Summary
Recent advances in photonic imaging and fluorescent protein technology offer unprecedented views of molecular space-time dynamics in living cells. At the same time, advances in computing hardware and software enable modeling of ever more complex systems, from global climate to cell division. As modeling and experiment become more closely integrated, we must address the issue of modeling cellular processes in 3-D. Here, we highlight recent advances related to 3-D modeling in cell biology. While some processes require full 3-D analysis, we suggest that others are more naturally described in 2-D or 1-D. Keeping the dimensionality as low as possible reduces computational time and makes models more intuitively comprehensible; however, the ability to test full 3-D models will build greater confidence in models generally and remains an important emerging area of cell biological modeling.
Which is best: 1-D, 2-D or 3-D?
There is a well-known story about a mathematical modeler giving a talk to biologists. The modeler starts by saying, “Let us assume that a cow has a spherical shape…” – the moral being that those silly modelers study abstractions that are lifeless. David McQueen, co-author of a famous 3-D mechanical model of the heart [1] once said: “2-D is too damn wrong, 3-D is too damn hard…” But is it actually so silly and wrong to consider a ‘ spherical cow’? Likewise, is it indeed too hard to go 3-D mathematically? The first rule of modeling is that a model should be as simple as possible and as complex as necessary to address a particular question. In this review, we argue that, following this principle, modeling in simplified 1-D or 2-D geometry is actually very useful in the vast majority of cases. There are, however, biological systems and phenomena that require 3-D simulations.
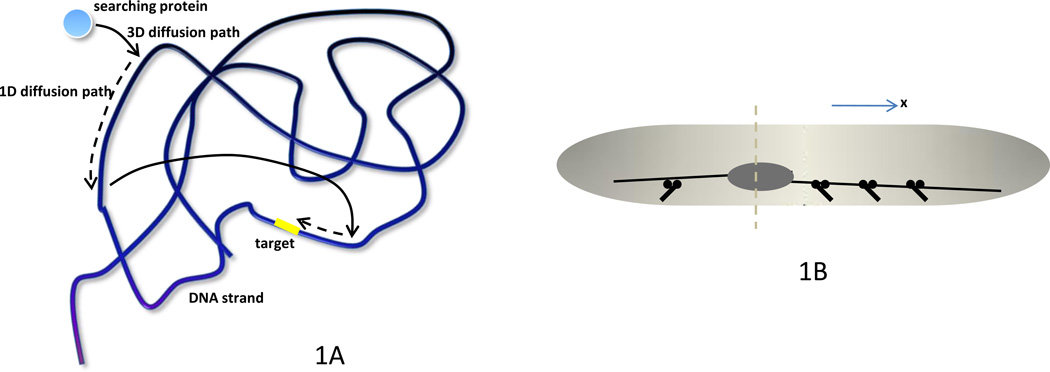
It is enough to glance at introductory textbook figures of animal cells to see that the 3-D shapes of cells, organelles and cytoskeletal structures are tremendously complex. Perhaps biological systems evolve to deal with this geometric complexity, and one strategy is to control the dimensionality of a particularly challenging process. A prime example that comes to mind is a protein searching for a target on a DNA molecule confined in a finite 3-D volume [2]. A naïve way to do the search would be to use ‘sliding’ – 1-D diffusion along the contour of the DNA. This way, the protein is constantly bound to the DNA and methodically tests all neighboring DNA sites, but such a process suffers from a slow, square-root-like, increase in the number of new positions probed as a function of time. Another possibility is to test just one site, then ‘jump’ – travel by 3-D diffusion to another site. This way, the number of new DNA sites probed grows linearly in time, but the protein misses the target, which can be very close to one of the tested sites, too often. Elegant mathematical models [2, 3] showed that the optimal strategy is to intertwine 1-D and 3-D search strategies (Figure 1A) – explore a DNA segment by the 1-D diffusion for a finite time, then jump in 3-D and explore another segment. Experimentation [4] demonstrated that, as the model predicted, the cell decreases the search time to a minimum by fine-tuning the average 3-D jump frequencies and 1-D diffusion rates suggesting that biological systems tend to organize and explore their space by sometimes reducing dimensionality of the process. This example illustrates also that modeling is crucial in understanding the role of geometry and dimensionality in cell life. To examine the emerging role of 3-D modeling in cell biology, we focus on three specific cellular processes: cell division, cell polarity and cell migration. For each process, we expose the logic behind choosing a model’s dimensionality, discuss complexities of 3-D modeling, cases when such models are needed, and examples of when they can be avoided in favor of simpler 1-D or 2-D models.
Figure 1. Search-and-capture and microtubule-mediated positioning mechanisms.
A. The optimal strategy of a search for a target on a DNA strand involves combining 3-D diffusive ‘jumps’ allowing sampling many DNA sites rapidly with 1-D diffusive ‘walks’ that enable local testing for the target site. B. In the elongated fission yeast, the cell division plane is along the short axis of the nucleus (dashed). The nucleus is elongated and positioned by opposing forces from the MTs growing to the left and right that are pulled outward by dynein motors at the cell cortex. If the nucleus is closer to the left end, then fewer motors are located at the left, and a greater number of opposing motors at the right pull the nucleus toward the cell center. The 1-D description of the process along the X-axis is adequate in this system. C. In the model for spindle oscillations in the lateral cross-section (Y-Z-plane) of the C. elegans embryo, MTs extending in dorsal and ventral directions (blue, along Z-axis) interact with dynein motors on the cortex that pull on the spindle and generate oscillations. A 1-D model is adequate if only those MTs are considered. However, the MTs extending in proximal-distal directions (red, along Y-axis), attaching to the cortex and bending are crucial to stabilize these oscillations, so the model becomes 2-D. D. Differential attachment rates of the MTs reaching the anterior and posterior half-cortices (black, along X-axis) cause asymmetric spindle movement, in which case a 3-D model is necessary. E. A minority of astral MTs (blue) growing from the spindle (black) poles toward the cell equator are stable, so molecular motors deliver myosin-activating molecular complexes (yellow) to the cell cortex, and contraction at the cell midline ensues. A majority of MTs (red) are unstable, do not reach the cell cortex and deplete the pool of motors and myosin-activating complexes (green) from the cortex, so the cell poles are relaxed. F. To accelerate the spindle assembly in prometaphase, microtubules (green) growing in 3-D from centrosomes located at the North and South poles of the cell search for kinetochores (blue) on the chromosomes (white/gray) that aggregate into a ‘bagel’-like volume along the cell equator.
Cell division
Position and orientation of the cell division plane is key to proper development of a multicellular organism. For example, a recent experimental/theoretical study of an epithelial sheet [5] established that the ‘long-axis rule known for a long time – that cells divide their long axis by forming a cleavage plane along the short axis – increases geometric regularity of the cells that are neighbors with the dividing cell. Here, we discuss a simpler question of the division geometry of a single, isolated cell. The plane of division in this case is often dictated by the cell shape [6]. On the other hand, it is known that the division plane usually passes through the middle and is perpendicular to the long axis of the nucleus or mitotic spindle (Figure 1B) [7]. Thus, two principal questions are: How do the nucleus and mitotic spindle read the cell geometry? How do they determine the division plane? There is no unique answer to this question – relevant mechanisms depend on the type of cells and conditions these cells are in – but in a few simpler cases the answer was obtained with the help of computational modeling. Briefly, long dynamic microtubules (MTs) spanning from the nucleus or spindle to the cell cortex and interacting with molecular motors are able to both sense and govern cell geometry.
This geometry can be characterized by its dimensionality. The system can be viewed as 1-D if the cell is elongated and events take place along its long axis; 2-D if we also consider phenomena taking place across the cell, but the cell is flat; or a cell can be 3- D, yet axisymmetric. Finally, in a fully 3-D system, there are significant differences along the anterior-posterior, dorsal-ventral and proximal-distal axes. In Box 1, we illustrate how scaling analysis can help to decide system dimensionality based on quantitative arguments. An excellent example of a 1-D system is seen in the elongated fission yeast Schizosaccharomyces pombe, where nuclear positioning aids in determining the cell division plane. One of the models of this process [8] is based on the observation that dynein motors on the cell cortex pull the MTs extending from the nucleus with their plus ends toward the cell poles (Figure 1B). The longer the MT, the more motors it can engage along its length, so if the nucleus is closer to one end of the cell, MTs at the other side are pulled with greater force. This servomechanism (length-dependent pulling force) centers the nucleus, or, if the motors detach faster under strain, leads to nuclear oscillations [8].
Box 1.
Partial differential equation (PDE) modeling is a traditional tool of Applied Mathematics widely used in modeling spatially explicit processes in biology [59]. When using this approach, continuous densities of key molecular players are introduced. For example, if a reaction-diffusion process involving a signaling molecule is modeled, then the molecular density, A(x, y, z,t), where x, y, z are spatial coordinates and t is time, is governed by the PDE:
Here, the left-hand side describes the local rate of density change, D is the diffusion coefficient, the terms in brackets are second spatial derivatives, F is a function describing the reaction, and k is the characteristic rate of reaction. Due to an enormous body of research on PDEs, they can often be solved approximately analytically (on paper, without a computer) providing invaluable insight. Usually though, PDEs are solved numerically, which is easy in 1-D, harder in 2-D and can be very challenging in 3-D – the amount of computer memory and time grows exponentially as the system’s dimensionality rises. The reason is the need to track densities at each grid point covering cell space; usually not less than ~ 100 grid points are necessary across the cell diameter, so if the system is 2-D, ~ 1002 grid points are needed, while in 3-D this number goes up to ~ 1003.
Scaling analysis procedure and non-dimensionalization [60] is crucial in simplifying PDEs and reducing the dimensionality. For example, here is how this procedure applies to the example above: let the characteristic cell dimensions in the x, y, z - directions be, Lx,Ly,Lz, respectively. We can introduce a characteristic reaction time scale τ = 1/k and non-dimensional time and distances:
In terms of these new variables, the PDE has the form:
If the cell is elongated in one of the directions, i.e. Lx ≫Ly,Lz, then terms with factors inside the brackets are much greater than the other terms, so approximately other terms can be neglected:
With appropriate boundary conditions, this last equation immediately suggests that on a fast time scale, the molecular density in Y- and Z-directions equilibrates to a constant, and on a longer time scale of interest, the density is only a function of X,T, but not Y,Z. Then, the PDE becomes 1-D on the longer time scale:
In a great number of cell systems and in more complex problems, a similar procedure – using characteristic scales to rescale and non-dimensionalize model variables – allows one to separate processes taking place on drastically different temporal and spatial scales – and thus to split the original problem into a few simpler ones.
Agent-based simulations are based on explicit tracking of all molecules in the cell involved in a studied mechanism so that based on the current configuration these molecules, all forces of interactions between them are computed, thermal and viscous forces are added, and then movements of every molecule are computed based on classical mechanics rules [61]. Usually, thousands of such molecule-agents are simulated, and the numerical code is quite involved, but when numerical codes are tuned in 2-D, their extension to 3-D does not require principally new numerical techniques, and the amount of memory and calculations increases only modestly. It is worth noting that there are other useful modeling techniques, such as cellular automata and Potts models, which are midway between the PDE and agent-based simulations in terms of complexity of 2-D to 3-D transition.
Homogenization is the practice of ignoring the microstructural details of a domain over which PDEs are solved or over which an agent-based simulation is run. For example, a rapidly diffusing molecule might bind reversibly to a slow-moving, large complex, and therefore has a fast-diffusing state and a slow-diffusing state. If the binding-unbinding rates are fast compared to the time scale of interest (e.g. morphogen gradient development), it is convenient to average over these two states and define an ‘effective’ diffusion coefficient (e.g. [22, 33]). The practice of homogenization has a long history in physics [34], and is used commonly in engineering to model the properties of composite materials, for example [36].
An early model of a similar positioning problem, the asymmetric positioning of the mitotic spindle during the first division of the Caenorhabditis elegans embryo, was also 1-D [9]. In this model, MTs extending in dorsal and ventral directions also interacted with dynein motors on the cortex (Figure 1C), with the important difference that the MT plus ends only, not the whole MT lengths, contacted the cortex. This system was revisited in [10], the authors of which convincingly demonstrated that the positioning problem in the C. elegans embryo is, in fact, 2-D. They discovered that MTs extending in dorsal and ventral directions reach the cortex, interact with dynein motors there and pull on the spindle causing spindle oscillations (Figure 1C), in agreement with [9]. However, the results of [10] indicated that MTs growing in proximal-distal directions are crucial to stabilize these oscillations – they attach to the cortex and bend elastically (Figure 1C). Thus, the forces in the ventral-dorsal direction are different from the forces in the proximal-distal direction, necessitating a 2-D model to mimic the spindle trajectory adequately. This study went even further because the 2-D model did not predict the different oscillation amplitudes of the anterior and posterior spindle poles (Figure 1B) that are seen experimentally. It turned out that asymmetric spindle movement could arise from a differential MT attachment rate on the posterior of the embryo compared to the anterior, and that full 3-D simulations were needed to quantitatively capture this asymmetry (Figure 1D). Also of note is that scale-up of the model from 2-D to 3-D was not that difficult because the authors used ‘agent-based’ simulation (see Box 1).
The models discussed above demonstrated that in simple cases the nucleus is positioned in the geometric center of the cell. A more complex, and essentially multi-dimensional, question is how the orientation of the nuclear long axis is determined. An elegant recent study [11] used controlled 2-D geometry by placing sea urchin eggs into micro-fabricated wells to manipulate cell geometry and to address this question. Surprisingly, the results challenged the ‘long-axis rule’: in a number of geometries, the cells divided at an angle different from that which is perpendicular to the cell’s longest axis. A 2-D computational model in which MTs from the two nuclear poles extended to the cell edges and were pulled outward by motor forces demonstrated that a balance of forces and torques elongate the nucleus and orient it in a unique way dependent on specific cell shape. The model correctly predicted these orientations for many cell shapes by using 2-D simulations; the authors suggest that cells divide along the ‘longest axis of symmetry’, a new rule that can replace the ‘long-axis rule’.
There is no complete answer yet to the second of the two questions posed in the beginning of this section – how does the mitotic spindle determine the division plane of the cell? Nevertheless, a significant contribution to solving this problem was made in a recent computational study [12]. The authors investigated the following hypothetical mechanism (Figure 1E): a minority of MTs reaching from the spindle poles to the cell ‘equator’ are stable, while the majority of the MTs are undergoing rapid dynamic instability and reach the cortex near the cell poles infrequently. Kinesin motors transport Rho-activating complex along the stable MTs to the future cleavage furrow site during anaphase and telophase, where eventually myosin is activated and contraction ensues. Kinesin motors also bind to MTs but rarely reach the cortex near the cell poles because the MTs growing in these directions usually catastrophe before contacting the cortex. As a result, the cortex near the cell poles is mechanically relaxed relative to the cell equator, which is necessary for proper cytokinesis. This agent-based model was simulated in 3-D and produced spectacular, life-like figures and movies. Other than these visual effects, it is not entirely clear whether 3-D simulations were needed in this case, especially taking into account that a single simulation took a few days of number-crunching on a powerful computer cluster. Consequently, it became too time-consuming to attempt a thorough search of the model parameter space. Likely, a 2-D model would lead to the same qualitative conclusions with less effort. On the other hand, a limited investigation of how sensitive the model results were to the parameter values was undertaken in 3-D, and it is an open question whether quantitative results of this investigation would be the same in 2-D.
Another recent study of mitotic spindle assembly illustrates very clearly that while qualitative answers can be obtained by reducing model dimensionality, quantitative conclusions sometime require full 3-D simulations. One of the mechanisms of spindle assembly is ‘search-and-capture’ in which centrosomal MTs grow and shrink until they capture kinetochores on chromosomes [13]. An influential modeling paper [14] showed that in order to assemble the spindle within the observed time, the MTs’ dynamic instability parameters should be optimized, so that MTs catastrophe often enough to not waste time growing in wrong directions, but not so often that they begin shortening before reaching a kinetochore. This model was essentially 1-D, considering only the centrosome-kinetochore distances, but not the actual chromosome distribution in the 3-D space of a mitotic cell. Recently, it was discovered that in some human cells, chromosomes briefly aggregate into an intricate ‘bagel’-like volume along the cell equator (Figure 1F), while the centrosomes stay at the ‘North and South poles’ of the cell [14]. Simulations in this paper revealed that there is a good reason for the cell to make this arrangement: all of the chromosomes are almost equidistant from the poles where the search originates, so the MT dynamic instability parameters can be tuned to the same pole-target distance. More importantly, as the MTs cannot penetrate chromosome arms, chromosomes that are too close to the pole could shield a large fraction of 3-D space from the searching MTs. The ‘bagel’-like configuration does not have such chromosomes, so the majority of targets are accessible from the poles. Only explicit 3-D simulations could estimate the time needed to assemble the spindle in this geometry and compare the result with the experimental measurement; 1-D and 2-D approximations would be grossly imprecise.
Cell polarity
In the previous section, we discussed how the cell reads its shape and imposes patterns by using MT-related force balances. Another group of mechanisms enabling cells to discriminate among all potential directions and to select a particular direction as being ‘special’ is based on reactions and diffusion of signaling molecules. This process, known as polarization, is fundamental to nearly all cells [15]. Theoreticians view this as an example of ‘symmetry-breaking’, a class of phenomena where a chemically reactive system spontaneously generates spatially heterogeneous patterns. Pioneering work by Alan Turing in the 1950s showed that simple sets of reactions coupled to finite rates of diffusion could lead to spontaneous symmetry breaking [16]. Subsequent theoretical studies in the 1960s and 1970s hypothesized that such reaction-diffusion mechanisms might underlie the morphogenesis of the early embryo [17, 18]. Fueled by recent advances in fluorescent protein imaging (i.e. confocal and 2-photon imaging, better digital cameras, and better fluorophores) and in digital technology (i.e. faster processors, larger memory capacity), we are witnessing a rapid growth in the integrated experimental-theoretical study of polarization. Here, we discuss how modeling in 3-D is shaping advances in our understanding of cell polarity by examining the specific case of embryo polarization.
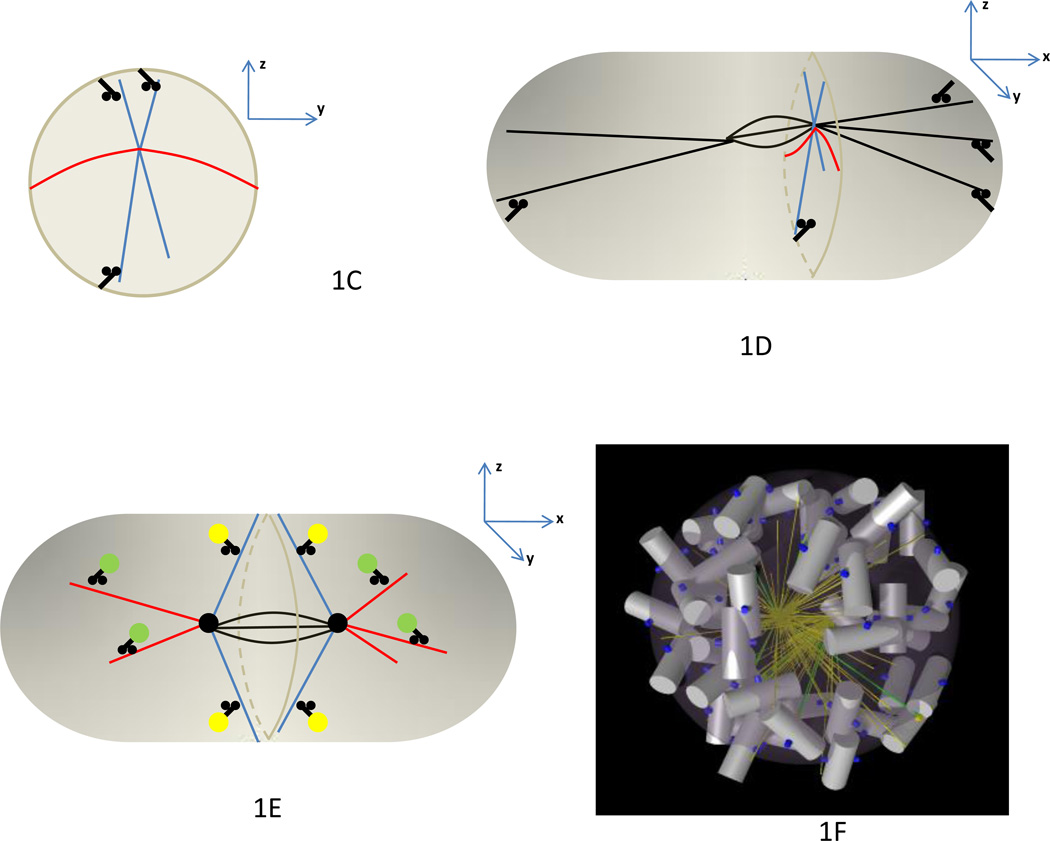
The polarization of early stage embryos is vital to determining the major axes of the mature organism: anterior-posterior, left-right, dorsal-ventral. Model organisms, such as C. elegans and Drosophila melanogaster, have been crucial to the study of embryo polarization, and decades of research in this field has facilitated mathematical modeling by providing a wealth of quantitative data within a consistent spatial-temporal framework (3-D spatially plus time reference frame, referred to hereafter as “3-D+T”). One of the best-studied polarity determinants is the protein bicoid, a transcriptional regulator that exists in an anterior-to-posterior concentration gradient in the early stages of D. melanogaster embryonic development [19]. Early work documented the existence of the bicoid gradient, and led to the synthesis-diffusion-degradation (SDD) model [19]. In this model, bicoid protein is synthesized from maternally inherited bicoid mRNA located near the anterior end of the embryo, diffuses, and is randomly degraded (Figure 2). Although the SDD was formulated without mathematics, it qualitatively described basic features of the bicoid gradient. Until recently, however, quantitative tests of the SDD model were lacking, so it was not clear whether this model was simply an attractive idea or a rigorous mechanistic explanation. For example, the SDD model predicts that at steady-state the gradient will decay according to an exponential distribution. Whether the bicoid gradient conforms to an exponential, or whether it even reaches a steady-state at all during developmentally relevant time scales were key questions left unanswered until recent years.
Figure 2. Cell polarity: Bicoid morphogen gradient in the Drosophila embryonic syncytium.
In the Synthesis-Diffusion-Degradation (SDD) model originally proposed, bicoid mRNA located in the anterior cortex serves a synthesis source for bicoid protein, which then diffuses away over time. Bicoid protein is eventually degraded, although whether degradation is occurring during the period when the gradient is ‘read-out’ by the nuclei is not clear. While 1-D models have been the norm and have been very informative, more recent 2-D and 3-D models are raising, and framing, new fundamental questions about this well-studied morphogen system. In addition, 2-D and 3-D models are facilitating more precise quantitative measurements needed for discrimination among competing models.
With the rapid advancement of digital cameras and fluorescent protein technology, the last few years have witnessed a flurry of new studies on bicoid, as well as other morphogens [20]. In the case of bicoid, detailed quantitative analysis confirmed an approximately exponential decay in the concentration of bicoid, as measured in 1-D along the contour of the embryo’s periphery, consistent with a simple diffusion-degradation mechanism [21]. For this simple model to work quantitatively, bicoid must diffuse at a rate sufficiently fast to allow the gradient to develop from a characteristic decay length of ~60 µm at 50 min after egg deposition to ~110 µm at 160 min [22]. Using photobleaching of bicoid-GFP via two-photon confocal microscopy, the first direct estimate of the bicoid diffusion coefficient was deemed too small to yield the observed extent of the gradient according to the SDD model [23]. This finding led to new modeling efforts to explain the additional missing convective transport mechanisms, perhaps via cytoskeleton-based transport [24]. One could regard this as an “SDD+” model, where the “+” refers to some extra transport component that is as yet undiscovered. Interpretation of photobleaching experiments is notoriously tricky [25, 26], however, and subsequent analysis using a full 3-D simulation of the bleaching-diffusion experiment re-estimated the measured value of the bicoid diffusion coefficient to be 3-fold higher than initially reported, and consistent with the SDD model after all [22]. It is important to note, however, that this measurement was made at a specific time and place in the developmental process: the anterior cytoplasm during mitotic cycle 13. To really test models, it will be vital to measure the diffusion coefficient in 3-D space and time (3D+T), both during mitosis and interphase (and inside and outside of the nucleus). It may turn out that the diffusion coefficient, rather than being a constant parameter, is actually a variable that depends on space and time, as recently suggested [27].
In fact, due to the combination of 3-D modeling and improved 3-D imaging, nearly all the basic assumptions of the SDD model have recently been thrown into question. For example, the first ‘D’ in the SDD model, ‘diffusion’, recently came into question when it was asserted, in the absence of mathematical modeling, that the bicoid protein gradient could be explained entirely by a pre-existing, maternally derived bicoid mRNA gradient [28]. In this case there would not be a requirement for bicoid protein diffusion at all, other than from the mRNA-ribosomal synthesis site to its target binding sites in the nucleus. However, it seems that this model would have difficulty explaining the increase in the decay length of the gradient over time, mentioned above. In addition, a recent report shows that protein diffusion is necessary to explain the different decay lengths in the mRNA and protein, as measured quantitatively [27]. An important aspect of this recent study is that it included a full 3-D model, using the experimentally measured embryo geometry as the domain over which the solution was obtained [29]. Combined with comprehensive 3-D quantitative analysis, they were able to show that the bicoid protein gradient is not simply a rescaled version of the bicoid mRNA gradient. The last ‘D’ in the SDD model, degradation, is also controversial because it is not clear that degradation is actually occurring during the period when bicoid influences polarity development. In fact, recent modeling analysis makes the case for bicoid not being degraded at all [30]. In this ‘nuclear-trapping’ model, net consumption of synthesized bicoid protein is provided by newly produced nuclei; as the number of nuclei increases the capacity of bicoid per unit volume of embryo also increases. Thus, the SDD model could in one respect be simpler than previously imagined in that no degradation is necessarily required, such that an ‘SD’ model (i.e. synthesis-diffusion) might be adequate.
In summary, it appears that understanding the dynamics of bicoid and other morphogens will require model predictions and measurements in 3-D+T to discriminate among various hypotheses and test long-held assumptions. For example, it may be important to incorporate even more realistic models of the cell geometry, such as the inward invaginations of the plasma membrane that would presumably limit diffusion on the 10–20 µm length scale [31]. However, as a full 3-D+T picture emerges, it may still be possible to mathematically homogenize the cytoplasm and thereby return to a simpler 1-D+T model [32, 33]. Such coarse-graining/homogenization methods are standard in engineering, and are commonly used to predict and simulate the electrical and diffusive transport properties of both naturally occurring and fabricated composite materials [34–36]. Reducing complexity by coarse-graining (i.e. from 3-D down to 2-D or 1-D) will make it easier to rapidly obtain in silico predictions of mutant phenotypes, and thereby gain a more complete molecular-level mechanistic understanding of morphogen systems.
Cell motility
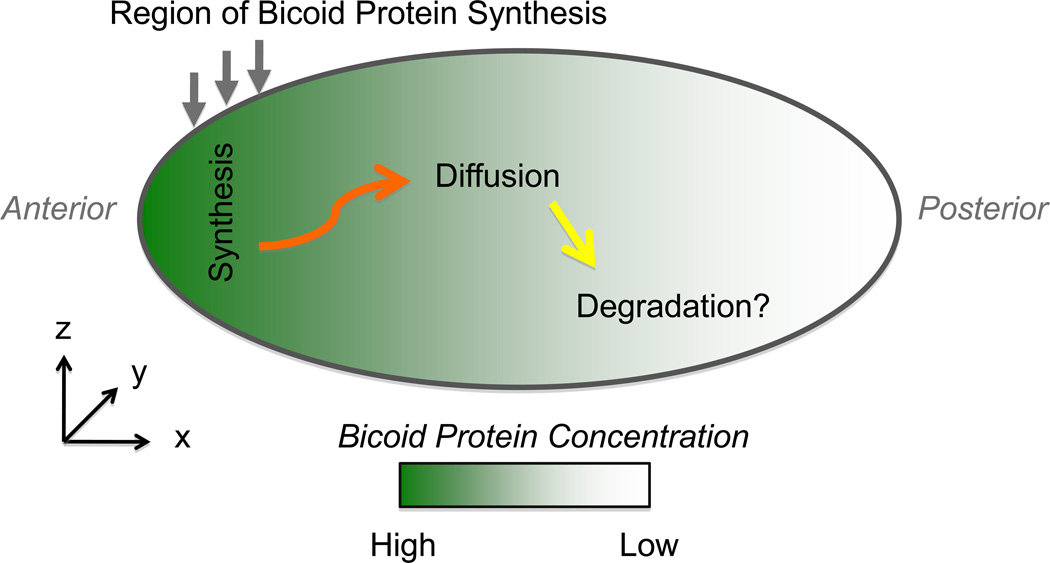
One particularly important cellular process that utilizes the reaction-diffusion processes that drive polarization is cell motility [37]. After morphogens determine the front, sides and rear, the cell moves by protruding the cell front through growth of actin networks, retracting the rear by myosin-driven contraction and adhering to the surface [38]. The whole phenomenon is so complex that the reductionist approach is essential. Thus, actin-propelled bacterial pathogens and plastic beads, which mimic cell protrusion but do not employ contraction or adhesion, have attracted much attention. Besides the reduction of biological complexity, dimensional reduction was widely used to model the protrusion. Thus, influential elastic propulsion theory [39] posits that actin gels grow at the bead surface and push away older parts of the actin network that surround the nascent actin filaments. These older actin gel layers stretch, and generate a squeezing stress propelling the bead forward. The first and simplest, 1-D formulation of this theory [39] explained successfully how the bead is propelled forward by the actin network growing at the bead’s rear (Figure 3A). The 1-D model, however, did not explain how the spherical bead, which is initially surrounded by a spherically symmetric ‘actin cloud’, breaks through this cloud as observed [40] and initiates unidirectional motility. This symmetry breaking phenomenon was explained by the 2-D model [41], which analyzed how radial compression of the inner layer of the actin gel equilibrated with the tangential stretching of the outer layer, so variations of the stress in two dimensions was explicitly considered. This model demonstrated that when the outer layer of the actin gel becomes thin in any location, then the stretching force per actin filament grows there and the gel starts to fail locally, making it possible for the bead to escape (Figure 3A). The exact geometry of the symmetry breaking, which in principle may not have axial symmetry, cannot be revealed by either 1-D or 2-D models, however. Recent experiments [42] indeed showed that the actin cloud breaks through an asymmetric linear crack [42] (Figure 3A). 3-D agent-based simulations faithfully reproduced this cracking process [42], providing the ultimate test for the actin elastic propulsion theory.
Figure 3. Dimensionality in actin protrusion and cell motility.
A. The actin-driven propulsion of a plastic bead can be, in the simplest case, modeled in 1-D by calculating gradients of actin density, deformation and stress along the long axis of the actin tail (left). The simplest model of the bead breaking through the initially symmetric ‘actin cloud’ can be formulated in 2-D (center): thinning of the actin gel (lighter region at the right) causes autocatalytic contraction of the gel at the opposite side of the bead and further breakage of the gel at the site of the initial thinning. Calculation of the elastic stress distribution in radial and tangential directions is required for modeling this process. In order to understand the exact geometry of the symmetry breaking, full 3-D simulation of the actin gel mechanics (right) is necessary. The figure at the right is reprinted from [42] with permission. B. Motile fish keratocyte cell is modeled in 3-D in the framework of the Virtual Cell software as the union of the cell body (half ellipsoid) and lamellipodium (thin ellipsoid) (left). Much insight can be obtained from 2-D models analyzing distributions of the key molecular players (actin, myosin, adhesion molecules, etc; in the figures, G-actin distribution is shown). These 2-D models can either address dynamics in the flat lamellipodium (bottom right), or in the dorsal-ventral plane in the middle of the cell (top right). Valuable answers can, in fact, be obtained even from 1-D models calculating front-to-rear dynamics along the dashed line at the center of the lamellipodium. Elements of the figure are reprinted from [58] with permission.
To elucidate how cells move on flat surfaces, understanding of actin-based protrusion has to be complemented by knowledge of contraction, adhesion and the role of the membrane enveloping the cell. One of the main mechanical questions about the migrating cell is how the cell manages to maintain its length such that the rear neither lags behind nor collapses onto the protruding front. This complicated question can be, in the first approximation, addressed in 1-D by considering the anterior-posterior slice across the moving cell. Pioneering 1-D modeling [43] illustrated under simple and natural assumptions that a simplified 1-D actomyosin strip protruding at one end and contracting and adhering to the surface across the cell indeed achieved stable size and crawling speed because the myosin contraction at the rear accelerated while the protrusion at the front slowed down as the cell was getting longer. Moreover, this simplistic model correctly predicted that the cell maximizes its speed at a moderate level of adhesion [44]; weaker adhesion causes the cell to treadmill in place while stronger adhesion makes the cell unable to pull forward the rear. The next step would be to understand how the migrating cell contains its lateral dimension, neither letting the sides expand too widely, nor allowing them to collapse to the middle. This question is answered only for simple, steadily crawling cells, such as fish keratocytes [45, 46] (Figure 3B). As this question concerns behavior in two dimensions, mathematical models were developed in 2-D based on experimental observations. These models proposed that the cell maintains low/high actin density at the sides/front of its edge, respectively. Lower-density actin networks at the sides is stalled by the outstretched cell membrane, while a higher-density actin gel at the front protrudes against the membrane tension, and myosin swept to the rear in the framework of the moving cell contracts pulling the rear forward. The 2-D models of the keratocyte are adequate because the motile appendage of the moving cell is very flat (Figure 3B) – its height is smaller than a micrometer compared to tens of micrometers in length and width. These models did not address this flatness. Interestingly, a limited 2-D model of the ventral-dorsal cross-section of the motile cell suggested that myosin contraction keeps the ventral and dorsal surfaces close together [47].
To fully understand the motile cell, which does not possess enough symmetry, a 3-D model is needed. The first attempt to tackle such a full 3-D model, albeit with somewhat artificial vertical force flattening the cell, was made in a recent pioneering study [48]. It remains to be seen if biological insight will come out of this model, however. Notably, unlike most 3-D computational models using the agent-based approach, the authors[48] employed computationally demanding numerical analysis of partial differential equations describing actin-myosin mechanics, so this traditional modeling tool is difficult, but not impossible, to use in 3-D. It is becoming clear that cell migration through a 3-D extracellular matrix in physiologically relevant systems employs mechanisms that are somewhat different from those on 2-D surfaces [49]. Respective 3-D modeling will undoubtedly be needed very soon. So far, lack of high quality data on the dynamics of actin, myosin and other key molecules in three spatial dimensions plus time, along with a lack of ideas for how to develop these 3-D models and computer limitations, hinder progress. However, recent success in high resolution imaging [50] will no doubt lead to more adequate simulations and understanding. One additional challenge of 3-D cell motility modeling is that, in addition to the cell mechanics, one has to model highly nontrivial deformations, stresses and breakage of the extracellular matrix surrounding the cell.
Concluding remarks
Computational modeling in cell biology generates quantitative hypotheses for experimental testing and provides rigorous framework for interpretation of the data. A model has to be as simple as possible for maximal qualitative insight, yet still capture essential features of the cell dynamics. Thus, if the symmetry and dimensions of the problem allow, there is no reason to develop a hyper-realistic 3-D model (other than to perhaps daze collaborators with life-like simulation movies!). A 3-D model is necessary when either i) a process is completely asymmetric and understanding its exact geometry is essential, or ii) a predicted quantity has to be compared with a more or less precise measurement. Clear illustrations of these two situations, in addition to the cases highlighted above, come from morphogenesis. For example, the developing vertebrate limb is highly asymmetric, and its multicellular pattern looks different in all three (anterior-posterior, dorsal-ventral and proximal-distal) dimensions. The most straightforward way to test our understanding of this process is to develop 3-D models, simulate them in 3-D and see if the simulations predict the observed pattern, as has been done [51, 52]. On the other hand, gastrulation – a fundamental morphogenetic behavior that transforms a flat cell sheet into a pit – has rotational symmetry, and so can be modeled successfully in 2-D [53, 54]. Both of these studies report insightful 2-D simulations of a frontal cross-section of cell sheets being buckled by asymmetric and heterogeneous actomyosin contractions. Notably, both studies contain exemplary discussions of the 2-D versus 3-D aspect of the problem and contend that though all qualitative model conclusions are valid in 3-D, the 2-D models ignore circumferential deformations and stresses along annular rings of cells around the symmetry axis. Thus, when accurate measurements of these strains and stresses become available and models have to be tested against those numbers, there will be no choice but to update the 2-D models to the full 3-D versions.
How hard is it to model in 3-D? It is not difficult to formulate a model in 3-D, but solving its partial differential equations, which are the most powerful and developed mathematical tool, is very taxing – the amount of memory, disk storage and processing time needed are enormous. With the increasing speed and bandwidth of modern computers, it is now possible to run 3-D models involving tens of millions of grid points and tens of thousands of time steps, but it takes terabytes of disk storage and several days to compute, even on the most powerful of supercomputers. This prevents exploratory use of modeling prohibiting frequent back and forth between models and experiments due to time constraints. While computer power is increasing rapidly, so is the known biological (i.e. interactome) complexity. There are techniques being developed to minimize computing time, such as parallel simulations (where the computation is spread across multiple machines) and graphical processing unit-based computation (developed for gaming applications). The latter is done on specialized processors that do not perform many generalized tasks, however, and are highly optimized to crunch data and equations for specific type of models [55]. In addition, as mentioned above, agent-based simulations in 3-D generally do not increase computational complexity significantly beyond 1-D or 2-D. Finally, hard thinking almost invariably makes a model, even a 3-D one, simple enough to simulate by using one of a growing number of user-friendly software interfaces, such as Virtual Cell [56] and CompuCell3D [57]. One has to always remember the second rule of modeling: computational time is inversely proportional to thinking time.
Acknowledgements
We thank R. Paul for donating Fig. 1D. This work was supported by NSF Grant DMS-0315782 and by NIH Grant GM-068952 to A.M. and by NIH Grants GM-071522, GM-076177, and CA-145044 to D.O.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.McQueen DM, Peskin CS. A three-dimensional computer model of the human heart for studying cardiac fluid dynamics. SIGGRAPH Comput. Graph. 2000;34(1):56–60. [Google Scholar]
- 2.Berg OG, Winter RB, von Hippel PH. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry. 1981;20(24):6929–6948. doi: 10.1021/bi00527a028. [DOI] [PubMed] [Google Scholar]
- 3.Sheinman M, Kafri Y. The effects of intersegmental transfers on target location by proteins. Phys Biol. 2009;6(1s):016003. doi: 10.1088/1478-3975/6/1/016003. [DOI] [PubMed] [Google Scholar]
- 4.Winter RB, Berg OG, von Hippel PH. Diffusion-driven mechanisms of protein translocation on nucleic acids. 3. The Escherichia coli lac repressor--operator interaction: kinetic measurements and conclusions. Biochemistry. 1981;20(24):6961–6977. doi: 10.1021/bi00527a030. [DOI] [PubMed] [Google Scholar]
- 5.Gibson WT, Veldhuis JH, Rubinstein B, Cartwright HN, Perrimon N, Brodland GW, Nagpal R, Gibson MC. Control of the mitotic cleavage plane by local epithelial topology. Cell. 2011;144(3):427–438. doi: 10.1016/j.cell.2010.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thery M, Bornens M. Cell shape and cell division. Curr Opin Cell Biol. 2006;18(6):648–657. doi: 10.1016/j.ceb.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 7.Reinsch S, Gonczy P. Mechanisms of nuclear positioning. J Cell Sci. 1998;111(Pt 16):2283–2295. doi: 10.1242/jcs.111.16.2283. [DOI] [PubMed] [Google Scholar]
- 8.Vogel SK, Pavin N, Maghelli N, Julicher F, Tolic-Norrelykke IM. Self-organization of dynein motors generates meiotic nuclear oscillations. PLoS Biol. 2009;7(4):e1000087. doi: 10.1371/journal.pbio.1000087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grill SW, Kruse K, Julicher F. Theory of mitotic spindle oscillations. Phys Rev Lett. 2005;94(10):108104. doi: 10.1103/PhysRevLett.94.108104. [DOI] [PubMed] [Google Scholar]
- 10.Kozlowski C, Srayko M, Nedelec F. Cortical microtubule contacts position the spindle in C. elegans embryos. Cell. 2007;129(3):499–510. doi: 10.1016/j.cell.2007.03.027. [DOI] [PubMed] [Google Scholar]
- 11.Minc N, Burgess D, Chang F. Influence of cell geometry on division-plane positioning. Cell. 2011;144(3):414–426. doi: 10.1016/j.cell.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Odell GM, Foe VE. An agent-based model contrasts opposite effects of dynamic and stable microtubules on cleavage furrow positioning. J Cell Biol. 2008;183(3):471–483. doi: 10.1083/jcb.200807129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirschner M, Mitchison T. Beyond self-assembly: From microtubules to morphogenesis. Cell. 1986;45:329–342. doi: 10.1016/0092-8674(86)90318-1. [DOI] [PubMed] [Google Scholar]
- 14.Magidson V, O'Connell CB, Loncarek J, Paul R, Mogilner A, Khodjakov A. The Spatial Arrangement of Chromosomes during Prometaphase Facilitates Spindle Assembly. Cell. 2011;146(4):555–567. doi: 10.1016/j.cell.2011.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li R, Gundersen GG. Beyond polymer polarity: how the cytoskeleton builds a polarized cell. Nat Rev Mol Cell Biol. 2008;9(11):860–873. doi: 10.1038/nrm2522. [DOI] [PubMed] [Google Scholar]
- 16.Turing AM. The chemical basis of morphogenesis. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 1952;237:37–72. [Google Scholar]
- 17.Wolpert L. Positional information and the spatial pattern of cellular differentiation. J Theor Biol. 1969;25(1):1–47. doi: 10.1016/s0022-5193(69)80016-0. [DOI] [PubMed] [Google Scholar]
- 18.Crick F. Diffusion in embryogenesis. Nature. 1970;225(5231):420–422. doi: 10.1038/225420a0. [DOI] [PubMed] [Google Scholar]
- 19.Driever W, Nusslein-Volhard C. A gradient of bicoid protein in Drosophila embryos. Cell. 1988;54(1):83–93. doi: 10.1016/0092-8674(88)90182-1. [DOI] [PubMed] [Google Scholar]
- 20.Grimm O, Coppey M, Wieschaus E. Modelling the Bicoid gradient. Development. 2010;137(14):2253–2264. doi: 10.1242/dev.032409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Houchmandzadeh B, Wieschaus E, Leibler S. Establishment of developmental precision and proportions in the early Drosophila embryo. Nature. 2002;415(6873):798–802. doi: 10.1038/415798a. [DOI] [PubMed] [Google Scholar]
- 22.Castle BT, Howard SA, Odde DJ. Assessment of Transport Mechanisms Underlying the Bicoid Morphogen Gradient. Cell Mol Bioeng. 2011;4(1):116–121. doi: 10.1007/s12195-010-0157-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gregor T, Wieschaus EF, McGregor AP, Bialek W, Tank DW. Stability and nuclear dynamics of the bicoid morphogen gradient. Cell. 2007;130(1):141–152. doi: 10.1016/j.cell.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hecht I, Rappel WJ, Levine H. Determining the scale of the Bicoid morphogen gradient. Proc Natl Acad Sci U S A. 2009;106(6):1710–1715. doi: 10.1073/pnas.0807655106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sprague BL, McNally JG. FRAP analysis of binding: proper and fitting. Trends Cell Biol. 2005;15(2):84–91. doi: 10.1016/j.tcb.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 26.Sprague BL, Pego RL, Stavreva DA, McNally JG. Analysis of binding reactions by fluorescence recovery after photobleaching. Biophys J. 2004;86(6):3473–3495. doi: 10.1529/biophysj.103.026765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Little SC, Tkacik G, Kneeland TB, Wieschaus EF, Gregor T. The formation of the Bicoid morphogen gradient requires protein movement from anteriorly localized mRNA. PLoS Biol. 2011;9(3):e1000596. doi: 10.1371/journal.pbio.1000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spirov A, Fahmy K, Schneider M, Frei E, Noll M, Baumgartner S. Formation of the bicoid morphogen gradient: an mRNA gradient dictates the protein gradient. Development. 2009;136(4):605–614. doi: 10.1242/dev.031195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fowlkes CC, Hendriks CL, Keranen SV, Weber GH, Rubel O, Huang MY, Chatoor S, DePace AH, Simirenko L, Henriquez C, Beaton A, Weiszmann R, Celniker S, Hamann B, Knowles DW, Biggin MD, Eisen MB, Malik J. A quantitative spatiotemporal atlas of gene expression in the Drosophila blastoderm. Cell. 2008;133(2):364–374. doi: 10.1016/j.cell.2008.01.053. [DOI] [PubMed] [Google Scholar]
- 30.Coppey M, Berezhkovskii AM, Kim Y, Boettiger AN, Shvartsman SY. Modeling the bicoid gradient: diffusion and reversible nuclear trapping of a stable protein. Dev Biol. 2007;312(2):623–630. doi: 10.1016/j.ydbio.2007.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mavrakis M, Rikhy R, Lippincott-Schwartz J. Plasma membrane polarity and compartmentalization are established before cellularization in the fly embryo. Dev Cell. 2009;16(1):93–104. doi: 10.1016/j.devcel.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sample C, Shvartsman SY. Multiscale modeling of diffusion in the early Drosophila embryo. Proc Natl Acad Sci U S A. 2010;107(22):10092–10096. doi: 10.1073/pnas.1001139107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Griffin EE, Odde DJ, Seydoux G. Regulation of the MEX-5 Gradient by a Spatially Segregated Kinase/Phosphatase Cycle. Cell. 2011;146(6):955–968. doi: 10.1016/j.cell.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maxwell JC. Treatise on Electricity and Magnetism. Clarendon Press; 1873. [Google Scholar]
- 35.Brosseau C. Modeling and simulation of dielectric heterostructures: a physica survey from an historical perspective. J Physics D: Applied Physics. 2006;39:1277–1294. [Google Scholar]
- 36.Toledo PG, Davis HT, Scriven LE. Transport properties of anisotropic porous media: effective medium theory. Chemical Engineering Science. 1991;47:391–405. [Google Scholar]
- 37.Weiner OD. Regulation of cell polarity during eukaryotic chemotaxis: the chemotactic compass. Curr Opin Cell Biol. 2002;14(2):196–202. doi: 10.1016/s0955-0674(02)00310-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112(4):453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 39.Gerbal F, Chaikin P, Rabin Y, Prost J. An elastic analysis of Listeria monocytogenes propulsion. Biophys J. 2000;79(5):2259–2275. doi: 10.1016/S0006-3495(00)76473-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cameron LA, Footer MJ, van Oudenaarden A, Theriot JA. Motility of ActA protein-coated microspheres driven by actin polymerization. Proc Natl Acad Sci U S A. 1999;96(9):4908–4913. doi: 10.1073/pnas.96.9.4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sekimoto K, Prost J, Jülicher F, Boukellal H, Bernheim-Grosswasser A. Role of tensile stress in actin gels and symmetry-breaking instability. European Physics Journal E Soft Matter. 2004;13:247–259. doi: 10.1140/epje/i2003-10073-y. [DOI] [PubMed] [Google Scholar]
- 42.Dayel MJ, Akin O, Landeryou M, Risca V, Mogilner A, Mullins RD. In silico reconstitution of actin-based symmetry breaking and motility. PLoS Biol. 2009;7(9):e1000201. doi: 10.1371/journal.pbio.1000201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.DiMilla PA, Barbee K, Lauffenburger DA. Mathematical model for the effects of adhesion and mechanics on cell migration speed. Biophys J. 1991;60(1):15–37. doi: 10.1016/S0006-3495(91)82027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Palecek SP, Loftus JC, Ginsberg MH, Lauffenburger DA, Horwitz AF. Integrin-ligand binding properties govern cell migration speed through cell-substratum adhesiveness. Nature. 1997;385(6616):537–540. doi: 10.1038/385537a0. [DOI] [PubMed] [Google Scholar]
- 45.Keren K, Pincus Z, Allen GM, Barnhart EL, Marriott G, Mogilner A, Theriot JA. Mechanism of shape determination in motile cells. Nature. 2008;453(7194):475–480. doi: 10.1038/nature06952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barnhart EL, Lee KC, Keren K, Mogilner A, Theriot JA. An adhesion-dependent switch between mechanisms that determine motile cell shape. PLoS Biol. 2011;9(5):e1001059. doi: 10.1371/journal.pbio.1001059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kruse K, Joanny JF, Julicher F, Prost J. Contractility and retrograde flow in lamellipodium motion. Phys Biol. 2006;3(2):130–137. doi: 10.1088/1478-3975/3/2/005. [DOI] [PubMed] [Google Scholar]
- 48.Herant M, Dembo M. Form and function in cell motility: from fibroblasts to keratocytes. Biophys J. 2010;98(8):1408–1417. doi: 10.1016/j.bpj.2009.12.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Even-Ram S, Yamada KM. Cell migration in 3D matrix. Curr Opin Cell Biol. 2005;17(5):524–532. doi: 10.1016/j.ceb.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 50.Soon L, Braet F, Condeelis J. Moving in the right direction-nanoimaging in cancer cell motility and metastasis. Microsc Res Tech. 2007;70(3):252–257. doi: 10.1002/jemt.20411. [DOI] [PubMed] [Google Scholar]
- 51.Chaturvedi R, Huang C, Kazmierczak B, Schneider T, Izaguirre JA, Glimm T, Hentschel HG, Glazier JA, Newman SA, Alber MS. On multiscale approaches to three-dimensional modelling of morphogenesis. J R Soc Interface. 2005;2(3):237–253. doi: 10.1098/rsif.2005.0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boehm B, Westerberg H, Lesnicar-Pucko G, Raja S, Rautschka M, Cotterell J, Swoger J, Sharpe J. The role of spatially controlled cell proliferation in limb bud morphogenesis. PLoS Biol. 2010;8(7):e1000420. doi: 10.1371/journal.pbio.1000420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sherrard K, Robin F, Lemaire P, Munro E. Sequential activation of apical and basolateral contractility drives ascidian endoderm invagination. Curr Biol. 2010;20(17):1499–1510. doi: 10.1016/j.cub.2010.06.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tamulonis C, Postma M, Marlow HQ, Magie CR, de Jong J, Kaandorp J. A cell-based model of Nematostella vectensis gastrulation including bottle cell formation, invagination and zippering. Dev Biol. 2011;351(1):217–228. doi: 10.1016/j.ydbio.2010.10.017. [DOI] [PubMed] [Google Scholar]
- 55.Christley S, Lee B, Dai X, Nie Q. Integrative multicellular biological modeling: a case study of 3D epidermal development using GPU algorithms. BMC Syst Biol. 2010;4:107. doi: 10.1186/1752-0509-4-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Slepchenko BM, Loew LM. Use of virtual cell in studies of cellular dynamics. Int Rev Cell Mol Biol. 2010;283:1–56. doi: 10.1016/S1937-6448(10)83001-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Swat MH, Hester SD, Balter AI, Heiland RW, Zaitlen BL, Glazier JA. Multicell simulations of development and disease using the CompuCell3D simulation environment. Methods Mol Biol. 2009;500:361–428. doi: 10.1007/978-1-59745-525-1_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Novak IL, Slepchenko BM, Mogilner A. Quantitative analysis of G-actin transport in motile cells. Biophys J. 2008;95(4):1627–1638. doi: 10.1529/biophysj.108.130096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Edelstein-Keshet L. Mathematical Models in Biology. New York: SIAM; 2008. [Google Scholar]
- 60.Logan JD. Applied Mathematics. Wiley; 2006. [Google Scholar]
- 61.Alberts JB, Odell GM. In silico reconstitution of Listeria propulsion exhibits nanosaltation. PLoS Biol. 2004;2(12):e412. doi: 10.1371/journal.pbio.0020412. [DOI] [PMC free article] [PubMed] [Google Scholar]