Abstract
2-Amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) is a heterocyclic aromatic amine that is formed during the cooking of meats and poultry. PhIP is a carcinogen in rodents and a potential human carcinogen. Several short-term biomarkers of PhIP have been established for human biomonitoring, but validated long-term biomarkers of the biologically effective dose of PhIP remain to be developed. Metabolites of PhIP have been reported to covalently bind to human serum albumin (SA), which is the most abundant protein in plasma; however, the chemical structures of PhIP-SA adducts are unknown. Cysteine34 is one of 35 conserved Cys residues in SA across species. Thirty-four of these Cys are involved in 17 disulfide bonds. The single unpaired Cys34 residue in SA is well known to react with carcinogenic metabolites and toxic electrophiles. 2-Nitro-1-methyl-6-phenylimidazo[4,5-b]pyridine (NO2-PhIP), 2-hydroxyamino-1-methyl-6-phenylimidazo[4,5-b]pyridine (HONH-PhIP), and 2-nitroso- 1-methyl-6-phenylimidazo[4,5-b]pyridine (NO-PhIP), three genotoxic metabolites of PhIP, were reacted with purified human SA or human plasma, and the SA adduction products, following enzymatic digestion, were separated by ultra performance liquid chromatography and characterized with a linear quadrupole ion trap mass spectrometer. The major adduct of NO2-PhIP was formed at the Cys34 of SA with bond formation occurring between the sulfhydryl group of Cys and the C-2 imidazole atom of PhIP. The major adducts formed between SA and HNOH-PhIP or NO-PhIP were identified as acid-labile sulfinamide linkages at Cys34. These PhIP-SA adducts represent a measure of bioactivation of PhIP and may serve as long-term biomarkers of the biologically effective dose of PhIP.
Introduction
Heterocyclic aromatic amines (HAAs) are carcinogens that are formed during the high-temperature cooking of meats.1 Thus far, more than 20 HAAs have been identified in cooked meats and poultry. 2-Amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) is the most mass-abundant HAA formed; concentrations can reach up to 480 parts per billion in well-done poultry.2 Many epidemiological investigations have examined the interrelationships among consumption of cooked red meat, its effect on human cancer risk of the digestive tract, the prostate gland, mammary gland, and the potential causal role of HAAs in the etiology of these cancers.3–5 A number of these studies have reported a positive association between cancer risk and well-done meat consumption, although some studies have shown no associations between well-done meat and cancer risk. A major limiting factor in many epidemiological studies is the uncertainty in quantitative estimates of chronic exposure to HAAs and, thus, the association of HAAs formed in cooked meat and cancer risk has been difficult to establish. There is a critical need to establish stable, long-term biomarkers of HAAs that can be implemented in molecular epidemioIogy studies.
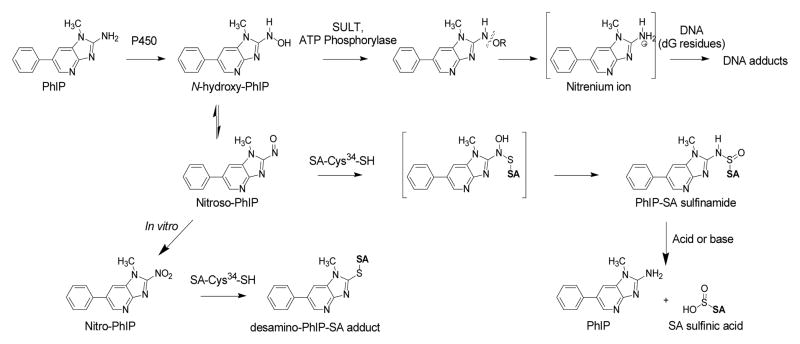
PhIP comprises about 70% of the daily mean intake of HAAs in the United States.6 The biomonitoring of PhIP and its metabolites in urine and the monitoring of PhIP in hair are promising approaches to assess exposure.7–9 However, PhIP requires metabolic activation to exert its genotoxic effects and long-term biomarkers of the biologically effective dose of PhIP have not been established. The bioactivaton of PhIP occurs by N-oxidation to form its N-hydroxylated derivative, 2-hydroxyamino-1-methyl-6-phenylimidazo[4,5-b]pyridine (HNOH-PhIP), by cytochrome P450 enzymes (Figure 1).10–12 Subsequently, HNOH-PhIP can undergo conjugation by phase II enzymes, to produce unstable esters, which undergo heterolytic cleavage to form the proposed nitrenium ion, that binds to DNA.7 HONH-PhIP can also undergo oxidation by P450s or transition metals to form 2-nitroso-1-methyl-6-phenylimidazo[4,5-b]pyridine (NO-PhIP);13,14 further oxidation of PhIP can occur to produce 2-nitro-1-methyl-6-phenylimidazo[4,5-b]pyridine(NO2-PhIP).15 Some N-oxidation products of PhIP are capable of binding to nucleophilic sites in proteins (Figure 1).16,17
Figure 1.
Chemical structures of PhIP, metabolites, and reaction products with SA.
The measurements of chemical specific DNA adducts in the target tissue are the most relevant findings for risk assessment of genotoxic carcinogens. Unfortunately, DNA adduct measurements in tissue are often precluded by the unavailability of biopsy samples in large scale human studies. Moreover, DNA adducts are often repaired and the adduct levels can be below the limit of detection, even when measured by sensitive tandem mass spectrometry techniques. Both hemoglobin (Hb) and serum albumin (SA) carcinogen adducts have been used as an alternative to DNA adducts for human biomarkers of several different classes of carcinogens.18–20 Stable carcinogen protein adducts are expected to accumulate and follow the kinetics of the lifetime of Hb or half-life of SA, during chronic exposure and augment the sensitivity of adduct detection.18,20
Carcinogen blood protein adducts of structurally related aromatic amines and HAAs are formed through their N-oxidized metabolites; such adduction products are a measure of the biologically effective dose.18,21 Many aromatic amines form adducts with Hb. The arylhydroxylamine metabolite can undergo oxidation to the arylnitroso intermediate within the erythrocyte and react with the Hb-Cys93β to form an arylamine-Hb sulfinamide adduct.22,23 Arylamine-Hb sulfinamide adduct formation occurs for a number of aromatic amines in experimental laboratory animals and in humans.21,23–25 Human biomonitoring studies have revealed presence of diverse arylamine-hemoglobin sulfinamide adducts in subjects, and elevated levels of this blood protein biomarker have been shown to be strongly associated with bladder cancer risk among smokers and nonsmokers.24,26,27 Several HAAs also form Hb-sulfinamide adducts in experimental animals; however, the level of adduct formation is very low.28–30 Preliminary findings from a human pilot study also have shown that the binding of HAAs to Hb is inefficient.31 The low level of HAA-Hb adduct formation will probably preclude the development of HAA-Hb adducts as biomarkers in humans.32
SA adducts have also served as means to biomonitor carcinogens and toxic electrophiles in humans.33 The scavenging properties of the single unpaired Cys34 residue in SA with numerous electrophiles are well documented.34 Mass spectrometric assays have been developed to measure SA adducts formed at the Cys34 with acrylamide,35 cyanide,36 nitrogen mustard,37 a,β-unsaturated aldehydes,38 the neurotoxin brevetoxin B,39 acetaminophen, 40 benzene,41 amongst other toxicants. Several HAAs also from adducts at the Cys34 residue of rodent SA or human SA, including: 2-amino-3-methylimidazo[4,5-f]quinoline (IQ),28 2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline (MeIQx),32 and PhIP.16,17 Chemical and spectroscopic analysis of the pronase digest recovered from rat SA modified with the N-hydroxylated metabolite of IQ proved the adduct was a tripeptide containing N2-cysteinsulfinyl-IQ.28 The adduct is formed by reaction of the nitroso derivative of IQ with Cys34 to form a semimercaptal, which undergoes rearrangement to form the sulfinamide adduct structure (Figure 1). Some of the SA adducts formed with the N-oxidized metabolites of MeIQx and PhIP may also exist as sulfinamides.32,42
There is one report on the employment of SA adducts of PhIP as a biomarker in humans42 The investigators indirectly measured a PhIP-SA adduct, following acid hydrolysis, which released PhIP from SA.42 Arylamine-sulfinamide protein adducts and arylamine-sulfinamide conjugates of glutathione are hydrolyzed by mildly acidic pH to generate the parent amines.21,23,28,43 The acid-labile character the PhIP-SA adduct suggests that a portion of the PhIP bound to SA exists as a sulfinamide linkage at Cys34.42 However, the investigators did not identify the reactive species responsible for adduct formation or characterize the structure of the intact PhIP-SA adduct. Thus, knowledge about the chemistry of PhIP-SA adduct formation is lacking. The goal of our current study is to identify and characterize the adduction products of N-oxidized metabolites of PhIP formed at the Cys34 residue of human SA, by mass spectrometry methods, as part of the validation process to establish a protein biomarker of this food carcinogen in humans.
Materials and Methods
Caution
PhIP is a carcinogen, which should be handled in a well-ventilated fume hood with the appropriate protective clothing.
Chemicals and Materials
PhIP was purchased from Toronto Research Chemicals (Toronto, ON, Canada) and PhIP-[2H5]-phenyl ([2H5]-PhIP, 99% isotopic purity) was a gift from Dr. Mark Knize and Dr. Kristen Kulp, (Lawrence Livermore National Laboratory). SA, cysteine, and 4-chloromercuribenzoic acid (4-CMB) were purchased from Sigma (St. Louis, MO). The peptide LQQCPF was obtained from Biosynthesis (Lewisville, TX), and human plasma was purchased from Bioreclamation LLC (Hicksville, NY). Sequence grade trypsin and chymotrypsin were purchased from Promega (Madison, WI). Pronase E, leucine aminopeptidase, and prolidase were purchased from Sigma (St. Louis, MO). All solvents were high-purity B & J Brand from Honeywell Burdick and Jackson (Muskegon, MI). ACS reagent-grade formic acid (88%) was purchased from J.T. Baker (Phillipsburg, NJ). Isolute C18 solid phase extraction (SPE) columns (25 mg) were from Biotage (Charlotte, NC). HiTrap Blue affinity and GE P10 columns were obtained from GE Healthcare (Piscataway, NJ). All other chemical reagents were ACS grade and purchased from Sigma-Aldrich.
Synthesis of PhIP N-Oxidized Intermediates and PhIP Adduction Products with LQQCPF
NO2-PhIP was synthesized by diazotization of PhIP as previously described.44 HNOH-PhIP was prepared by reduction of NO2-PhIP with hydrazine and Pd/C serving as a catalyst.44,45 2-Thio-1-methyl-6- phenylimidazo[4,5-b]pyridine (HS-PhIP) was prepared by reaction of NO2-PhIP (50 μg, 200 nmol) with (NH4)2S (20% solution, 0.1 mL) in DMF (0.9 mL) at room temperature for 18 h. The desired reaction product was concentrated to dryness by vacuum centrifugation and redissolved in H2O:CH3OH (1:1) containing 0.1% HCO2H. The product was characterized by ESI/MS. NO2-PhIP was quantitatively converted to HS-PhIP ([M+H]+ at m/z 242.1): the product ion spectrum confirmed the structure (vide infra). NO-PhIP was prepared by the oxidation of HNOH-PhIP (0.7 μmol in 100 μL C2H5OH) with K3Fe(CN)6 (84 μmol in 500 μL H2O), under ambient conditions for 3 h. The solution was diluted with H2O (2 mL) and applied to a C18 SPE column, washed with 10% CH3OH in H2O and NO-PhIP was subsequently eluted with CH3OH (1 mL). The product was characterized by UV/vis spectroscopy and mass spectrometry (vide infra).
N-Oxidized PhIP-modified peptides were prepared by the following reactions: cysteine (Cys) (10 μg, 82 nmol) in 50 mM ammonium bicarbonate buffer, pH 8.5 (1 mL) with NO2-PhIP (28 μg, 110 nmol in DMSO (8 μL)); LQQCPF (30 μg, 41 nmol) in 50 mM ammonium bicarbonate buffer, pH 8.5 (1 mL) with NO2-PhIP (31 μg, 122 nmol in DMSO (10 μL)); or LQQCPF (20 μg, 27 nmol) in H2O (1 mL) with NO-PhIP (6.5 g, 27 nmol in CH3OH (40 μL) ([2H5]-NO2-PhIP and [2H5]-NO-PhIP) were reacted at 37ºC for 18 h, followed by enrichment with C18 SPE as described above. Modified peptides were purified by HPLC with Agilent 1100 HPLC system (Palo Alto, CA) and an Agilent Eclipse XDB-C18 column (4.6 × 250 mm). A linear gradient was employed, starting from 100% A solvent (0.1% HCO2H) and reaching 100% B solvent (95% CH3CN containing, 4.9% H2O, and 0.1% HCO2H) at 20 min, at a flow rate of 1 mL/min. The wavelength was monitored at 210 and 320 nm. The desired products were collected product and vacuum centrifuged to dryness and resuspended in H2O. The concentration was determined by UV measurement, utilizing the same molar extinction coefficient of PhIP. The identities of the PhIP-modified peptides were characterized by LC-ESI/MS/MS (vide infra). The approximate yield of the NO2-PhIP-modified LQQC*PF (C-desamino-PhIP) was ~ 60%, and the yield of the LQQC*PF (C-[S=O]-PhIP) sulfinamide was ~ 10%, based on the recovery of the modified peptides.
Synthesis of LQQCPF Sulfinic Acid
LQQC(SO2H)PF was prepared by oxidation of LQQCPF (73 μg, 100 nmol) with Fenton's reagent (Fe2+/H2O2) as described in literature.46 The reaction mixture was desalted with a C18 SPE cartrdige as described above, and LQQC(SO2H)PF was eluted from the SPE with CH3OH and assayed directly by mass spectrometry.
Pretreatment of Cys34 in SA
Reduction of mixed disulfides formed at Cys34 of commerical human SA was done by treatment of SA (50 mg, 750 nmol) in 0.1 M potassium phosphate buffer, pH 7.4 (2 mL) with β-mercaptoethanol (40 μmol) at room temperature for 90 min.28 These mild reducing conditions do not open internal disulfide bonds of SA.47 Excess β-mercaptoethanol was removed from SA by gel filtration chromatography. The Cys34 was blocked by titration of the thiol residue to its endpoint with 4-chloromecuribenzoic acid (4-CMB),28,48 followed gel filtration chromatography. The reduced sulfhydryl content of SA was determined using Ellman’s reagent.49
Modification of SA and Plasma with NO2-PhIP, HNOH-PhIP and NO-PhIP
A solution of NO2-PhIP (30 nmol in 2.2 μL DMSO), HNOH-PhIP (30 nmol in 4.2 μL C2H5OH) or NO-PhIP (30 nmol in 50 μL C2H5OH) was reacted with commercial SA reduced with β-mercaptoethanol or reacted with reduced SA and that had been pretreated with 4-CMB (2 mg SA, 30 nmol/1 mL 10 mM potassium phosphate buffer, pH 7.4) The reaction of N-oxidized PhIP derivatives with SA was performed at 37 ºC for 18 h. Unbound PhIP derivatives were removed from SA by solvent extraction 3 times with equal volumes of dichloromethane. The PhIP-modified SA was then subjected to gel-filtration chromatography with the PD-10 column in 10 mM potassium phosphate buffer (pH 7.4).
Human plasma (200 μL) was diluted with PBS to a final volume of 1 mL. This solution was reacted with HNOH-PhIP (150 nmol in 27 μL C2H5OH) or NO-PhIP (150 nmol in 40 μL C2H5OH) at 37 ºC for 18 h. The plasma was diluted with 4 mL buffer A (50 mM KH2PO4, pH 7.0) and centrifuged at 3000 g for 5 min, to remove particulates, before it was applied to a HiTrap Blue affinity column. The resin was washed with buffer A (10 mL) to remove plasma proteins, and then SA was eluted from the affinity column with 50 mM KH2PO4 buffer (pH 7.0) containing, 1.5 M KCl (3 mL). Thereafter, any remaining unbound PhIP derivatives were removed by solvent extraction with dichloromethane (3 mL, 3 times), followed by size exclusion chromatography of SA with the P10 column. The amount of SA was determined by UV absorption at 280 nm.50
Proteolytic Digestion
Trypsin was prepared at a concentration of 0.2 mg/mL in 50 mM CH3CO2H, and chymotrypsin was prepared at a concentration of 0.2 mg/mL in 1 mM HCl. Pronase, leucine aminopeptidase and prolidase were dissolved in H2O at concentrations of 1, 0.5 and 0.25 mg/mL, respectively. 1) Trypsin/chymotrypsin digestion: To a solution of PhIP-modified protein (5 μg, 75 pmol, in 10 mM potassium phosphate buffer, pH 7.4 (10 μL)) containing [2H5]-PhIP LQQC*PF internal standard [5 pmol LQQC*PF (C-desamino-[2H5]-PhIP) or 1.35 pmol LQQC*PF (C-[S=O]-[2H5]-PhIP) sulfinamide] in 50 mM ammonium bicarbonate buffer (35 μL, pH 8.5) containing CaCl2 (1 mM) was added trypsin at a protease:protein ratio of 1:10 (w/w) and chymotrypsin at a protease:protein ratio of 1:12.5 (w/w). The mixture (total digestion volume = 50 μL) was incubated at 37º C for 20 h. Thereafter, the protein digest was diluted with H2O (1 mL) and applied to a C18 SPE column and processed as previously described. The CH3OH eluents were concentrated to dryness by vacuum centrifugation and resuspended in H2O (200 μL) for subsequent UPLC-ESI/MS/MSn analysis. 2) Pronase E/leucine aminopeptidase/prolidase digestion: This three enzyme digestion mixture is a modification of the procedure reported in the literature.51,52 To a solution of modified protein (5 μg, 75 pmol) spiked with [2H5]-PhIP-modified LQQC*PF (5 pmol LQQC*PF (C-desamino-[2H5]-PhIP) or 1.35 pmol LQQC*PF (C-[S=O]-[2H5]-PhIP) sulfinamide) in 50 mM ammonium bicarbonate buffer (pH 8.5) containing 1 mM MnCl2 was added pronase E at a protease:protein ratio of 1:2 (w/w), leucine aminopeptidase at a protease:protein ratio of 1:30 (w/w) and prolidase at a protease:protein ratio of 1:8 (w/w). The mixture was incubated at 37º C for 20 h, followed by the same SPE purification procedure described above.
Acid Hydrolysis of HNOH-PhIP and NO-PhIP-Modified SA
PhIP-modified SA (5 μg, 75 pmol) with LQQC*PF (C-[S=O]-[2H5]-PhIP) (1.35 pmol) added as an internal standard in H2O (300 μL) was hydrolyzed with 0.16 N HCl (12 N, 4 μL). Samples were incubated at 37 ºC for 1 h and subjected to SPE as described above.
UPLC-ESI/MS/MSn Analyses of Peptides
The interpretation of the fragment ions of non-modified and PhIP-modified peptides generated by tandem MS was done manually and facilitated by online software (Protein Prospector, Univ. of California, San Francisco, http://us.expasy.org/proteomics). LC separation of peptides was performed with a NanoAcquity UPLC system (Waters Corp., Milford, MA) equipped with an Vydac Everest C18 column (0.3 × 150 mm) from Grace (Deerfield, IL). Digests (3 μL) were injected and peptides were resolved with a linear gradient starting from 100% A solvent (0.01% HCO2H) to 100% B solvent (95% CH3CN containing, 4.99% H2O, and 0.01% HCO2H) over 11 min, at a flow rate of 5 μL/min. MS spectra were acquired with a linear quadrupole ion trap mass spectrometer (LTQ, Thermo Fisher, San Jose, CA). Xcalibur version 2.07 software was used for data manipulations. All analyses were conducted in the positive ionization mode and employed an Advance CaptiveSpray™ source from Michrom Bioresource Inc. (Auburn, CA). The temperature of capillary tube was set at 200 °C; the spray voltage was 1.5 kV; and the in-source fragmentation was 10 V. There was no sheath or auxilliary gas. Helium was used as the collision and damping gas in the ion trap and was set at a pressure of 1 mTorr. One μscan was used for data acquisition. The automatic gain control (AGC) settings were full MS target 30,000 and MSn target 10,000, and the maximum injection time was 10 ms. Tandem mass spectrometry (MS/MS) was employed to monitor and characterize PhIP-SA adducts. The transitions monitored were as follows: LQQC*PF (C-desamino-PhIP) ([M+2H]2+ at m/z 472.0 → 242.1, 684.3 and 701.3); LQQC*PF (C-desamino-[2H5]-PhIP) ([M+2H]2+ at m/z 474.5 → 247.1, 689.3 and 706.3); C*-desamino-PhIP ([M+H]+ m/z 329.2 → 242.1) C-desamino-[2H5]-PhP ([M+H]+ m/z 334.2 → 247.1); LQQC*PF (C-[S=O]-PhIP) sulfinamide ([M+2H]2+ m/z 487.2 → 225.1, 749.3); LQQC*PF (C-[S=O]-[2H5]-PhIP) sulfinamide ([M+2H]2+ m/z 489.7 → 230.1, 749.3); C*PF (C- [S=O]-PhIP) sulfinamide ([M+H]+ at m/z 604.2 → 225.1, 380.2); C*PF (C-[S=O]-[2H5]-PhIP) sulfinamide ([M+H]+ m/z 609.2 → 230.1, 380.2); Several of these fragment ions were selected for characterization at the MS/MS3 scan stage. MS/MS was conducted on PhIP (m/z 225.2 → 210.1), [2H5]-PhIP (m/z 230.2 → 215.1). The activation Q was 0.35 and activation time was 10 msec. The normalized collision energies and isolation widths were optimized for each compound.
Product ion spectra of N-oxidized PhIP derivatives were acquired by infusion with a Finnigan Quantum Ultra triple stage quadrupole mass spectrometer interfaced with the CaptiveSpray™ source from Michrom Bioresource Inc. Typical instrument tuning parameters were: capillary temperature 200 °C, source spray voltage 1.5 kV, tube lens offset 95 V, capillary offset 35 V, and source fragmentation 10 V. Argon, set at 1.5 mTorr, was used as the collision gas, and the collision energy was set between 15 to 30 V.
Results
Characterization of PhIP N-Oxidation Products
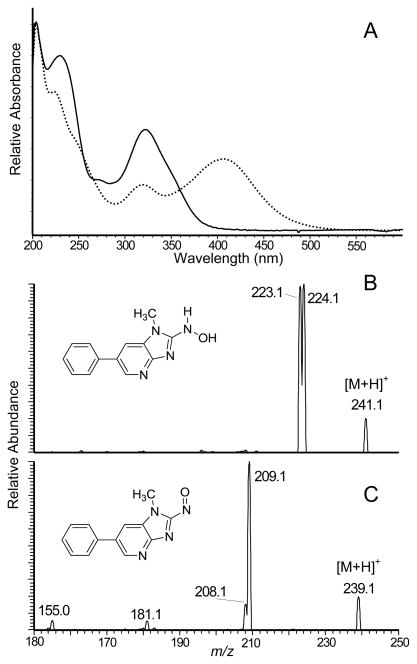
The oxidation of HNOH-PhIP by treatment with K3Fe(CN)6 produced NO-PhIP. The UV-vis spectra of HNOH-PhIP and NO-PhIP are shown in Figure 2A. The spectrum of HONH-PhIP displayed a prominent maximum at 320 nm, whereas a new absorption band appeared at 405 nm in the spectrum of NO-PhIP. The product ion mass spectra of HONH-PhIP and NO-PhIP are presented in Figures 2B and 2C. The protonated molecules [M+H]+ of HONH-PhIP and NO-PhIP are observed respectively at m/z 241.1 and m/z 239.1. HONH-PhIP undergoes fragmentation to form product ions at m/z 224.1 and 223.1, consistent with losses of OH• and H2O,53 whereas NO-PhIP undergoes fragmentation to produce a principal ion at m/z 209.1, which is attributed to the loss of the NO group.
Figure 2.
(A) UV spectra of HNOH-PhIP (solid curve) and NO-PhIP (dash curve), (B) ESI/MS/MS product ion spectra of HONH-PhIP and NO-PhIP were acquired with a triple quadrupole mass spectrometer.
UV Spectral Characterization of SA Modified with NO2-PhIP, HNOH-PhIP and NO-PhIP
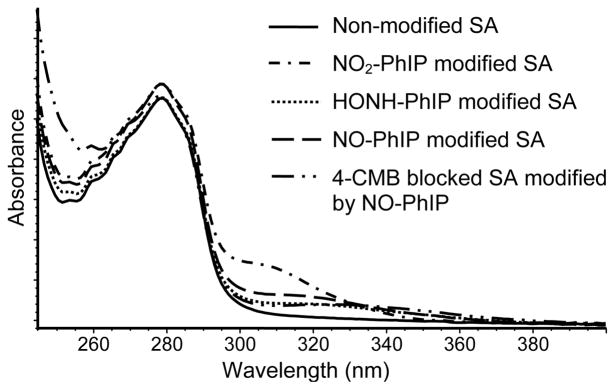
The adduction products of N-oxidized derivatives of PhIP formed with peptides and SA were initially characterized by UV spectroscopy. PhIP and PhIP-modified LQQC*PF peptides display maxima at ~ 310 – 330 nm, whereas non-modified LQQCPF and SA do not possess a chromophore around these wavelengths. A major portion of the Cys34 in the commercial SA is present as mixed disulfides (0.25 nmol reduced -SH/nmol SA);28,54 mild reduction of the SA with β-mercaptoethanol increased the reduced sulfhydryl content of Cys34 to 0.95 nmol -SH/nmol SA. We examined the reactivity of PhIP-N-oxidized derivatives with SA that had been reduced with β-mercaptoethanol without or with treatment with 4-CMB, a reagent that is highly selective for modification of sulfhydryl groups.48 The extent of non-covalent binding of PhIP to SA was assessed by incubation of equimolar concentrations of PhIP with SA for 18 h, followed by solvent extraction with dichloromethane, and then by gel-filtration chromatography of SA, to remove PhIP. There was no increase in the chromophore of SA around 310 – 330 nm, thereby demonstrating that this tandem clean-up procedure was efficient at removal of non-bound PhIP from SA. The UV spectra of commercial SA reacted with equimolar concentrations of NO2-PhIP, HONH-PhIP or NO-PhIP, followed by the tandem clean-up procedure, are shown in (Figure 3). NO2-PhIP-modified SA displayed the highest level of absorbance at 310 nm, corresponding to ~0.29 nmol of PhIP bound per nmol of SA. The binding of HONH-PhIP and NO-PhIP to SA was several fold lower. The estimate in the level of adduct formation was based on the assumption that PhIP-SA adducts possess molar absorption coefficients that are comparable to that of PhIP (315 nm; e (M−1 cm−1) at 22,220). It is noteworthy that the UV spectrum of SA that was reacted with 4-CMB, prior to treatment with NO-PhIP, also displayed a chromophore at 320 nm, suggesting that nucleophilic sites other than Cys34 have reacted with NO-PhIP.
Figure 3.
UV spectra of N-oxidized derivatives of PhIP bound to SA.
Mass Spectrometric Characterization of SA Adducts
Both glutathione and the Cys34 residue in rat SA have been reported to react with N-oxidized metabolites of PhIP in vitro.15,16 One stable adduct was reported to form between rat SA and NO2-PhIP, where the thiol group of Cys34 displaced the NO2 moiety of PhIP, by nucleophilic substitution, to form a sulfur-carbon linked adduct.16 This adduct was stable, whereas adduction products formed between rat SA or glutathione and HONH-PhIP derivatives that presumably contained sulfur-nitrogen linked adducts were unstable15,16 Therefore, we examined the reaction products formed between human SA with NO2-PhIP to determine if a sulfur-carbon linked adduct at Cys34 had been formed with NO2-PhIP. A stable PhIP-SA adduct could serve as a guide and allow us to optimize proteolytic digestion conditions for other N-oxidized PhIP metabolites bound to SA.
NO2-PhIP-Modified-SA
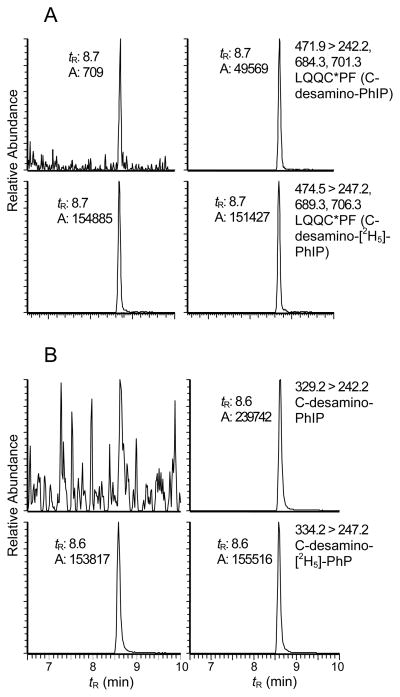
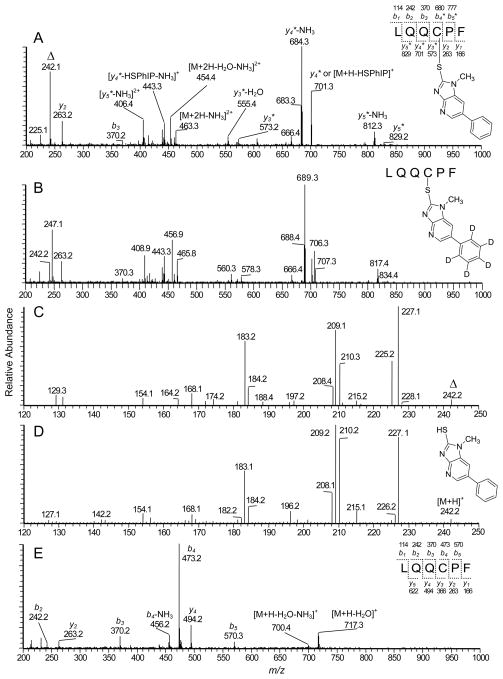
The target sequence LQQCPF (L31-F36) contains Cys34, which is recovered from SA digested with chymotrypsin. Reconstructed ion chromatograms of the peptide obtained from NO2-PhIP-modified SA digested with a mixture trypsin and chymotrypsin and the LQQC*PF (C- desamino-[2H5]-PhIP) internal standard are shown in Figure 4A. A peak (tR = 8.7 min) was detected as a doubly charged species [M+2H]2+ at m/z 471.9. This mass is consistent with the molecular weight of a reaction product formed between LQQCPF (734.3 Da) and NO2-PhIP (254.1 Da), where the thiol group of Cys34 displaced the nitro moiety of PhIP, to form a peptide adduct containing desamino-PhIP. The product ion spectra support the proposed assignment of the adduct structure (Figure 5). The b2 (m/z 242.1) and b3 (m/z 370.3) ions are present, and the proposed y4* (m/z 701.3), y5* (m/z 829.2) and y5*-NH3 (m/z 812.3) ions contain an additional mass of 208 Da, which corresponds to desamino-PhIP; these product ions are consistent with the predicted fragmentation and the site of PhIP adduction occurring at Cys34.
Figure 4.
Reconstructed ion chromatograms of A) PhIP-LQQC*PF adducts recovered from NO2-PhIP- modified SA following digestion with trypsin/chymotrypsin: NO2-PhIP-modified SA (right panel) and non-modified SA (left panel); and B) PhIP-C* adducts obtained from NO2-PhIP-modified SA by digestion with Pronase E, leucine aminopeptidase, and prolidase: NO2-PhIP modified SA (right panel) and non-modified SA (left panel).
Figure 5.
Product ion mass spectra of A) LQQC*PF recovered from NO2-PhIP-modified SA, B) LQQC*PF modified with NO2-[2H5]-PhIP, C) Second generation product ion spectrum of m/z 242.2 (denoted with a triangle symbol) from LQQC*PF, D) Synthetic HS-PhIP, and E) LQQCPF. The symbol * denotes the PhIP-adducted amino acid in the peptide.
The proposed b2 ion at m/z 242.1 is observed in the product ion spectrum of NO2-PhIP-modified LQQC*PF (Figure 5A) and in the spectrum of [2H5]-PhIP-modified LQQC*PF (Figure 5B); an ion also arises at m/z 247.1 in the spectrum of [2H5]-PhIP-modified LQQC*PF. We note that a cleavage of the side chain of Cys with retention of the sulfur group on the PhIP (m/z 242.1) or [2H5]-PhIP (m/z 247.1) moieties could explain the creation of these fragment ions. The second generation product ion spectrum (MS3) of m/z 242.1 (Figure 5C) derived from LQQC*PF was very similar to the product ion spectrum obtained with synthetic HS-PhIP ([M+H]+ at m/z 242.1) (Figure 5D). Fragment ions at m/z 227.1, 210.3, 209.1, and 208.1 correspond to, respectively, the loss of the CH3· moiety of PhIP, and losses of S, HS·, H2S, and the ion at m/z 183.2 corresponds to the loss of thiocyanic acid, HSCN. The product ion observed at m/z 225.2 (Figure 5C) is ascribed to the loss of NH3 from the b2 ion of LQQC*PF; this ion is not observed in product ion spectrum of HS-PhIP (Figure 5D). The MS3 product ion spectrum of the proposed HS-[2H5]-PhIP ion at m/z 247.1 displayed fragment ions at m/z 232.1, 215.3, 214.3, 213.1, 188.2, showing that the major fragment ions were increased by 5 Da over HS-PhIP (Figure S-1, Supporting Information). The product ion spectrum of the naturally occurring H[34S]-PhIP isotope displayed the same pattern of fragmentation that was observed in the spectrum of H[32S]-PhIP (Figure S-1, Supporting Information). These mass spectral data confirm that bond formation occurred between the sulfur atom of Cys34 and the C-2 imidazole atom of NO2-PhIP.
Proteolytic digestion of NO2-PhIP-modified SA with a 3-enzyme mixture containing pronase E, leucine aminopeptidase, and prolidase resulted in an apparent complete digestion of the NO2-PhIP bound to Cys34 of SA, to produce the monoamino acid adduct C*-desamino-PhIP (tR = 8.6 min) (Figure 4B). The digestion of the PhIP-SA adduct with the 3-enzymes was more efficient than the digestion with trypsin/chymotrypsin, and increased the recovery of C*-desamino-PhIP by ~5-fold. PhIP-modified penta, tetra-, tri- or dipeptides were not detected with the 3-enzyme mixture, whereas the digestion with Pronase E alone produced several different peptides containing the C*-desamino-PhIP- adduct. (Figure S-2, Supporting Information).
HNOH-PhIP- and NO-PhIP-Modified SA
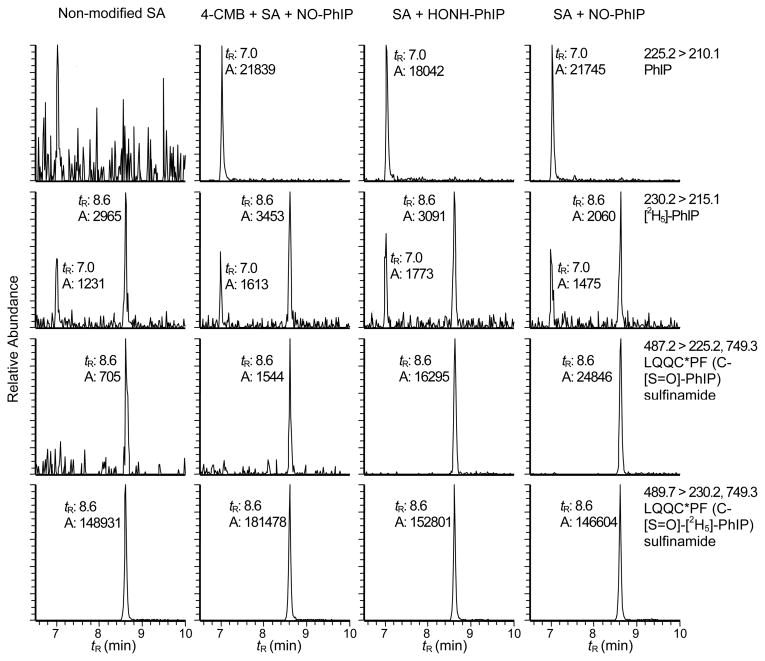
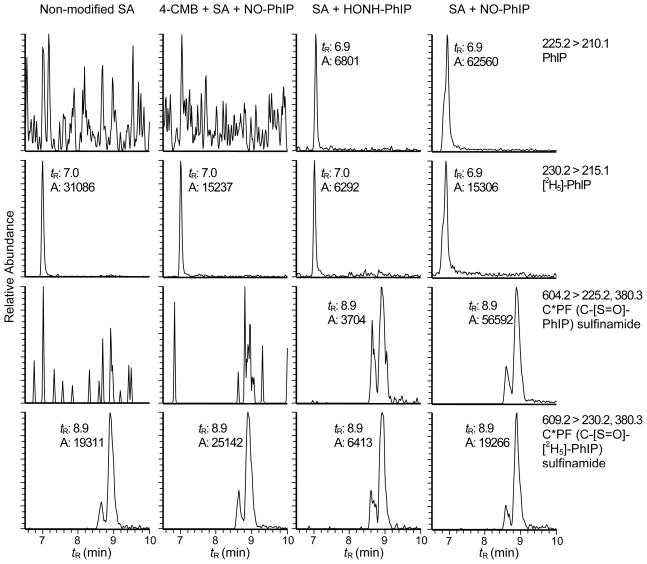
The reaction of SA with HONH-PhIP and NO-PhIP produced identical peptide adducts at Cys34. The reconstructed ion chromatograms presented in Figures 6A - 6D depict the LQQC*PF adduction products (tR = 8.6 min) recovered from PhIP-modified SA that was digested with trypsin/chymotrypsin. A peak corresponding to PhIP is also observed at tR = 7.0 min. The pretreatment of SA with 4-CMB, prior to reaction with N-oxidized PhIP, decreased the amount of the LQQC*PF adduct by greater than 97% (Figure 6B).
Figure 6.
Reconstructed ion chromatograms of PhIP and LQQC*PF (C-[S=O]-PhIP) sulfinamide, which formed by reaction HONH-PhIP or NO-PhIP with SA, followed by digestion with trypsin/chymotrypsin. LQQC*PF (C-[S=O]-[2H5]-PhIP served as an internal standard. Non-modified SA and SA pretreated with 4-CMBA prior to reaction with NO-PhIP served as negative controls.
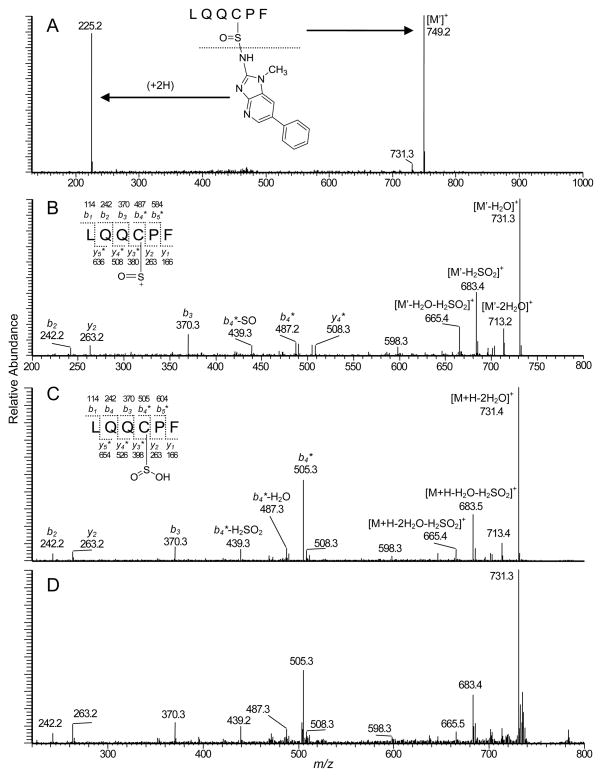
The full scan spectrum of LQQC*PF (tR = 8.6 min) adducted with HONH-PhIP or NO-PhIP displayed a predominant doubly charged species [M+2H]2+ at m/z 487.4 (data not shown), which is an addition mass of 239 Da to the protonated peptide LQQCPF (m/z 735.3). The 239 Da increase in mass corresponds to PhIP (223 Da) and an oxygen atom (16 Da). The adducted peptides coeluted with the synthetically [2H5]-NO-PhIP-modified LQQC*PF (C-[S=O]-[2H5]-PhIP) sulfinamide adduct. The product ion spectrum of the proposed LQQC*PF (C-[S=O]-PhIP) sulfinamide adduct ([M+2H]2+ at m/z 487.4) is shown in Figure 7. Two major fragment ions were observed at m/z 225.2 and 749.2, whereas the product ion spectrum of the [2H5]-NO-PhIP-modified LQQC*PF peptide ([M+2H]2+ at m/z 489.7) displayed ions at m/z 230.2 and 749.2. The ion at m/z 225.2 was identified as the protonated PhIP molecule. The second generation product ion spectrum of m/z 225.2 contained prominent fragment ions at m/z 210.1 and 208.1, indicative of loss of the CH3· and NH3 groups from PhIP (data not shown). These findings indicate that the extra oxygen atom is associated with the peptide rather than the PhIP moiety. The mass difference of 14 Da observed between LQQC*PF product ion at m/z 749.2 and the protonated peptide LQQCPF (m/z 735.3) is consistent with the presence of a sulfur-oxygen double bond on Cys34. The second generation product ion spectrum of the ion [M']+ at m/z 749.2 (Figure 7B) produced b- and y- ions, which are consistent with the predicted fragmentation. The appearance of the b2 (m/z 242.1) and b3 (m/z 370.3) ions, the proposed modified b4 ions at m/z 487.2 (addition of 14 Da) and m/z 439.3 [b4- SO]+, and the ion at m/z 683.4 [M+H-PhIP-H2SO2]+ support the proposed peptide adduct structure with the site of NO-PhIP modification occurring at the sulfhydryl group of Cys34 in the form of sulfinamide linkage. The losses of H2O+SO under these collision-induced dissociation conditions are characteristic of sulfinamide and sulfinic acid derivatives.46,55 Thus, the most likely structure of the ion at m/z 749.2 would be oxygenated LQQC*PF (LQQC[S=O]PF), as shown in the Figure 7.
Figure 7.
Product ion mass spectra of A) LQQC*PF (C-[S=O]-PhIP) sulfinamide adduct formed by reaction NO-PhIP with SA and digested with trypsin/chymotrypsin, B) Second generation product ion spectrum of the ion at m/z 479.2 in the spectrum of LQQC*PF (C-[S=O]-PhIP) sulfinamide, C) Product ion spectrum of LQQC*PF (C-[S=O]-PhIP) sulfinamide prepared synthetically and hydrolyzed with 0.16 N HCl, D) Product ion spectrum of synthetic LQQC(SO2H)PF obtained oxidation of LQQCPF with Fenton's reagent.
Noticeable amounts of PhIP were recovered from the proteolytic digest of SA that had been pretreated with 4-CMB prior to reaction with NO-PhIP (Figure 6B); PhIP was also present in the digest of HONH-PhIP- and NO-PhIP-modified SA (Figures 6C and 6D). Since the tandem solvent extraction and gel filtration procedure quantitatively removed unbound PhIP from SA, the PhIP appears to arise during proteolysis. The internal standard LQQC*PF (C-[S=O]-[2H5]-PhIP) sulfinamide was stable under these enzymatic digestion conditions, and only traces of [2H5]-PhIP were detected in the proteolytic digests. We surmise that a labile adduct of PhIP was formed at a site other than the Cys34 in SA and underwent hydrolysis during proteolytic digestion of the PhIP-modified SA to regenerate PhIP.
Acid Hydrolysis of NO-PhIP-modified LQQC*PF (C-[S=O]-PhIP) Sulfinamide to Form the LQQC(SO2H)PF Sulfinic Acid
Arylamine-sulfinamide protein adducts and arylamine-sulfinamide glutathione conjugates are hydrolyzed under mildly acidic pH conditions to form the parent amines and the corresponding sulfinic acids.23,28,43 The synthetic NO-PhIP-modified LQQC*PF peptide was incubated in 0.16 N HCl at 37 °C, and the hydrolysis product was characterized by MS. The full scan spectrum displayed an ion at m/z 225.1, which was attributed to protonated PhIP. A second ion was observed at m/z 767.3, a mass 32 Da greater than LQQCPF, which is consistent with the appearance of the sulfinic acid LQQC(SO2H)PF ([M+H]+ m/z 767.3) (Figure S-3, Supporting Information). The product ion mass spectra of the acid-hydrolyzed LQQC*PF (C-[S=O]-PhIP) sulfinamide and the synthetic sulfinic acid LQQC(SO2H)PF, obtained by reacting LQQCPF with Fenton's reagent (Fe2+/H2O2),46 were in excellent agreement (Figures 7C and 7D). The b- and y-ions, and characteristic fragment ions attributed to the sulfinic acid moiety46,55 are present in the product ion spectrum. The generation of the LQC(SO2H)PF peptide by acid hydrolysis of NO-PhIP-modified LQQC*PF is additional proof that the intact LQQC*PF PhIP adduct contains the N2-cysteinsulfinyl-PhIP linkage.
The reconstructed ion chromatograms presented in Figure 8 depict the PhIP-sulfinamide adducts recovered from SA modified with HONH-PhIP or NO-PhIP, following digestion with a 3-enzyme system containing Pronase E, leucine aminopeptidase, and prolidase. The LQQC*PF (C-[S=O]-[2H5]-PhIP) sulfinamide was employed as an internal standard. Unlike NO2-PhIP-modified-SA, where the enzymatic digestion with the 3-enzymes produced the monoamino acid C*-desamino-PhIP adduct in high yield (Figure 4B), the monoamino acid C*-PhIP (C-[S=O]-PhIP) sulfinamide adduct was not produced by the enzyme mixture. The digestion stopped at the tripeptide C*PF (C-[S=O]-PhIP) sulfinamide (tR = 8.9 min). Moreover, the sulfur-nitrogen bond of the sulfinamide linkage in the PhIP- modified SA and the LQQC*PF (C-[S=O]-[2H5]-PhIP) sulfinamide internal standard underwent hydrolysis and elevated levels of PhIP and [2H5]-PhIP (tR = 7.0 min) were recovered, following digestion. The 3-enzyme mixture either catalyzed the hydrolysis of the sulfinamide linkage to produce PhIP, or the sulfinamide linkage in C*PF (C-[S=O]-PhIP) was labile and underwent hydrolysis. The tripeptide adduct often appeared as two partially resolved peaks, which exist as a pair of diastereomers, because of the new chiral center introduced at the sulfur atom in the sulfinamide adduct structure.28 The product ion spectrum of C*PF (C-[S=O]-PhIP) sulfinamide [M+H]+ at m/z 604.3 displayed fragment ions attributed to PhIP (m/z 225.1) with retention of the oxgyen atom on the sulfur containing peptide [M+H]+ at m/z 380.2 (Figure S-4, Supporting Information). Surprisingly, the digest of the 4-CMB-modified SA reacted with NO-PhIP (Figure 8B) did not contain any PhIP, as was observed in the digest done with trypsin/chymotrypsin (Figure 6B).
Figure 8.
Reconstructed ion chromatograms of PhIP and C*PF (C-[S=O]-PhIP) sulfinamide obtained from HONH-PhIP and NO-PhIP-modified SA digested with pronase E, leucine aminopeptidase, and prolidase. C*PF (C-[S=O]-[2H5]-PhIP) served as an internal standard. Non-modified SA and SA pretreated with 4-CMBA prior to reaction with NO-PhIP served as negative controls.
UPLC/ESI/MS/MS Analyses of PhIP-SA from Human Plasma Reacted with HONH-PhIP and NO-PhIP
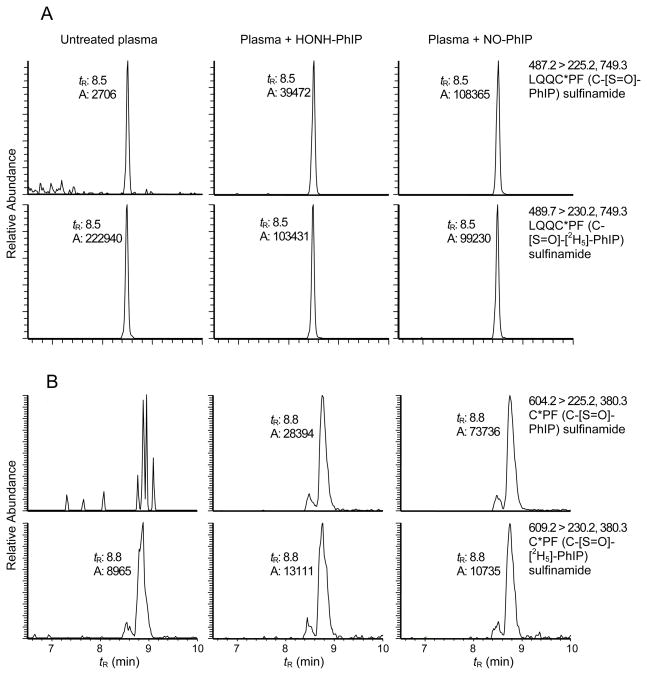
The formation of PhIP-SA adduction products was also examined in human plasma treated with HNOH-PhIP and NO-PhIP because plasma proteins, fatty acids, drugs, and other organic compounds that are associated with SA can induce structural changes in the conformation of SA and affect the reactivity of the Cys34 residue.54,56 SA was purified from PhIP-treated plasma by the HiTrap Blue affinity column and digested by trypsin/chymotrypsin or the 3-enzyme system. The reconstructed ion chromatograms of PhIP-SA adduction products are shown in Figure 8. It is evident that LQQC*PF (C-[S=O]-PhIP) sulfinamide was formed at Cys34 of SA in plasma. A small amount of the NO2-PhIP- modified LQQC*PF (C-desamino-PhIP) accounting for ~several percent of the ion counts observed for LQQC*PF (C-[S=O]-PhIP) was found in the NO-PhIP modified SA in plasma (data not shown).
Efficacy of Proteolytic Digestion of PhIP-Modified SA and Estimates of Cys34 Sulfinamide Formation
The levels of covalent binding of NO2-PhIP, HOHN-PhIP, and NO-PhIP to commercial SA were based upon UV spectroscopy, and the amounts of PhIP adducts formed at Cys34 were determined by LC/MS measurements. The data are summarized in Table 1. Under the assumption that the increase in UV chromophore of SA centered around 310 – 330 nm is attributed to covalently bound PhIP adducts, approximately 30% of the NO2-PhIP bound to SA, 13% of the NO-PhIP and 8% of the HONH-PhIP had bound to SA. The quantity of PhIP-SA-Cys34 adducts recovered from SA with a digestion mixture of pronase E, leucine aminopeptidase, and prolidase are 4 to 15-fold greater than the amounts of adduct recovered with a digestion mixture containing trypsin and chymotrypsin. The C*PF (C-desamino-PhIP) adduct accounted for approximately 50% of the NO2-PhIP bound to SA, and the C*PF (C-[S=O]-PhIP) sulfinamide accounted for about 45% of the NO-PhIP bound to SA. The amount of C*PF (C-[S=O]-PhIP) sulfinamide corresponded to ~85% of the adduction products recovered as PhIP following acid treatment of NO-PhIP-modified SA. The remaining portion of the acid-labile material recovered as PhIP appears to be derived from another adduct. The 4-CMB-modified SA contained very little free Cys34 and only traces of LQQC*PF or C*PF (C-[S=O]-PhIP) sulfinamide were present in the digests of the NO-PhIP-modified SA that had been pretreated with 4-CMB (Figure 6 and Figure 8). However, about 20% of the NO-PhIP bound to the 4-CMB-modified SA was recovered as PhIP following treatment with acid (Table 1). The structure(s) of this labile adduct remains to be identified. The amount of LQQC*PF and C*PF (C-[S=O]-PhIP) sulfinamide adducts recovered from the HONH-PhIP-modified SA was more variable than the amount of sulfinamide adducts recovered from NO-PhIP-modified SA, particularly with the 3-enzyme system. However, the amounts of PhIP recovered by acid treatment of HONH-PhIP- and NO-PhIP-modified SA were comparable. On the basis of adduct formation estimated by the UV spectra of PhIP-modified SA, there appear to be adduction products of SA formed by NO2-PhIP and NO-PhIP through nucleophiles other than Cys34. These sites of adduction remain to be determined.
Table 1.
UV Measurement of PhIP-N -Oxidized Metabolites Bound to Commercial Human Serum Albumin and LC-ESI/MS/MS Estimates of Adducts Formed at Cys34
| pmol PhIP adduct/nmol HSA1 | ||||
|---|---|---|---|---|
| Assay method | NO2-PhIP-modified HSA | HNOH-PhIP-modified HSA | NO-PhIP-modified HSA | NO-PhIP-modified HSA pretreated with 4-CMBA |
| UV Estimate | 293 ± 23 | 82 ± 11 | 131 ± 19 | 71 ± 7 |
| pmol PhIP-Cys 34 adduct/nmol HSA1 | ||||
|---|---|---|---|---|
| LC-ESI/MS/MS | NO2-PhIP modified SA | HNOH-PhIP-modified HSA | NO-PhIP-modified HSA | NO-PhIP-modified HSA pretreated with 4-CMBA5 |
| Acid hydrolysis2 | - | 72.5 ± 8.0 | 69.5 ± 11.1 | 13.4± 1.6 |
| Trypsin/chymotrypsin3 | 21.5 ± 1.0 | 2.2 ± 0.1 | 3.5 ± 0.5 | 0.1 ± 0.0 |
| 3-enzyme digestion4 | 121 ± 31.6 | 11.8 ± 1.7 | 59.0 ± 6.1 | 0.1 ± 0.0 |
| 3-enzyme digestion4 | 85.9 ± 3.5 | 69.4 ± 8.1 | 49.1 ± 6.3 | Not assayed |
Each value represents the average and standard deviation of 3 experiments.
Measured as PhIP
Measured as LQQC*PF adduct following digestion with trypsin/chymotrypsin
Measured as C*PF following digestion with pronase E, leucine aminopeptidase, and prolidase; two digests done on different days
The amount of titratable Cys in β-mercaptoethanol reduced HSA was 0.95 nmol Cys/nmol SA and <0.02 nmol Cys/nmol SA in 4-CMBA-treated HSA
The amounts of PhIP sulfinamide adduct formed at Cys34 of SA, following reaction of plasma with either HONH-PhIP or NO-PhIP are summarized in Table 2. The quantity of C*PF (C-[S=O]-PhIP) sulfinamide formed with either HONH-PhIP- or NO-PhIP-modified SA corresponds to ~90% or more of the amount of PhIP recovered following acid hydrolysis of PhIP-SA. The levels of C*PF (C-[S=O]-PhIP) sulfinamide formed with SA in plasma are several fold lower than adduct levels produced by comparable reactions of N-oxidized PhIP metabolites with commercial SA. However, plasma contains other proteins and low molecular weight nucleophiles that can potentially react with N-oxidized PhIP derivatives. Moreover, approximately 30% of the Cys34 of SA in plasma is present as mixed disulfides;54 these factors likely decrease the yields of PhIP-SA adducts formed in plasma. Our data demonstrate that the PhIP sulfinamide (C-[S=O]-PhIP) at Cys34 represents the major acid-labile adduct of PhIP formed with SA in human plasma.
Table 2.
LC-ESI/MS/MS Estimates of PhIP-N -Oxidized Metabolites Bound to Cys34 of Human Serum Albumin in Plasma
| pmol PhIP-Cys 34 adduct/nmol HSA1 | ||
|---|---|---|
| Assay | HNOH-PhIP-modified HSA | NO-PhIP-modified HSA |
| Acid hydrolysis2 | 11.2 ± 1.0 | 27.1 ± 2.2 |
| Trypsin/chymotrypsin digestion3 | 2.1 ± 0.4 | 4.1 ± 0.9 |
| 3-Enzyme digestion4 | 10.0 ± 1.1 | 34.8 ± 3.5 |
Each value represents the average and standard deviation of 3 experiments.
Measured as PhIP
Measured as LQQC*PF following digestion with trypsin/chymotrypsin
Measured as C*PF following digestion with pronase E, leucine aminopeptidase, and prolidase
Discussion
The results presented here demonstrate that the Cys34 residue of human SA is an important nucleophilic site for binding of PhIP-N-oxidized metabolites. The Cys34 residue of SA has an unusually low pKa value, 6.5 compared to about 8.0 – 8.5 in many other proteins or peptides, and it predominantly exists as the thiolate anion at physiological pH.38,56 The covalent binding of electrophiles to Cys residues is enhanced when the thiol exists as the thiolate anion.20 Crystallography and NMR studies with human SA have shown that the Cys34 resides in a shallow crevice of the protein.56–58 However, computational studies have shown that the thiolate anion is accessible to solvent,38 and Cys34 exists in equilibrium between buried and exposed conformations.56–58 These molecular and structural features of SA can help to explain the higher reactivity of Cys34 towards many low molecular weight electrophiles than other nucleophilic sites in SA or other proteins.34 The Cys34 of human SA is well known to covalently bind to many carcinogens and electrophilic toxicants of diverse structures.33,34
The amounts of PhIP-SA-Cys34 adducts recovered by digestion with trypsin and chymotrypsin were 5- to 10-fold lower than the amounts of PhIP-SA-Cys34 adducts obtained from a protease mixture containing pronase E, leucine aminopeptidase, and prolidase. These findings are not surprising. The 3-enzyme mixture is expected to hydrolyze SA nearly to complete digestion,51,52 whereas efficient digestion by trypsin and chymotrypsin requires denaturation of SA to unfold the protein, followed by chemical reduction of disulfide bonds and alkylation of the newly formed sulfhydryl groups to prevent formation of mixed disulfides, to expose as many sites as possible for proteolysis.59 Human SA contains 17 intrachain disulfide bonds and its stability or resistance to denaturation is well known.54 The denaturation of PhIP-modified SA with heat, dithiothreitol, followed by alkylation of the unfolded protein with iodoacetamide prior to proteolytic digestion,59 resulted in complete hydrolysis of the LQQC*PF (C-[S=O]-PhIP) sulfinamide linkage and also destroyed ~50% of the LQQC*PF (C-desamino-PhIP) adduct (unpublished observations L.P and R.J.T.). Thus, the digestion and recovery of PhIP adducts from the LQQC*PF sequence in native SA is incomplete, when trypsin and chymotrypsin are employed for enzymatic digestion.
The rates of protein digestion by trypsin in organic-aqueous mixtures were reported to be superior and resulted in a higher amino acid sequence coverage than a tryptic digestion conducted in aqueous solution.60 We explored several different conditions of denaturation of PhIP-modified SA, however, the amounts of LQQC*PF (C-[S=O]-PhIP) recovered from NO-PhIP-modified SA digested with trypsin/chymotrypsin in CH3CN-aqueous mixtures, or in the presence of other denaturing reagents, were significantly lower than the proteolysis of PhIP-modified SA in ammonium bicarbonate buffer (pH 8.5) (Figure S-5, Supporting Information). The activity of chymotrypsin decreases with increasing concentration of water-miscible polar organic solvents;61 the use of organic solvents or urea as denaturing reagents also adversely affected the efficacy of digestion of PhIP-SA-Cys34 adducts.
The adduct of NO2-PhIP formed at Cys34 of SA underwent complete digestion by the proteolytic mixture of pronase E, leucine aminopeptidase, and prolidase to produce the monoamino acid C*-desamino PhIP adduct. Pronase E alone was less efficient at digestion, and the C*P (C*-desamino PhIP dipeptide adduct was recovered in high abundance (Figure S-2, Supporting Information). Prolidase, an Xaa-Pro dipeptidase, which hydrolyzes dipeptides with proline at the carboxy terminus, was critical for the complete digestion of this NO2-PhIP-SA adduct. However, the digestion of the PhIP sulfinamide formed at Cys34 did not proceed beyond the C*PF (C-[S=O]-PhIP) tripeptide with this 3-enzyme mixture. Morever, noticeable amounts of PhIP were recovered from the digest. The presence of PhIP may be due to the inherent instability of the C*PF (C-[S=O]-PhIP) sulfinamide linkage, or the proteases may have catalyzed hydrolysis of the sulfinamide linkage. Thus far, we have not been able to synthesize the C*PF (C-[S=O]-PhIP) sulfinamide to examine its chemical stability. The Cys34 adduct containing N2-cysteinsulfinyl-IQ in rat SA,28 and the adducts derived from the reactive N-acetyl-p-benzoquinoneimine metabolite acetaminophen,40 sulfur mustard,62 and acrylamide35 formed with the Cys34 of human SA also did not undergo digestion beyond the tripeptide stage with pronase. Thus, the efficacy of proteoloytic digestion of toxicant-modified SA is highly dependent upon structure of the toxicant and the nature of the bond formed between the toxicant and the sulfhydryl group of Cys34.
There are two reports on the binding of PhIP to SA in vivo. In one pilot study with human volunteers undergoing colorectal cancer surgery, the amount of 14C-PhIP bound to SA was estimated by accelerator mass spectrometry at ~500 pg PhIP/g SA (2.2 fmol/mg SA), 24 h after an oral dose of 14C-PhIP (75 μg/person).31 In a study with healthy human subjects on a non-controlled diet, an acid-labile PhIP-SA adducts(s) was (were) detected at 10-fold higher levels in meat-eaters than in vegetarians (6.7 ± 1.6 vs 0.7 ± 0.3 fmol PhIP/mg protein; mean ± SE).42 The differences in levels of PhIP-SA adducts reported between these studies may be explained by the different dose regimen. PhIP-SA adduct was obtained following a single dose, in the accelerator mass spectrometry study, whereas the adduct level in the population-based study represents chronic exposure. The PhIP-SA adduct would be present at a ~28-fold higher level than the level of adduct formed after a single dose, assuming the adduct is stable over the lifespan of SA.20 The structure(s) of any of the PhIP adducts bound to SA in vivo are unknown. It seems likely that some portion of the acid-labile adduct of PhIP bound to human SA occurred as a sulfinamide linkage at the Cys34 residue.
The levels of PhIP-SA adducts at Cys34 characterized in our study are about 5,000-fold greater than the levels of PhIP-SA adducts reported in humans.31,42 Our ongoing studies are focused on the optimization of enzymatic digestion conditions and selective enrichment of PhIP-SA adducts, prior to UPLC-ESI/MS/MSn, to measure PhIP adducts formed at the Cys34 residue or possibly other sites of SA in humans.
Supplementary Material
Figure 9.
Reconstructed ion chromatograms of LQQC*PF (C-[S=O]-PhIP) and C*PF (C-[S=O]-PhIP) sulfinamide adducts obtained from HONH-PhIP- and NO-PhIP-modified plasma, followed by HiTrap Blue affinity purification of SA. A) LQQC*PF (C-[S=O]-PhIP) obtained by trypsin/chymotrypsin digestion of SA, and B) C*PF (C-[S=O]-PhIP) obtained by pronase E, leucine aminopeptidase, and prolidase digestion.
Acknowledgments
Funding Support: This research was supported by grant R01 CA122320 (L.P. and R.J.T.) from the National Cancer Institute.
Abbreviations
- 4-CMB
4-chloromercuribenzoic acid
- PhIP
2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine
- HONH-PhIP
2-hydroxyamino-1-methyl-6-phenylimidazo[4,5-b]pyridine
- NO-PhIP
2-nitroso-1-methyl-6-phenylimidazo[4,5-b]pyridine
- NO2-PhIP
2-nitro-1-methyl-6-phenylimidazo[4,5-b]pyridine
- HS-PhIP
2-thio-1-methyl-6-phenylimidazo[4,5-b]pyridine
- MeIQx
2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline
- IQ
2-amino-3-methylimidazo[4,5-f]quinoline
- Hb
hemoglobin
- HAA
heterocyclic aromatic amine
- LC-ESI/MS/MS
liquid chromatography-electrospray ionization/tandem mass spectrometry
- SA
serum albumin
- SPE
solid phase extraction
- UPLC
ultra performance liquid chromatography
Footnotes
Supporting Information Available: Additional information as noted in text (Figure S-1 – Figure S-4). This material is available free of charge via the Internet at http://pubs.acs.org.
Reference List
- 1.Sugimura T, Wakabayashi K, Nakagama H, Nagao M. Heterocyclic amines: Mutagens/carcinogens produced during cooking of meat and fish. Cancer Sci. 2004;95:290–299. doi: 10.1111/j.1349-7006.2004.tb03205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Felton JS, Jagerstad M, Knize MG, Skog K, Wakabayashi K. Contents in foods, beverages and tobacco. In: Nagao M, Sugimura T, editors. Food Borne Carcinogens Heterocyclic Amines. John Wiley & Sons Ltd; Chichester, England: 2000. pp. 31–71. [Google Scholar]
- 3.Sinha R. An epidemiologic approach to studying heterocyclic amines. Mutat Res. 2002;506–507:197–204. doi: 10.1016/s0027-5107(02)00166-5. [DOI] [PubMed] [Google Scholar]
- 4.Knize MG, Felton JS. Formation and human risk of carcinogenic heterocyclic amines formed from natural precursors in meat. Nutr Rev. 2005;63:158–165. doi: 10.1111/j.1753-4887.2005.tb00133.x. [DOI] [PubMed] [Google Scholar]
- 5.Alaejos MS, Gonzalez V, Afonso AM. Exposure to heterocyclic aromatic amines from the consumption of cooked red meat and its effect on human cancer risk: a review. Food Addit Contam Part A Chem Anal Control Expo Risk Assess. 2008;25:2–24. doi: 10.1080/02652030701474235. [DOI] [PubMed] [Google Scholar]
- 6.Keating GA, Bogen KT. Estimates of heterocyclic amine intake in the US population. J Chromatogr B Analyt Technol Biomed Life Sci. 2004;802:127–133. doi: 10.1016/j.jchromb.2003.10.047. [DOI] [PubMed] [Google Scholar]
- 7.Turesky RJ, Le Marchand L. Metabolism and biomarkers of heterocyclic aromatic amines in molecular epidemiology studies: lessons learned from aromatic amines. Chem Res Toxicol. 2011;24:1169–1214. doi: 10.1021/tx200135s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alexander J, Reistad R, Hegstad S, Frandsen H, Ingebrigtsen K, Paulsen JE, Becher G. Biomarkers of exposure to heterocyclic amines: approaches to improve the exposure assessment. Food Chem Toxicol. 2002;40:1131–1137. doi: 10.1016/s0278-6915(02)00053-4. [DOI] [PubMed] [Google Scholar]
- 9.Kobayashi M, Hanaoka T, Hashimoto H, Tsugane S. 2-Amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) level in human hair as biomarkers for dietary grilled/stir-fried meat and fish intake. Mutat Res. 2005;588:136–142. doi: 10.1016/j.mrgentox.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 10.Zhao K, Murray S, Davies DS, Boobis AR, Gooderham NJ. Metabolism of the food derived mutagen and carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) by human liver microsomes. Carcinogenesis. 1994;15:1285–1288. doi: 10.1093/carcin/15.6.1285. [DOI] [PubMed] [Google Scholar]
- 11.Turesky RJ, Constable A, Richoz J, Varga N, Markovic J, Martin MV, Guengerich FP. Activation of heterocyclic aromatic amines by rat and human liver microsomes and by purified rat and human cytochrome P450 1A2. Chem Res Toxicol. 1998;11:925–936. doi: 10.1021/tx980022n. [DOI] [PubMed] [Google Scholar]
- 12.Crofts FG, Sutter TR, Strickland PT. Metabolism of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine by human cytochrome P4501A1, P4501A2 and P4501B1. Carcinogenesis. 1998;19:1969–1973. doi: 10.1093/carcin/19.11.1969. [DOI] [PubMed] [Google Scholar]
- 13.Kim D, Kadlubar FF, Teitel CH, Guengerich FP. Formation and reduction of aryl and heterocyclic nitroso compounds and significance in the flux of hydroxylamines. Chem Res Toxicol. 2004;17:529–536. doi: 10.1021/tx034267y. [DOI] [PubMed] [Google Scholar]
- 14.Lindeke B. The Non- and postenzymatic chemistry of N-oxygenated molecules. Drug Metab Rev. 1982;13:71–121. doi: 10.3109/03602538209002232. [DOI] [PubMed] [Google Scholar]
- 15.Alexander J, Wallin H, Rossland OJ, Solberg KE, Holme JA, Becher G, Andersson R, Grivas S. Formation of a glutathione conjugate and a semistable transportable glucuronide conjugate of N2-oxidized species of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) in rat liver. Carcinogenesis. 1991;12:2239–2245. doi: 10.1093/carcin/12.12.2239. [DOI] [PubMed] [Google Scholar]
- 16.Reistad R, Frandsen H, Grivas S, Alexander J. In vitro formation and degradation of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) protein adducts. Carcinogenesis. 1994;15:2547–2552. doi: 10.1093/carcin/15.11.2547. [DOI] [PubMed] [Google Scholar]
- 17.Chepanoske CL, Brown K, Turteltaub KW, Dingley KH. Characterization of a peptide adduct formed by N-acetoxy-2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP), a reactive intermediate of the food carcinogen PhIP. Food Chem Toxicol. 2004;42:1367–1372. doi: 10.1016/j.fct.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 18.Skipper PL, Tannenbaum SR. Protein adducts in the molecular dosimetry of chemical carcinogens. Carcinogenesis. 1990;11:507–518. doi: 10.1093/carcin/11.4.507. [DOI] [PubMed] [Google Scholar]
- 19.Skipper PL, Peng X, SooHoo CK, Tannenbaum SR. Protein adducts as biomarkers of human carcinogen exposure. Drug Metab Rev. 1994;26:111–124. doi: 10.3109/03602539409029787. [DOI] [PubMed] [Google Scholar]
- 20.Tornqvist M, Fred C, Haglund J, Helleberg H, Paulsson B, Rydberg P. Protein adducts: quantitative and qualitative aspects of their formation, analysis and applications. J Chromatogr B Analyt Technol Biomed Life Sci. 2002;778:279–308. doi: 10.1016/s1570-0232(02)00172-1. [DOI] [PubMed] [Google Scholar]
- 21.Neumann HG. Biomonitoring of aromatic amines and alkylating agents by measuring hemoglobin adducts. Int Arch Occup Environ Health. 1988;60:151–155. doi: 10.1007/BF00378690. [DOI] [PubMed] [Google Scholar]
- 22.Kiese M, Taeger K. The fate of phenylhydroxylamine in human red cells. Naunyn Schmiedebergs Arch Pharmacol. 1976;292:59–66. doi: 10.1007/BF00506490. [DOI] [PubMed] [Google Scholar]
- 23.Green LC, Skipper PL, Turesky RJ, Bryant MS, Tannenbaum SR. In vivo dosimetry of 4-aminobiphenyl in rats via a cysteine adduct in hemoglobin. Cancer Res. 1984;44:4254–4259. [PubMed] [Google Scholar]
- 24.Bryant MS, Vineis P, Skipper PL, Tannenbaum SR. Hemoglobin adducts of aromatic amines: associations with smoking status and type of tobacco. Proc Natl Acad Sci U S A. 1988;85:9788–9791. doi: 10.1073/pnas.85.24.9788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones CR, Sabbioni G. Identification of DNA adducts using HPLC/MS/MS following in vitro and in vivo experiments with arylamines and nitroarenes. Chem Res Toxicol. 2003;16:1251–1263. doi: 10.1021/tx020064i. [DOI] [PubMed] [Google Scholar]
- 26.Skipper PL, Tannenbaum SR, Ross RK, Yu MC. Nonsmoking-related arylamine exposure and bladder cancer risk. Cancer Epidemiol Biomarkers Prev. 2003;12:503–507. [PubMed] [Google Scholar]
- 27.Gan J, Skipper PL, Gago-Dominguez M, Arakawa K, Ross RK, Yu MC, Tannenbaum SR. Alkylaniline-hemoglobin adducts and risk of non-smoking-related bladder cancer. J Natl Cancer Inst. 2004;96:1425–1431. doi: 10.1093/jnci/djh274. [DOI] [PubMed] [Google Scholar]
- 28.Turesky RJ, Skipper PL, Tannenbaum SR. Binding of 2-amino-3-methylimidazo[4,5-f]quinoline to hemoglobin and albumin in vivo in the rat. Identification of an adduct suitable for dosimetry. Carcinogenesis. 1987;8:1537–1542. doi: 10.1093/carcin/8.10.1537. [DOI] [PubMed] [Google Scholar]
- 29.Lynch AM, Murray S, Boobis AR, Davies DS, Gooderham NJ. The measurement of MeIQx adducts with mouse haemoglobin in vitro and in vivo: implications for human dosimetry. Carcinogenesis. 1991;12:1067–1072. doi: 10.1093/carcin/12.6.1067. [DOI] [PubMed] [Google Scholar]
- 30.Dingley KH, Freeman SP, Nelson DO, Garner RC, Turteltaub KW. Covalent binding of 2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline to albumin and hemoglobin at environmentally relevant doses. Comparison of human subjects and F344 rats. Drug Metab Dispos. 1998;26:825–828. [PubMed] [Google Scholar]
- 31.Dingley KH, Curtis KD, Nowell S, Felton JS, Lang NP, Turteltaub KW. DNA and protein adduct formation in the colon and blood of humans after exposure to a dietary-relevant dose of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine. Cancer Epidemiol Biomarkers Prev. 1999;8:507–512. [PubMed] [Google Scholar]
- 32.Lynch AM, Murray S, Zhao K, Gooderham NJ, Boobis AR, Davies DS. Molecular dosimetry of the food-borne carcinogen MeIQx using adducts of serum albumin. Carcinogenesis. 1993;14:191–194. doi: 10.1093/carcin/14.2.191. [DOI] [PubMed] [Google Scholar]
- 33.Rubino FM, Pitton M, Di FD, Colombi A. Toward an “omic” physiopathology of reactive chemicals: thirty years of mass spectrometric study of the protein adducts with endogenous and xenobiotic compounds. Mass Spectrom Rev. 2009;28:725–784. doi: 10.1002/mas.20207. [DOI] [PubMed] [Google Scholar]
- 34.Rappaport SM, Li H, Grigoryan H, Funk WE, Williams ER. Adductomics: Characterizing exposures to reactive electrophiles. Toxicol Lett. 2011 doi: 10.1016/j.toxlet.2011.04.002. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Noort D, Fidder A, Hulst AG. Modification of human serum albumin by acrylamide at cysteine-34: a basis for a rapid biomonitoring procedure. Arch Toxicol. 2003;77:543–545. doi: 10.1007/s00204-003-0484-5. [DOI] [PubMed] [Google Scholar]
- 36.Fasco MJ, Iii CR, Stack RF, O'hehir C, Barr JR, Eadon GA. Cyanide adducts with human plasma proteins: albumin as a potential exposure surrogate. Chem Res Toxicol. 2007;20:677–684. doi: 10.1021/tx6003425. [DOI] [PubMed] [Google Scholar]
- 37.Noort D, Hulst AG, Jansen R. Covalent binding of nitrogen mustards to the cysteine-34 residue in human serum albumin. Arch Toxicol. 2002;76:83–88. doi: 10.1007/s00204-001-0318-2. [DOI] [PubMed] [Google Scholar]
- 38.Aldini G, Regazzoni L, Orioli M, Rimoldi I, Facino RM, Carini M. A tandem MS precursor-ion scan approach to identify variable covalent modification of albumin Cys34: a new tool for studying vascular carbonylation. J Mass Spectrom. 2008;43:1470–1481. doi: 10.1002/jms.1419. [DOI] [PubMed] [Google Scholar]
- 39.Wang Z, Ramsdell JS. Analysis of interactions of brevetoxin-B and human serum albumin by liquid chromatography/mass spectrometry. Chem Res Toxicol. 2011;24:54–64. doi: 10.1021/tx1002854. [DOI] [PubMed] [Google Scholar]
- 40.Damsten MC, Commandeur JN, Fidder A, Hulst AG, Touw D, Noort D, Vermeulen NP. Liquid chromatography/tandem mass spectrometry detection of covalent binding of acetaminophen to human serum albumin. Drug Metab Dispos. 2007;35:1408–1417. doi: 10.1124/dmd.106.014233. [DOI] [PubMed] [Google Scholar]
- 41.Bechtold WE, Willis JK, Sun JD, Griffith WC, Reddy TV. Biological markers of exposure to benzene: S-phenylcysteine in albumin. Carcinogenesis. 1992;13:1217–1220. doi: 10.1093/carcin/13.7.1217. [DOI] [PubMed] [Google Scholar]
- 42.Magagnotti C, Orsi F, Bagnati R, Celli N, Rotilio D, Fanelli R, Airoldi L. Effect of diet on serum albumin and hemoglobin adducts of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) in humans. Int J Cancer. 2000;88:1–6. doi: 10.1002/1097-0215(20001001)88:1<1::aid-ijc1>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 43.Neumann HG. Analysis of hemoglobin as a dose monitor for alkylating and arylating agents. Arch Toxicol. 1984;56:1–6. doi: 10.1007/BF00316343. [DOI] [PubMed] [Google Scholar]
- 44.Turesky RJ, Lang NP, Butler MA, Teitel CH, Kadlubar FF. Metabolic activation of carcinogenic heterocyclic aromatic amines by human liver and colon. Carcinogenesis. 1991;12:1839–1845. doi: 10.1093/carcin/12.10.1839. [DOI] [PubMed] [Google Scholar]
- 45.Westra JG. A rapid and simple synthesis of reactive metabolites of carcinogenic aromatic amines in high yield. Carcinogenesis. 1981;2:355–357. doi: 10.1093/carcin/2.4.355. [DOI] [PubMed] [Google Scholar]
- 46.Men L, Wang Y. Fragmentation of the deprotonated ions of peptides containing cysteine, cysteine sulfinic acid, cysteine sulfonic acid, aspartic acid, and glutamic acid. Rapid Commun Mass Spectrom. 2006;20:777–784. doi: 10.1002/rcm.2374. [DOI] [PubMed] [Google Scholar]
- 47.Peters T., Jr . All about Albumin Biochemistry, Genetics, and Medical applications. Academic Press; San Diego: 1996. [Google Scholar]
- 48.Boyer PD. Spectrophotometric Study of the Reaction of Protein Sulfhydryl Groups with Organic Mercurials. J Am Chem Soc. 1954;76:4331–4337. [Google Scholar]
- 49.Riener CK, Kada G, Gruber HJ. Quick measurement of protein sulfhydryls with Ellman's reagent and with 4,4'-dithiodipyridine. Anal Bioanal Chem. 2002;373:266–276. doi: 10.1007/s00216-002-1347-2. [DOI] [PubMed] [Google Scholar]
- 50.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsao M, Otter DE. Quantification of glutamine in proteins and peptides using enzymatic hydrolysis and reverse-phase high-performance liquid chromatography. Anal Biochem. 1999;269:143–148. doi: 10.1006/abio.1998.3091. [DOI] [PubMed] [Google Scholar]
- 52.Baxter JH, Lai CS, Phillips RR, Dowlati L, Chio JJ, Luebbers ST, Dimler SR, Johns PW. Direct determination of methionine sulfoxide in milk proteins by enzyme hydrolysis/high-performance liquid chromatography. J Chromatogr A. 2007;1157:10–16. doi: 10.1016/j.chroma.2007.04.035. [DOI] [PubMed] [Google Scholar]
- 53.Turesky RJ, Parisod V, Huynh-Ba T, Langouët S, Guengerich FP. Regioselective differences in C(8)- and N-oxidation of 2-amino-3,8- dimethylimidazo[4,5-f]quinoxaline by human and rat liver microsomes and cytochromes P450 1A2. Chem Res Toxicol. 2001;14:901–911. doi: 10.1021/tx010035s. [DOI] [PubMed] [Google Scholar]
- 54.Peters T., Jr Serum albumin. Adv Protein Chem. 1985;37:161–245. doi: 10.1016/s0065-3233(08)60065-0. [DOI] [PubMed] [Google Scholar]
- 55.Fu X, Mueller DM, Heinecke JW. Generation of intramolecular and intermolecular sulfenamides, sulfinamides, and sulfonamides by hypochlorous acid: a potential pathway for oxidative cross-linking of low-density lipoprotein by myeloperoxidase. Biochemistry. 2002;41:1293–1301. doi: 10.1021/bi015777z. [DOI] [PubMed] [Google Scholar]
- 56.Stewart AJ, Blindauer CA, Berezenko S, Sleep D, Tooth D, Sadler PJ. Role of Tyr84 in controlling the reactivity of Cys34 of human albumin. FEBS J. 2005;272:353–362. doi: 10.1111/j.1742-4658.2004.04474.x. [DOI] [PubMed] [Google Scholar]
- 57.Carter DC, Ho JX. Structure of serum albumin. Adv Protein Chem. 1994;45:153–203. doi: 10.1016/s0065-3233(08)60640-3. [DOI] [PubMed] [Google Scholar]
- 58.Christodoulou J, Sadler PJ. 1H NMR of albumin in human blood plasma: drug binding and redox reactions at Cys34. FEBS Lett. 1995;376:1–5. doi: 10.1016/0014-5793(95)01231-2. [DOI] [PubMed] [Google Scholar]
- 59.Kinter M, Sherman NE. Protein Sequencing and Identification Using Tandem Mass Spectrometry. Wiley-Interscience; New York: 2000. [Google Scholar]
- 60.Russell WK, Park ZY, Russell DH. Proteolysis in mixed organic-aqueous solvent systems: applications for peptide mass mapping using mass spectrometry. Anal Chem. 2001;73:2682–2685. doi: 10.1021/ac001332p. [DOI] [PubMed] [Google Scholar]
- 61.Simon LM, László K, Vértesi A, Bagi K, Szájani B. Stability of hydrolytic enzymes in water-organic solvent systems. J Mol Catal B: Enzymatic. 1998;4:41–45. [Google Scholar]
- 62.Noort D, Hulst AG, de Jong LP, Benschop HP. Alkylation of human serum albumin by sulfur mustard in vitro and in vivo: mass spectrometric analysis of a cysteine adduct as a sensitive biomarker of exposure. Chem Res Toxicol. 1999;12:715–721. doi: 10.1021/tx9900369. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.