SUMMARY
Porphyromonas gingivalis is a low-abundance oral anaerobic bacterium implicated in periodontitis, a polymicrobial inflammatory disease, and the associated systemic conditions. However, the mechanism by which P. gingivalis contributes to inflammation and disease has remained elusive. Here we show that P. gingivalis, at very low colonization levels, triggers changes to the amount and composition of the oral commensal microbiota leading to inflammatory periodontal bone loss. The commensal microbiota and the complement pathway were both required for P. gingivalis-induced bone loss as germ-free mice or conventionally raised C3a and C5a receptor deficient mice did not develop bone loss after inoculation with P. gingivalis. These findings demonstrate that a single, low-abundance species can disrupt host-microbial homeostasis to cause inflammatory disease. The identification and targeting of similar low-abundance pathogens with community-wide impact may be important for treating inflammatory diseases of polymicrobial etiology.
INTRODUCTION
Despite considerable recent attention to the composition of the human microbiome, the mechanisms underlying complex host-bacterial and inter-bacterial interactions that lead to polymicrobial inflammatory diseases remain poorly defined. An attractive model to address this issue is periodontitis, an oral inflammatory disease induced by a multispecies biofilm that is readily accessible for investigation (Pihlstrom et al., 2005).
Porphyromonas gingivalis, a low-abundance oral anaerobic bacterium, is implicated in periodontitis and associated systemic conditions (Desvarieux et al., 2005; Genco and Van Dyke, 2010; Lundberg et al., 2010; Pihlstrom et al., 2005; Tonetti et al., 2007). However, the fundamental mechanism by which P. gingivalis contributes to this polymicrobial disease has remained elusive. Using a murine periodontal model, we show here that P. gingivalis, even in low numbers, can orchestrate inflammatory periodontitis through obligatory interactions with the complement system and the oral commensal microbiota.
Complement is centrally involved in immunity and inflammation through direct effects on immune cells or via crosstalk and regulation of Toll-like receptor and other pathways (Hajishengallis and Lambris, 2010). Although an important component of host immunosurveillance, complement becomes a major link between infection and inflammatory pathology when overactivated or deregulated (Ricklin et al., 2010; Zipfel and Skerka, 2009). The triggering of complement proceeds via distinct cascade mechanisms (classical, lectin, or alternative) which converge at the third complement component (C3) and lead to the generation of effector molecules that mediate recruitment and activation of inflammatory cells (via the anaphylatoxins C3a and C5a), microbial opsonization and phagocytosis (via opsonins such as C3b and iC3b), and direct lysis of targeted microbes (through the C5b-9 membrane attack complex) (Ricklin et al., 2010).
In addition to its established involvement in a number of local or systemic inflammatory or autoimmune diseases (e.g., ischemia/reperfusion injury, systemic lupus erythematosus, and asthma) (Ricklin et al., 2010), complement may also be implicated in periodontitis, as suggested by clinical and histological observations: Chronically inflamed gingiva display significantly elevated complement activity, whereas induction of experimental gingival inflammation in human volunteers causes progressive elevation of complement cleavage products correlating with increased clinical indices of inflammation (Hajishengallis, 2010; Nikolopoulou-Papaconstantinou et al., 1987; Patters et al., 1989; Schenkein and Genco, 1977). However, the precise roles of the various complement pathways in periodontitis have not been defined, and is thus uncertain which ones mediate destructive inflammation, or which ones might conversely mediate protective host responses. In this regard, we have recently shown that C5a receptor-deficient (C5aR−/−) mice, in contrast to wild-type controls, are resistant to experimental periodontitis induced after oral inoculation with P. gingivalis (Liang et al., 2011).
In that study, however, it was reasonable to assume that the inflammatory bone loss was caused directly by P. gingivalis and, moreover, we did not address its possible interactions with the oral commensal microbiota or requirements for its colonization. Here we show that P. gingivalis transiently inhibits chemokine induction, subverts complement, and stably colonizes the murine oral cavity, albeit at very low levels; strikingly, however, low-level colonization by P. gingivalis causes changes to the oral commensal microbiota through mechanisms that depend on the anaphylatoxin receptors (C3aR and C5aR). Collectively, these actions lead to destructive inflammatory disease that requires the presence of the commensal microbiota and intact complement pathways, since P. gingivalis failed to cause periodontitis in germ-free mice or conventionally raised mice deficient in C3aR or C5aR. Thus our data show that a single low-abundance bacterium can instigate pathogenic host-polymicrobial interactions through a normally benign microbiota. These findings may have implications for disorders at intestinal and other mucosal or skin surfaces, where similar locally active immune defenses maintain homeostasis in the presence of a large microbial burden (Darveau, 2010; Grice and Segre, 2011; Hooper and Macpherson, 2010; Slack et al., 2009).
RESULTS
P. gingivalis triggers changes to the amount and composition of the oral microbiota
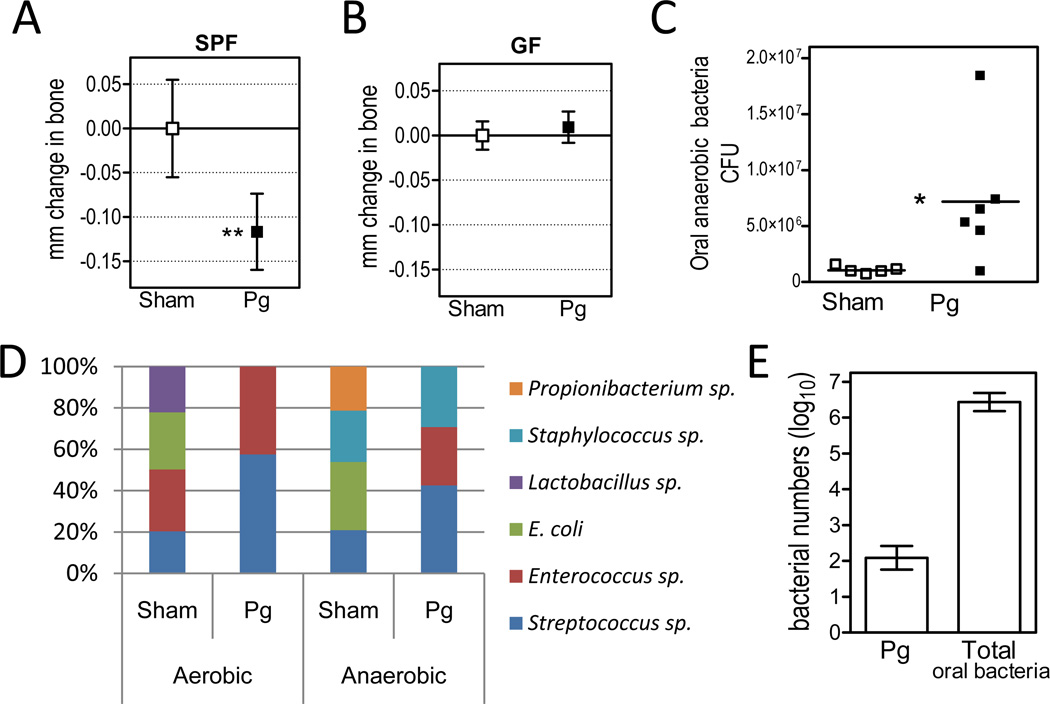
A common approach to study periodontitis in animals is to orally inoculate specific-pathogen-free (SPF) mice with P. gingivalis and after 6 weeks measure periodontal bone loss, the hallmark of this disease (Baker et al., 2000; Graves et al., 2008). Similar to these studies, we found that P. gingivalis caused significant periodontal bone loss compared to sham-treated controls (Figure 1A). In contrast, colonization of germ-free (GF) mice by P. gingivalis failed to induce bone loss (Figure 1B). Furthermore, introduction of P. gingivalis into SPF mice led to elevation of the total cultivatable commensal bacterial load (Figure 1C) and changes to the qualitative composition of this microbiota (Figure 1D). Importantly, the numbers of P. gingivalis were estimated to be 4 to 5 log10 units lower than the total bacterial counts (i.e., <0.01% of the total) on the basis of quantitative real-time PCR (Figure 1E). Accordingly, P. gingivalis was not routinely detectable by culture, although it could be visualized by immunofluorescence microscopy of oral swabs (Figure S1). Hence, although a very minor constituent of the total microbiota, P. gingivalis significantly altered the numbers and community organization of the commensal bacteria, the presence of which were essential for P. gingivalis-induced bone loss.
Figure 1. Oral inoculation of SPF (but not GF) mice with P. gingivalis causes periodontal bone loss and elevation of the commensal bacterial load.
Ten- to twelve-week-old specific-pathogen-free (SPF) or germ-free (GF) C3H mice were orally inoculated with P. gingivalis (Pg) or vehicle only (Sham). Six weeks later, the mice were assessed for periodontal bone loss (A, SPF; B, GF), levels of total cultivatable oral anaerobic bacteria (C), and changes to the qualitative composition of the microbiota detected by aerobic or anaerobic culture (D). (E) Pg and total bacteria were enumerated in the periodontal tissue of Pg-inoculated mice by real-time PCR of the ISPg1 gene (Pg) or the 16S rRNA gene (total oral bacteria). ISPg1 was selected to increase the sensitivity of Pg detection since this gene is present in 31 copies in the Pg genome (the gene copy numbers were thus divided by 31 to obtain genome equivalents). Negative ‘mm change in bone’ values indicate bone loss relative to bone levels of sham mice at the end of the experiment. Lactobacilli were detected in similar numbers following aerobic and anaerobic culture of samples from sham-treated mice, but as they represented a very low percentage of the total anaerobic counts (<0.01%), they do not appear in the anaerobic bar chart in D. In A, B, and E, data are means ± SD (n ≥ 5 mice per group). In C, CFU data are shown for each individual mouse with horizontal lines denoting mean values. In D, each organism was expressed as log10 CFU and shown as a proportion of the total cultured organisms. *p <0.05; **p <0.01 versus corresponding sham control.
Transmission of the commensal oral microbiota to co-caged GF mice causes bone loss
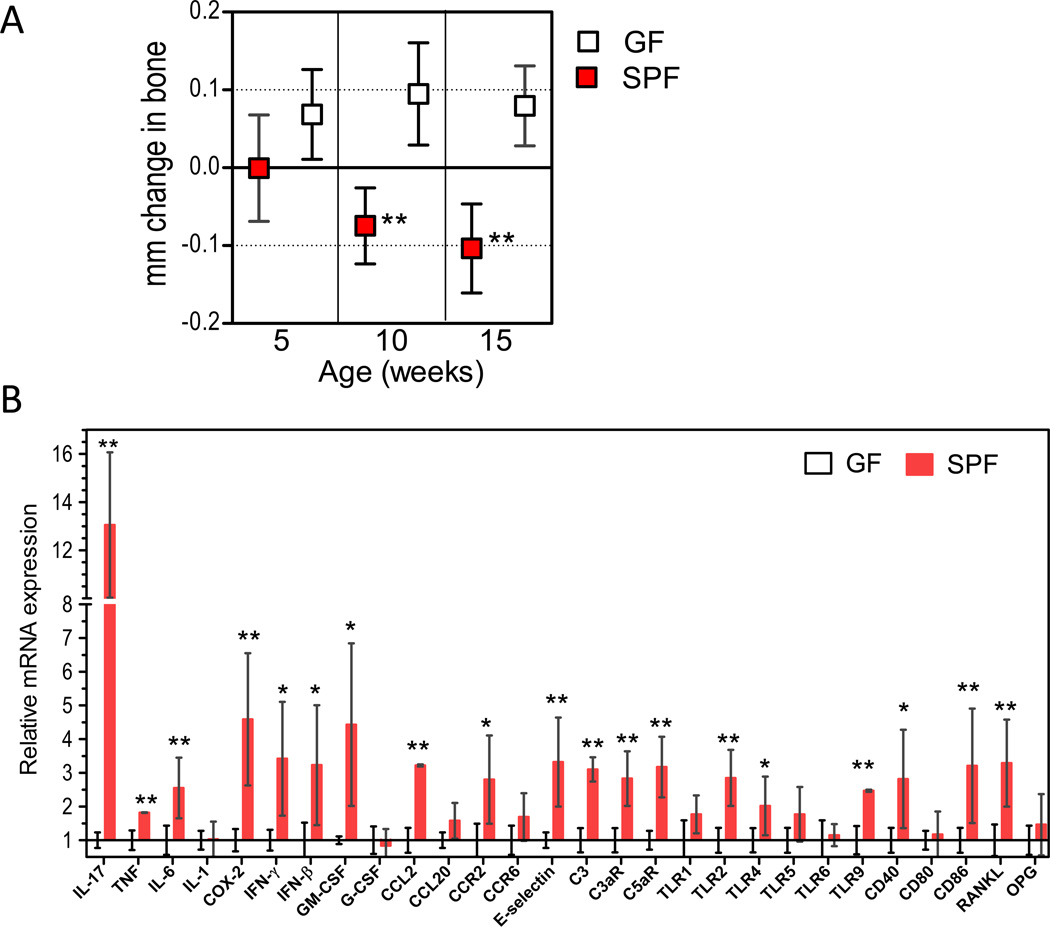
Interestingly, the normal homeostatic relationship between periodontal tissue and the commensal oral microbiota may result in bone loss, albeit at a slower rate. Indeed, periodontal bone measurements revealed modest but statistically significant bone loss in SPF mice compared to GF controls (Figure 2A), accompanied by relatively higher gingival expression of inflammatory mediators, including molecules implicated in bone resorption, such as interleukin (IL)-17, IL-6, cyclooxygenase-2 (COX-2), and receptor activator of nuclear factor-κB ligand (RANKL) (Figure 2B). To firmly conclude that commensal bacteria were responsible for naturally occurring bone loss, we performed co-caging experiments, which allowed the monitoring of the transmission of the oral microbiota. There was a complete transmission of the cultivatable aerobic and anaerobic commensal microbiota from SPF to GF mice within 14 days (Figure 3, A and B). Moreover, 16 weeks after co-caging (but not earlier, at 6 weeks) the conventionalized GF mice developed periodontal bone loss, similar to that seen in age-matched SPF controls (Figure 3C). Therefore, these data, together with the increased expression of inflammatory mediators in SPF compared to GF animals (Figure 2B), suggest that the commensal microbiota can cause modest bone loss through a low-grade inflammatory process. The introduction of P. gingivalis leads to an acceleration of this process such that bone loss seen in 16-week-old mice after 6 weeks colonization by P. gingivalis is equivalent to the bone loss seen in untreated SPF mice at 18 months (Figure S2; ‘18-month’ panel). Taken together, the data in figures 1, 3, and S2 suggest that P. gingivalis causes periodontitis through alterations to the oral commensal microbiota that amplify the intrinsic non-pathologic bone loss elicited by this community.
Figure 2. Natural periodontal bone loss in specific-pathogen-free (SPF) but not germ-free (GF) mice.
(A) SPF and GF mice at the indicated ages were assessed for periodontal bone levels. Negative values indicate bone loss relative to 5-week-old SPF mice. (B) Gingiva were dissected from 10-week-old SPF or GF mice and gingival mRNA levels of the indicated molecules were determined by quantitative real-time PCR (normalized against GAPDH mRNA and expressed as fold change in SPF transcript levels relative to GF transcripts levels, which were assigned an average value of 1). Data are means ± SD (n = 5 to 6 mice per group). *p <0.05; **p <0.01 between age-matched SPF and GF mice.
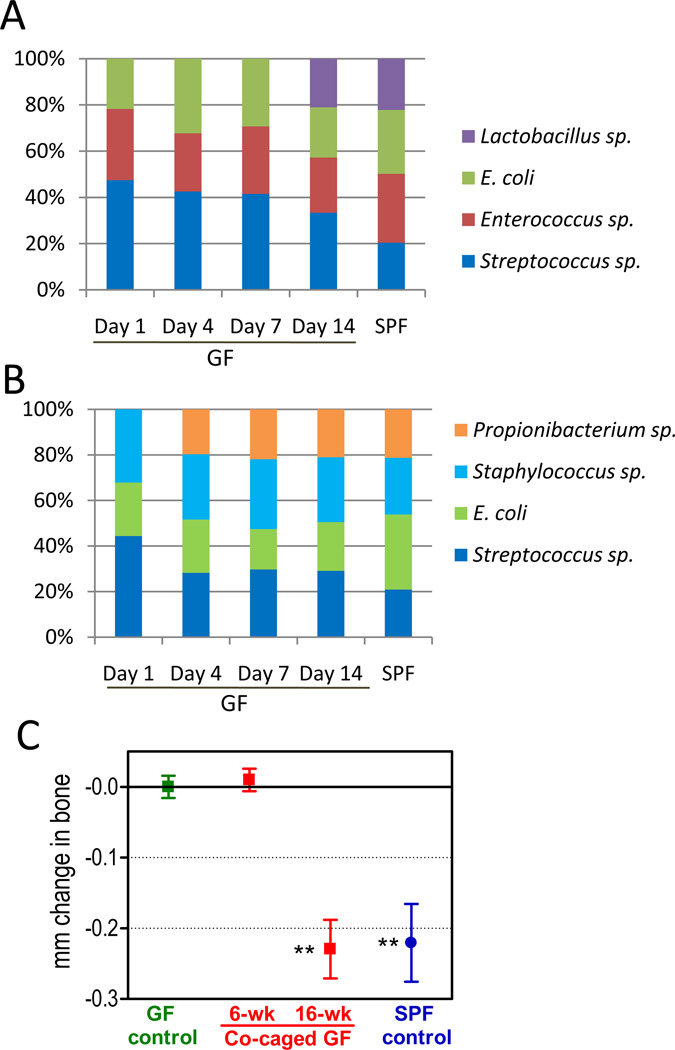
Figure 3. Transmission of the commensal oral microbiota to co-caged GF mice and development of periodontal bone loss after an extended period.
(A–B) Eight- to ten-week-old GF C3H mice were co-caged with SPF C3H mice. Oral swabs were taken at the indicated timepoints and the bacteria were cultured aerobically (A) and anaerobically (B). By day 14, there was complete transmission of the cultivatable oral microbiota from the SPF to the GF mice. (C) GF mice were co-caged with SPF mice and were assessed for periodontal bone levels at 6 and 16 weeks after co-caging. Negative values indicate bone loss relative to bone levels of mice maintained under GF conditions for the entire period (GF control); data are means ± SD (n = 5 to 8 mice per group). **p <0.01 versus GF control.
P. gingivalis exploits complement to elevate the oral bacterial load and instigate bone loss
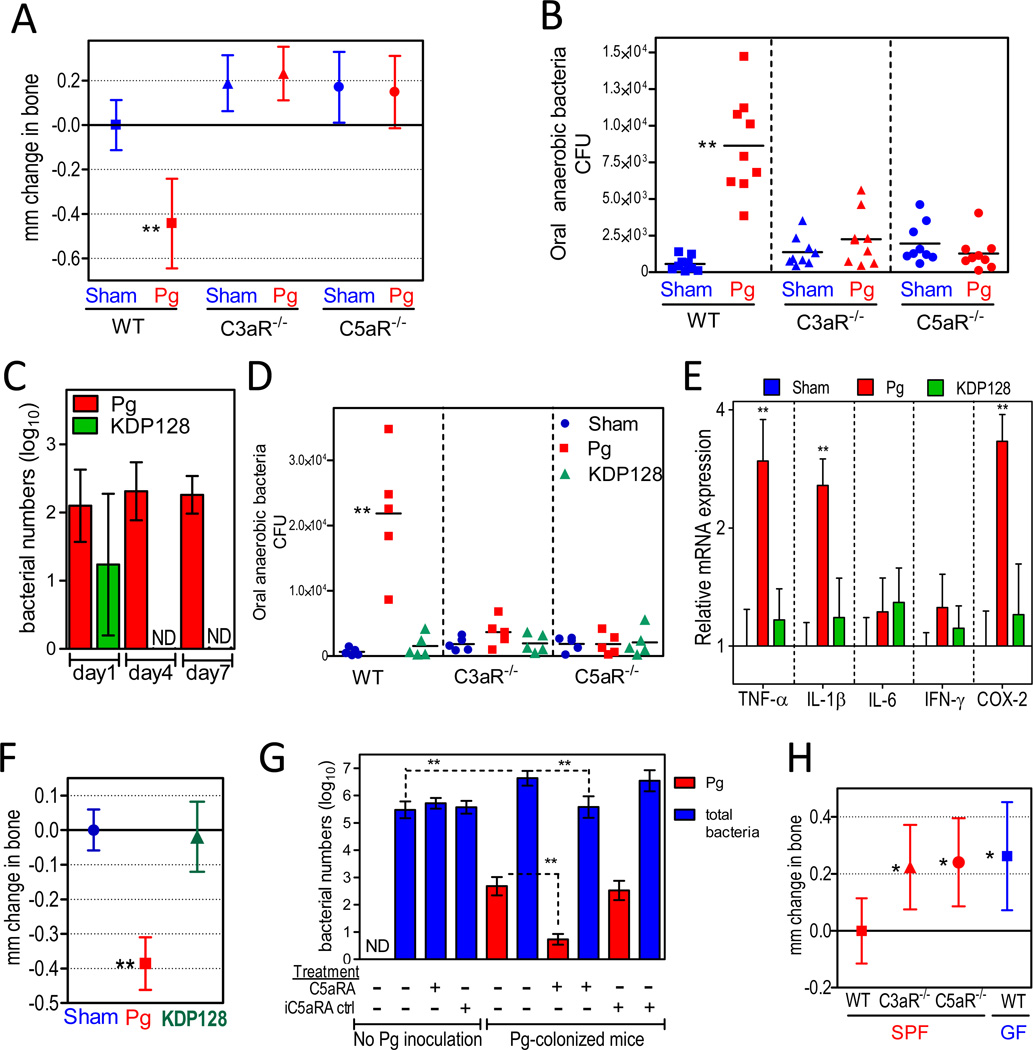
In contrast to normal SPF mice, complement C3a or C5a receptor knock-out mice (C3aR−/− or C5aR−/−) did not develop bone loss after inoculation with P. gingivalis (Figure 4A). Furthermore, no change was observed in the oral commensal microbiota in these knock-out mice after P. gingivalis inoculation (Figure 4B), consistent with the hypothesis that an altered commensal microbiota is required for P. gingivalis-associated bone loss. The inability of P. gingivalis to alter the oral microbiota in C5aR−/− mice may be explained by our demonstration that this pathogen exploits C5aR to inhibit the killing capacity of leukocytes (Liang et al., 2011; Wang et al., 2010). P. gingivalis-affected leukocytes with impaired killing function would likely allow uncontrolled growth of other bacterial species in the same biofilm, which may account for the observed elevation of the total bacterial numbers following P. gingivalis colonization of SPF animals (Figures 1C and 4B). This C5aR-dependent subversive activity of P. gingivalis depends on its expression of cysteine proteases (gingipains), which cleave C5 and generate high local concentrations of C5a (Liang et al., 2011; Wang et al., 2010). Consistent with the importance of gingipains in C5aR-dependent subversion of leukocytes and promotion of bacterial survival (Liang et al., 2011), a gingipain-deficient isogenic mutant (KDP128) was not detectable by 4 days post-inoculation (Figure 4C) and failed to elevate the oral bacterial load even in C5aR-expressing wild-type mice (Figures 4D and S3A). Moreover, mice inoculated with the mutant did not demonstrate increased expression of inflammatory mediators in the periodontal tissue (Figure 4E) nor did they develop bone loss relative to sham-treated controls (Figure 4F).
Figure 4. Complement-dependent elevation of the oral bacterial load and induction of bone loss.
(A–B) Ten- to twelve-week-old BALB/c mice, either wild-type (WT) or deficient in C3aR (C3aR−/−) or C5aR (C5aR−/−), were orally inoculated with P. gingivalis (Pg) or vehicle only (Sham) and assessed for bone loss (A) and levels of oral anaerobic bacteria (B). (C) Quantitative detection, by real-time PCR of the ISPg1 gene, of wild-type Pg or a gingipain-deficient isogenic mutant (KDP128) in the periodontal tissue of inoculated WT BALB/c mice. ND, not detected. (D) WT, C3aR−/−, or C5aR−/− mice were orally inoculated with Pg or KDP128 and assessed for levels of oral anaerobic bacteria seven days later. (E–F) WT mice were inoculated with Pg or KDP128 and euthanized six weeks post-inoculation: Gingiva were dissected and processed for quantitative real-time PCR to determine gingival mRNA expression of the indicated molecules (normalized against GAPDH mRNA levels and presented as fold change relative to sham mice) (E), whereas defleshed maxillae were assessed for periodontal bone loss (F). (G) Effects of C5aR antagonist (C5aRA), or inactive analog control (iC5aRA ctrl), on the numbers of P. gingivalis or total bacteria in the periodontal tissue of mice with or without previous inoculation with P. gingivalis (for details see Experimental Procedures). ND, Pg not detected. (H) Periodontal bone levels in C3aR−/− and C5aR−/− mice relative to WT SPF or GF mice. Data are means ± SD (A, n = 9; C, E, F, and G, n = 5; H, n = 8 mice per group). In A, F, and H, negative values indicate bone loss relative to bone levels of the indicated controls (zero baseline), whereas positive values indicate increased bone levels. CFU counts are shown for each individual mouse with horizontal lines denoting mean values. *p <0.05; **p <0.01 versus corresponding control.
The C3aR requirement for the P. gingivalis effect on the oral microbiota and bone loss may be related to its synergistic interactions with C5aR that include reciprocal augmentation of receptor expression (Ricklin et al., 2010). Accordingly, P. gingivalis-inoculated C3aR−/− mice displayed significantly reduced expression of C5aR (and certain other, but not all, inflammatory receptors examined) compared to P. gingivalis-inoculated wild-type mice (Figure S3B). Therefore, in the absence of C5aR or C3aR (the absence of which diminishes C5aR expression), P. gingivalis may not be able to impair innate immunity, elevate the oral bacterial burden, and promote disease.
Since C5aR is required for the in vivo survival of P. gingivalis (Liang et al., 2011; Wang et al., 2010), we reasoned that local administration of a C5aR antagonist would block the persistence of P. gingivalis, leading to its removal from the periodontal tissue of initially colonized mice. If so, this would also establish whether the experimental removal of P. gingivalis is able to reverse the increase in the total microbial load induced by colonization by this organism (Figures 1C and 4B). As hypothesized, 2 days following local administration of a specific and potent C5aR antagonist (C5aRA; Ac-F[OP(d)Cha-WR]), but not of an inactive analog (iC5aRA; Ac-F[OP(d)Cha-A(d)R]), there was a 2 × log10 reduction in the numbers of P. gingivalis in the periodontal tissue of previously colonized mice (Figure 4G). In essence, the pathogen was almost eliminated (99% reduction). Strikingly, the loss of P. gingivalis was accompanied by significant reduction in the total numbers of oral anaerobic bacteria, which returned to their original levels prior to P. gingivalis colonization (Figure 4G). This reduction in the total microbiota was not a direct effect on the commensal bacteria by C5aRA since the antagonist failed to reduce the total oral bacterial numbers in mice not colonized with P. gingivalis (Figure 4G). These data clearly indicate that the experimental removal of P. gingivalis from the periodontal tissue, similar to its introduction, exerts a major influence on the oral microbiota. In both cases, the effects of P. gingivalis are strictly dependent on complement pathways.
Intriguingly, complement may play additional roles in P. gingivalis-induced periodontitis that are not related to immune subversion. In this regard, we observed that young C3aR−/− and C5aR−/− SPF mice had similar periodontal bone levels to those of age-matched normal GF mice, (Figure 4H), indicating that complement contributes to the commensal microbiota-induced bone loss seen in clinically healthy SPF mice. This complement-dependent process continues throughout the mouse life and by 18 months, mice develop very considerable bone loss relative to age-matched GF mice or C3aR−/− and C5aR−/− SPF mice (Figure S2; ‘18-month’ panel). The failure of P. gingivalis to induce bone loss in C3aR−/− or C5aR−/− SPF mice demonstrates that, in normal SPF mice, P. gingivalis exploits the existing complement-commensal bacteria interaction, a form of host immuno-inflammatory surveillance, to promote disease progression.
Defective leukocyte recruitment disrupts host-microbial homeostasis and causes bone loss
Besides complement exploitation, additional mechanisms of immune subversion may contribute to the ability of P. gingivalis to compromise host defense and promote commensal bacteria-mediated disease. One putative mechanism may involve ‘local chemokine paralysis’ whereby P. gingivalis inhibits IL-8 induction by gingival epithelial cells (Bainbridge et al., 2010; Darveau et al., 1998). IL-8 inhibition may be instigated by P. gingivalis early in the infection process to suppress or delay the recruitment of neutrophils into the oral tissues, thereby facilitating P. gingivalis colonization.
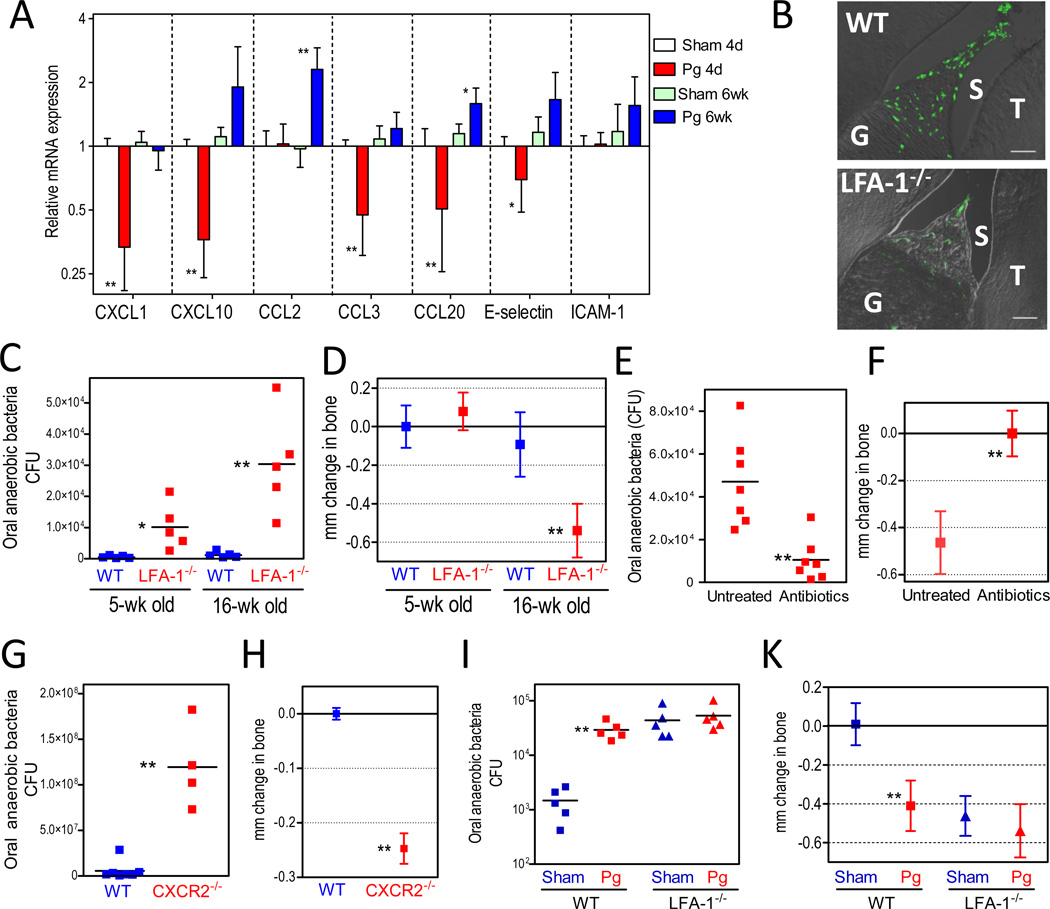
In the present study, the gingival expression of the murine CXCL1 (a functional mouse ortholog of human IL-8) was downregulated– along with certain other chemokines or pertinent adhesion molecules– soon after P. gingivalis oral inoculation (Figure 5A). Chemokine expression returned to baseline or higher levels about 6 weeks later (Figure 5A). Hence we reasoned that deregulation of leukocyte recruitment may have contributed to P. gingivalis-induced periodontitis in this murine model. We therefore examined whether mice genetically deficient in leukocyte recruitment would also display increased total numbers of oral bacteria and bone loss equivalent to that observed in P. gingivalis-colonized normal mice. Mice lacking leukocyte function-associated antigen-1 (LFA-1) had reduced neutrophil infiltration (Figure 5B), harbored a higher commensal bacterial load (Figure 5C), and developed significant periodontal bone loss relative to wild type controls (Figure 5D). The spontaneous periodontitis seen in LFA-1−/− mice mirrors the increased prevalence and severity of periodontitis in individuals with leukocyte adhesion deficiency type 1 (Darveau, 2010). Oral antibiotic treatment of LFA-1−/− mice resulted in significant reduction of the commensal bacterial load (Figure 5E) and prevented the development of periodontal bone loss (Figure 5F), confirming the requirement of the commensal microbiota in this process. Furthermore, and similar to LFA-1−/− mice, mice deficient in CXCR2, the chemokine receptor for CXCL1, also displayed elevated levels of commensal bacteria and periodontal bone loss relative to wild-type controls (Figure 5, G and H), confirming the key role played by an intact leukocyte recruitment machinery in protection of the periodontal tissue.
Figure 5. Compromised homeostasis in the periodontal tissue.
(A) BALB/c mice were orally inoculated with P. gingivalis (Pg) or vehicle only (sham) and sacrificed 4 days or 6 weeks post-inoculation. Dissected gingiva were processed for real-time PCR to determine mRNA expression of the indicated molecules (normalized against GAPDH mRNA and shown as fold change relative to 4-day sham-infected mice). Data are means ± SD (n = 5 mice per group). (B) Diminished recruitment of neutrophils to the gingiva of LFA-1−/− mice. Sagittal sections of periodontal tissue from 5-week-old wild-type (WT) or LFA-1−/− mice were stained for the neutrophil marker LyG6. Shown are representative overlays of differential interference contrast and fluorescent confocal images. G, gingiva; S, sulcus; T, teeth. (C–D) WT and LFA-1−/− mice were assessed for levels of oral anaerobic bacteria (C) and periodontal bone loss (D) as they aged from 5 to 16 weeks. (E–F) LFA-1−/− mice given antibiotics-containing drinking water or plain water (control) were assessed for levels of oral anaerobic bacteria (E) and bone loss (F). (G–H) Eighteen- to twenty-week-old WT and CXCR2−/− mice were assessed for oral anaerobic bacteria (G) and periodontal bone levels (H). (I–K) Ten-week-old WT or LFA-1−/− mice were orally inoculated with P. gingivalis (Pg) or vehicle only (Sham) and six weeks later were assessed for levels of oral anaerobic bacteria (I) and bone loss (K). CFU are shown for each mouse and horizontal lines denote mean values. Bone loss data are means ± SD (n = 5 to 7 mice per group). *p <0.05; **p <0.01 versus corresponding control.
Finally, P. gingivalis inoculation of LFA-1−/− mice had no additive effects on either the total microbial count or bone loss compared to unchallenged LFA-1−/− mice (Figure 5, I and K). Hence, together with the earlier findings (Figures 1 and 4), these observations indicate that pathogenic bone loss is caused by the commensals when the host defenses are compromised by either genetic deficiencies or by the deregulatory effects of P. gingivalis.
DISCUSSION
In this paper, we show that a single pathogen of low abundance can instigate pathogenic host-polymicrobial interactions through the normally benign oral microbiota. Specifically, oral colonization by P. gingivalis can exploit or modify the host response in ways that alter the amount and composition of the oral microbial community, leading to destructive inflammatory disease. The fundamental mechanism by which P. gingivalis causes periodontitis in this model therefore involves disruption of host homeostasis. When the homeostasis is already compromised (e.g., due to congenital immunodeficiencies), the role of P. gingivalis becomes minimal.
Although the ability of P. gingivalis to initiate periodontitis was established in a study using a nonhuman primate model (Holt et al., 1988), the findings were interpreted as evidence for a specific microbial etiology. However, retrospective analysis of another study in the same model is consistent with our findings that P. gingivalis exerts growth-enhancing effects on the oral microbiota: Immunization of nonhuman primates (Macaca fascicularis; a natural host for P. gingivalis) with a gingipain-based vaccine reduced both P. gingivalis and the total subgingival bacterial load and protected against bone loss (Page et al., 2007). Moreover, exogenous infection of these animals with P. gingivalis led to an elevation in the total subgingival bacterial load (Page et al., 2007). Here, taking advantage of the analytical tools available in the mouse model, we elucidated the mechanism underlying the role of P. gingivalis in periodontal pathogenesis. Specifically, we demonstrated that this pathogen exploits complement and compromises leukocyte recruitment to induce inflammatory periodontal bone loss through an altered oral microbiota.
In this regard, P. gingivalis uses C5aR for intercepting, through subversive crosstalk, otherwise protective Toll-like receptor−mediated antimicrobial signaling pathways, thereby leading to impairment of the capacity of leukocytes to control bacteria (Hajishengallis and Lambris, 2011; Wang et al., 2010). Consistent with this, we now showed that C5aR was essential for the ability of P. gingivalis to induce elevation of the total numbers of the oral microbiota. Such changes to the microbiota were not observed when mice were inoculated with a gingipain-deficient mutant, which cannot cleave C5 to generate locally high levels of C5a for C5aR activation (Liang et al., 2011).
Intriguingly, P. gingivalis promoted disease by exploiting the proinflammatory properties of complement, through which the altered commensal microbiota caused pathogenic bone loss in young mice. In the absence of P. gingivalis, the oral commensal microbiota induced modest, complement-dependent bone loss in young mice under a controlled inflammatory state: a state which also characterizes clinically healthy human periodontal tissue (Darveau, 2010) and is analogous to the immuno-inflammatory surveillance state described for the healthy intestine (Hooper and Macpherson, 2010; Slack et al., 2009). However, the altered composition and increased numbers of the oral microbiota in P. gingivalis-colonized mice appeared to amplify complement-dependent inflammation and accelerate the bone loss. Indeed, the bone loss induced by P. gingivalis in 6 weeks was comparable to that induced by the oral microbiota alone over a period of 18 months. The bone loss seen in 18-month-old SPF mice was significantly more than in age-matched C3aR−/− and C5aR−/− SPF mice or normal GF mice, the bone loss of which may be due to age-related hormonal changes that influence bone metabolism (Syed and Hoey, 2010).
Periodontal health in humans is critically dependent upon leukocyte surveillance of the oral tissues and a coordinated gradient of IL-8 characterizes clinically healthy but not diseased gingival tissue (Darveau, 2010; Tonetti et al., 1998). The ability of P. gingivalis to inhibit the induction of gingival IL-8 or IL-8-like chemokines in humans and rodents (Bainbridge et al., 2010; Darveau et al., 1998; this study, Figure 5A)– even transiently– would be anticipated to delay the recruitment of neutrophils and, thereby, facilitate its initial colonization of the periodontal tissues. This inhibitory effect of P. gingivalis on IL-8 is mediated by a secreted serine phosphatase (SerB) and, accordingly, a SerB-deficient isogenic mutant induces higher neutrophil recruitment to the periodontal tissue (Bainbridge et al., 2010). Following these initial events, the ability of P. gingivalis to persist in the periodontal tissue may depend upon expression of the gingipain proteases and exploitation of C5aR, as outlined above, in order to escape the clearance mechanisms of neutrophils that will eventually be recruited in response to the increased microbial burden. Consistent with this scheme of events, a gingipain-deficient mutant, which is nevertheless proficient in SerB expression, appeared able to colonize but was subsequently eliminated from the periodontal tissue.
Following establishment of P. gingivalis, we envision the following cascade of events: colonization at low levels impairs innate immunity and leads to increased numbers of oral commensal bacteria and thereby enhanced inflammation. The inflammatory environment is favorable to further bacterial growth since the gingival inflammatory exudate is a rich source of nutrients, such as degraded host proteins and hemin, a source of essential iron (Darveau, 2010; Hajishengallis, 2009). This, moreover, can alter the composition of the oral microbiota favoring those bacteria (e.g., proteolytic and asaccharolytic organisms) that can better exploit these environmental changes. These changes result in even higher inflammation and bone resorption, leading to increased niche space (deeper periodontal pockets) for the bacteria.
In contrast to our findings, previous studies by an independent group have shown that P. gingivalis could cause bone loss in germ-free rats (Evans et al., 1992; Malek et al., 1994). In those studies, however, the P. gingivalis infecting dose was approximately 1000-fold higher than we used in the mouse model (0.75 × 1012 bacterial cells vs. 109 in our study). It is possible that such high doses of P. gingivalis may outweigh any requirement for cooperation with the commensal microbiota to induce bone loss. However, at the low levels of P. gingivalis observed in this investigation, and indeed frequently in human periodontitis, a combined action of this bacterium and other bacteria may be required to induce bone loss.
P. gingivalis was not detectable in stable/healthy clinical states and comprised only 0.8% of total clones in active human periodontitis (Kumar et al., 2006), although its presence has been significantly correlated with progressive bone loss (Chaves et al., 2000). Similarly low levels of P. gingivalis have been reported in another human study (predominantly found at ≤ 0.03 of the total microbiota) (Doungudomdacha et al., 2000) and in the mouse periodontitis model (Pathirana et al., 2007), consistent with our findings that P. gingivalis was detected in the murine periodontal tissue at least 4 log10 units lower than the total oral bacteria.
By analogy to the ‘keystone species’ concept in macro-ecology, i.e., low-abundance species with a major supporting role for an entire ecological community (Brown et al., 2001; Ebenman and Jonsson, 2005; Power et al., 1996), P. gingivalis may be regarded as such a species by fulfilling the criteria of low abundance and major influence on the microbial community. This notion is further substantiated by the demonstrated impact on the oral microbiota caused by the experimental removal of P. gingivalis from the biofilm. Indeed, local administration of a C5aR antagonist in the gingiva of P. gingivalis-colonized mice resulted in essentially complete elimination of the pathogen and a >10-fold suppression of the oral microbiota, which returned to its original numbers prior to P. gingivalis colonization. This suggests that the continuous presence of low colonization levels of P. gingivalis is required for maintaining high numbers of the oral microbiota which are directly correlated with enhanced inflammatory bone loss. Hence, P. gingivalis appears to meet the definition of a ‘keystone species’ in that its supportive impact on the community is ‘disproportionately large relative to its abundance’ (Brown et al., 2001; Ebenman and Jonsson, 2005; Power et al., 1996). This role contrasts with that of dominant species which normally provide the major energy flow in an ecosystem. Moreover, P. gingivalis could be characterized as a ‘keystone pathogen’, defined as a keystone species which supports and shapes a microbial community in ways that also promote disease pathogenesis.
Enteric pathogens have also been shown to exert global changes on the gut microbiota in rodent models of inflammatory disease (Garrett et al., 2010; Lupp et al., 2007; Stecher et al., 2007). However, in these examples, the infecting organism normally becomes established as the dominant component of the gut microbiota and causes suppression of the commensals. Conversely, in the case of P. gingivalis, its presence at low levels appears to exert a stimulatory effect on the growth of the oral commensal microbiota. Therefore, P. gingivalis exhibits some unique characteristics which support its characterization as a keystone pathogen. Importantly, our identification of specific host receptors required for the subversive effects of P. gingivalis not only provide a mechanistic basis for our observations, but also suggest approaches for inhibiting the disease by targeted intervention (e.g., C5aR antagonists).
Although the composition of the human microbiome has received considerable attention in recent years, the precise mechanisms whereby the microbial communities mediate disease or protection from it remain largely uncertain. Our findings demonstrate that an inflammatory disease can be caused through dysregulation of host-polymicrobial interactions instigated by a single species that appears to act as a keystone pathogen. Changes in the composition of the gut microbiota have been implicated in inflammatory bowel disease, colon cancer, obesity, diabetes, and coronary heart disease (Bloom et al., 2011; Kinross et al., 2011; Saleh and Trinchieri, 2011). The identification and targeting of possible keystone pathogens may help treat or prevent inflammatory diseases of polymicrobial etiology.
EXPERIMENTAL PROCEDURES
Mice
All animal procedures described in this study were approved by the institutional animal care and use committees, in compliance with established federal and state policies. Germ-free (GF) C3H/Orl mice (Charles River Laboratories International) or BALB/c mice (The Jackson Laboratory) were maintained in isolators at the Royal Veterinary College, University of London, or the Germ-free Animal Research Facility at the Center for Oral Health Research in the Medical University of South Carolina, respectively. The sterility of GF animals was examined by aerobic and anaerobic culture of oral swabs and fecal pellets on non-selective media and by PCR using universal 16S primers. Specific-pathogen-free (SPF) mice (below) were maintained in individually ventilated cages at the animal care facilities of Queen Mary University of London and the University of Louisville. C3aR−/−, C5aR−/−, CXCR2−/− and wild-type mice on BALB/c background were from The Jackson Laboratory. LFA-1−/− mice on C57BL/6 background were generously provided by Dr. C.M. Ballantyne (Baylor College of Medicine) (Ding et al., 1999). C3H/HeNcrl mice were from Charles River Laboratories International. In co-caging experiments, SPF and GF mice were co-caged at a ratio of 1:3.
Periodontitis model
Periodontal bone loss was induced in mice by oral inoculation with P. gingivalis, as originally described by Baker (Baker et al., 2000) with slight modifications (Wang et al., 2007). Briefly, by means of a ball-ended feeding needle, mice were orally inoculated three times at 2-day intervals with 109 CFU P. gingivalis (ATCC 33277 or W50) suspended in 2% carboxy-methylcellulose vehicle. Sham controls received vehicle alone. The mice were euthanized six weeks after the last oral inoculation. Real-time PCR of the ISPg1 gene (Naito et al., 2008) was used to quantify the levels of P. gingivalis in harvested periodontal tissue. P. gingivalis was also detected in oral swabs by immunofluorescence using MAb-1B5 (Paramonov et al., 2005). Assessment of periodontal bone loss in defleshed maxillae was performed under a dissecting microscope (x40) fitted with a video image marker measurement system (VIA-170K; Boeckeler Instruments). Specifically, the distance from the cementoenamel junction (CEJ) to the alveolar bone crest (ABC) was measured on 14 predetermined points on the buccal surfaces of the maxillary molars. To calculate bone loss, the 14-site total CEJ-ABC distance for each mouse was subtracted from the mean CEJ-ABC distance of sham-infected mice (Baker et al., 2000). The results were expressed in mm and negative values indicated bone loss relative to sham controls. The same method was used to measure the bone levels in GF versus SPF mice or in wild-type versus gene-knockout mice. In certain experiments, bone loss was measured in mice which were administered antibiotics in their drinking water (sulfamethoxazole and trimethoprim at a final concentration of 800 µg/ml and 400 µg/ml, respectively). To assess the oral microbial burden, the murine oral cavity was sampled for 30 seconds using sterile swabs or paperpoints. Serial dilutions of the extracts were plated onto blood agar plates for aerobic and anaerobic growth and CFU determination. In certain experiments, the predominant cultivatable organisms were purified by subculture and identified by 16S ribosomal RNA sequencing.
C5aR antagonist intervention
A specific and potent C5aR antagonist (C5aRA), the cyclic hexapeptide Ac-F[OP(d)Cha-WR] (acetylated phenylalanine–[ornithine-proline-(d)cyclohexylalanine-tryptophan-arginine]), and an inactive analog (iC5aRA), Ac-F[OP(d)Cha-A(d)R] (acetylated phenylalanine–[ornithine-proline-(d)cyclohexylalanine-alanine-(d)arginine]), were synthesized in the laboratory of one of the coauthors (JDL) as previously described (Finch et al., 1999; Markiewski et al., 2008). Groups of mice were orally inoculated or not with P. gingivalis (as described above) and 7 days later were injected with C5aRA, or iC5aRA control, into the palatal gingiva, on the mesial of the first molar and in the papillae between first and second and third molars on both sides of the maxilla (1 µl of 1 µg per site; total of six sites). Two days later (day 9), the mice were sacrificed and maxillary periodontal tissue was harvested to determine the levels of P. gingivalis colonization and the number of total oral bacteria, using quantitative real-time PCR of the ISPg1 gene (P. gingivalis) or the 16S rRNA gene (total oral bacteria). Two groups of mice, one of which was inoculated with P. gingivalis, were not treated with C5aRA or iC5aRA at day 7 and were sacrificed the same day for determining the levels of P. gingivalis and of total oral bacteria prior to the C5aRA or iC5aRA interventions.
Quantitative real-time PCR
Gingival tissue was excised from around the maxillary molars. For performing real-time PCR of host genes, total RNA was extracted by using the PerfectPure RNA cell kit (5 Prime, Fisher) and quantified by spectrometry at 260 and 280 nm. The RNA was reverse-transcribed using the High-Capacity cDNA Archive kit (Applied Biosystems) and real-time PCR with cDNA was performed using the ABI 7500 Fast System, according to the manufacturer's protocol (Applied Biosystems). For performing real-time PCR of bacterial genes, genomic DNA was isolated from maxillary periodontal tissue (including both soft and hard tissue) using the DNeasy kit (Qiagen) and was quantified by spectrometry at 260 and 280 nm. TaqMan probes, sense primers, and antisense primers for detection and quantification of host or bacterial genes by real-time PCR were purchased from Applied Biosystems.
Immunohistochemistry
Upper jaws (maxillae) with intact surrounding tissue were fixed in 4% paraformaldehyde, decalcified in Immunocal Solution (Decal Chemical Corp.) for 15 days, and embedded in OCT compound. Serial mesio-distal sections (7- to 8-µm thick) parallel to the long axis of the teeth (sagittal) were stained with FITC-conjugated monoclonal antibody to mouse Ly6G (LifeSpan BioSciences), a specific neutrophil marker (Daley et al., 2008). The specificity of staining was confirmed by using FITC-conjugated isotype control (rat IgG2b). Images were captured using a laser-scanning confocal microscope (Olympus FV1000).
Statistical analysis
Data were evaluated by analysis of variance and the Dunnett multiple-comparison test using the InStat program (GraphPad Software, San Diego, CA). Where appropriate (comparison of two groups only), two-tailed t tests were performed. P < 0.05 was taken as the level of significance. All experiments were performed at least twice for verification.
HIGHLIGHTS.
P. gingivalis induces changes to the amount and composition of the oral microbiota
P. gingivalis causes periodontitis in conventional but not germ-free mice
Complement is also required for P. gingivalis-induced periodontitis
Defective leukocyte recruitment disrupts host-microbial homeostasis and causes bone loss
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the NIH (DE015254, DE018292, and DE021580 to GH; DE018274 to RPD; GM062134 and AI068730 to JDL; P20RR017696 to KLK), the U.S. Department of Defense (W81XWH-07-P-0481 to RPD), and the Medical Research Council (UK) (G0900408 to MAC). We thank Stephen Lory (Harvard Medical School) and Thomas T. MacDonald (Barts and the London School of Medicine) for critical review of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bainbridge B, Verma RK, Eastman C, Yehia B, Rivera M, Moffatt C, Bhattacharyya I, Lamont RJ, Kesavalu L. Role of Porphyromonas gingivalis phosphoserine phosphatase enzyme SerB in inflammation, immune response, and induction of alveolar bone resorption in rats. Infect. Immun. 2010;78:4560–4569. doi: 10.1128/IAI.00703-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker PJ, Dixon M, Roopenian DC. Genetic control of susceptibility to Porphyromonas gingivalis-induced alveolar bone loss in mice. Infect. Immun. 2000;68:5864–5868. doi: 10.1128/iai.68.10.5864-5868.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom SM, Bijanki VN, Nava GM, Sun L, Malvin NP, Donermeyer DL, Dunne WM, Jr, Allen PM, Stappenbeck TS. Commensal Bacteroides species induce colitis in host-genotype-specific fashion in a mouse model of inflammatory bowel disease. Cell Host Microbe. 2011;9:390–403. doi: 10.1016/j.chom.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JH, Whitham TG, Morgan Ernest SK, Gehring CA. Complex species interactions and the dynamics of ecological systems: long-term experiments. Science. 2001;293:643–650. doi: 10.1126/science.293.5530.643. [DOI] [PubMed] [Google Scholar]
- Chaves ES, Jeffcoat MK, Ryerson CC, Snyder B. Persistent bacterial colonization of Porphyromonas gingivalis, Prevotella intermedia, and Actinobacillus actinomycetemcomitans in periodontitis and its association with alveolar bone loss after 6 months of therapy. J. Clin. Periodontol. 2000;27:897–903. doi: 10.1034/j.1600-051x.2000.027012897.x. [DOI] [PubMed] [Google Scholar]
- Daley JM, Thomay AA, Connolly MD, Reichner JS, Albina JE. Use of Ly6G-specific monoclonal antibody to deplete neutrophils in mice. J. Leukoc. Biol. 2008;83:64–70. doi: 10.1189/jlb.0407247. [DOI] [PubMed] [Google Scholar]
- Darveau RP. Periodontitis: a polymicrobial disruption of host homeostasis. Nat. Rev. Microbiol. 2010;8:481–490. doi: 10.1038/nrmicro2337. [DOI] [PubMed] [Google Scholar]
- Darveau RP, Belton CM, Reife RA, Lamont RJ. Local chemokine paralysis, a novel pathogenic mechanism for Porphyromonas gingivalis. Infect. Immun. 1998;66:1660–1665. doi: 10.1128/iai.66.4.1660-1665.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desvarieux M, Demmer RT, Rundek T, Boden-Albala B, Jacobs DR, Jr, Sacco RL, Papapanou PN. Periodontal microbiota and carotid intima-media thickness: the Oral Infections and Vascular Disease Epidemiology Study (INVEST) Circulation. 2005;111:576–582. doi: 10.1161/01.CIR.0000154582.37101.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding ZM, Babensee JE, Simon SI, Lu H, Perrard JL, Bullard DC, Dai XY, Bromley SK, Dustin ML, Entman ML, et al. Relative contribution of LFA-1 and Mac-1 to neutrophil adhesion and migration. J. Immunol. 1999;163:5029–5038. [PubMed] [Google Scholar]
- Doungudomdacha S, Rawlinson A, Douglas CW. Enumeration of Porphyromonas gingivalis, Prevotella intermedia and Actinobacillus actinomycetemcomitans in subgingival plaque samples by a quantitative-competitive PCR method. J. Med. Microbiol. 2000;49:861–874. doi: 10.1099/0022-1317-49-10-861. [DOI] [PubMed] [Google Scholar]
- Ebenman B, Jonsson T. Using community viability analysis to identify fragile systems and keystone species. Trends Ecol. Evol. 2005;20:568–575. doi: 10.1016/j.tree.2005.06.011. [DOI] [PubMed] [Google Scholar]
- Evans RT, Klausen B, Sojar HT, Bedi GS, Sfintescu C, Ramamurthy NS, Golub LM, Genco RJ. Immunization with Porphyromonas (Bacteroides) gingivalis fimbriae protects against periodontal destruction. Infect. Immun. 1992;60:2926–2935. doi: 10.1128/iai.60.7.2926-2935.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch AM, Wong AK, Paczkowski NJ, Wadi SK, Craik DJ, Fairlie DP, Taylor SM. Low-molecular-weight peptidic and cyclic antagonists of the receptor for the complement factor C5a. J. Med. Chem. 1999;42:1965–1974. doi: 10.1021/jm9806594. [DOI] [PubMed] [Google Scholar]
- Garrett WS, Gallini CA, Yatsunenko T, Michaud M, DuBois A, Delaney ML, Punit S, Karlsson M, Bry L, Glickman JN, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genco RJ, Van Dyke TE. Prevention: Reducing the risk of CVD in patients with periodontitis. Nat. Rev. Cardiol. 2010;7:479–480. doi: 10.1038/nrcardio.2010.120. [DOI] [PubMed] [Google Scholar]
- Graves DT, Fine D, Teng YT, Van Dyke TE, Hajishengallis G. The use of rodent models to investigate host-bacteria interactions related to periodontal diseases. J. Clin. Periodontol. 2008;35:89–105. doi: 10.1111/j.1600-051X.2007.01172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice EA, Segre JA. The skin microbiome. Nat Rev Microbiol. 2011;9:244–253. doi: 10.1038/nrmicro2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G. Porphyromonas gingivalis-host interactions: open war or intelligent guerilla tactics? Microbes Infect. 2009;11:637–645. doi: 10.1016/j.micinf.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G. Complement and periodontitis. Biochem. Pharmacol. 2010;80:1992–2001. doi: 10.1016/j.bcp.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Lambris JD. Crosstalk pathways between Toll-like receptors and the complement system. Trends Immunol. 2010;31:154–163. doi: 10.1016/j.it.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Lambris JD. Microbial manipulation of receptor crosstalk in innate immunity. Nat. Rev. Immunol. 2011;11:187–200. doi: 10.1038/nri2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt SC, Ebersole J, Felton J, Brunsvold M, Kornman KS. Implantation of Bacteroides gingivalis in nonhuman primates initiates progression of periodontitis. Science. 1988;239:55–57. doi: 10.1126/science.3336774. [DOI] [PubMed] [Google Scholar]
- Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat. Rev. Immunol. 2010;10:159–169. doi: 10.1038/nri2710. [DOI] [PubMed] [Google Scholar]
- Kinross JM, Darzi AW, Nicholson JK. Gut microbiome-host interactions in health and disease. Genome Med. 2011;3:14. doi: 10.1186/gm228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar PS, Leys EJ, Bryk JM, Martinez FJ, Moeschberger ML, Griffen AL. Changes in periodontal health status are associated with bacterial community shifts as assessed by quantitative 16S cloning and sequencing. J. Clin. Microbiol. 2006;44:3665–3673. doi: 10.1128/JCM.00317-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang S, Krauss JL, Domon H, McIntosh ML, Hosur KB, Qu H, Li F, Tzekou A, Lambris JD, Hajishengallis G. The C5a receptor impairs IL-12-dependent clearance of Porphyromonas gingivalis and is required for induction of periodontal bone loss. J. Immunol. 2011;186:869–877. doi: 10.4049/jimmunol.1003252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg K, Wegner N, Yucel-Lindberg T, Venables PJ. Periodontitis in RA-the citrullinated enolase connection. Nat. Rev. Rheumatol. 2010;6:727–730. doi: 10.1038/nrrheum.2010.139. [DOI] [PubMed] [Google Scholar]
- Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, Finlay BB. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2:119–129. doi: 10.1016/j.chom.2007.06.010. [DOI] [PubMed] [Google Scholar]
- Malek R, Fisher JG, Caleca A, Stinson M, van Oss CJ, Lee JY, Cho MI, Genco RJ, Evans RT, Dyer DW. Inactivation of Porphyromonas gingivalis fimA gene blocks periodontal damage in gnotobiotic rats. J. Bacteriol. 1994;176:1052–1059. doi: 10.1128/jb.176.4.1052-1059.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markiewski MM, DeAngelis RA, Benencia F, Ricklin-Lichtsteiner SK, Koutoulaki A, Gerard C, Coukos G, Lambris JD. Modulation of the antitumor immune response by complement. Nat. Immunol. 2008;9:1225–1235. doi: 10.1038/ni.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito M, Hirakawa H, Yamashita A, Ohara N, Shoji M, Yukitake H, Nakayama K, Toh H, Yoshimura F, Kuhara S, et al. Determination of the genome sequence of Porphyromonas gingivalis strain ATCC 33277 and genomic comparison with strain W83 revealed extensive genome rearrangements in P. gingivalis. DNA Res. 2008;15:215–225. doi: 10.1093/dnares/dsn013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolopoulou-Papaconstantinou AA, Johannessen AC, Kristoffersen T. Deposits of immunoglobulins, complement, and immune complexes in inflamed human gingiva. Acta Odontol. Scand. 1987;45:187–193. doi: 10.3109/00016358709098858. [DOI] [PubMed] [Google Scholar]
- Page RC, Lantz MS, Darveau R, Jeffcoat M, Mancl L, Houston L, Braham P, Persson GR. Immunization of Macaca fascicularis against experimental periodontitis using a vaccine containing cysteine proteases purified from Porphyromonas gingivalis. Oral Microbiol. Immunol. 2007;22:162–168. doi: 10.1111/j.1399-302X.2007.00337.x. [DOI] [PubMed] [Google Scholar]
- Paramonov N, Rangarajan M, Hashim A, Gallagher A, Aduse-Opoku J, Slaney JM, Hounsell E, Curtis MA. Structural analysis of a novel anionic polysaccharide from Porphyromonas gingivalis strain W50 related to Arg-gingipain glycans. Mol. Microbiol. 2005;58:847–863. doi: 10.1111/j.1365-2958.2005.04871.x. [DOI] [PubMed] [Google Scholar]
- Pathirana RD, O'Brien-Simpson NM, Brammar GC, Slakeski N, Reynolds EC. Kgp and RgpB, but not RgpA, are important for Porphyromonas gingivalis virulence in the murine periodontitis model. Infect. Immun. 2007;75:1436–1442. doi: 10.1128/IAI.01627-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patters MR, Niekrash CE, Lang NP. Assessment of complement cleavage in gingival fluid during experimental gingivitis in man. J. Clin. Periodontol. 1989;16:33–37. doi: 10.1111/j.1600-051x.1989.tb01609.x. [DOI] [PubMed] [Google Scholar]
- Pihlstrom BL, Michalowicz BS, Johnson NW. Periodontal diseases. Lancet. 2005;366:1809–1820. doi: 10.1016/S0140-6736(05)67728-8. [DOI] [PubMed] [Google Scholar]
- Power ME, Tilman D, Estes JA, Menge BA, Bond WJ, Mills LS, Daily G, Castilla JC, Lubchenco J, Paine RT. Challenges in the quest for keystones. BioScience. 1996;46:609–620. [Google Scholar]
- Ricklin D, Hajishengallis G, Yang K, Lambris JD. Complement: a key system for immune surveillance and homeostasis. Nat. Immunol. 2010;11:785–797. doi: 10.1038/ni.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh M, Trinchieri G. Innate immune mechanisms of colitis and colitis-associated colorectal cancer. Nat. Rev. Immunol. 2011;11:9–20. doi: 10.1038/nri2891. [DOI] [PubMed] [Google Scholar]
- Schenkein HA, Genco RJ. Gingival fluid and serum in periodontal diseases. II. Evidence for cleavage of complement components C3, C3 proactivator (factor B) and C4 in gingival fluid. J. Periodontol. 1977;48:778–784. doi: 10.1902/jop.1977.48.12.778. [DOI] [PubMed] [Google Scholar]
- Slack E, Hapfelmeier S, Stecher B, Velykoredko Y, Stoel M, Lawson MA, Geuking MB, Beutler B, Tedder TF, Hardt WD, et al. Innate and adaptive immunity cooperate flexibly to maintain host-microbiota mutualism. Science. 2009;325:617–620. doi: 10.1126/science.1172747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecher B, Robbiani R, Walker AW, Westendorf AM, Barthel M, Kremer M, Chaffron S, Macpherson AJ, Buer J, Parkhill J, et al. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 2007;5:2177–2189. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed FA, Hoey KA. Integrative physiology of the aging bone: insights from animal and cellular models. Ann. N. Y. Acad.Sci. 2010;1211:95–106. doi: 10.1111/j.1749-6632.2010.05813.x. [DOI] [PubMed] [Google Scholar]
- Tonetti MS, D'Aiuto F, Nibali L, Donald A, Storry C, Parkar M, Suvan J, Hingorani AD, Vallance P, Deanfield J. Treatment of periodontitis and endothelial function. N. Engl. J. Med. 2007;356:911–920. doi: 10.1056/NEJMoa063186. [DOI] [PubMed] [Google Scholar]
- Tonetti MS, Imboden MA, Lang NP. Neutrophil migration into the gingival sulcus is associated with transepithelial gradients of interleukin-8 and ICAM-1. J. Periodontol. 1998;69:1139–1147. doi: 10.1902/jop.1998.69.10.1139. [DOI] [PubMed] [Google Scholar]
- Wang M, Krauss JL, Domon H, Hosur KB, Liang S, Magotti P, Triantafilou M, Triantafilou K, Lambris JD, Hajishengallis G. Microbial hijacking of complement-toll-like receptor crosstalk. Sci. Signal. 2010;3:ra11. doi: 10.1126/scisignal.2000697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Shakhatreh M-AK, James D, Liang S, Nishiyama S-i, Yoshimura F, Demuth DR, Hajishengallis G. Fimbrial proteins of Porphyromonas gingivalis mediate in vivo virulence and exploit TLR2 and complement receptor 3 to persist in macrophages. J. Immunol. 2007;179:2349–2358. doi: 10.4049/jimmunol.179.4.2349. [DOI] [PubMed] [Google Scholar]
- Zipfel PF, Skerka C. Complement regulators and inhibitory proteins. Nat. Rev. Immunol. 2009;9:729–740. doi: 10.1038/nri2620. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.