Abstract
Background & Aims
Expression of the netrin-1 dependence receptor UNC5C is reduced in many colorectal tumors; mice with the UNC5C mutations have increased progression of intestinal tumors. We investigated whether specific variants in UNC5C increase risk for colorectal cancer (CRC).
Methods
We analyzed the sequence of UNC5C in blood samples from 1801 patients with CRC and 4152 controls from 3 cohorts (France, USA, and Finland). Almost all cases from France and the USA had familial CRC; of the Finnish cases, 92/984 were familial. We analyzed whether CRC segregates with the UNC5C variant A628K in 3 families with histories of CRC. We also performed haplotype analysis, to determine the origin of this variant.
Results
Of 817 patients with familial CRC, 14 had 1 of 4 different, unreported missense variants in UNC5C. The variants p.Asp353Asn (encodes D353N), p.Arg603Cys (encodes R603C), and p.Gln630Glu (encodes Q630E) did not occur significantly more often in cases than controls. The variant p.Ala628Lys (A628K) was detected in 3 families in the French cohort (odds ratio [OR], 8.8; Wald’s 95% confidence interval [CI], 1.47–52.93; P=.03) and in 2 families the US cohort (OR, 1.9; P=.6), but was not detected in the Finnish cohort; UNC5C A628K segregated with CRC in families. Three families with A628K had a 109 kb identical haplotype that spanned most of UNC5C, indicating recent origin of this variant in Caucasians (14 generations; 95% CI, 6–36 generations). Transfection of HEK293T cells with UNC5C-A628K significantly reduced apoptosis compared to wild-type UNC5C, measured in an assay of active caspase-3.
Conclusion
Inherited mutations in UNC5C prevent apoptosis and increase risk for CRC.
Keywords: Colon cancer, tumor suppression, tumorigenesis, neoplasm, UNC5H3
Introduction
The commonly accepted scheme for transmembrane receptor-mediated signal transduction relies on the fact that the receptor becomes active upon ligand binding. However, over the last decade a new functional family of receptors has been identified. These receptors named dependence receptors share the ability to be active in the absence of their respective ligand and in this setting trigger apoptosis. Such receptors thus create cellular states of dependence on their respective ligands 1, 2. The prototypical dependence receptors are the netrin-1 receptors. Netrin-1, a diffusible laminin-related protein, has been shown to play a major role in the control of neuronal navigation during the development of the nervous system, by interacting with its main receptors, DCC (Deleted in Colorectal Cancer) 3–5 and UNC5H 6, 7. However, netrin-1 has rapidly emerged as a multifunctional protein implicated in multiple functions beyond the brain 8, 9. We and others have shown that a large part of its activity is associated with the fact that netrin-1 regulates endothelial and epithelial cell survival by inhibiting the pro-apoptotic activity of the dependence receptors DCC and UNC5H, -i.e., UNC5H1, UNC5H2, UNC5H3, UNC5H4 also called UNC5A,B,C,D- 9–15.
The pro-apoptotic activity of these unbound receptors has been suggested to act as a mechanism to eliminate tumor cells that would develop in settings of ligand unavailability such as proliferation of tumor cells in an environment with constant and limited ligand presence or migration of metastatic tumor cells in tissues with no or low ligand expression. This hypothesis fits with the observation that the DCC and UNC5H genes are down-regulated in tumors, hence suggesting that the loss expression of these genes represents a selective advantage for tumor development 9, 16–18. Along this line, it has been shown in different animal models that the balance between netrin-1 and its receptors is important in the regulation of cancer progression. On the one hand a gain of netrin-1 has been shown to block tumor cell death in vitro and to be associated with intestinal tumor initiation and progression in mice 19, 20. On the other hand, inactivation of UNC5C (UNC5H3/rcm) in mice is associated with intestinal tumor progression 21, thus demonstrating per se that UNC5C acts as a tumor suppressor.
Based on these observations, we speculated that genetic abnormalities of UNC5C might be involved in human colorectal carcinogenesis. In order to identify germline mutations in the human UNC5C gene that may increase predisposition to cancer, we analyzed genomic DNA from human blood samples of unrelated individuals with family histories of CRC. We report here the detection of several missense mutations, one of which leads to significant loss of pro-apoptotic activity of UNC5C and co-segregates with the disease.
Results and Conclusion
The UNC5C gene was analyzed in three independent cohorts. A first cohort- from France- included 328 unrelated individuals affected with CRC from French families and 1911 controls (1031 cancer-free individuals from the Calvados region in France, 780 blood donors from the Rhône-Alpes region in France, 100 Portuguese control individuals; Portuguese controls were included because one UNC5C variant was detected in a French family of Portuguese origin). A second cohort from the US included 489 unrelated individuals affected with CRC from Ohio families and 1416 Ohio controls. A third cohort was composed of 984 unrelated individuals affected with CRC from Finnish families and 825 blood donors.
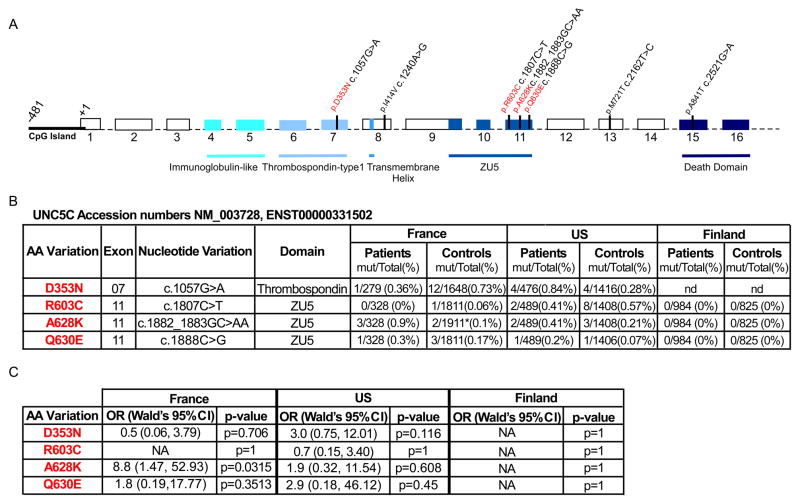
We first sequenced all 16 exons of the UNC5C gene (Fig. 1A) in a panel of 235 unrelated individuals affected with CRC from French or North American families. We identified 20 different nontruncating variants within the UNC5C coding sequence, including 13 synonymous (8 not yet described in databases), and 7 non synonymous changes (Suppl. Table 1). We focused our attention on 4 of the 7 detected missense variants; p.Asp353Asn (D353N), p.Arg603Cys (R603C), p.Ala628Lys (A628K) and p.Gln630Glu (Q630E) (Fig. 1). Two of the other missense variations (p.Met721Thr in exon 13 and p.Ala841Thr in exon 15) were already described as polymorphic in dbSNP and were found at the same frequency in the 235 cases of CRCs and in our controls (Suppl. Table 1). One other variant p.Iso414Val in exon 8 was found only once in a patient and not in 500 controls. However, the change of an Isoleucine to Valine should not be associated with any significant change in the protein folding (non-polar side-chain to hydrophobic amino acid) and consistent with this assumption we failed to detect any functional change in UNC5C carrying this variation (not shown and Fig. 4E).
Figure 1. Genomic features of the UNC5C gene.
A, Schematic representation of the UNC5C gene showing exon-intron structure, location of CpG island, protein domains, and sequence changes observed in this study. Above the sequence are 7 missense changes. Those 4 not already described in databases are depicted in color. B, Table recapitulating the different newly identified missense variations. AA : Amino acid. C, Table showing the conditional maximum likelihood estimate of the odds ratio (OR), associated Wald’s 95% confidence interval (CI), and p-values as obtained with Fisher’s exact text.
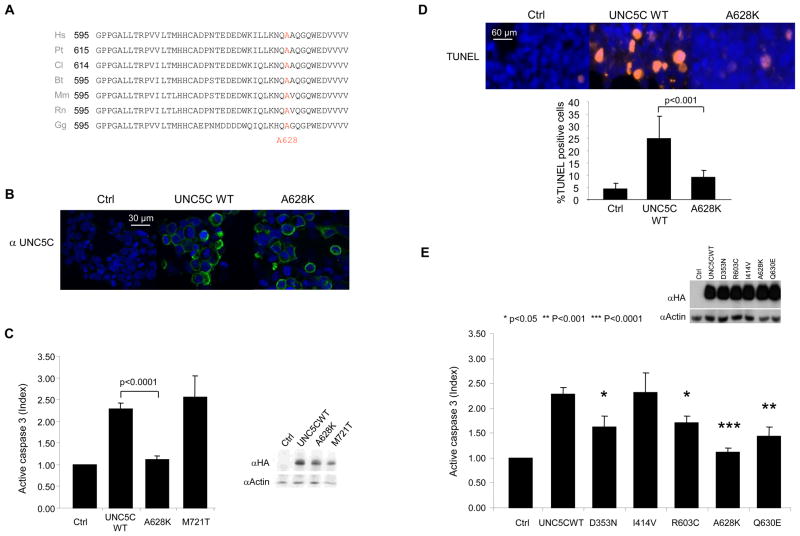
Figure 4. Loss of proapoptotic function of UNC5C A628K variant.
A, Sequence alignment of the UNC5C regions containing the A628 residue among species. Hs: Homo sapiens, Pt: Pan troglodytes, Cl: Canis lupus familiaris, Bt: Bos taurus, Rn: Rattus norvegicus, Mm: Mus musculus, Gg: Gallus gallus. B, Immunostaining using UNC5C antibody and confocal microscopy after transient transfection of HEK293T cells with mock vector (control) or constructs containing the missense variation (UNC5C A628K) or wild-type UNC5C (UNC5C WT) allele. C, Apoptosis quantification by active caspase-3 activity assay after transient transfection of HEK293T cells using mock vector (control) or constructs containing missense mutation (UNC5C A628K or UNC5C M721T) or wild-type UNC5C (UNC5C WT) allele. Data are mean ± SEM. Statistical analysis, student test, p values are indicated. Upper panel: UNC5C expression is controlled by immunoblot using anti-HA (Hemagglutinin epitope) antibodies (all UNC5C proteins were HA-tagged). Immunoblot on actin is shown as a control for loading. D. Analysis of A628K variant pro-apoptotic activity after transient transfection in HEK293T cells of UNC5C wild-type (WT) or A628K (A628K). Apoptosis quantification by TUNEL staining. Data are mean ± standard deviation (lower panel). E. Analysis of UNC5C D353N, R603C and Q630E pro-apoptotic activity after transient transfection in HEK293T cells. Apoptosis quantification by active caspase-3 activity assay. Inset: HA-immunoblot of the different mutants expressed in HEK293T (all UNC5C proteins were HA-tagged). Data are mean ± SEM. Statistical analysis, student test, p values are indicated.
One of the missense mutations, p.Asp353Asn (D353N) is located in exon 7 encoding an extracellular thrombospondin domain and the 3 others, p.Arg603Cys (R603C), p.Ala628Lys (A628K) and p.Gln630Glu (Q630E), are located in exon 11 coding for the intracellular ZU-5 domain that has been shown to be involved in the pro-apoptotic activity of UNC5C 11, 22,23 (Fig. 1B). We extended our search for variants in exons 7 and 9–11 (exons 9 to 11 encode the ZU-5 domain) to a larger panel of individuals from families with colorectal cancer from France and the US (Suppl. Table 1, Fig. 2, Suppl. Fig. 1). The changes leading to the above amino acid substitutions were detected in 14/817 (1.7%) independently ascertained individuals with familial cancer from France and North America (Fig. 1B). As exon 11 encoding the ZU5 domain appeared to show most changes (9/817 (1.1%)), we extended the analysis of exon 11 to a panel of individuals from families with colorectal cancer from Finland. Pedigrees of the 14 families with colorectal cancer with UNC5C changes are shown, in Fig. 2 for the A628K change and in Suppl. Fig. 1 for the other 3 changes.
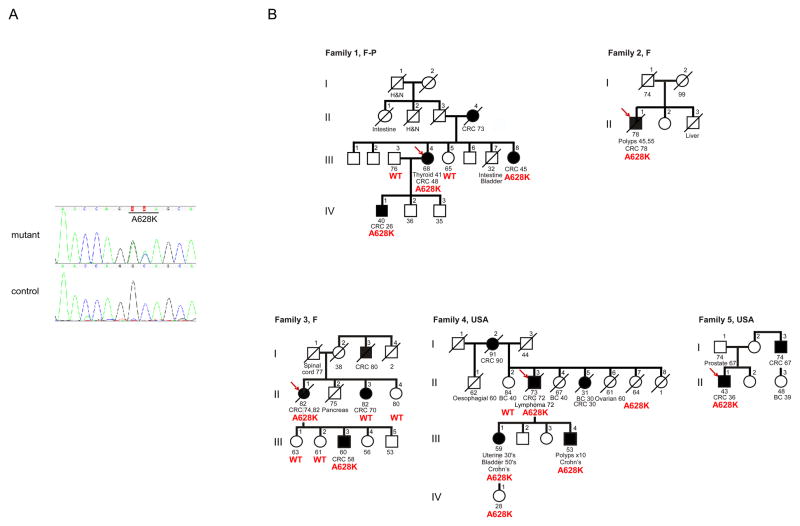
Figure 2. Pedigrees of the 5 families in which the proband had a deleterious A628K mutation in UNC5C.
A, Four-color-sequencing chromatograms from an automated DNA sequencer showing the germline missense mutation in the UNC5C gene from a patient (Family 1, III.3), and a control sequence (Caucasian control population). The coding strand is shown. M: C and A, R: G and A. B, The probands screened for UNC5C mutations are indicated by red arrows. Individuals with cancer are shown as filled circles/squares. Different cancers are indicated beneath the relevant individuals, with age at diagnosis next to the cancer type. WT, normal sequence observed; The A628K mutation in UNC5C gene is indicated beneath individuals. Absence of a genotype indicates that DNA samples could not be obtained. BC (Breast Cancer). CaSU (Cancer Site Unknown). Crohn s (Crohn s disease). CRC (Colorectal Cancer). H&N (Head&Neck cancer). Roman numerals refer to generations, and individuals within a generation are numbered from left to right. Families origin: USA (USA), France (F), France and Portugal (F-P).
Figure 1B summarizes the proportion of the four detected mutations (D353N, R603C, A628K and Q630E) in the three independent cohorts of patients and controls. As shown in Figure 1C, the Finnish cohort was not informative as none of the four mutations were detected in patients or controls. The French cohort and the US cohort showed no significant differences between probands and controls for the mutations D353N, R603C, and Q630E. The A628K mutation was significantly more frequent in the probands than in the controls in the French cohort resulting in an Odds Ratio (OR) of 8.8 (Wald’s 95%CI: 1.47–52.93) for risk of CRC (p=0.03 by Fisher’s exact test) (Fig. 1C). A similar trend was seen for the A628K mutation in the US cohort, but the difference was not statistically significant (OR 1.9; p 0.608).
In an effort to ascertain the genetic transmission of the probands’ A628K mutation and determine its cosegregation with CRC in family members, samples that were available for other members of families 1, 3 and 4 (Figure 2) were analyzed. Including the probands, 12 individuals in the five families were A628K mutation carriers; of these, 8 had CRC (age at diagnosis, mean 52 years; median 48 years). Only 4 mutation carriers did not have CRC but 2 of them had either other cancers (uterine cancer, bladder cancer) or/and Crohn’s disease (family 4, III.1 and III.4 respectively). The last two carriers were either young (IV.1, age 28) or died without diagnosed cancer at age 64 (II.7).
Finally, in these 3 families there were 7 individuals who tested negative for the mutation; none had CRC except individual II.3 in family 3 who developed CRC at 70 years of age, suggesting the possibility of a phenocopy.
These observations, albeit of limited coverage, suggest dominant inheritance with high CRC penetrance conferred by this mutation. A closer look at the pedigrees discloses a wide array of different cancers in several individuals belonging to many families. As only few of these could be studied for the mutations, it is too early to tell whether mutations in UNC5C might predispose to other cancers in addition to CRC.
We analyzed whether, as expected for a tumor suppressor gene mutated in the germline in one allele, UNC5C gene expression is switched off in tumors in individuals carrying the A628K mutation. We did not observe loss of heterozygosity (LOH) in any of the probands tested (families 1, 4, 5 and families 11, 12 and 14). However, in agreement with recent data 21, 24, we detected strong DNA methylation at the UNC5C promoter in every case tested (probands of families 4, 5, 11, 12 and 14) suggesting that both UNC5C alleles are silenced in the tumors (Suppl. Figure 2).
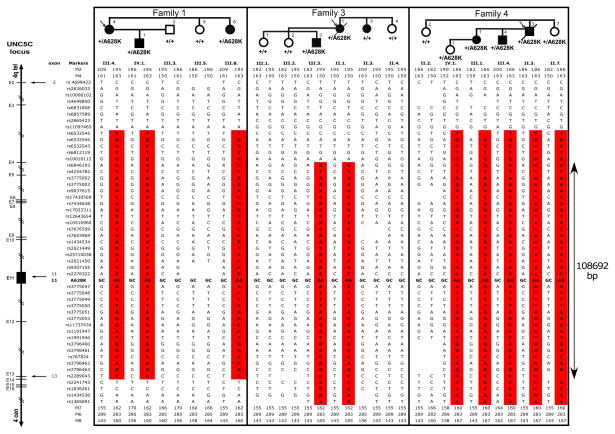
Because the A628K mutation was common and occurred both in France (with one family of Portuguese origin) and in the United States, we asked whether this might be a recurrent de novo hot-spot mutation or a founder mutation inherited over many generations. A haplotype analysis was conducted with 55 polymorphic SNPs and microsatellite markers spanning about 388 kbp within the UNC5C locus in families 1, 3 and 4. We identified a shared region encompassing 108 692 bp around the A628K mutation suggesting that the mutation probably emanated from a common founder. The haplotype is shown in Fig. 3. Using the likelihood based method developed by Genin and colleagues 25, the most recent common ancestor of the A628K mutation was estimated to have lived 14 generations ago (95% confidence interval (CI) = 6–36). Assuming that one generation is 25 years, this corresponds to 350 years (95% CI = 150–900).
Figure 3. Haplotyping of individuals in families 1, 3 and 4.
Left, Map of the UNC5C locus indicating the approximate position of the different SNP markers in this region. Most are intronic. The position of the markers is indicated. Right, Pedigrees of individuals genotyped in families 1, 3 and 4. Individual identifiers as in Figure 2B. Location of markers and the A628K mutation in exon 11 are given on the left. Arrows indicate exonic markers. A common disease-associated haplotype (red) covering 108692 bp and comprising the A628K mutation is shared by all tested mutation-positive individuals.
Because at least one of the identified UNC5C variants appears to be associated with the development of CRC and because it was recently proposed that UNC5C negatively regulates tumor progression via its intrinsic ability to trigger apoptosis, we next analysed whether these allelic variants lead to the loss or reduction of UNC5C s proapoptotic function. UNC5C sequence analysis among species reveals that the amino-acid A628, located in the ZU5 domain, a domain known to be implicated in UNC5H pro-apoptotic activity 22, 23, is conserved among species from Gallus gallus to Homo sapiens (Fig 4A). We then assessed whether the A628K mutation in UNC5C modulates its dependence receptor activity i.e., its ability to trigger apoptosis when expressed in the absence of its ligand. HEK293T cells were transiently transfected with constructs encoding wild-type or A628K mutant UNC5C (Fig. 4BCD). Both wild-type UNC5C and UNC5C A628K were normally expressed and located at the plasma membrane after transfection, as shown by immunohistochemistry (Fig. 4B) and immunoblot (Fig. 4C). Cell death was then first analysed by measuring caspase activation (Fig. 4C). We observed that while wild-type UNC5C receptor is pro-apoptotic in the absence of its ligand, the A628K mutant is no longer effective in triggering apoptosis. As a further control, a UNC5C variant with the already described innocuous polymorphism p.Met721Thr (M721T) was assessed. As shown in Fig. 4C, this variant failed to modulate the receptor pro-apoptotic activity. Cell death was also analysed by TUNEL assay (Fig. 4D). Similarly, while wild-type UNC5C receptor is pro-apoptotic in the absence of its ligand, the UNC5C A628K mutant was not effective in triggering apoptosis.
Together with the observation that in mice, inactivation of UNC5C (UNC5H3/rcm) is associated with intestinal tumor progression 21, and that UNC5C expression has already been shown to be down-regulated in a large fraction of human colorectal cancers mainly via promoter methylation 21,24, the data presented here support the view that individuals with a heterozygous loss-of-apoptotic-function constitutional “germline” mutation in the UNC5C gene have an increased likelihood of developing colorectal cancer, and possibly other cancers, and this feature is hereditary. Interestingly transfection of an half proportion of UNC5C A628K together with an half proportion of UNC5C WT is sufficient to reduce apoptosis induction (not shown), suggesting that the mutation in one UNC5C allele may provide an initial survival advantage that is further amplified via a general UNC5C promoter methylation in the tumor. Whether other mutations than A628K also confer a predisposition to CRC remains to be determined by studying further cases and controls. Indeed, even though our present study failed to show a significantly increased proportion of the D353N, R603C, and Q630E mutations in probands over controls (Fig. 1C), we observed that these mutations all affect UNC5C pro-apoptotic activity significantly (Fig. 4E).
Taken together these data suggest that one or several germline mutations in UNC5C may have a role in CRC susceptibility. Three main findings support the germline A628K mutation as a predisposing change: its effect on the pro-apoptotic function of the receptor, its co-segregation with disease in three families, and the fact that it is more frequent in cases than in controls. Nevertheless, the relative weakness of the association and the fact that the OR reached statistical significance only in the French cohort calls for further studies in geographically and ethnically different patients. It also calls for experiments to look for changes in other genes implicated in the netrin-1 dependence receptor pathway, such as DAPK1 and PR65β, recently described as mediators of UNC5H-induced apoptosis 26, 27. Our results further support the notion that these receptors participate in the control of tumor development.
Methods
Individuals and clinical samples
We obtained genomic DNA from blood from 1801 individuals with colorectal cancer (984 from the Department of Medical Genetics at the University of Helsinki, Helsinki, Finland, 489 from the Division of Human Genetics at the Ohio State University, Columbus USA, 179 from the Centre Leon Bérard-HEH, Lyon France, 60 from the CHU Strasbourg, France, and 10 from the previously described CCREF cohort 28), representing unrelated families. Institutional review board approval and informed consent for cancer gene predisposition research was obtained from all patients. Almost all families fulfil the criteria of revised Bethesda guidelines 29 or had at least two cases of CRC (except families 2 and 9 who show one case of CRC associated with another type of cancer). Family 10 is an exception as the proband had kidney cancer at 37 years of age and developed colon polyps at 43 years of age but was the only living member of the family with 4 cases of CRC. The samples from Finland are mostly sporadic CRC but included 92 familial cases 30, 31. Cases of familial polyposis or hamartomatous polyposis syndromes or with mismatch repair gene mutations were excluded.
The tumors of probands have been checked for a mismatch repair (MMR) genes deficiency (i.e., MLH1, MSH2 and MSH6) 32. All the individuals from Ohio had microsatellite stable tumors. Additionally, for the probands of the families 4, 7, 9, 11–12 and 14, the presence of the 3 MMR proteins was verified by immunohistochemistry. All the individuals from the Molecular Oncology Unit at Centre Léon Bérard (probands of family 1–3, 10 and 13) have been sequenced for MLH1, MSH2, and MSH2 (exons and intron/exon junctions) and no mutations were detected.
The control samples were obtained after approval by the appropriate Internal Review Boards. Every donor signed an approved consent form for genetic studies. The control cohorts were derived as follows. The 1416 individuals from the metropolitan region of Columbus, Ohio, USA belonged to a series of randomly chosen volunteers representing the same geographic, ethnic and social background as the CRC patients from Ohio 33). The 1031 individuals from the Calvados colorectal Family (CCREF) study 28 were randomly derived relatives of probands with CRC. The 780 anonymous blood donors were from the Etablissement Français du Sang Rhône-Alpes34. Because one of the probands with the A628K mutation was of Portuguese ethnicity, 100 randomly chosen Portuguese controls were also studied (a kind gift of Professor J. Sequeiros, University of Porto, Portugal). The 825 Finnish anonymous blood donors were obtained from the Finnish Red Cross Blood Transfusion Service. All the blood donors were healthy and cancer-free at the time the blood was drawn.
Conditional maximum likelihood estimate of the odds ratio (OR), associated 95% confidence interval (CI) and p-values were obtained with Fisher’s exact text.
UNC5C DNA sequencing and haplotype analysis
Genomic DNA (50ng) was amplified by PCR using primers encompassing all the exons of UNC5C with HotStarTaq (Qiagen) or AmpliTaq Gold DNA Polymerase (Applied Biosystems). PCR products were screened using endo-1 to detect heteroduplex 35, or by single-strand conformation polymorphism (SSCP) gel electrophoresis 36 or by High Resolution Melting (Roche Applied Science, Basel, CH) 37 and then sequenced in both directions by Cogenics (Grenoble), or GATC Biotech (Konstanz) or by using the ABI Prism BigDye Terminator Cycle Sequencing Kit version 3.1 and the Applied Biosystems 3730 DNA Analyzer (PE Applied Biosystems, Foster City, CA). SNP Markers were designed with Hapmap and UCSC databases, and microsatellite markers by using Repeat Masker (http://www.repeatmasker.org) for haplotype and LOH analysis. All primers and PCR conditions are available upon request.
Loss of heterozygosity (LOH) assessment and UNC5C promoter methylation analysis
Microsatellites and SNPs were used as markers to compare normal (blood) and tumor tissues, using the SNaPshot (PE Applied Biosystems, Foster City, CA) technique as described previously 38 or by using fluorescently labeled primers for the microsatellites 39. All primers and PCR conditions are available upon request.
The UNC5C promoter region, amplified from bisulfite-modified DNA by four different PCRs, was cloned into a TA vector (pCR2.1 Invitrogen, Carlsbad, CA). Ten positive recombinants were isolated and sequenced by using the ABI Prism BigDye Terminator Cycle Sequencing Kit version 3.1 and the Applied Biosystems 3730 DNA Analyzer (PE Applied Biosystems, Foster City, CA)
Plasmid constructs
pcDNA3-UNC5C-HA containing human UNC5C full-length coding sequence was used as template for insertion of the different mutant alleles using the Quick-change strategy. All primers are available upon request. HA : Haemagglutinin tag.
Cell line, transfection procedure, cell death assays
Transient transfections of human embryonic kidney cells (HEK293T) were performed using Lipofectamine reagent according to manufacturer’s instructions (Invitrogen). For detection of DNA fragmentation, Terminal deoxynucleodityl transferase mediated dUTP-biotin Nick End Labelling (TUNEL) was performed with 300U/mL TUNEL enzyme (300U/mL) and 6 μM biotinylated dUTP (Roche Diagnostics), and detected with avidine coupled Cy3. Caspase-3 activity was measured using Caspase-3/CPP32 Fluorometric Assay Kit from Gentaur Biovision (Belgium) 40, 41.
Immunohistochemistry and immunoblot
HEK293T cultures were fixed in 4% paraformaldehyde. Coverslips were then hybridized one hour at room temperature with an antibody against human UNC5C (1/500, MAB1005, R&D System). After rinsing in Phosphate Buffer Saline, slides were incubated with an Alexa-488-Donkey anti-Mouse antibody (1/500, Molecular Probes) and nuclei were stained with Hoescht. Slides were analysed using confocal microscopy (Leica TCS SP2). Immunoblot was performed as described previously using anti-HA antibody 26.
Supplementary Material
Acknowledgments
Grant support: This work was supported by the Ligue Contre le Cancer (to PM), the NIH (NS45093 to PM; CA67941 and CA16058 to ADLC), the FRM (PM), the ARC (PM), the Region Rhone-Alpes (to PM, JYS and JCS) and the Institut National du Cancer (INCA) (to PM and JYS).
We wish to thank Dr. F. Calheiros and Dr. A. Calheiros and Laboratório de Análises “Dr. Maria Paulo Lda” (Porto-Portugal) for the precious help regarding the family 3, Pr. J. Sequeiros (University of Porto-Portugal) and Dr F. Camoin (EFS Rhône-Alpes) for DNA controls, Dr. O. Caron (Strasbourg-France) for samples, K.L Guan for UNC5C construct, M. Leone, C. Navarro, C. Lafaye, G. Montmain, E. Ruano, James D. Perko, Nidhi Shah and Melissa Wu for technical help, S. Lyonnet and D. Stoppa-Lyonnet for precious advice and comments and M. Susag for text correction.
Footnotes
Author Contributions: MMC, MC, JT, LM have made experiments, NA, IC, YD, CL, SL, AP, JCS, JYS, QW, HH, ST, LA have provided materials, MMC, LA, SMT, AdlC, AB, PM designed experiments and analyzed data, SMT, AdlC, AB and PM have written the manuscript.
The authors declare to have no conflict of interest in this study
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mehlen P, Thibert C. Dependence receptors: between life and death. Cell Mol Life Sci. 2004;61:1854–66. doi: 10.1007/s00018-004-3467-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bredesen DE, Mehlen P, Rabizadeh S. Receptors that mediate cellular dependence. Cell Death Differ. 2005;12:1031–43. doi: 10.1038/sj.cdd.4401680. [DOI] [PubMed] [Google Scholar]
- 3.Serafini T, Colamarino SA, Leonardo ED, Wang H, Beddington R, Skarnes WC, Tessier-Lavigne M. Netrin-1 is required for commissural axon guidance in the developing vertebrate nervous system. Cell. 1996;87:1001–14. doi: 10.1016/s0092-8674(00)81795-x. [DOI] [PubMed] [Google Scholar]
- 4.Keino-Masu K, Masu M, Hinck L, Leonardo ED, Chan SS, Culotti JG, Tessier-Lavigne M. Deleted in Colorectal Cancer (DCC) encodes a netrin receptor. Cell. 1996;87:175–85. doi: 10.1016/s0092-8674(00)81336-7. [DOI] [PubMed] [Google Scholar]
- 5.Forcet C, Stein E, Pays L, Corset V, Llambi F, Tessier-Lavigne M, Mehlen P. Netrin-1-mediated axon outgrowth requires deleted in colorectal cancer-dependent MAPK activation. Nature. 2002;417:443–7. doi: 10.1038/nature748. [DOI] [PubMed] [Google Scholar]
- 6.Ackerman SL, Kozak LP, Przyborski SA, Rund LA, Boyer BB, Knowles BB. The mouse rostral cerebellar malformation gene encodes an UNC-5-like protein. Nature. 1997;386:838–42. doi: 10.1038/386838a0. [DOI] [PubMed] [Google Scholar]
- 7.Hong K, Hinck L, Nishiyama M, Poo MM, Tessier-Lavigne M, Stein E. A ligand-gated association between cytoplasmic domains of UNC5 and DCC family receptors converts netrin-induced growth cone attraction to repulsion. Cell. 1999;97:927–41. doi: 10.1016/s0092-8674(00)80804-1. [DOI] [PubMed] [Google Scholar]
- 8.Cirulli V, Yebra M. Netrins: beyond the brain. Nat Rev Mol Cell Biol. 2007;8:296–306. doi: 10.1038/nrm2142. [DOI] [PubMed] [Google Scholar]
- 9.Mehlen P, Delloye-Bourgeois C, Chedotal A. Novel roles for Slits and netrins: axon guidance cues as anticancer targets? Nat Rev Cancer. 2011;11:188–97. doi: 10.1038/nrc3005. [DOI] [PubMed] [Google Scholar]
- 10.Mehlen P, Rabizadeh S, Snipas SJ, Assa-Munt N, Salvesen GS, Bredesen DE. The DCC gene product induces apoptosis by a mechanism requiring receptor proteolysis. Nature. 1998;395:801–4. doi: 10.1038/27441. [DOI] [PubMed] [Google Scholar]
- 11.Llambi F, Causeret F, Bloch-Gallego E, Mehlen P. Netrin-1 acts as a survival factor via its receptors UNC5H and DCC. Embo J. 2001;20:2715–22. doi: 10.1093/emboj/20.11.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang H, Ozaki T, Shamim Hossain M, Nakamura Y, Kamijo T, Xue X, Nakagawara A. A newly identified dependence receptor UNC5H4 is induced during DNA damage-mediated apoptosis and transcriptional target of tumor suppressor p53. Biochem Biophys Res Commun. 2008;370:594–8. doi: 10.1016/j.bbrc.2008.03.152. [DOI] [PubMed] [Google Scholar]
- 13.Tanikawa C, Matsuda K, Fukuda S, Nakamura Y, Arakawa H. p53RDL1 regulates p53-dependent apoptosis. Nat Cell Biol. 2003;5:216–23. doi: 10.1038/ncb943. [DOI] [PubMed] [Google Scholar]
- 14.Arakawa H. Netrin-1 and its receptors in tumorigenesis. Nat Rev Cancer. 2004;4:978–87. doi: 10.1038/nrc1504. [DOI] [PubMed] [Google Scholar]
- 15.Castets M, Coissieux MM, Delloye-Bourgeois C, Bernard L, Delcros JG, Bernet A, Laudet V, Mehlen P. Inhibition of endothelial cell apoptosis by netrin-1 during angiogenesis. Dev Cell. 2009;16:614–20. doi: 10.1016/j.devcel.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 16.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–67. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 17.Grady WM. Making the case for DCC and UNC5C as tumor-suppressor genes in the colon. Gastroenterology. 2007;133:2045–9. doi: 10.1053/j.gastro.2007.10.034. [DOI] [PubMed] [Google Scholar]
- 18.Mehlen P, Guenebeaud C. Netrin-1 and its dependence receptors as original targets for cancer therapy. Curr Opin Oncol. 2010;22:46–54. doi: 10.1097/CCO.0b013e328333dcd1. [DOI] [PubMed] [Google Scholar]
- 19.Mazelin L, Bernet A, Bonod-Bidaud C, Pays L, Arnaud S, Gespach C, Bredesen DE, Scoazec JY, Mehlen P. Netrin-1 controls colorectal tumorigenesis by regulating apoptosis. Nature. 2004;431:80–4. doi: 10.1038/nature02788. [DOI] [PubMed] [Google Scholar]
- 20.Paradisi A, Maisse C, Coissieux MM, Gadot N, Lepinasse F, Delloye-Bourgeois C, Delcros JG, Svrcek M, Neufert C, Flejou JF, Scoazec JY, Mehlen P. Netrin-1 up-regulation in inflammatory bowel diseases is required for colorectal cancer progression. Proc Natl Acad Sci U S A. 2009;106:17146–51. doi: 10.1073/pnas.0901767106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bernet A, Mazelin L, Coissieux MM, Gadot N, Ackerman SL, Scoazec JY, Mehlen P. Inactivation of the UNC5C Netrin-1 receptor is associated with tumor progression in colorectal malignancies. Gastroenterology. 2007;133:1840–8. doi: 10.1053/j.gastro.2007.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williams ME, Strickland P, Watanabe K, Hinck L. UNC5H1 induces apoptosis via its juxtamembrane domain through an interaction with NRAGE. J Biol Chem. 2003;278:17483–17490. doi: 10.1074/jbc.M300415200. [DOI] [PubMed] [Google Scholar]
- 23.Wang R, Wei Z, Jin H, Wu H, Yu C, Wen W, Chan LN, Wen Z, Zhang M. Autoinhibition of UNC5b revealed by the cytoplasmic domain structure of the receptor. Mol Cell. 2009;33:692–703. doi: 10.1016/j.molcel.2009.02.016. [DOI] [PubMed] [Google Scholar]
- 24.Shin SK, Nagasaka T, Jung BH, Matsubara N, Kim WH, Carethers JM, Boland CR, Goel A. Epigenetic and genetic alterations in Netrin-1 receptors UNC5C and DCC in human colon cancer. Gastroenterology. 2007;133:1849–57. doi: 10.1053/j.gastro.2007.08.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Genin E, Tullio-Pelet A, Begeot F, Lyonnet S, Abel L. Estimating the age of rare disease mutations: the example of Triple-A syndrome. J Med Genet. 2004;41:445–9. doi: 10.1136/jmg.2003.017962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Llambi F, Lourenco FC, Gozuacik D, Guix C, Pays L, Del Rio G, Kimchi A, Mehlen P. The dependence receptor UNC5H2 mediates apoptosis through DAP-kinase. Embo J. 2005;24:1192–201. doi: 10.1038/sj.emboj.7600584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guenebeaud C, Goldschneider D, Castets M, Guix C, Chazot G, Delloye-Bourgeois C, Eisenberg-Lerner A, Shohat G, Zhang M, Laudet V, Kimchi A, Bernet A, Mehlen P. The dependence receptor UNC5H2/B triggers apoptosis via PP2A-mediated dephosphorylation of DAP kinase. Mol Cell. 40:863–76. doi: 10.1016/j.molcel.2010.11.021. [DOI] [PubMed] [Google Scholar]
- 28.Andrieu N, Launoy G, Guillois R, Ory-Paoletti C, Gignoux M. Familial relative risk of colorectal cancer: a population-based study. Eur J Cancer. 2003;39:1904–11. doi: 10.1016/s0959-8049(03)00420-9. [DOI] [PubMed] [Google Scholar]
- 29.Rustgi AK. The genetics of hereditary colon cancer. Genes Dev. 2007;21:2525–38. doi: 10.1101/gad.1593107. [DOI] [PubMed] [Google Scholar]
- 30.Aaltonen LA, Salovaara R, Kristo P, Canzian F, Hemminki A, Peltomaki P, Chadwick RB, Kaariainen H, Eskelinen M, Jarvinen H, Mecklin JP, de la Chapelle A. Incidence of hereditary nonpolyposis colorectal cancer and the feasibility of molecular screening for the disease. N Engl J Med. 1998;338:1481–7. doi: 10.1056/NEJM199805213382101. [DOI] [PubMed] [Google Scholar]
- 31.Salovaara R, Loukola A, Kristo P, Kaariainen H, Ahtola H, Eskelinen M, Harkonen N, Julkunen R, Kangas E, Ojala S, Tulikoura J, Valkamo E, Jarvinen H, Mecklin JP, Aaltonen LA, de la Chapelle A. Population-based molecular detection of hereditary nonpolyposis colorectal cancer. J Clin Oncol. 2000;18:2193–200. doi: 10.1200/JCO.2000.18.11.2193. [DOI] [PubMed] [Google Scholar]
- 32.Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN, Srivastava S. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–57. [PubMed] [Google Scholar]
- 33.Hampel H, Frankel WL, Martin E, Arnold M, Khanduja K, Kuebler P, Nakagawa H, Sotamaa K, Prior TW, Westman J, Panescu J, Fix D, Lockman J, Comeras I, de la Chapelle A. Screening for the Lynch syndrome (hereditary nonpolyposis colorectal cancer) N Engl J Med. 2005;352:1851–60. doi: 10.1056/NEJMoa043146. [DOI] [PubMed] [Google Scholar]
- 34.He H, Liyanarachchi S, Akagi K, Nagy R, Li J, Dietrich RC, Li W, Sebastian N, Wen B, Xin B, Singh J, Yan P, Alder H, Haan E, Wieczorek D, Albrecht B, Puffenberger E, Wang H, Westman JA, Padgett RA, Symer DE, de la Chapelle A. Mutations in U4atac snRNA, a component of the minor spliceosome, in the developmental disorder MOPD I. Science. 2011;332:238–40. doi: 10.1126/science.1200587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Triques K, Piednoir E, Dalmais M, Schmidt J, Le Signor C, Sharkey M, Caboche M, Sturbois B, Bendahmane A. Mutation detection using ENDO1: application to disease diagnostics in humans and TILLING and Eco-TILLING in plants. BMC Mol Biol. 2008;9:42. doi: 10.1186/1471-2199-9-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liechti-Gallati S, Schneider V, Neeser D, Kraemer R. Two buffer PAGE system-based SSCP/HD analysis: a general protocol for rapid and sensitive mutation screening in cystic fibrosis and any other human genetic disease. Eur J Hum Genet. 1999;7:590–8. doi: 10.1038/sj.ejhg.5200338. [DOI] [PubMed] [Google Scholar]
- 37.Rouleau E, Lefol C, Bourdon V, Coulet F, Noguchi T, Soubrier F, Bieche I, Olschwang S, Sobol H, Lidereau R. Quantitative PCR high-resolution melting (qPCR-HRM) curve analysis, a new approach to simultaneously screen point mutations and large rearrangements: application to MLH1 germline mutations in Lynch syndrome. Hum Mutat. 2009;30:867–75. doi: 10.1002/humu.20947. [DOI] [PubMed] [Google Scholar]
- 38.Valle L, Serena-Acedo T, Liyanarachchi S, Hampel H, Comeras I, Li Z, Zeng Q, Zhang HT, Pennison MJ, Sadim M, Pasche B, Tanner SM, de la Chapelle A. Germline allele-specific expression of TGFBR1 confers an increased risk of colorectal cancer. Science. 2008;321:1361–5. doi: 10.1126/science.1159397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Canzian F, Salovaara R, Hemminki A, Kristo P, Chadwick RB, Aaltonen LA, de la Chapelle A. Semiautomated assessment of loss of heterozygosity and replication error in tumors. Cancer Res. 1996;56:3331–7. [PubMed] [Google Scholar]
- 40.Delloye-Bourgeois C, Brambilla E, Coissieux MM, Guenebeaud C, Pedeux R, Firlej V, Cabon F, Brambilla C, Mehlen P, Bernet A. Interference with netrin-1 and tumor cell death in non-small cell lung cancer. J Natl Cancer Inst. 2009;101:237–47. doi: 10.1093/jnci/djn491. [DOI] [PubMed] [Google Scholar]
- 41.Delloye-Bourgeois C, Fitamant J, Paradisi A, Cappellen D, Douc-Rasy S, Raquin MA, Stupack D, Nakagawara A, Rousseau R, Combaret V, Puisieux A, Valteau-Couanet D, Benard J, Bernet A, Mehlen P. Netrin-1 acts as a survival factor for aggressive neuroblastoma. J Exp Med. 2009;206:833–47. doi: 10.1084/jem.20082299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.