases.
Drosophila offers several advantages for modeling human diseases. Current Drosophila techniques allow for cell-type specific manipulation of gene expression with single cell precision in vivo within the CNS and other tissues (reviewed in St Johnston 2002). Commonly used Drosophila genetic tools provide the ability to dissect multigene interactions and identify new gene functions: the fly genome is sequenced and well-annotated; thousands of loss-of-function mutants for the majority of genes in the genome are publicly available; genes can be knocked-out with targeted mutagenesis, insertional mutagenesis, or RNA interference (RNAi); tissue-specific gene overexpression can be carried out with spatial and temporal precision using establishing techniques; binary transgene expression systems, such as the Gal4/UAS system, allow for combination of multiple transgenes in the same animal to achieve transgene activation or misexpression under specific promoters; and numerous reagents exist for cell-type specific genetic and phenotypic analysis in vivo, especially in the CNS (Bellen et al. 2004; Brand and Perrimon 1993; Lee and Luo 1999). This includes transgenic fly collections with genetically encoded conditional RNAi constructs that allow for in vivo knock-down of almost all genes in the genome (Dietzl et al. 2007). Other advantages offered by the fly system are the short life span, large number of progeny, well-described anatomy, and ease of dissection and visualization of complex tissues.
Drosophila neuroblasts and glia within the central nervous system
The Drosophila CNS is comprised of a bilaterally symmetrical brain and nerve cord composed of neurons and glia. Numerous studies have shown a remarkable evolutionary conservation of neurodevelopmental mechanisms between flies and humans. Indeed, many genes that regulate mammalian neurodevelopment were first discovered in Drosophila neurodevelopment.
Like the mammalian CNS, the Drosophila CNS arises from ectodermal cells that produce self-renewing neural stem-like cells, called neuroblasts (which are not formally classified as glia). In Drosophila, anatomical characteristics and marker gene expression are used to clearly distinguish between neurons, neural stem/progenitor cells, and glia, which are a diverse group of non-neuronal cells in the CNS. Similarities between mammalian and Drosophila neural stem/progenitor cells and neurons are well established. More recently, striking similarities have been observed between Drosophila glia and their vertebrate counterparts in terms of their function, development, and gene expression, (reviewed in Freeman et al. 2003; Freeman and Doherty 2006; Jones 2001). Specific subtypes of fly glia are homologous to specialized subtypes of vertebrate glia; for example, Drosophila neuropil glia share many characteristics with vertebrate astrocytes (Doherty et al. 2009). Furthermore, Drosophila glia express genes indicative of their roles in the nervous system, including amino acid and ion transporters, and cell-type specific transcription factors that also show conservation in mammalian glia (Egger et al. 2002; Freeman et al. 2003; Soustelle et al. 2002).
Drosophila neurogenesis takes place in two phases, primary embryonic neurogenesis and secondary post-embryonic neurogenesis. Neuroblasts generally divide asymmetrically to renew themselves and make ganglion mother cells (GMCs), which are intermediate neural progenitor cells (reviewed in Doe 2008; Neumuller and Knoblich 2009). During secondary neurogenesis, neuronal and glial lineages bifurcate, with many postembryonic neuroblasts generating only neurons that contribute to the adult brain, while glia arise from both neuroblasts and lineage-specific glial progenitor cells (unpublished data and Colonques et al. 2007; Schmid et al. 1999; Stork et al. 2008). Following secondary neurogenesis, neuroblasts differentiate or die and few if any new neurons are born in the adult (reviewed in Sousa-Nunes et al. 2010), and glia maintain a proliferative capacity into adulthood (unpublished data and Kato et al. 2009).
In studies of Drosophila neurogenesis, several groups have discovered loss-of-function mutations that cause tumor-like growths in the fly brain. These mutations disrupt asymmetric cell division of neuroblasts and/or cell fate specification of their daughters such that daughter cells adopt a neuroblast-like fate (reviewed in Neumuller and Knoblich 2009). These mutant neuroblast-like cells fail to respond to cues that restrain normal proliferation and continue to divide, resulting in accumulation of abnormal proliferative cells at the expense of neurons (Betschinger et al. 2006; Caussinus and Gonzalez 2005; Lee et al. 2006a). Mutant neuroblast-derived cells create invasive, genetically unstable malignant tumors upon transplantation into wild-type hosts, demonstrating their similarities to human cancer cells (Caussinus and Gonzalez 2005; Ryo et al. 1984; Woodhouse et al. 2003). Drosophila genes that yield neuroblast-derived tumors have mammalian orthologs, some of which regulate mammalian neural stem cell self-renewal and/or become deleted in malignant gliomas, such as the regulators of asymmetric cell division lgl (Lgl1) and brat (TRIM32, TRIM3) (Boulay et al. 2009; Klezovitch et al. 2004; Schwamborn et al. 2009). This is consistent with conserved mechanisms of neurogenesis and tumor suppression between Drosophila and mammals. Yet, unlike malignant gliomas, Drosophila neuroblast-derived brain tumors do not accumulate ectopic glia or express glial markers. Instead, neuroblast-derived brain tumors may be more similar to other human brain cancers. For example, medulloblastoma, the most common childhood brain tumor, has a similar etiology: medulloblastomas can arise from transformed neuronal progenitor cells that have acquired mutations that prevent their exit from the cell cycle, block neuronal differentiation of their progeny, and promote accumulation of abnormal immature progenitor cells that inappropriately proliferate to form aggressive tumors (reviewed in Emmenegger and Wechsler-Reya 2008). Continued studies of genes that regulate Drosophila neurogenesis and tumorigenesis in the brain may bring new insights into the genetic, cellular, and molecular basis of human brain cancers, including both medulloblastoma and GBM. To facilitate this process, we and others have embarked on creating overt Drosophila brain cancer models, as described below (Read et al. 2009; Witte et al. 2009).
A DROSOPHILA GLIOBLASTOMA MODEL
The similarities between the fly and human CNS and the advantages of fly genetics make Drosophila an attractive system in which to model gliomas, particularly GBMs. While a Drosophila model cannot address all aspects of GBM or other gliomas, such a model could recapitulate important features, either at the phenotypic or genotypic level (or both), and therefore be extremely useful in understanding glial pathogenesis. In the process of creating Drosophila models for GBM, several criteria must be considered. The Drosophila brain is not the human brain, and although the two share numerous properties, important species-specific differences exist between the two. Thus, analysis of fly GBM model and other brain cancer models must necessarily be focused on specific phenotypic and genetic issues that can be appropriately addressed in the fly, such as proliferation, invasion, inappropriate differentiation, and interactions between genetic pathways and pathway components.
The first, and most crucial, step to creating any Drosophila disease model is to ask whether a fly model can recapitulate key aspects of the human disease and relevant mouse models, both genetically and phenotypically. Given the importance of the EGFR-Ras and PI3K pathways in GBM and the high degree of conservation of these pathways and their biological functions in flies, overt Drosophila GBM models logically focused on these pathways. Since co-activation of EGFR-Ras and PI3K signaling induces GBM tumors in mouse models (Holland et al. 2000; Marumoto et al. 2009), co-activation of these pathways was tested in Drosophila for the ability to induce phenotypes consistent with tumorigenesis. Drosophila have single orthologs each for EGFR(dEGFR), and other pathway components Raf(dRaf), PIK3CA(dp110), PTEN(dPTEN), and Akt(dAkt), and one relevant ortholog for Ras(dRas1) and Rb(Rbf1). Overexpression assays were performed using cell-type specific transcriptional drivers for glia, neuroblasts, and GMCs.
Co-activation of EGFR-Ras and PI3K in Drosophila glia induces neoplastic tumors
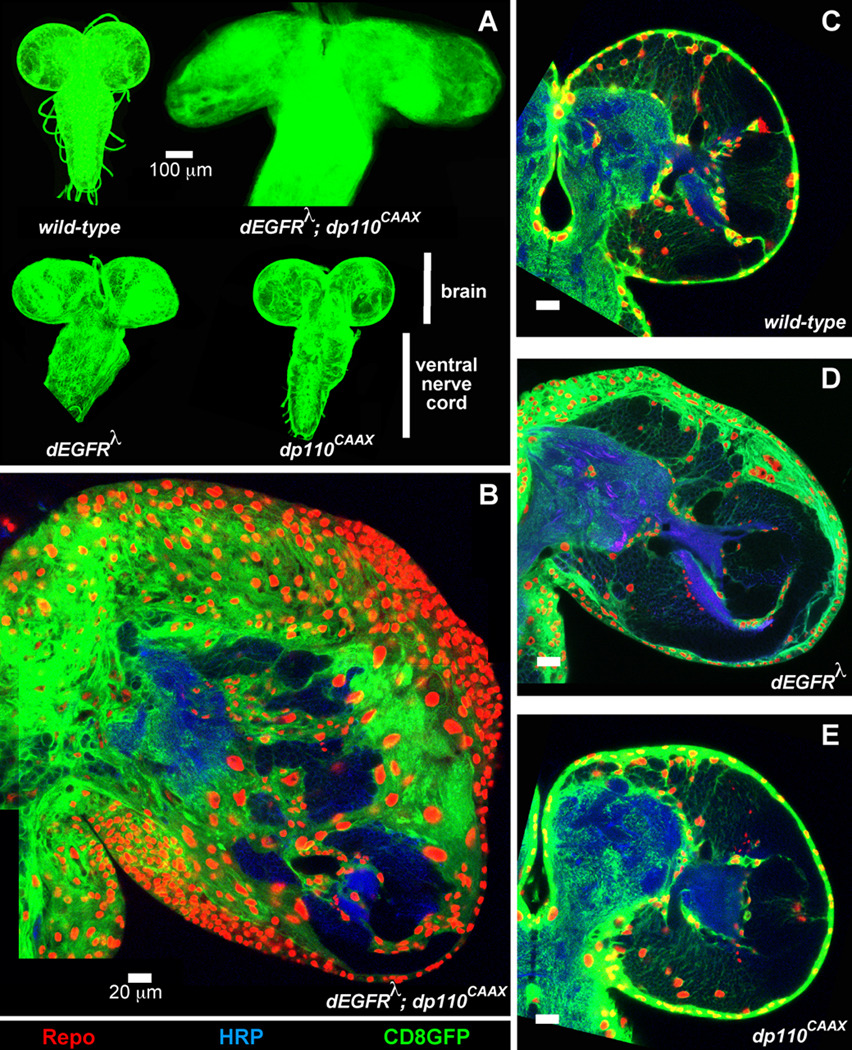
In the Drosophila larval brain, glial-specific co-activation of EGFR-Ras and PI3K synergize to stimulate glial neoplasia (Read et al. 2009; Witte et al. 2009). Co-overexpression of a constitutively activated dEGFR variant (dEGFRλ) and dp110 (dp110CAAX) in glia, using the repo-Gal4 glial-specific transcriptional driver, induced progressive accumulation of 50–100-fold excess glia (Figure 1) (Read et al. 2009). Neoplastic dEGFRλ;dp110CAAX glia arise during post-embryonic stages, disrupt the normal cellular architecture of the larval brain, are highly proliferative, lose normal stellate glial morphologies, and generate multilayered aggregations of abnormal glia throughout the brain; in these ways dEGFRλ;dp110CAAX glia are neoplastic (Read et al. 2009). Neoplastic overgrowth and inappropriate migration also occurred upon co-overexpression of other core pathway components, such as activated dRas1 (dRas1V12), dAkt, or a dPTENRNAi as well as activated forms of other RTKs such as PDGFRalpha and FGFR orthologs (Read et al. 2009; Witte et al. 2009). The effects of EGFR and PI3K co-activation are strongly enhanced by loss of Rbf1 (Read et al. 2009), which is consistent with co-occurrence of Rb pathway mutations with EGFR and PI3K pathway mutations in GBM. In contrast, glial-specific activation of the EGFR-Ras pathway alone induced only 5–10 fold excess glia in the larval brain, and PI3K alone gave viable animals with fairly normal brains (Figure 1). Glial neoplasia is not simply caused by promotion of cell cycle entry and progression; glial-specific expression of the G1 cyclins-cdks (dCyclinD-dCdk4 or dCyclinE-dCdk2), Rbf1 knock-down, or co-overexpression of dCyclinE-dCdk2 with Stg (Cdc25) all fail to induce neoplasia (Read et al. 2009). Yet, blocking cell-cycle entry and progression via overexpression of Dap, the single Drosophila cdk-inhibitor, strongly suppressed glial overproliferation and also partially prevented the morphogenesis defects observed in dEGFRλ;dp110CAAX glia, implying that these defects are secondary to overproliferation (Read et al. 2009).
Figure 1. Co-activation of EGFR-Ras and PI3K in Drosophila glia causes neoplasia.
(A) Optical projections of whole brain-ventral nerve cord complexes from larvae displayed at the same scale. Dorsal view; anterior up. Glia are labeled with CD8GFP (green) driven by the repo-Gal4 transcriptional driver. Each brain is composed of 2 symmetrical hemispheres attached to the ventral nerve cord (VNC). In dEGFRλ;dp110CAAX larvae, both brain hemispheres and the VNC are greatly enlarged relative to other genotypes due to excess glial cells. (B–E) Optical sections of larval brain hemispheres, displayed at the same scale. 20 mm scale bars. Frontal sections, midway through brains. Anterior up; midline to left. Glial cell nuclei are labeled with the Repo nuclear marker (red). Glial cell bodies and membranes are labeled with membrane-bound GFP (green) driven by the repo-Gal4 transcriptional driver. Brains are counter-stained with anti-HRP (blue), which reveals neuronal fiber tracts (neuropil) at high intensity and some cell bodies of neurons and neuronal precursors at low intensity. Dark areas within brains contain unstained neural progenitor cells. In dEGFRλ;dp110CAAX brains (B) there is a dramatic increase in number of glial nuclei relative to wild-type (C), dEGFRλ alone, or (D) dp110CAAX alone (E). Adapted from (Read et al. 2009).
The proliferative and tumorigenic potential of mutant glia has been assessed in the abdominal transplant assay, a classic test of tumorigenicity (Woodhouse et al. 2003). Larval brain fragments with GFP-labeled dEGFRλ;dp110CAAX and wild-type glia were transplanted into young adult hosts. Wild-type transplants grew and survived over 1–6 weeks, but produced few glia, whereas dEGFRλ;dp110CAAX mutant glia continued to proliferate into massive and invasive tumors that filled hosts’ abdomens, causing premature death (Read et al. 2009). Tumors were embedded with trachea, which are Drosophila oxygen delivery tubules, suggesting that tumors stimulated growth of new trachea or enveloped existing trachea perhaps in a process akin to tumor angiogenesis (Read et al. 2009).
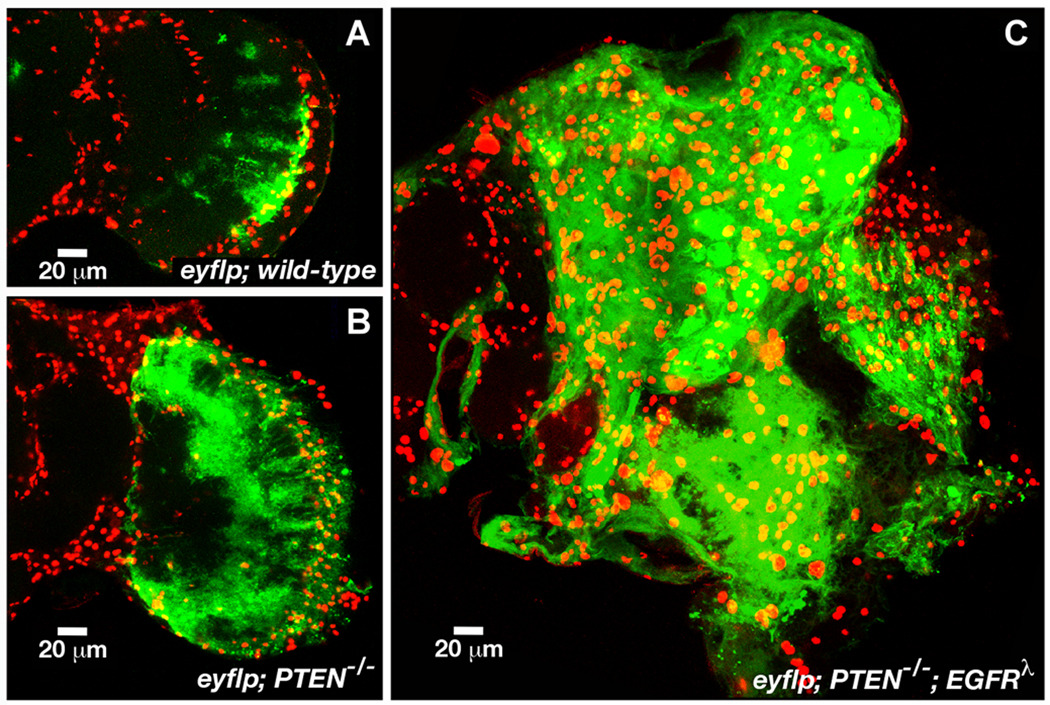
To explore the invasive potential of mutant glia within the CNS, FLP-FRT clonal analysis has been used, a technique in which discrete clones of mutant cells are induced in an otherwise normal CNS, a situation analogous to somatic tumorigenesis. With this technique, FLP-recombinase was used to create GFP-labeled mutant glial cells from mitotic progenitors such that only mutant cells (and all of their clonal progeny) initiated expression of transgenes (Lee and Luo 1999). Random clones induced from mitotic founder cells late in development or in the young adult brain were examined in the adult brain: wild-type control clones consisted of 1–3 cells, single mutant dPTEN−/− or dEGFRλ mutant glial clones typically contained 2-fold more cells than wild-type controls, and dEGFRλ;dPTEN−/− or dRas1V1;dPTEN−/− clones created tumor-like and invasive overgrowths in the brain that were sometimes associated with trachea (Figure 2) (Read et al. 2009). Since neoplastic larval glia derived from random clones were concentrated in brain regions that normally contain proliferating glia, neoplastic glia may be derived from these cells (Chotard et al. 2005; Colonques et al. 2007; Stork et al. 2008). The ey-FLP recombinase, which is active in ey-expressing glial progenitors (Chotard et al. 2005), was used to create clones from a subpopulation of glial progenitors. Single mutant ey-FLP clones contained a modest number of excess glia relative to wild-type, whereas double mutant dRas1V12-dEGFRλ;dPTEN−/− clones became large invasive tumors in adult brains (Figure 2) (Read et al. 2009).
Figure 2. Co-activation of EGFR and PI3K in glial progenitors creates invasive tumors.
(A–B) FLP/FRT clones in adult brains derived from a population of mitotic glial cells that express the ey-FLP recombinase and the repo-Gal4 transcriptional driver. Membrane bound GFP (green) marks cell bodies and membranes of glial clones derived by FLP/FRT mitotic recombination (see text). The Repo nuclear protein (red) marks all glial cell nuclei, in both clones and surrounding normal tissue. Optical sections through brains of similarly aged adults matched to scale. 20 mm scale bars. Each panel shows half brains. dPTEN−/− (B) clones are composed of 2–5-fold more cells than wild-type controls (A). In contrast, dEGFRλ;dPTEN−/− (C) double mutant clones form large tumors visible in adult brains. Adapted from (Read et al. 2009).
EGFR-Ras and PI3K and their effectors in neuroblasts
The phenotype triggered by co-activation of EGFR and PI3K in glia is distinct from other Drosophila brain-overgrowth and brain tumor mutants, which are characterized by accretion of abnormal neuroblast-like cells as previously discussed (Betschinger et al. 2006; Caussinus and Gonzalez 2005). dEGFRλ;dp110CAAX neoplastic glial cells do not express transcription factors or cell-fate determinants characteristic of neuroblasts (unpublished data and Read et al. 2009) and, therefore, are not converted into neuroblasts. Moreover, constitutive EGFR-Ras and PI3K signaling does not elicit neoplastic overgrowth from neuroblasts or intermediate neuronal progenitor cells/GMCs, as assessed with overexpression assays using several defined neuroblast-specific promoters (unpublished data and Lavery et al. 2007; Read et al. 2009). Thus, co-activation EGFR-Ras and PI3K is not sufficient to transform all proliferative neural cell types, despite the fact that neuroblasts are capable of undergoing neoplastic transformation in response to several other genetic mutations. Yet, the Tor, Myc, and Pnt/Stg pathways required for the development of EGFR and PI3K dependent glial neoplasia (see below, Read et al. 2009) are involved in the development of normal neuroblasts and GMCs (unpublished data and Orian et al. 2007; Siegrist et al. 2010). This implies that other genes modulate these pathways in neuroblasts and control competence to undergo neoplastic transformation, and these genes must act differentially in glial and neuronal lineages. One such gene may be Dap, which encodes the single p21/p27 cdk-inhibitor in the fly genome. As they normally exit the cell cycle, neuroblasts and/or GMCs normally express Dap, whereas glial cells rarely express Dap (Read et al. 2009; Wallace et al. 2000). Dap is highly regulated by numerous signaling pathways including Ras (Liu et al. 2002; Wallace et al. 2000). Studies of Dap regulation in glia and neuroblasts may illuminate the genetic origins of glioblastoma, especially given that loss p21 expression may underlie the tumorigenic response of mammalian neuro-glial progenitor cells to EGFR (Ligon et al. 2007).
EGFR-PI3K drives glial neoplasia through a combinatorial genetic network
Co-activation of EGFR-Ras and PI3K signaling in Drosophila glia gives rise to neoplastic, proliferative, invasive cells that create transplantable tumors, mirroring the overproliferation and inappropriate invasion observed in human GBM. This fly GBM model was created to facilitate forward genetic screens to identify other genes that suppress (improve) or enhance (worsen) disease phenotypes upon mutation or inhibition. Within the fly community, extensive and successful use of genetic modifier screens has verified their power for identifying novel genes and gene functions in signaling pathways and cellular processes (Karim et al. 1996; Read et al. 2005; Yoshikawa et al. 2003). Such enhancer-suppressor screens have identified both core components and cell-type specific regulators of numerous signaling pathways (Karim et al. 1996; Simon 2000; Voas and Rebay 2004), especially RTK signaling pathways. A key advantage of this genetic approach is that it is in vivo, screening within a living organism for gene function, which sets it apart from many correlative molecular and biochemical screening methods, such as microarray analysis or genomic sequencing, which do not directly assess gene function. Virtually all signal transduction pathways present in flies are highly conserved in mammals, even to the extent that mouse and human components can functionally replace their Drosophila orthologs (Halder et al. 1995). Thus, gene products identified as modifiers of Drosophila GBM-like phenotypes represent excellent candidates for orthologous genes involved in pathogenesis of human GBM. In particular, genes that suppress glial neoplasia when their function is reduced may represent promising targets for pharmacological intervention in human GBM.
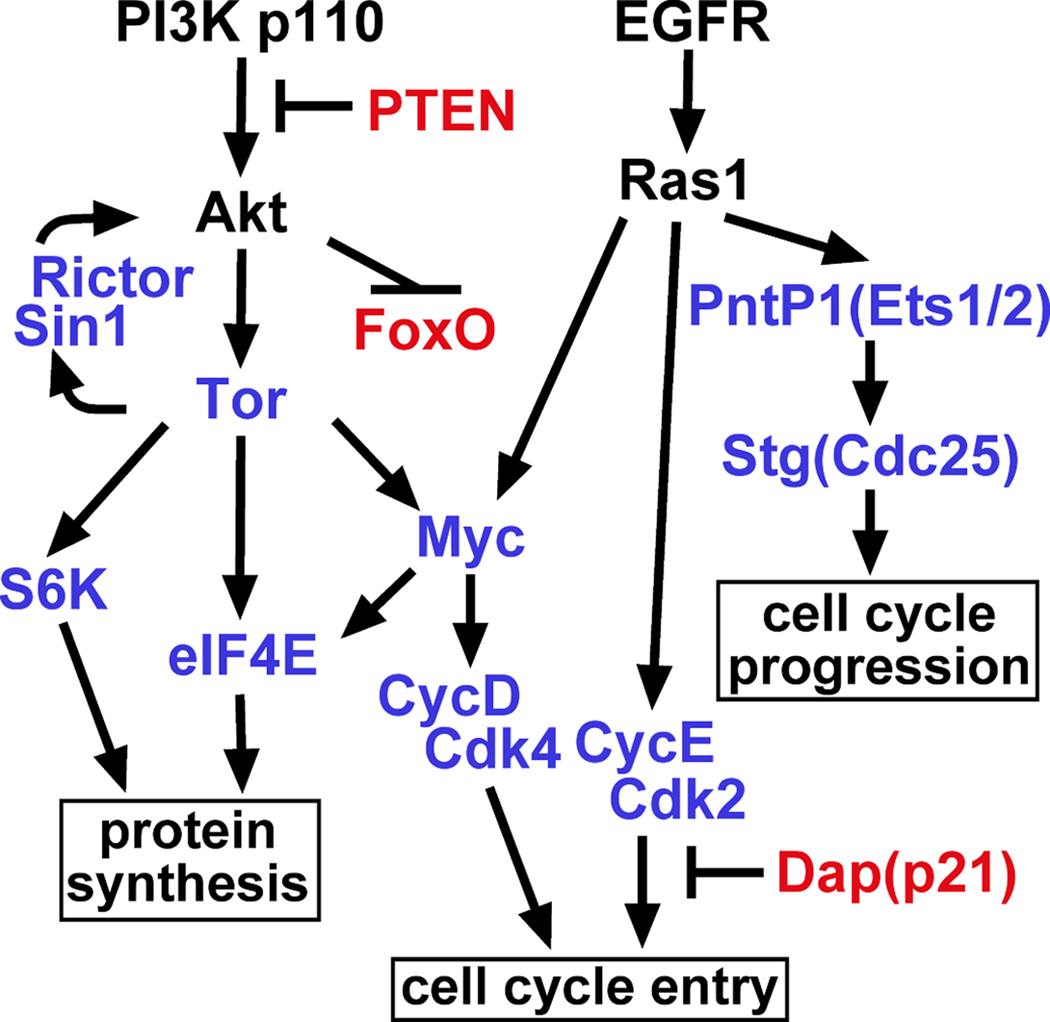
To date, many loci have been tested for their ability to modify EGFR-PI3K-driven glial neoplasia when overexpressed, mutated, and/or knocked-down. Initial genetic tests to validate this Drosophila GBM model show that at least four pathway circuits are required for EGFR and PI3K- driven glial neoplasia in Drosophila, including the Myc pathway to drive cell cycle entry and protein translation, an Erk-ETS-Cdc25 circuit to promote cell cycle progression, and the Tor pathway which provides protein translational capacity necessary for proliferation and growth (Figure 3) (Read et al. 2009). In glia, these pathway components act synergistically, rather than additively, in a combinatorial network downstream of EGFR and PI3K signaling. Therefore, Drosophila shows evolutionary conservation of oncogene cooperation. Orthologs for many of the pathway components presented below are implicated in human GBM, although specific roles for some, such as theTORC2 complex, have not been defined despite their expression in GBM tumors (Masri et al. 2007).
Figure 3. EGFR-PI3K drives glial neoplasia through a combinatorial genetic network.
Pathway diagram of key effectors involved in glial neoplasia initiated by EGFR and PI3K in Drosophila, showing the pathway circuits driving cell cycle entry and progression, and protein translation. Positive regulators are indicated in blue and negative regulators are indicated in red. Arrows indicate pathway connections, although these connections are not necessarily direct. Adapted from (Read et al. 2009).
Akt and Tor
Tor kinase is a key target of PI3K and Akt (Hu et al. 2005; Pelloski et al. 2006). In Drosophila, genetic reduction of Tor activity reduced dEGFλ;dp110CAAX-dependent glial overgrowth. Tor resides in two complexes, TORC1 and TORC2. TORC1, which includes the Raptor protein, drives growth and protein synthesis (Guertin and Sabatini 2007). Glial-specific RNAi of the Drosophila orthologs of Raptor and the TORC1 effectors S6-kinase (dS6K) and eIF4E (deIF4E) all significantly reduced accumulation of dEGFRλ;dp110CAAX glia; glial-specific RNAi of dTSC1, a TORC1 repressor, enhanced dEGFRλ;dp110CAAX phenotypes (Read et al. 2009). However, dTSC1 RNAi in wild-type glia, which can mimic TORC1 activation (Guertin and Sabatini 2007; Teleman et al. 2008), did not produce neoplasia (Read et al. 2009). Thus, TORC1 activity, which is necessary for normal glial development (Read et al. 2009), is also necessary for glial neoplasia, but is not sufficient.
TORC2, which includes the Sin1 and Rictor proteins, directly phosphorylates and fully activates Akt (Guertin and Sabatini 2007). dSin1−/−; dEGFRλ;dp110CAAX and dRictor−/−; dEGFRλ;dp110CAAX triple mutants showed no glial neoplasia (Read et al. 2009). Alone, dSin1−/− and dRictor−/− mutant flies had no detectable glial defects (Read et al. 2009). Thus, TORC2 is dispensable for normal glial development, but is necessary for glial neoplasia. GBMs show elevated rictor expression and TORC2 activity, although little is known about their function or regulation in GBM (Masri et al. 2007). Our results suggest that TORC2 may be a promising therapeutic target specific to EGFR-PI3K transformed glia.
Myc and CyclinD-Cdk4
Co-activation of EGFR and PI3K signaling upregulates expression of the dMyc bHLH transcription factor in glia. This is consistent with findings that, in flies and mammals, EGFR-Ras, PI3K, and Tor signaling upregulate Myc protein levels (Gera et al. 2004; Moberg et al. 2004; Pelengaris et al. 2002; Prober and Edgar 2002; Teleman et al. 2008). dMyc is also highly expressed in neuroblasts, suggesting that it promotes a stem/progenitor cell fate in both neoplastic and normal proliferative cells in the CNS, much like mammalian Myc (Betschinger et al. 2006). Reduction of Myc activity rescued glial neoplasia in Drosophila (Read et al. 2009), a result also recently documented in a mouse model of PTEN-dependent GBM (Zheng et al. 2008). Thus, Myc activity is rate-limiting: in human GBMs, c-myc is overexpressed and sometimes amplified (Wiedemeyer et al. 2008), and perhaps amplification and/or overexpression is selected for this reason.
In mammals, Myc oncogenes are well-known to cooperate with RTK-Ras signaling to drive neoplastic transformation (reviewed in Pelengaris et al. 2002), and this property of Myc is conserved in flies: co-overexpression of dMyc with dEGFRλ produced a neoplastic phenotype (Read et al. 2009), indicating that dMyc can substitute for PI3K when combined with EGFR, consistent with recent data demonstrating the Myc upregulation is a key output of downstream of PI3K (Teleman et al. 2008). dMyc target genes include dCyclinD (Orian et al. 2005). We observed dMyc-dependent dCyclinD overexpression in dEGFRλ;dp110CAAX mutant glia, and a requirement for dCyclinD-dCdk4 in neoplasia, although dCdk4 itself is not required for normal glial proliferation (Read et al. 2009). D-cyclins and Cdk4 are commonly amplified and/or overexpressed in GBMs, but, as in Drosophila, Cdk4 is not required for normal proliferation in most mouse tissues (Kozar and Sicinski 2005; McLendon et al. 2008; Parsons et al. 2008). Thus, Cdk4, like TORC2, may be a key output of pathogenic EGFR and PI3K signaling in human GBM, indicating that it may be an important therapeutic target. Consistent with this, a Cdk4 inhibitor can effectively suppress the growth of GBM xenografts in mouse (Michaud et al.).
Pnt (Ets 1/2) and Stg (Cdc25)
Relative to wild-type glia, dEGFRλ;dp110CAAX glia show high levels of nuclear, activated Erk and expression of PntP1, an ETS-family transcription factor induced by Erk activity (Baonza et al. 2002; Klaes et al. 1994; Read et al. 2009). PntP1 was also detected wild-type neuroblasts, suggesting that it promotes a proliferative progenitor state in neural cells. In developing eye tissue, Pnt proteins directly upregulate Stg (Cdc25) expression to stimulate G2-M cell cycle progression (Baonza et al. 2002). In neoplastic glia, RNAi of PntP1 reduced Stg expression and completely suppressed proliferation. Stg itself is rate limiting for glial EGFR-PI3K dependent neoplasia: genetic reduction of Stg partially suppressed neoplasia, whereas Stg overexpression synergistically enhanced neoplasia. Thus, dEGFRλ;dp110CAAX induces neoplasia via coordinated stimulation of G1-S entry, via CyclinD-cdk4, and G2-M progression through Stg. Yet, co-overexpression of Stg with the G1 cyclins-cdks fails to induce glial neoplasia, despite the fact that this can induce chronic proliferation in other Drosophila cell types (Buttitta et al. 2007).
Ets1, Ets2, and Cdc25B (human orthologs of PntP1 and Stg, respectively) are highly overexpressed in GBMs according to gene expression microarrays performed by the TCGA, although these genes do not have established roles in GBM (unpublished data and Furnari et al. 2007; Kitange et al. 1999; Masri et al. 2007). Notably, Cdc25B is the only Cdc25 ortholog highly expressed in GBM, implying that it may be tumor-specific (unpublished data). Cdc25B, which also regulates cell cycle progression, may be a promising therapeutic target since it is not apparently expressed in normal adult neural progenitors and is not required for normal mammalian cell proliferation (Ferguson et al. 2005; Lein et al. 2007).
CONCLUSION, IMPLICATIONS, AND FUTURE DIRECTIONS
Drosophila provides an experimental organismal model system geared towards probing the frontiers of biology in a systematic manner in which phenotypic and genetic readouts are paramount. The results discussed here have established Drosophila as a robust model system for the study of the genetic and cellular origins of human glioblastoma (GBM), which are lethal primary brain tumors caused by genetic mutations that perturb essential developmental and homeostatic processes in glial cells. This model represents an important organotypic and cell-type specific Drosophila neurological disease model in which malignant neural cells are created by mutations in the signature genes and pathways thought to be driving forces in a homologous human disease. Further studies of the Drosophila glial cells prone to neoplastic transformation may illuminate the cellular origins of human gliomas, which are thought to arise from glial stem and progenitor cells, and facilitate informative comparisons to human GBM stem/propagating cells.
In the future, Drosophila will also be useful for creating models for different genetic subtypes of GBM. This may be especially useful for distinguishing those combinations of genetic mutations and pathways that drive glial tumorigenesis from the large number of genes that show mutations and altered expression in GBMs as uncovered by recent genomic analyses of patient samples (McLendon et al. 2008; Parsons et al. 2008). Furthermore, Drosophila have emerged as a pliable model for readily evaluating therapeutic drugs for neurological diseases (Choi et al. 2010; McBride et al. 2005), and this EGFR-PI3K dependent GBM model may also aid in the identification and design of improved therapeutic agents that target gliomagenic pathways. Indeed, we and others have already used the Drosophila GBM model to successfully test known and potential therapeutics for GBM (unpublished data and Witte et al. 2009). This larval model has several advantages for drug testing in that compounds can be directly fed to animals, tumorous brains rapidly develop over 5 days, inhibition of neoplastic proliferation can be readily observed in live animals with fluorescence microscopy; drug targets can be independently verified genetically; and drug and drug targets can then be verified in corresponding mammalian model systems.
Through genetic and phenotypic analysis of the EGFR-PI3K model, crucial downstream effectors of EGFR and PI3K that drive neoplasia have been identified; many of these genes are mutated and/or activated in human GBM. This firmly validates the relevance of this model to human GBM. Notably, when activated individually or in combination, these downstream effector pathways are not sufficient to induce glial neoplasia, implying a requirement for coordinated stimulation of multiple effector pathways, some of which must still remain unidentified. Thus, large-scale forward genetic screens can now be performed in vivo in the Drosophila CNS to identify new genes involved in glial neoplasia that may reveal new mechanisms and molecules underlying glial transformation that may represent new therapeutic opportunities for human GBM.
ACKNOWLEDGEMENTS
I thank Drs. Webster Cavenee and Frank Furnari for critical reading of this manuscript, and Dr. John Thomas for his mentorship. R.D. Read is supported by awards from the National Institute of Neurological Disorders and Stroke and the American Brain Tumor Association.
REFERENCES
- Aguirre A, Dupree JL, Mangin JM, Gallo V. A functional role for EGFR signaling in myelination and remyelination. Nat Neurosci. 2007;10(8):990–1002. doi: 10.1038/nn1938. [DOI] [PubMed] [Google Scholar]
- Akai H, Gateff E, Davis LE, Schneiderman HA. Virus-like particles in normal and tumorous tissues of Drosophila. Science. 1967;157(3790):810–813. doi: 10.1126/science.157.3790.810. [DOI] [PubMed] [Google Scholar]
- Bachoo RM, Maher EA, Ligon KL, Sharpless NE, Chan SS, You MJ, Tang Y, DeFrances J, Stover E, Weissleder R, et al. Epidermal growth factor receptor and Ink4a/Arf: convergent mechanisms governing terminal differentiation and transformation along the neural stem cell to astrocyte axis. Cancer Cell. 2002;1(3):269–277. doi: 10.1016/s1535-6108(02)00046-6. [DOI] [PubMed] [Google Scholar]
- Baeza N, Weller M, Yonekawa Y, Kleihues P, Ohgaki H. PTEN methylation and expression in glioblastomas. Acta Neuropathol (Berl) 2003;106(5):479–485. doi: 10.1007/s00401-003-0748-4. [DOI] [PubMed] [Google Scholar]
- Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- Baonza A, Murawsky CM, Travers AA, Freeman M. Pointed and Tramtrack69 establish an EGFR-dependent transcriptional switch to regulate mitosis. Nat Cell Biol. 2002;4(12):976–980. doi: 10.1038/ncb887. [DOI] [PubMed] [Google Scholar]
- Bellen HJ, Levis RW, Liao G, He Y, Carlson JW, Tsang G, Evans-Holm M, Hiesinger PR, Schulze KL, Rubin GM, et al. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics. 2004;167(2):761–781. doi: 10.1534/genetics.104.026427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betschinger J, Mechtler K, Knoblich JA. Asymmetric segregation of the tumor suppressor brat regulates self-renewal in Drosophila neural stem cells. Cell. 2006;124(6):1241–1253. doi: 10.1016/j.cell.2006.01.038. [DOI] [PubMed] [Google Scholar]
- Bilder D. Epithelial polarity and proliferation control: links from the Drosophila neoplastic tumor suppressors. Genes Dev. 2004;18(16):1909–1925. doi: 10.1101/gad.1211604. [DOI] [PubMed] [Google Scholar]
- Boockvar JA, Kapitonov D, Kapoor G, Schouten J, Counelis GJ, Bogler O, Snyder EY, McIntosh TK, O'Rourke DM. Constitutive EGFR signaling confers a motile phenotype to neural stem cells. Mol Cell Neurosci. 2003;24(4):1116–1130. doi: 10.1016/j.mcn.2003.09.011. [DOI] [PubMed] [Google Scholar]
- Boulay JL, Stiefel U, Taylor E, Dolder B, Merlo A, Hirth F. Loss of heterozygosity of TRIM3 in malignant gliomas. BMC Cancer. 2009;9:71. doi: 10.1186/1471-2407-9-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118(2):401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Brennan C, Momota H, Hambardzumyan D, Ozawa T, Tandon A, Pedraza A, Holland E. Glioblastoma subclasses can be defined by activity among signal transduction pathways and associated genomic alterations. PLoS One. 2009;4(11):e7752. doi: 10.1371/journal.pone.0007752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broderick DK, Di C, Parrett TJ, Samuels YR, Cummins JM, McLendon RE, Fults DW, Velculescu VE, Bigner DD, Yan H. Mutations of PIK3CA in anaplastic oligodendrogliomas, high-grade astrocytomas, and medulloblastomas. Cancer Res. 2004;64(15):5048–5050. doi: 10.1158/0008-5472.CAN-04-1170. [DOI] [PubMed] [Google Scholar]
- Brumby AM, Richardson HE. Using Drosophila melanogaster to map human cancer pathways. Nat Rev Cancer. 2005;5(8):626–639. doi: 10.1038/nrc1671. [DOI] [PubMed] [Google Scholar]
- Buffo A, Rite I, Tripathi P, Lepier A, Colak D, Horn AP, Mori T, Gotz M. Origin and progeny of reactive gliosis: A source of multipotent cells in the injured brain. Proc Natl Acad Sci U S A. 2008;105(9):3581–3586. doi: 10.1073/pnas.0709002105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows RC, Wancio D, Levitt P, Lillien L. Response diversity and the timing of progenitor cell maturation are regulated by developmental changes in EGFR expression in the cortex. Neuron. 1997;19(2):251–267. doi: 10.1016/s0896-6273(00)80937-x. [DOI] [PubMed] [Google Scholar]
- Buttitta LA, Katzaroff AJ, Perez CL, de la Cruz A, Edgar BA. A double-assurance mechanism controls cell cycle exit upon terminal differentiation in Drosophila. Dev Cell. 2007;12(4):631–643. doi: 10.1016/j.devcel.2007.02.020. [DOI] [PubMed] [Google Scholar]
- Caric D, Raphael H, Viti J, Feathers A, Wancio D, Lillien L. EGFRs mediate chemotactic migration in the developing telencephalon. Development. 2001;128(21):4203–4216. doi: 10.1242/dev.128.21.4203. [DOI] [PubMed] [Google Scholar]
- Caussinus E, Gonzalez C. Induction of tumor growth by altered stem-cell asymmetric division in Drosophila melanogaster. Nat Genet. 2005 doi: 10.1038/ng1632. [DOI] [PubMed] [Google Scholar]
- Chen R, Nishimura MC, Bumbaca SM, Kharbanda S, Forrest WF, Kasman IM, Greve JM, Soriano RH, Gilmour LL, Rivers CS, et al. A hierarchy of self-renewing tumor-initiating cell types in glioblastoma. Cancer Cell. 2010;17(4):362–375. doi: 10.1016/j.ccr.2009.12.049. [DOI] [PubMed] [Google Scholar]
- Cheng L, Bao S, Rich JN. Potential therapeutic implications of cancer stem cells in glioblastoma. Biochem Pharmacol. 2010;80(5):654–665. doi: 10.1016/j.bcp.2010.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi CH, Schoenfeld BP, Bell AJ, Hinchey P, Kollaros M, Gertner MJ, Woo NH, Tranfaglia MR, Bear MF, Zukin RS, et al. Pharmacological reversal of synaptic plasticity deficits in the mouse model of Fragile X syndrome by group II mGluR antagonist or lithium treatment. Brain Res. 2010 doi: 10.1016/j.brainres.2010.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotard C, Leung W, Salecker I. glial cells missing and gcm2 cell autonomously regulate both glial and neuronal development in the visual system of Drosophila. Neuron. 2005;48(2):237–251. doi: 10.1016/j.neuron.2005.09.019. [DOI] [PubMed] [Google Scholar]
- Cloughesy TF, Yoshimoto K, Nghiemphu P, Brown K, Dang J, Zhu S, Hsueh T, Chen Y, Wang W, Youngkin D, et al. Antitumor activity of rapamycin in a Phase I trial for patients with recurrent PTEN-deficient glioblastoma. PLoS Med. 2008;5(1):e8. doi: 10.1371/journal.pmed.0050008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman H, Zhang L, Sulman EP, McDonald JM, Shooshtari NL, Rivera A, Popoff S, Nutt CL, Louis DN, Cairncross JG, et al. A multigene predictor of outcome in glioblastoma. Neuro Oncol. 2010;12(1):49–57. doi: 10.1093/neuonc/nop007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colognato H, ffrench-Constant C. Mechanisms of glial development. Curr Opin Neurobiol. 2004;14(1):37–44. doi: 10.1016/j.conb.2004.01.009. [DOI] [PubMed] [Google Scholar]
- Colonques J, Ceron J, Tejedor FJ. Segregation of postembryonic neuronal and glial lineages inferred from a mosaic analysis of the Drosophila larval brain. Mech Dev. 2007;124(5):327–340. doi: 10.1016/j.mod.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Comings DE. A general theory of carcinogenesis. Proc Natl Acad Sci U S A. 1973;70(12):3324–3328. doi: 10.1073/pnas.70.12.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448(7150):151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Doe CQ. Neural stem cells: balancing self-renewal with differentiation. Development. 2008;135(9):1575–1587. doi: 10.1242/dev.014977. [DOI] [PubMed] [Google Scholar]
- Doetsch F, Caille I, Lim DA, Garcia-Verdugo JM, Alvarez-Buylla A. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell. 1999;97(6):703–716. doi: 10.1016/s0092-8674(00)80783-7. [DOI] [PubMed] [Google Scholar]
- Doherty J, Logan MA, Tasdemir OE, Freeman MR. Ensheathing glia function as phagocytes in the adult Drosophila brain. J Neurosci. 2009;29(15):4768–4781. doi: 10.1523/JNEUROSCI.5951-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger B, Leemans R, Loop T, Kammermeier L, Fan Y, Radimerski T, Strahm MC, Certa U, Reichert H. Gliogenesis in Drosophila: genome-wide analysis of downstream genes of glial cells missing in the embryonic nervous system. Development. 2002;129(14):3295–3309. doi: 10.1242/dev.129.14.3295. [DOI] [PubMed] [Google Scholar]
- Ekstrand AJ, Sugawa N, James CD, Collins VP. Amplified and rearranged epidermal growth factor receptor genes in human glioblastomas reveal deletions of sequences encoding portions of the N- and/or C-terminal tails. Proc Natl Acad Sci U S A. 1992;89(10):4309–4313. doi: 10.1073/pnas.89.10.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmenegger BA, Wechsler-Reya RJ. Stem cells and the origin and propagation of brain tumors. J Child Neurol. 2008;23(10):1172–1178. doi: 10.1177/0883073808321062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endersby R, Baker SJ. PTEN signaling in brain: neuropathology and tumorigenesis. Oncogene. 2008;27(41):5416–5430. doi: 10.1038/onc.2008.239. [DOI] [PubMed] [Google Scholar]
- Ferguson AM, White LS, Donovan PJ, Piwnica-Worms H. Normal cell cycle and checkpoint responses in mice and cells lacking Cdc25B and Cdc25C protein phosphatases. Mol Cell Biol. 2005;25(7):2853–2860. doi: 10.1128/MCB.25.7.2853-2860.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fomchenko EI, Holland EC. Mouse models of brain tumors and their applications in preclinical trials. Clin Cancer Res. 2006;12(18):5288–5297. doi: 10.1158/1078-0432.CCR-06-0438. [DOI] [PubMed] [Google Scholar]
- Fraser MM, Zhu X, Kwon CH, Uhlmann EJ, Gutmann DH, Baker SJ. Pten loss causes hypertrophy and increased proliferation of astrocytes in vivo. Cancer Res. 2004;64(21):7773–7779. doi: 10.1158/0008-5472.CAN-04-2487. [DOI] [PubMed] [Google Scholar]
- Freeman MR, Delrow J, Kim J, Johnson E, Doe CQ. Unwrapping glial biology: Gcm target genes regulating glial development, diversification, and function. Neuron. 2003;38(4):567–580. doi: 10.1016/s0896-6273(03)00289-7. [DOI] [PubMed] [Google Scholar]
- Freeman MR, Doherty J. Glial cell biology in Drosophila and vertebrates. Trends Neurosci. 2006;29(2):82–90. doi: 10.1016/j.tins.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21(21):2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- Furnari FB, Lin H, Huang HS, Cavenee WK. Growth suppression of glioma cells by PTEN requires a functional phosphatase catalytic domain. Proc Natl Acad Sci U S A. 1997;94(23):12479–12484. doi: 10.1073/pnas.94.23.12479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Neufeld TP, Pan D. Drosophila PTEN regulates cell growth and proliferation through PI3K-dependent and -independent pathways. Dev Biol. 2000;221(2):404–418. doi: 10.1006/dbio.2000.9680. [DOI] [PubMed] [Google Scholar]
- Gateff E. Malignant neoplasms of genetic origin in Drosophila melanogaster. Science. 1978;200(4349):1448–1459. doi: 10.1126/science.96525. [DOI] [PubMed] [Google Scholar]
- Gateff E, Schneiderman HA. Neoplasms in mutant and cultured wild-tupe tissues of Drosophila. Natl Cancer Inst Monogr. 1969;31:365–397. [PubMed] [Google Scholar]
- Gera JF, Mellinghoff IK, Shi Y, Rettig MB, Tran C, Hsu JH, Sawyers CL, Lichtenstein AK. AKT activity determines sensitivity to mammalian target of rapamycin (mTOR) inhibitors by regulating cyclin D1 and c-myc expression. J Biol Chem. 2004;279(4):2737–2746. doi: 10.1074/jbc.M309999200. [DOI] [PubMed] [Google Scholar]
- Giese A, Bjerkvig R, Berens ME, Westphal M. Cost of migration: invasion of malignant gliomas and implications for treatment. J Clin Oncol. 2003;21(8):1624–1636. doi: 10.1200/JCO.2003.05.063. [DOI] [PubMed] [Google Scholar]
- Groszer M, Erickson R, Scripture-Adams DD, Lesche R, Trumpp A, Zack JA, Kornblum HI, Liu X, Wu H. Negative regulation of neural stem/progenitor cell proliferation by the Pten tumor suppressor gene in vivo. Science. 2001;294(5549):2186–2189. doi: 10.1126/science.1065518. [DOI] [PubMed] [Google Scholar]
- Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12(1):9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Haas-Kogan D, Shalev N, Wong M, Mills G, Yount G, Stokoe D. Protein kinase B (PKB/Akt) activity is elevated in glioblastoma cells due to mutation of the tumor suppressor PTEN/MMAC. Curr Biol. 1998;8(21):1195–1198. doi: 10.1016/s0960-9822(07)00493-9. [DOI] [PubMed] [Google Scholar]
- Halder G, Callaerts P, Gehring WJ. Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science. 1995;267(5205):1788–1792. doi: 10.1126/science.7892602. [DOI] [PubMed] [Google Scholar]
- Hirth F. Drosophila melanogaster in the study of human neurodegeneration. CNS Neurol Disord Drug Targets. 2010;9(4):504–523. doi: 10.2174/187152710791556104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoelzinger DB, Demuth T, Berens ME. Autocrine factors that sustain glioma invasion and paracrine biology in the brain microenvironment. J Natl Cancer Inst. 2007;99(21):1583–1593. doi: 10.1093/jnci/djm187. [DOI] [PubMed] [Google Scholar]
- Holland EC, Celestino J, Dai C, Schaefer L, Sawaya RE, Fuller GN. Combined activation of Ras and Akt in neural progenitors induces glioblastoma formation in mice. Nat Genet. 2000;25(1):55–57. doi: 10.1038/75596. [DOI] [PubMed] [Google Scholar]
- Holland EC, Hively WP, DePinho RA, Varmus HE. A constitutively active epidermal growth factor receptor cooperates with disruption of G1 cell-cycle arrest pathways to induce glioma-like lesions in mice. Genes Dev. 1998;12(23):3675–3685. doi: 10.1101/gad.12.23.3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Holland EC. Applications of mouse glioma models in preclinical trials. Mutat Res. 2005;576(1–2):54–65. doi: 10.1016/j.mrfmmm.2004.08.023. [DOI] [PubMed] [Google Scholar]
- Hu X, Pandolfi PP, Li Y, Koutcher JA, Rosenblum M, Holland EC. mTOR promotes survival and astrocytic characteristics induced by Pten/AKT signaling in glioblastoma. Neoplasia. 2005;7(4):356–368. doi: 10.1593/neo.04595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HS, Nagane M, Klingbeil CK, Lin H, Nishikawa R, Ji XD, Huang CM, Gill GN, Wiley HS, Cavenee WK. The enhanced tumorigenic activity of a mutant epidermal growth factor receptor common in human cancers is mediated by threshold levels of constitutive tyrosine phosphorylation and unattenuated signaling. J Biol Chem. 1997;272(5):2927–2935. doi: 10.1074/jbc.272.5.2927. [DOI] [PubMed] [Google Scholar]
- Inda MD, Bonavia R, Mukasa A, Narita Y, Sah DW, Vandenberg S, Brennan C, Johns TG, Bachoo R, Hadwiger P, et al. Tumor heterogeneity is an active process maintained by a mutant EGFR-induced cytokine circuit in glioblastoma. Genes Dev. 2010;24(16):1731–1745. doi: 10.1101/gad.1890510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones BW. Glial cell development in the Drosophila embryo. Bioessays. 2001;23(10):877–887. doi: 10.1002/bies.1129. [DOI] [PubMed] [Google Scholar]
- Karim FD, Chang HC, Therrien M, Wassarman DA, Laverty T, Rubin GM. A screen for genes that function downstream of Ras1 during Drosophila eye development. Genetics. 1996;143(1):315–329. doi: 10.1093/genetics/143.1.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K, Awasaki T, Ito K. Neuronal programmed cell death induces glial cell division in the adult Drosophila brain. Development. 2009;136(1):51–59. doi: 10.1242/dev.023366. [DOI] [PubMed] [Google Scholar]
- Kitange G, Kishikawa M, Nakayama T, Naito S, Iseki M, Shibata S. Expression of the Ets-1 proto-oncogene correlates with malignant potential in human astrocytic tumors. Mod Pathol. 1999;12(6):618–626. [PubMed] [Google Scholar]
- Klaes A, Menne T, Stollewerk A, Scholz H, Klambt C. The Ets transcription factors encoded by the Drosophila gene pointed direct glial cell differentiation in the embryonic CNS. Cell. 1994;78(1):149–160. doi: 10.1016/0092-8674(94)90581-9. [DOI] [PubMed] [Google Scholar]
- Klezovitch O, Fernandez TE, Tapscott SJ, Vasioukhin V. Loss of cell polarity causes severe brain dysplasia in Lgl1 knockout mice. Genes Dev. 2004;18(5):559–571. doi: 10.1101/gad.1178004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblum HI, Hussain R, Wiesen J, Miettinen P, Zurcher SD, Chow K, Derynck R, Werb Z. Abnormal astrocyte development and neuronal death in mice lacking the epidermal growth factor receptor. J Neurosci Res. 1998;53(6):697–717. doi: 10.1002/(SICI)1097-4547(19980915)53:6<697::AID-JNR8>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Kozar K, Sicinski P. Cell cycle progression without cyclin D-CDK4 and cyclin D-CDK6 complexes. Cell Cycle. 2005;4(3):388–391. doi: 10.4161/cc.4.3.1551. [DOI] [PubMed] [Google Scholar]
- Laks DR, Masterman-Smith M, Visnyei K, Angenieux B, Orozco NM, Foran I, Yong WH, Vinters HV, Liau LM, Lazareff JA, et al. Neurosphere formation is an independent predictor of clinical outcome in malignant glioma. Stem Cells. 2009;27(4):980–987. doi: 10.1002/stem.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavery W, Hall V, Yager JC, Rottgers A, Wells MC, Stern M. Phosphatidylinositol 3-kinase and Akt nonautonomously promote perineurial glial growth in Drosophila peripheral nerves. J Neurosci. 2007;27(2):279–288. doi: 10.1523/JNEUROSCI.3370-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CY, Wilkinson BD, Siegrist SE, Wharton RP, Doe CQ. Brat is a Miranda cargo protein that promotes neuronal differentiation and inhibits neuroblast self-renewal. Dev Cell. 2006a;10(4):441–449. doi: 10.1016/j.devcel.2006.01.017. [DOI] [PubMed] [Google Scholar]
- Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, Pastorino S, Purow BW, Christopher N, Zhang W, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006b;9(5):391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- Lee LA, Orr-Weaver TL. Regulation of cell cycles in Drosophila development: intrinsic and extrinsic cues. Annu Rev Genet. 2003;37:545–578. doi: 10.1146/annurev.genet.37.110801.143149. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22(3):451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445(7124):168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Lemke G. Glial control of neuronal development. Annu Rev Neurosci. 2001;24:87–105. doi: 10.1146/annurev.neuro.24.1.87. [DOI] [PubMed] [Google Scholar]
- Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, Puc J, Miliaresis C, Rodgers L, McCombie R, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275(5308):1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- Ligon KL, Huillard E, Mehta S, Kesari S, Liu H, Alberta JA, Bachoo RM, Kane M, Louis DN, Depinho RA, et al. Olig2-regulated lineage-restricted pathway controls replication competence in neural stem cells and malignant glioma. Neuron. 2007;53(4):503–517. doi: 10.1016/j.neuron.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu TH, Li L, Vaessin H. Transcription of the Drosophila CKI gene dacapo is regulated by a modular array of cis-regulatory sequences. Mech Dev. 2002;112(1–2):25–36. doi: 10.1016/s0925-4773(01)00626-8. [DOI] [PubMed] [Google Scholar]
- Marino S, Krimpenfort P, Leung C, van der Korput HA, Trapman J, Camenisch I, Berns A, Brandner S. PTEN is essential for cell migration but not for fate determination and tumourigenesis in the cerebellum. Development. 2002;129(14):3513–3522. doi: 10.1242/dev.129.14.3513. [DOI] [PubMed] [Google Scholar]
- Marumoto T, Tashiro A, Friedmann-Morvinski D, Scadeng M, Soda Y, Gage FH, Verma IM. Development of a novel mouse glioma model using lentiviral vectors. Nat Med. 2009;15(1):110–116. doi: 10.1038/nm.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masri J, Bernath A, Martin J, Jo OD, Vartanian R, Funk A, Gera J. mTORC2 activity is elevated in gliomas and promotes growth and cell motility via overexpression of rictor. Cancer Res. 2007;67(24):11712–11720. doi: 10.1158/0008-5472.CAN-07-2223. [DOI] [PubMed] [Google Scholar]
- McBride SM, Choi CH, Wang Y, Liebelt D, Braunstein E, Ferreiro D, Sehgal A, Siwicki KK, Dockendorff TC, Nguyen HT, et al. Pharmacological rescue of synaptic plasticity, courtship behavior, and mushroom body defects in a Drosophila model of fragile X syndrome. Neuron. 2005;45(5):753–764. doi: 10.1016/j.neuron.2005.01.038. [DOI] [PubMed] [Google Scholar]
- McLendon R, Friedman A, Bigner D, Van Meir EG, Brat DJ, Mastrogianakis M, Olson JJ, Mikkelsen T, Lehman N, Aldape K, et al. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeill H, Craig GM, Bateman JM. Regulation of neurogenesis and epidermal growth factor receptor signaling by the insulin receptor/target of rapamycin pathway in Drosophila. Genetics. 2008;179(2):843–853. doi: 10.1534/genetics.107.083097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellinghoff IK, Wang MY, Vivanco I, Haas-Kogan DA, Zhu S, Dia EQ, Lu KV, Yoshimoto K, Huang JH, Chute DJ, et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N Engl J Med. 2005;353(19):2012–2024. doi: 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- Michaud K, Solomon DA, Oermann E, Kim JS, Zhong WZ, Prados MD, Ozawa T, James CD, Waldman T. Pharmacologic inhibition of cyclin-dependent kinases 4 and 6 arrests the growth of glioblastoma multiforme intracranial xenografts. Cancer Res. 70(8):3228–3238. doi: 10.1158/0008-5472.CAN-09-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moberg KH, Mukherjee A, Veraksa A, Artavanis-Tsakonas S, Hariharan IK. The Drosophila F box protein archipelago regulates dMyc protein levels in vivo. Curr Biol. 2004;14(11):965–974. doi: 10.1016/j.cub.2004.04.040. [DOI] [PubMed] [Google Scholar]
- Nagane M, Coufal F, Lin H, Bogler O, Cavenee WK, Huang HJ. A common mutant epidermal growth factor receptor confers enhanced tumorigenicity on human glioblastoma cells by increasing proliferation and reducing apoptosis. Cancer Res. 1996;56(21):5079–5086. [PubMed] [Google Scholar]
- Neumuller RA, Knoblich JA. Dividing cellular asymmetry: asymmetric cell division and its implications for stem cells and cancer. Genes Dev. 2009;23(23):2675–2699. doi: 10.1101/gad.1850809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orian A, Delrow JJ, Rosales Nieves AE, Abed M, Metzger D, Paroush Z, Eisenman RN, Parkhurst SM. A Myc-Groucho complex integrates EGF and Notch signaling to regulate neural development. Proc Natl Acad Sci U S A. 2007;104(40):15771–15776. doi: 10.1073/pnas.0707418104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orian A, Grewal SS, Knoepfler PS, Edgar BA, Parkhurst SM, Eisenman RN. Genomic binding and transcriptional regulation by the Drosophila Myc and Mnt transcription factors. Cold Spring Harb Symp Quant Biol. 2005;70:299–307. doi: 10.1101/sqb.2005.70.019. [DOI] [PubMed] [Google Scholar]
- Pan D. The hippo signaling pathway in development and cancer. Dev Cell. 2010;19(4):491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelengaris S, Khan M, Evan G. c-MYC: more than just a matter of life and death. Nat Rev Cancer. 2002;2(10):764–776. doi: 10.1038/nrc904. [DOI] [PubMed] [Google Scholar]
- Pelloski CE, Lin E, Zhang L, Yung WK, Colman H, Liu JL, Woo SY, Heimberger AB, Suki D, Prados M, et al. Prognostic associations of activated mitogen-activated protein kinase and Akt pathways in glioblastoma. Clin Cancer Res. 2006;12(13):3935–3941. doi: 10.1158/1078-0432.CCR-05-2202. [DOI] [PubMed] [Google Scholar]
- Perera RM, Zoncu R, Johns TG, Pypaert M, Lee FT, Mellman I, Old LJ, Toomre DK, Scott AM. Internalization, intracellular trafficking, and biodistribution of monoclonal antibody 806: a novel anti-epidermal growth factor receptor antibody. Neoplasia. 2007;9(12):1099–1110. doi: 10.1593/neo.07721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, Misra A, Nigro JM, Colman H, Soroceanu L, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9(3):157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- Pong WW, Gutmann DH. The ecology of brain tumors: lessons learned from neurofibromatosis-1. Oncogene. 2010 doi: 10.1038/onc.2010.519. [DOI] [PubMed] [Google Scholar]
- Prigent SA, Nagane M, Lin H, Huvar I, Boss GR, Feramisco JR, Cavenee WK, Huang HS. Enhanced tumorigenic behavior of glioblastoma cells expressing a truncated epidermal growth factor receptor is mediated through the Ras-Shc-Grb2 pathway. J Biol Chem. 1996;271(41):25639–25645. doi: 10.1074/jbc.271.41.25639. [DOI] [PubMed] [Google Scholar]
- Prober DA, Edgar BA. Interactions between Ras1, dMyc, and dPI3K signaling in the developing Drosophila wing. Genes Dev. 2002;16(17):2286–2299. doi: 10.1101/gad.991102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raizer JJ, Abrey LE, Lassman AB, Chang SM, Lamborn KR, Kuhn JG, Yung WK, Gilbert MR, Aldape KA, Wen PY, et al. A phase II trial of erlotinib in patients with recurrent malignant gliomas and nonprogressive glioblastoma multiforme postradiation therapy. Neuro Oncol. 2010;12(1):95–103. doi: 10.1093/neuonc/nop015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read RD, Cavenee WK, Furnari FB, Thomas JB. A drosophila model for EGFR-Ras and PI3K-dependent human glioma. PLoS Genet. 2009;5(2):e1000374. doi: 10.1371/journal.pgen.1000374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read RD, Goodfellow PJ, Mardis ER, Novak N, Armstrong JR, Cagan RL. A Drosophila model of multiple endocrine neoplasia type 2. Genetics. 2005;171(3):1057–1081. doi: 10.1534/genetics.104.038018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter LT, Bier E. Using Drosophila melanogaster to uncover human disease gene function and potential drug target proteins. Expert Opin Ther Targets. 2002;6(3):387–399. doi: 10.1517/14728222.6.3.387. [DOI] [PubMed] [Google Scholar]
- Ryo H, Shiba T, Fukunaga A, Kondo S, Gateff E. Chromosomal aberrations and retrovirus-like particles produced by in vivo transplantation in neoplastic brain cells of a Drosophila mutant strain. Gann. 1984;75(1):22–28. [PubMed] [Google Scholar]
- Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304(5670):554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- Sanai N, Tramontin AD, Quinones-Hinojosa A, Barbaro NM, Gupta N, Kunwar S, Lawton MT, McDermott MW, Parsa AT, Manuel-Garcia Verdugo J, et al. Unique astrocyte ribbon in adult human brain contains neural stem cells but lacks chain migration. Nature. 2004;427(6976):740–744. doi: 10.1038/nature02301. [DOI] [PubMed] [Google Scholar]
- Sarkaria JN, Carlson BL, Schroeder MA, Grogan P, Brown PD, Giannini C, Ballman KV, Kitange GJ, Guha A, Pandita A, et al. Use of an orthotopic xenograft model for assessing the effect of epidermal growth factor receptor amplification on glioblastoma radiation response. Clin Cancer Res. 2006;12(7 Pt 1):2264–2271. doi: 10.1158/1078-0432.CCR-05-2510. [DOI] [PubMed] [Google Scholar]
- Schmid A, Chiba A, Doe CQ. Clonal analysis of Drosophila embryonic neuroblasts: neural cell types, axon projections and muscle targets. Development. 1999;126(21):4653–4689. doi: 10.1242/dev.126.21.4653. [DOI] [PubMed] [Google Scholar]
- Schwamborn JC, Berezikov E, Knoblich JA. The TRIM-NHL protein TRIM32 activates microRNAs and prevents self-renewal in mouse neural progenitors. Cell. 2009;136(5):913–925. doi: 10.1016/j.cell.2008.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegrist SE, Haque NS, Chen CH, Hay BA, Hariharan IK. Inactivation of both Foxo and reaper promotes long-term adult neurogenesis in Drosophila. Curr Biol. 2010;20(7):643–648. doi: 10.1016/j.cub.2010.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon MA. Receptor tyrosine kinases: specific outcomes from general signals. Cell. 2000;103(1):13–15. doi: 10.1016/s0092-8674(00)00100-8. [DOI] [PubMed] [Google Scholar]
- Simon MA, Bowtell DD, Dodson GS, Laverty TR, Rubin GM. Ras1 and a putative guanine nucleotide exchange factor perform crucial steps in signaling by the sevenless protein tyrosine kinase. Cell. 1991;67(4):701–716. doi: 10.1016/0092-8674(91)90065-7. [DOI] [PubMed] [Google Scholar]
- Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432(7015):396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- Sousa-Nunes R, Cheng LY, Gould AP. Regulating neural proliferation in the Drosophila CNS. Curr Opin Neurobiol. 2010;20(1):50–57. doi: 10.1016/j.conb.2009.12.005. [DOI] [PubMed] [Google Scholar]
- Soustelle L, Besson MT, Rival T, Birman S. Terminal glial differentiation involves regulated expression of the excitatory amino acid transporters in the Drosophila embryonic CNS. Dev Biol. 2002;248(2):294–306. doi: 10.1006/dbio.2002.0742. [DOI] [PubMed] [Google Scholar]
- St Johnston D. The art and design of genetic screens: Drosophila melanogaster. Nat Rev Genet. 2002;3(3):176–188. doi: 10.1038/nrg751. [DOI] [PubMed] [Google Scholar]
- Stark MB. An hereditary tumor in the fruit fly, Drosophila. Journal of Cancer Research. 1918;3:279–301. [Google Scholar]
- Stark MB. A Benign Tumor that is Hereditary in Drosophila. Proc Natl Acad Sci U S A. 1919;5(12):573–580. doi: 10.1073/pnas.5.12.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steck PA, Pershouse MA, Jasser SA, Yung WK, Lin H, Ligon AH, Langford LA, Baumgard ML, Hattier T, Davis T, et al. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet. 1997;15(4):356–362. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- Steffan JS, Bodai L, Pallos J, Poelman M, McCampbell A, Apostol BL, Kazantsev A, Schmidt E, Zhu YZ, Greenwald M, et al. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature. 2001;413(6857):739–743. doi: 10.1038/35099568. [DOI] [PubMed] [Google Scholar]
- Stork T, Engelen D, Krudewig A, Silies M, Bainton RJ, Klambt C. Organization and function of the blood-brain barrier in Drosophila. J Neurosci. 2008;28(3):587–597. doi: 10.1523/JNEUROSCI.4367-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawa N, Ekstrand AJ, James CD, Collins VP. Identical splicing of aberrant epidermal growth factor receptor transcripts from amplified rearranged genes in human glioblastomas. Proc Natl Acad Sci U S A. 1990;87(21):8602–8606. doi: 10.1073/pnas.87.21.8602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh H, Deng W, Gage FH. Signaling in adult neurogenesis. Annu Rev Cell Dev Biol. 2009;25:253–275. doi: 10.1146/annurev.cellbio.042308.113256. [DOI] [PubMed] [Google Scholar]
- Sulis ML, Parsons R. PTEN: from pathology to biology. Trends Cell Biol. 2003;13(9):478–483. doi: 10.1016/s0962-8924(03)00175-2. [DOI] [PubMed] [Google Scholar]
- Sun Y, Goderie SK, Temple S. Asymmetric distribution of EGFR receptor during mitosis generates diverse CNS progenitor cells. Neuron. 2005;45(6):873–886. doi: 10.1016/j.neuron.2005.01.045. [DOI] [PubMed] [Google Scholar]
- Teleman AA, Hietakangas V, Sayadian AC, Cohen SM. Nutritional control of protein biosynthetic capacity by insulin via Myc in Drosophila. Cell Metab. 2008;7(1):21–32. doi: 10.1016/j.cmet.2007.11.010. [DOI] [PubMed] [Google Scholar]
- Uhrbom L, Dai C, Celestino JC, Rosenblum MK, Fuller GN, Holland EC. Ink4a-Arf loss cooperates with KRas activation in astrocytes and neural progenitors to generate glioblastomas of various morphologies depending on activated Akt. Cancer Res. 2002;62(19):5551–5558. [PubMed] [Google Scholar]
- Uhrbom L, Kastemar M, Johansson FK, Westermark B, Holland EC. Cell type-specific tumor suppression by Ink4a and Arf in Kras-induced mouse gliomagenesis. Cancer Res. 2005;65(6):2065–2069. doi: 10.1158/0008-5472.CAN-04-3588. [DOI] [PubMed] [Google Scholar]
- Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voas MG, Rebay I. Signal integration during development: insights from the Drosophila eye. Dev Dyn. 2004;229(1):162–175. doi: 10.1002/dvdy.10449. [DOI] [PubMed] [Google Scholar]
- Wallace K, Liu TH, Vaessin H. The pan-neural bHLH proteins DEADPAN and ASENSE regulate mitotic activity and cdk inhibitor dacapo expression in the Drosophila larval optic lobes. Genesis. 2000;26(1):77–85. doi: 10.1002/(sici)1526-968x(200001)26:1<77::aid-gene10>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- Wiedemeyer R, Brennan C, Heffernan TP, Xiao Y, Mahoney J, Protopopov A, Zheng H, Bignell G, Furnari F, Cavenee WK, et al. Feedback circuit among INK4 tumor suppressors constrains human glioblastoma development. Cancer Cell. 2008;13(4):355–364. doi: 10.1016/j.ccr.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson CW, Chuang PT. Mechanism and evolution of cytosolic Hedgehog signal transduction. Development. 2010;137(13):2079–2094. doi: 10.1242/dev.045021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte HT, Jeibmann A, Klambt C, Paulus W. Modeling glioma growth and invasion in Drosophila melanogaster. Neoplasia. 2009;11(9):882–888. doi: 10.1593/neo.09576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhouse EC, Fisher A, Bandle RW, Bryant-Greenwood B, Charboneau L, Petricoin EF, 3rd, Liotta LA. Drosophila screening model for metastasis: Semaphorin 5c is required for l(2)gl cancer phenotype. Proc Natl Acad Sci U S A. 2003;100(20):11463–11468. doi: 10.1073/pnas.2031202100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa S, McKinnon RD, Kokel M, Thomas JB. Wnt-mediated axon guidance via the Drosophila Derailed receptor. Nature. 2003;422(6932):583–588. doi: 10.1038/nature01522. [DOI] [PubMed] [Google Scholar]
- Zheng H, Ying H, Yan H, Kimmelman AC, Hiller DJ, Chen AJ, Perry SR, Tonon G, Chu GC, Ding Z, et al. p53 and Pten control neural and glioma stem/progenitor cell renewal and differentiation. Nature. 2008;455(7216):1129–1133. doi: 10.1038/nature07443. [DOI] [PMC free article] [PubMed] [Google Scholar]