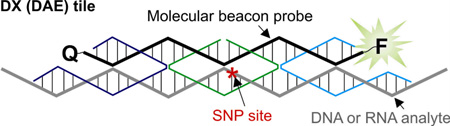
Abstract
A sensor that fluoresces in the presence of specific nucleic acids was designed and characterized. The sensor uses a molecular beacon probe and three adaptor strands to form five-stranded associate, a DX-tile, with a specific analyte. The new sensor can be used as a highly selective and affordable tool for real-time analysis of DNA and RNA.
Keywords: Molecular beacon probe, double crossover motif, DNA nanotechnology, multicomponent DNA sensors, SNP analysis
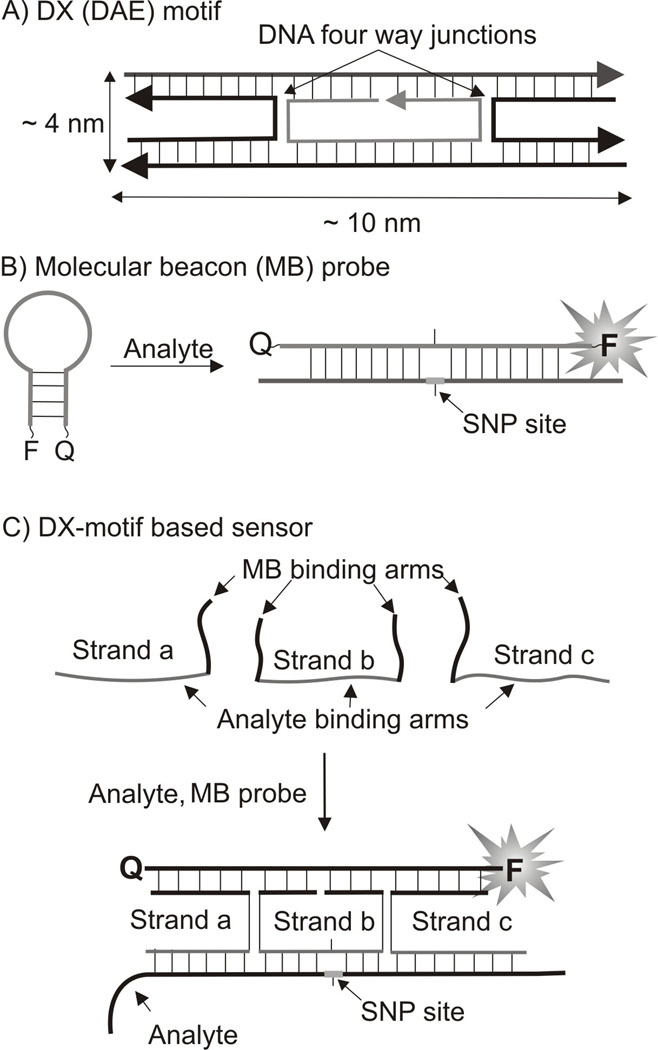
DNA nanotechnology takes advantage of the molecular recognition properties of nucleic acids to assemble DNA strands into predicted structures and shapes[1]. Double crossover DNA associates, also known as DX motifs (or DX tiles),[2] are non-naturally occurring structural elements introduced by DNA nanotechnology. DX tiles are the associates of 4 or 5 DNA strands constituting two DNA helixes linked to each other by two DNA four-way junctions (DNA crossovers). The size of classical tile may vary from 9 × 4 nm to 13 × 4 nm. It was demonstrated that such structures are roughly twice as stiff as regular double stranded DNA.[2b,c] DX motifs are classified based on parallel (DP) and antiparallel (DA)[2a] depending on relative orientation of the two double stranded helical domains. Furthermore, the two crossover points can be separated by the odd (O) or even (E) number of half-helical turns. Here, we explore the potential of the DX structure, namely double-crossover antiparallel even (DAE) motif (Figure 1A) to serve as a sensor for the analysis of specific nucleic acids. The sensor has the following advantages (i) it allows fluorescent detection of specific DNA and RNA analytes in real time; (ii) it is sensitive to single nucleotide differences in analytes at ambient temperatures; (iii) it significantly reduces the cost of multiplex assays.
Figure 1.
The design of a DX motif-based sensor. A) Double-crossover antiparallel even (DAE) DX tile developed by DNA nanotechnology. B) General scheme for hybridization of MB probe to an analysed nucleic acid. C) DX motif–forming sensor for nucleic acid analysis suggested in this study. The adaptor strands are unmodified oligodeoxyribonucleotides. The position of an SNP site is shown in red.
Over the last four decades, nucleic acid hybridization techniques have been widely explored for the detection of specific nucleic acid sequences in such formats as Southern and Northern blots, fluorescent in situ hybridization, polymerase chain reaction (PCR), and DNA microarrays. As a rule, most of the techniques have been using the formation of regular DNA duplexes between probe and the analyzed DNA or RNA. Rear exceptions from this rule include probes that form G-quadruplexes, three-or four-way junctions,[3] all are representatives of naturally occurring structural motifs. In this study we report how an artificially structure developed by DNA nanotechnology, DX motif, can be efficiently utilized for nucleic acid analysis.
The introduction of real-time hybridization probes[4] was an important development in nucleic acid analysis, since it enabled instant fluorescent signal readout after formation of a probe-analyte complex, thus avoiding the need to separate probe-analyte hydride from the excess amount of the unbound probe. A state-of-the-art tool for real-time nucleic acid analysis is molecular beacon (MB) probe.[5] MB probe is a fluorophore- and a quencher-conjugated DNA hairpin (Figure 1B). Binding to a specific analyte separates the fluorophore from the quencher, thus leading to the fluorescence increase. In this study we demonstrate how integration of MB probe into DAE motif improves analysis of specific DNA and RNA sequences.
The DX motif-forming sensor suggested here takes advantage of the three adaptor strands a, b and c, which cooperatively hybridize to both the analyzed nucleic acid and the MB probe and form the DAE structure (Figure 1C). In the resultant DX motif-containing complex, the MB probe is fixed in the elongated conformation, providing the high fluorescent signal. Importantly, the analyte-binding arm of strand b is relatively short, only 10 nucleotides. This enables strand b to form stable hybrid only with a fully complementary analyte. Therefore, if the analyte contains a single base mispairing, the fluorescent signal of the sensor is reduced: this provides the basis for high selectivity of the approach and makes it applicable for molecular diagnostics of single nucleotide polymorphisms (SNP sites).[6] Furthermore, in the case of DX tile-based sensor, the MB probe does not directly hybridize to the analyte. Indirect reporter binding makes it possible to use the same expensive MB probe for the analysis of many nucleic acid sequences, just the sequences of three unmodified and affordable adaptor DNA strands a, b, and c need to be re-designed to recognize new analytes.
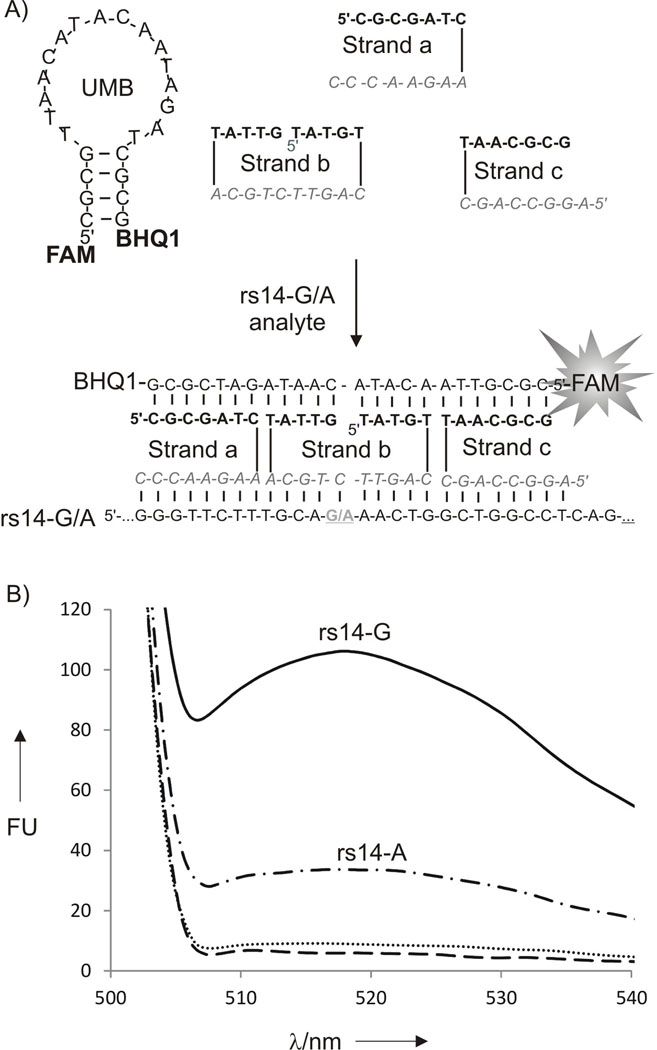
Human polymorphic site rs1490413 (abbreviated as rs14) was chosen as a model analyte (Table S1, Supporting information). This site has been selected earlier as a useful molecular marker for human identification.[7] Figure 2A demonstrates the structure of the tetracomponent probe in the absence (top) or in the presence (bottom) of rs14. Indeed, addition of the fully complementary analyte rs14-G to the tetracomponent probe triggered about 12-fold fluorescence increase (Figure 2B). At the same time, addition of the mismatched rs14-A resulted in a significantly lower fluorescence signal. The discrimination factor, which was calculated according to the formula (Frs14G-F0)/(Frs14A-F0), was about 4. The selectivity of the probe was also confirmed by polyacrilamide gel electrophoresis (Figure S1). The low mobility band corresponding to the DX tile complex was clearly visible in the presence of rs14-G, while only faint band appeared in the presence of rs14-A. The DX motif forming sensor was able to detect as low as ~ 5 nM (Figure S2), which is in the range of that found for regular MB probes.[5,8]
Figure 2.
The DX-tile based sensor for the analysis of human polymorphic site rs1490413 (rs14). A) The probe’s design. Stands a, b and c have analyte-binding arms (bold) and MB-binding arms (italic). The SNP site is underlined. B) Fluorescence response of DX tile–forming to the presence of analyte rs14 A (500 nM) and rs14G (500 nM). Strands a, b and c (500 nM) and UMB (20 nM) were incubated in the absence (dotted line) or presence of 500 nM rs14-A (dash-dotted line) or 500 nM rs14-A (solid line). Control sample contained 20 nM UMB (dashed line) for 15 min at 22°C, and the fluorescence of the sample was measured.
In comparison with the conventional MB approach, however, the DX tile-based sensor is more complex: there are four oligonucleotide strands instead of just one. Such complexity may raise concerns about the rational of using multicomponent sensors. In order to emphasize the advantages of the new approach, we studied the performance of a traditional MB probe, which was designed to recognize the same rs14-G analyte (Figure 3).
Figure 3.
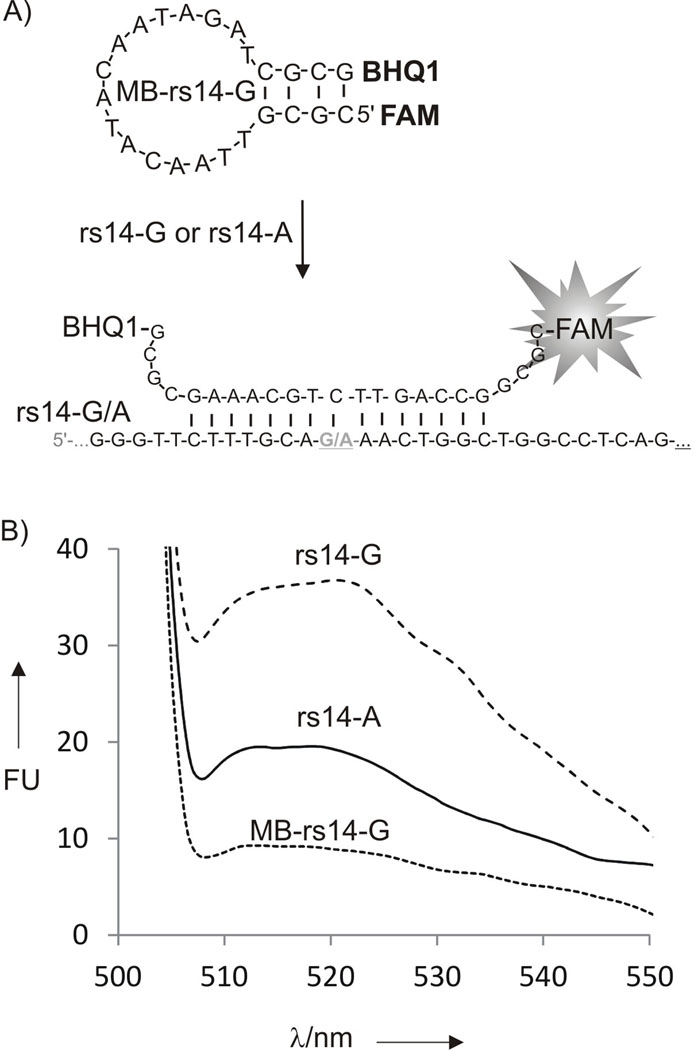
Molecular beacon probe for the analysis of human rs1490413 polymorphic site. A) Hybridization of MB probe to rs14 analyte. B) Fluorescent response of MB-rs14-G (20 nM) in the absence (dotted curve) or presence of complementary 500 nM rs14-G (dashed curve) or single base mismatched 500 nM rs14-A (solid curve) analytes. The fluorescent spectra were taken after 15 min of incubation at 22°C. All experiment have been done in optimal conditions for MB probe performance (concentrations and buffer) with exception that the temperature (22°C) was not optimized for SNP discrimination by MB probe.
MB-rs14 probe contained 15-nucleotide loop complementary to the SNP site of rs14-G analyte and a short but stable stem (Figure 3A). Fully matched rs14-G increased fluorescence less than four times (Figure 3B). The fluorescence signal was twice as high as the background in the presence of the mismatched rs14-A. The calculated discrimination factor was 2.6. Clearly, both signal-to-background ratio and the discrimination factor were lower than that of DX tile-based sensor. It has to be emphasized that the two approaches were compared under different but optimal conditions for each sensor. The optimized conditions included concentrations for each DNA strand and the buffers. One can argue that further optimization of MB probe can result in the improved performance. It is possible, but never guaranteed, since for some DNA or RNA fragments MB probe cannot be designed at all.[9] Moreover, such optimization requires synthesis of a series of expensive MB probes. On the contrary, in DX motif-based approach, the adaptor strands connect the MB probe reporter to the analyzed nucleic acid, thus making MB sequence analyte-independent. Therefore, MB probe can be optimized only once and used for the analysis of almost any RNA or DNA sequences.
To demonstrate this advantage, we designed adaptor strands a, b and c complementary to a different analyte, hepatitis C virus RNA (Hep C RNA, Table S1). The design of the adaptor strands was straightforward: the MB-binding arms were unchanged; the sequences of the analyte-binding arms were of the same length as for rs14 analyte, but made complementary to an arbitrary region of Hep C RNA. The probe detected the specific RNA sequence with the signal-to-background ratio of ~ 37 (Figure S3). Importantly, the specificity of the probe was excellent, as revealed by the addition of ten-fold access of a non-complementary RNA (Figure S3).
Earlier we have explored the advantage of indirect binding of MB probe to analyte by designing a self-assembling probe that uses two adaptor strands to form a DNA four-way junction structure.[8–10] The approach offered an exceptional selectivity in SNP analysis at room temperature. However, such sensor required chemically modified adaptor strands. Out of two possible conformations of the DNA four way junction, the modifications in adaptor strands stabilized the one that contained MB probe in elongated conformation thus maximizing the fluorescence of the DNA associate. Chemical modifications significantly increase the cost of the multiplex method for nucleic acid analysis. Here we took advantage of artificially developed DNA motif in order to reduce the cost of multicomponent sensor.
The important advantage of multicomponent probes that have several analyte binding arms is high specificity towards single nucleotide polymorphisms.[3] Such probes can be designed to recognize ≥20 nucleotide analyte fragment (26 nucleotide in design reported here), which provides excellent specificity in binding of a unique fragment in DNA the size of a genome. At the same time one analyte binding arm of the probe is short enough to be sensitive to a single base miss-match. Indeed, a short hybrid is destabilized by a single mismatch greater than a long one.[11] Thus the split design of the analyte binding arm allows binding of extended region in analyzed DNA/RNA without compromising the high accuracy of SNP analysis.
The purpose of nanotechnology is ‘to put specific molecular species where we want them, when we want them there’.[12] In the present study, we demonstrated how an MB probe can be placed at the edge of DX motif in its elongated conformation only when a specific analyte is present in the solution. The formation of the DX motif is detected in real time by fluorescence without the need to separate the sensor-analyte complex from excess amount of unbound sensor. It this study we provide the evidences that DX motif-based probe is advantageous over a state-of-the art sensor, an MB probe. In particular: (i) the sensor utilizes as a signal reporter the optimal MB probe with the analyte-independent sequence, thus eliminating the need for optimization of the MB probe for each new analyte; (ii) the sensor is more specific to SNP sites at ambient (22°C) temperatures, which opens an opportunity for SNP genotyping without using elevated temperatures or precise temperature control; (iii) the sensor uses unmodified DNA adaptor strands, which makes it affordable in case of analysis of multiple DNA or RNA sequences. This last advantage opens a possibility of significant cost reduction for conventional real-time assays, such as SNP-specific qPCR. Moreover, the approach is adoptable for SNP genotyping in microarray format, if one of the adaptor strands is attached to the solid substrate. Another possible advantage, which was left out of the scope of this study, is the possibility to increase the signal turn on ratio of MB probe incorporated in DX tile due to greater stiffness of the DX tile structure.[2b,c]
In conclusion, this study demonstrates how the developments of DNA nanotechnology can add to the conventional approach for nucleic acid analysis. Other DNA structures and approaches developed by DNA nanotechnology recently,[1] may further fulfill the growing demands in efficient tools for molecular diagnostics.
Experimental Section
All oligonucleotides were custom-made by Integrated DNA Technologies, Inc (Coralville, IA). The complete sequences of all oligonucleotides are shown in Table S1 (Supporting information). For fluorescence assay, UMB (20 nM), strand b (500 nM), strands a and c (600 nM), and the indicated concentrations (1–500 nM) of the analytes were mixed in a buffer containing MgCl2 (50 mM), TrisHCl (50 mM), pH 7.4. For MB-rs14-G assay, the probe (20 nM) was incubated the presence or albescence of rs-14-G or rs14-A oligonucleotides (500 nM) for 15 min the buffer previously optimized for MB probe[14] (100 mM KCl, 1 mM Mg). Fluorescence spectra were recorded on a Perkin-Elmer (San Jose, CA) LS-55 Luminescence Spectrometer with a Hamamatsu xenon lamp (excitation at 485 nm; emission 517 nm) after 15 min of incubation at room temperature (22°C).
Supplementary Material
Acknowledgements
Support from NHGRI R21HG004060 and NSF CCF Division of Computer and Communication Foundations (Award Number 1117205) is greatly appreciated.
Glossary
- SNP
single-nucleotide polymorphism
- MB
molecular beacon
Footnotes
Supporting information for this article is available on the WWW under http://www.angewandte.org or from the author.
References
- 1.a) Seeman NC. Annu. Rev. Biochem. 2010;79:65–87. doi: 10.1146/annurev-biochem-060308-102244. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Krishnan Y, Simmel FC. Angew. Chem. 2011;123:3180–3215. doi: 10.1002/anie.200907223. [DOI] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. Engl. 2011;50:3124–3156. doi: 10.1002/anie.200907223. [DOI] [PubMed] [Google Scholar]; c) Bath J, Turberfield AJ. Nat. Nanotechnol. 2007;2:275–284. doi: 10.1038/nnano.2007.104. [DOI] [PubMed] [Google Scholar]; d) Becerril HA, Woolley AT. Chem. Soc. Rev. 2009;38:329–337. doi: 10.1039/b718440a. [DOI] [PubMed] [Google Scholar]; e) Endo M, Sugiyama H. Chembiochem. 2009;10:2420–2443. doi: 10.1002/cbic.200900286. [DOI] [PubMed] [Google Scholar]; f) Tørring T, Voigt NV, Nangreave J, Yan H, Gothelf KV. Chem. Soc. Rev. 2001 doi: 10.1039/c1cs15057j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.a) Fu TJ, Seeman NC. Biochemistry. 1993;32:3211–3220. doi: 10.1021/bi00064a003. [DOI] [PubMed] [Google Scholar]; b) Kim S, Kim J, Qian P, Shin J, Amin R, Ahn SJ, Labean TH, Kim MK, Park SH. Nanotechnology. 2011;22:245706. doi: 10.1088/0957-4484/22/24/245706. [DOI] [PubMed] [Google Scholar]; c) Sa-Ardyen P, Vologodskii AV, Seeman NC. Biophys J. 2003;84:3829–3837. doi: 10.1016/S0006-3495(03)75110-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kolpashchikov DM. Chem. Rev. 2010;110:4709–4723. doi: 10.1021/cr900323b. [DOI] [PubMed] [Google Scholar]
- 4.a) Morrison LE, Halder TC, Stols LM. Anal. Biochem. 1989;183:231–244. doi: 10.1016/0003-2697(89)90473-9. [DOI] [PubMed] [Google Scholar]; b) Ebata K, Masuko M, Ohtani H, Kashiwasake-Jibu M. Photochem Photobiol. 1995;62:836–839. doi: 10.1111/j.1751-1097.1995.tb09144.x. [DOI] [PubMed] [Google Scholar]; b) Oser A, Valet G. Angew. Chem. Int. Ed. Engl. 1990;29:1167–1168. [Google Scholar]; c) Kolpashchikov DM. J. Am. Chem. Soc. 2005;127 doi: 10.1021/ja0529788. 12442-1243. [DOI] [PubMed] [Google Scholar]
- 5.a) Tyagi S, Kramer FR. Nat. Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]; b) Marras SA, Tyagi S, Kramer FR. Clin. Chim. Acta. 2006;363:48–60. doi: 10.1016/j.cccn.2005.04.037. [DOI] [PubMed] [Google Scholar]; c) Wang K, Tang Z, Yang CJ, Kim Y, Fang X, Li W, Wu Y, Medley CD, Cao Z, Li J, Colon P, Lin H, Tan W. Angew. Chem. 2009;121:870–885. doi: 10.1002/anie.200800370. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. Engl. 2009;48:856–870. doi: 10.1002/anie.200800370. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Li Y, Zhou X, Ye D. Biochem. Biophys. Res. Commun. 2008;373:457–461. doi: 10.1016/j.bbrc.2008.05.038. [DOI] [PubMed] [Google Scholar]; e) Venkatesan N, Seo YJ, Kim BH. Chem. Soc. Rev. 2008;37:648–663. doi: 10.1039/b705468h. [DOI] [PubMed] [Google Scholar]
- 6.a) Stankiewicz P, Lupski JR. Annu. Rev. Med. 2010;61:437–455. doi: 10.1146/annurev-med-100708-204735. [DOI] [PubMed] [Google Scholar]; b) Foley SL, Lynne AM, Nayak R. Infect. Genet. Evol. 2009;9:430–440. doi: 10.1016/j.meegid.2009.03.004. [DOI] [PubMed] [Google Scholar]; c) Bier FF, von Nickisch-Rosenegk M, Ehrentreich-Förster E, Reiss E, Henkel J, Strehlow R, Andresen D. Adv. Biochem. Eng. Biotechnol. 2008;109:433–453. doi: 10.1007/10_2007_087. [DOI] [PubMed] [Google Scholar]; d) Wu CC, Rubinelli P, Lin TL. Avian Dis. 2007;51:515–526. doi: 10.1637/0005-2086(2007)51[515:MDADOI]2.0.CO;2. [DOI] [PubMed] [Google Scholar]; e) Tan JC, Miller BA, Tan A, Patel JJ, Cheeseman IH, Anderson TJ, Manske M, Maslen G, Kwiatkowski DP, Ferdig MT. Genome Biol. 2011;12:R35. doi: 10.1186/gb-2011-12-4-r35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanchez JJ, Phillips C, Børsting C, Balogh K, Bogus M, Fondevila M, Harrison CD, Musgrave-Brown E, Salas A, Syndercombe-Court D, Schneider PM, Carracedo A, Morling N. Electrophoresis. 2006;27:1713–1724. doi: 10.1002/elps.200500671. [DOI] [PubMed] [Google Scholar]
- 8.Gerasimova YV, Hayson A, Ballantyne J, Kolpashchikov DM. Chembiochem. 2010;11:1762–1768. doi: 10.1002/cbic.201000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.a) Browne KA. J. Am. Chem. Soc. 2005;127:1989–1994. doi: 10.1021/ja046369w. [DOI] [PubMed] [Google Scholar]; b) Kim Y, Yang CJ, Tan W. Nucleic Acids Res. 2007;35:7279–7287. doi: 10.1093/nar/gkm771. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Grimes J, Gerasimova YV, Kolpashchikov DM. Angew. Chem. 2010;122:9134–9137. doi: 10.1002/anie.201004475. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. Engl. 2010;49:8950–8953. [Google Scholar]; d) Nguyen C, Grimes J, Gerasimova YV, Kolpashchikov DM. Chemistry: A European J. 2011 doi: 10.1002/chem.201101987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.a) Gerasimova YV, Peck S, Kolpashchikov DM. Chem. Commun. 2010;46:8761–8763. doi: 10.1039/c0cc03248d. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Kolpashchikov DM. J. Am. Chem. Soc. 2006;128:10625–10628. doi: 10.1021/ja0628093. [DOI] [PubMed] [Google Scholar]
- 11.Demidov VV, Frank-Kamenetskii MD. Trends Biochem. Sci. 2004;29:62–71. doi: 10.1016/j.tibs.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 12.Gu H, Chao J, Xiao S-J, Seeman NC. Nature Nanotech. 2009;4:245–248. doi: 10.1038/nnano.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.