Abstract
The Candida albicans cell wall provides an architecture that allows for the organism to survive environmental stresses as well as interaction with host tissues. Previous work has focused on growing C. albicans on media such as Sabouraud or YPD at 30 °C. Because C. albicans normally colonizes a host, we hypothesized that cultivation on blood or serum at 37 °C would result in structural changes in cell wall mannan. C. albicans SC5314 was inoculated onto YPD, 5% blood, or 5% serum agar media three successive times at 30 °C and 37 °C, then cultivated overnight at 30 °C in YPD. The mannan was extracted and characterized using 1D and 2D 1H NMR techniques. At 30 °C cells grown in blood and serum contain less acid-stable terminal β-(1→2)-linked D-mannose and α-(1→2)-linked D-mannose-containing side chains, while the acid-labile side chains of mannan grown in blood and serum contain fewer β-Man-(1→2)-α-Man-(1→ side chains. The decrement in acid-stable mannan side chains is greater at 37 °C than at 30 °C. Cells grown on blood at 37 °C show fewer →6)-α-Man-(1→ structural motifs in the acid-stable polymer backbone. The data indicate that C. albicans, grown on media containing host derived components, produces less complex mannan. This is accentuated when the cells are cultured at 37 °C. This study demonstrates that the C. albicans cell wall is a dynamic and adaptive organelle, which alters its structural phenotype in response to growth in host-derived media at physiological temperature.
Keywords: Mannan, NMR, Growth conditions
1. Introduction
The opportunistic pathogen Candida albicans is one of the most commonly isolated organisms as a hospital-acquired infection in critical care wards.1,2 C. albicans is a commensal organism readily isolated from human skin and has several virulence traits, including the production of hydrolytic enzymes, and adhesins as well as the ability to undergo a morphological shift from yeast to hyphal morphologies, 3–5 which allow it to be pathogenic in immunocompromised individuals. In addition, one of the major contributors to C. albicans virulence is its cell wall, which serves a major function in protecting the organism from environmental stress as well as acting as a support structure for attachment of adhesion molecules and other virulence-associated proteins. In addition, the fungal cell wall serves as the interface with the environment in which the cell grows, such as a human host. Recent studies have demonstrated that the fungal cell wall is recognized by a number of host specific receptors that allow for an appropriate immune response.6,7
The C. albicans cell wall is composed of chitin (a polymer of N-acetylglucosamine) that is attached to a matrix of β-D-(1→3)-glucan. The β-D-(1→3)-glucan polymer has a number of β-D-(1→6)-glucan branch points, which serve as linkage sites via a glycophosphatidylinositol remnant and internal repeat (PIR) moieties for N- and O-linked mannosylated protein attachment.8 It has been suggested that C. albicans masks the underlying cell wall β-D-(1→3)-glucan with a dense layer of mannan and/or mannoprotein.9 As β-D-glucan is the primary fungal pathogen associated molecular pattern (PAMP),10–12 this “masking” or covering the glucan with mannan/mannoprotein is thought to reduce recognition of the yeast by anti-fungal innate immune mechanisms, such as via recognition by Dectin-1.6 The Candida cell wall has been extensively studied, but most of these investigations have focused on defining cell wall structure following cultivation in medium such as YPD or Sabouraud agar.13–21 While these data have advanced our knowledge, they have not addressed the question of what structural changes occur in the Candida cell wall in response to cultivation in complex biological media, such as blood or serum.
Indeed, little is known about the changes that occur in the cell wall composition and architecture as a result of cultivating fungi on different media and under different environmental conditions, such as growth at physiologic temperature (37 °C). In this study, we compared and contrasted the structure of mannan/mannoprotein in the cell wall of C. albicans strain SC5314 grown in blood, serum, or YPD. In addition, we also compared and contrasted the effect of cultivation temperature on mannan/mannoprotein structure. To the best of our knowledge, this is the first in depth investigation of how growth conditions, i.e. temperature and medium, impact the structural phenotype of mannan/mannoprotein in the C. albicans cell wall. When considered as a whole, our results indicate that the C. albicans cell wall mannan structure is complex, dynamic and highly adaptable. We speculate that changing the structural phenotype of the cell wall may confer a survival advantage to the organism.
2. Results and Discussion
Over more than a decade, several groups 18,22–29 have made great strides in understanding the structure of mannan isolated from fungal cell walls of several Candida species. By using elegant 2D NMR experiments (up to 600 MHz) these investigators examined the structural details of mannan side chains after carefully degrading the mannan and isolating the side chain fragments. Vinogradov and coworkers30 used 750 MHz NMR to extend these structural studies on intact mannan isolated from Saccharomyces cerevisiae with and without degradation followed by isolation of the fragmented side chains. Maes and coworkers 31 used solid-state magic-angle-spinning NMR at 800 MHz to examine mannan structures in intact cell walls of live C. albicans cells. Using solid-state NMR, Maes and coworkers observed the anomeric proton resonance for α-Man-1→PO4, i.e., a single mannosyl repeat unit attached to the phosphodiester linkage. However, Maes and coworkers31 and others18,22–30,32 did not report identification of this linkage in previous solution-state NMR studies.
2.1 Growth on different culture media at different temperatures alters mannan structure
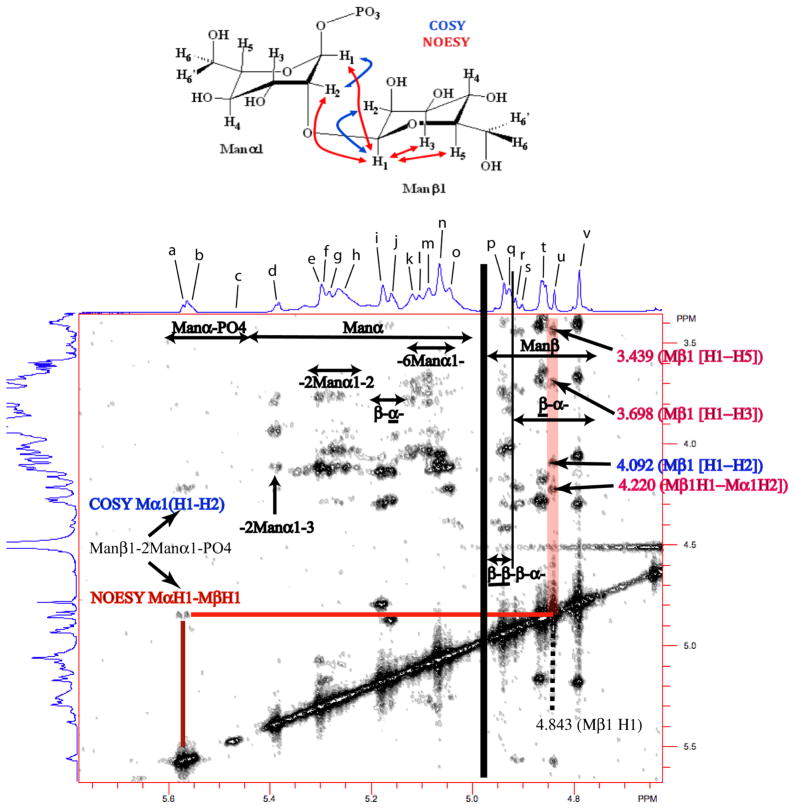
In this study we utilized 1D and 2D COSY and NOESY NMR experiments at 950 MHz (Figs. S1–S3) as well as previously published chemical shift assignments characteristic of individual mannosyl motifs in specific side chain fragments based on 2D NMR studies of isolated side chain fragments22–28,30–32 to assign proton NMR resonances to structural motifs of non-degraded, intact mannans. Specifically we correlated the unique chemical shifts of the anomeric proton, H-1, and its neighboring proton, H-2, in specific mannosyl repeat units of isolated mannan side chain fragments to the chemical shifts of mannosyl repeat units in similar chemical environments in non-degraded, intact mannans. By this approach, we provide structural assignments for acid-stable and acid-labile mannan side chains without the time-consuming degradation and isolation of individual side chain fragments and detailed 2D NMR side chain structural characterization studies. Based upon these assignments, detailed structural changes in isolated cell wall mannan from C. albicans SC5314 strain as a function of temperature and growth medium conditions were made based upon 600 MHz proton 1D NMR spectra. Tables 1 and 2 show chemical shift assignments for H-1 and H-2 protons from specific mannosyl repeat units indicated in Bold within the structural motifs indicated as a to v based upon COSY and NOESY NMR data. From the 2D COSY (Fig. S2) and NOESY spectra (Fig. 2), various structural features can be discerned. For example, for the structural motif β-Man-(1→2)-α-Man-1→PO4 (Fig. 2, a), the anomeric proton, H-1, of α-Man-1→PO4 resonates at 5.572 ppm while H-2 resonates at 4.219 ppm (Fig. 2 and peak a, Tables 1 and 2). The NOESY correlation across the glycosidic linkage of β-Man-(1→2)-α-Man-1→PO4 between anomeric protons (α-Man1 [5.572 ppm] →β-Man1 [4.840 ppm], (peak u, Tables 1 and 2)) is clearly evident. The β-Man1 anomeric proton resonance showed the expected correlation to α-Man1 H-2 (4.219 ppm) across the glycosidic link and the expected intra residue NOEs with H-2 (4.088 ppm), H-3 (3.698 ppm), and H-5 (3.439 ppm). Taken together these data define the β-Man-(1→2)-α-Man-(1→PO4 structural motif in the acid-labile portion of mannan. Similarly, other spectral regions can be defined for structural motifs containing α-Man and β-Man in subregions →2)-α-Man-(1→, →2)-α-Man-(1→3, →6)-α-Man-(1→, →2)-β-Man-(1→2)-β-Man-(1→2)-β-Man-(1→ (β-β-β-), and →2)-β-Man-(1→2)-α-Man-(1→ (β-α-) as well as α-Man-PO4 (Fig. 2). A few of the resonance assignments in Tables 1–3 show multiple side chain assignments associated with particular resonances, such as the assignments for side chains e and f. The structural fragments assigned to these resonances are very similar and exhibit similar H-1 and H-2 chemical shifts for the indicated mannosyl motifs. Changes in the intensity of the overlapping resonances indicated by peaks e and f indicate changes in side chain composition involving →3)-α-Man-(1→2)-αMan-(1→3)-[α-Man-(1→6]α-Man-(1→2 structural motifs and are examined together for changes in side chain composition. The three overlapping resonances indicated by peak t are assigned to terminal and internal β-(1→2)-linked D-mannosyl repeat units in either acid-stable or the acid-labile side chains of the mannan isolate. Different intensities for peaks q and t reflect changes in side chains that contain β-Man-(1→ [2)-β-Man(1→]n2)-α-Man-(1→ (n = 1 for peak t and = 2 for peak q) structural motifs primarily which may also be reflected in peaks b, p, r, and s. The H-1 resonance at 4.927 ppm for α-Man-(1→6) motifs overlaps resonances for other structural motifs assigned as peak q and therefore is difficult to analyze in terms of structural changes to the mannan isolate.
Table 1.
Changes in mannan structural motifs as a function of growth in either blood or serum and growth temperature during the three passages at either 30 °C or 37 °C and final growth at either 30 °C or 37 °C relative to growth in YPD under the same conditions. Peak intensity differences are based upon spectral normalization using peak n.
| Label | 1H Chemical Shifts (ppm) | Structural Motifs | 30 °C/30 °Ca | 37 °C/30 °C | 37 °C/37 °C | ||||
|---|---|---|---|---|---|---|---|---|---|
| H-1 | H-2 | - | Bloodb | Serumb | Bloodb | Serumb | Bloodb | Serumb | |
| a | 5.572 | 4.219 | β-Man-(1→2)-α-Man-(1→PO4 | ⇓c | ⇓ | = | ⇑ | ⇑ | ⇑ |
| b | 5.560 | 4.207 | β-Man-(1→ [2β-Man-(1→]n2)-β-Man-(1→2)-α-Man- (1→PO4 | = | = | = | ⇔ | ⇑ | ⇑ |
| c | 5.471 | 4.033 | α-Man-(1→PO4 | = | ⇑ | ⇑ | ⇔ | ⇑ | ⇔ |
| d | 5.385 | 4.109 | α-Man-(1→2)-α-Man-(1→3)-α-Man-(1→ | ⇓ | = | ⇓ | ⇓ | = | ⇓ |
| e | 5.303 | 4.120 | α-Man-(1→3?)-α-Man-(1→2)-α-Man-(1→3)-(α-Man- (1→6)-α-Man-(1→2) | ⇓ | ⇓ | ⇓ | ⇓ | ⇓ | ⇔ |
| f | 5.297 | 4.135 | α-Man-(1→3)-α-Man-(1→6)-α-Man-(1→2)-α-Man- (1→3)-α-Man-(1→6)-α-Man-(1→2) | ⇓ | ⇓ | ⇓ | ⇓ | ⇓ | ⇔ |
| g | 5.286 | 4.132 | α-Man-(1→2)-α-Man-(1→2) | ⇓ | ⇓ | ⇓ | ⇓ | ⇑ | ⇑ |
| h | 5.266 | 4.110 | α-Man-(1→2)-α-Man-(1→2) | ⇓ | ⇓ | ⇓ | ⇓ | ⇑ | ⇔&⇑ |
| i | 5.179 | 4.290 | β-Man-(1→2)-α-Man-(1→ | ⇓ | ⇓ | ⇓ | ⇓ | ⇑ | ⇑ |
| j | 5.162 | 4.271 | β-Man-(1→2)-α-Man-(1→2) | ⇓ | ⇓ | ⇓ | ⇓ | ⇑ | ⇑ |
| k | 5.120 | 4.048 | →6)-α-Man-(1→ d | ⇓ | ⇓ | ⇓ | ⇓ | -e | -e |
| l | 5.107 | -e | →6)-α-Man-(1→ d | ⇓ | ⇓ | ⇓ | ⇓ | -e | -e |
| m | 5.088 | 4.089 | →6)-α-Man-(1→ d | ⇓ | ⇓ | ⇓ | ⇓ | -e | -e |
| n | 5.067 | 4.083 | α-Man-(1→2 →6)-α-Man-[α-Man-(1→2)-α-Man]-(1→ | = | = | = | = | = | = |
| o | 5.045 | 4.222 | α-Man-(1→3)-α-Man-(1→2)-α-Man-(1→ | = | ⇓ | ⇓ | ⇓ | ⇑ | ⇑ |
| p | 4.940 | 4.171 | β-Man-(1→2)-[β-Man-(1→2)]n-α-Man-(1→f β-Man-(1→2)-β-Man-(1→2)-β-Man-(1→2)-α-Man- (1→PO4 | = | = | = | = | ⇑ | ⇑ |
| q | 4.933 | 4.414 | β-Man-(1→2)-β-Man-(1→2)-β-Man-(1→2)-α-Man- (1→β-Man-(1→2)-β-Man-(1→2)-β -Man-(1→2)-α-Man- (1→PO4 | ⇓ | ⇓ | ⇓ | ⇓ | ⇔ | ⇓ |
| q | 4.930 | 4.163 | β-Man-(1→2)-β-Man-(1→2)-β-Man-(1→2)-α-Man- (1→PO4 | ⇓ | ⇓ | ⇓ | ⇓ | ⇔ | ⇓ |
| q | 4.927 | 4.016 | α-Man-(1→6) | -g | -g | -g | -g | -e | -e |
| r | 4.917 | 4.309 | β-Man-(1→ [β-Man-(1→2)]n2)-β-Man-(1→2)-α-Man- (1→PO4 | ⇔ | ⇔ | ⇓ | ⇔ | ⇔ | ⇓ |
| s | 4.904 | 4.293 | β-Man-(1→ [β-Man-(1→2)]n2)-β-Man-(1→2)-α-Man- (1→PO4 | = | = | ⇔ | ⇑ | ⇔ | ⇔ |
| t | 4.866 | 4.276 | β-Man-(1→2)-β-Man-(1→2)-α-Man-(1→ | ⇓ | ⇓ | ⇓ | ⇓ | ⇑ | ⇑ |
| t | 4.862 | 4.168 | β-Man-(1→2)-β-Man-(1→2)-α-Man-(1→PO4 | ⇓ | ⇓ | ⇓ | ⇓ | ⇑ | ⇑ |
| t | 4.857 | 4.168 | β-Man-(1→2)-β-Man-(1→2)-α-Man-(1→2(3) β-Man-(1→2)-β-Man-(1→2)-α-Man-(1→PO4 | ⇓ | ⇓ | ⇓ | ⇓ | ⇑ | ⇑ |
| u | 4.840 | 4.088 | β-Man-(1→2)-αMan-(1→PO4 | ⇓ | ⇓ | ⇔ | = | ⇑ | ⇑ |
| v | 4.791 | 4.056 | β-Man-(1→2)-αMan-(1→ | ⇓ | ⇓ | ⇓ | ⇓ | ⇑ | ⇑ |
Growth temperatures are shown as follows: growth temperature during three passages, slash separator, then final growth temperature.
Relative to presence of the structural feature in mannan isolated from YPD with the resonance for →6α-Man-(1→ at 5.07 ppm set to the same height for both spectra.
Symbols: =: no difference; ⇔: very slight difference; ⇑: increase; ⇓: decrease
Repeat unit along the backbone with different side chains attached.
COSY crosspeak not observed.
Chemical shift for the anomeric proton of Man next to terminal β–Man-(1→
See text for discussion.
Table 2.
Mannan structural changes as a function of temperature for growth under 30 °C/37 °C and 37 °C/37 °C (constant 37 °C) growth conditions relative to 30 °C/30 °C (constant 30 °C) growth conditions as a function of growth medium. Peak intensity differences are based upon spectral normalization using peak n.
| Label | 1H Chemical Shifts (ppm) | Structural Motifs | 37 °C/30 °Ca | 37 °C/37 °C | |||||
|---|---|---|---|---|---|---|---|---|---|
| H-1 | H-2 | YPDb | Bloodb | Serumb | YPDb | Bloodb | Serumb | ||
| a | 5.572 | 4.219 | β-Man-(1→2)-α-Man-(1→PO4 | ⇓c | = | ⇓ | ⇓ | ⇓ | ⇓ |
| b | 5.560 | 4.207 | β-Man-(1→ [β-Man-(1→2]n2)-β-Man-(1→2)-α-Man- (1→PO4 | = | = | = | ⇓ | ⇓ | ⇓ |
| c | 5.471 | 4.033 | α-Man-(1→PO4 | = | = | ⇓ | ⇓ | ⇓ | ⇓ |
| d | 5.385 | 4.109 | α-Man-(1→2)-α-Man-(1→3)-α-Man-(1→ | ⇑ | ⇑ | ⇓ | ⇔ | ⇔ | ⇔ |
| e | 5.303 | 4.120 | α-Man-(1→3)[? →]-α-Man-(1→2)-α-Man-(1→3)-α-Man-(1→6)-α-Man-(1→2) | ⇓ | ⇑ | ⇓ | ⇓ | ⇓ | ⇓ |
| f | 5.297 | 4.135 | α-Man-(1→3)-[α-Man-(1→6)]α-Man-(1→2)-α-Man- (1→3)[αMan-(1→6)]-α-Man-(1→2) | ⇓ | ⇑ | ⇓ | ⇓ | ⇓ | ⇓ |
| g | 5.286 | 4.132 | α-Man-(1→2)-α-Man-(1→2) | ⇔ | ⇔ | ⇓ | ⇓ | ⇓ | ⇓ |
| h | 5.266 | 4.110 | α-Man-(1→2)-α-Man-(1→2) | ⇑ | ⇑ | ⇓ | ⇓ | ⇓ | ⇓ |
| i | 5.179 | 4.290 | β-Man-(1→2)-α-Man-(1→ | ⇓ | ⇑ | ⇓ | ⇓ | ⇓ | ⇓ |
| j | 5.162 | 4.271 | β-Man-(1→2)-α-Man-(1→2) | ⇑ | ⇑ | ⇓ | ⇓ | ⇓ | ⇓ |
| k | 5.120 | 4.048 | →6)-α-Man-(1→ d | = | = | ⇓ | -e | -e | -e |
| l | 5.107 | -e | →6)-α-Man-(1→ d | ⇓ | = | ⇓ | -e | -e | -e |
| m | 5.088 | 4.089 | →6)-α-Man-(1→ d | ⇓ | ⇑ | ⇓ | -e | -e | -e |
| n | 5.067 | 4.083 | α-Man-(1→2) →6)-α-Man(α-Man-(1→2)α-Man)-(1→ | = | = | = | = | = | = |
| o | 5.045 | 4.222 | α-Man-(1→3)-α-Man-(1→2)-α-Man-(1→ | ⇔ | = | ⇓ | ⇓ | ⇓ | ⇓ |
| p | 4.940 | 4.171 | β-Man-(1→2)[β-Man-(1→2)]n-α-Man-(1→f β-Man-(1→2)-β-Man-(1→2)-β-Man-(1→2)-α-Man- (1→PO4 | = | = | ⇔ | ⇓ | ⇓ | ⇓ |
| q | 4.933 | 4.414 | β-Man-(1→2)-β-Man-(1→2)-β-Man-(1→2)-α-Man- (1→β-Man-(1→2)-β-Man-(1→2)-β-Man-(1→2)-α-Man- (1→PO4 | = | ⇑ | ⇓ | ⇓ | ⇓ | ⇓ |
| q | 4.930 | 4.163 | β-Man-(1→2)-β-Man-(1→2)-β-Man-(1→2)-α-Man- (1→PO4 | = | ⇑ | ⇓ | ⇓ | ⇓ | ⇓ |
| q | 4.927 | 4.016 | α-Man-(1→6) | -g | -g | -g | -g | -e | -e |
| r | 4.917 | 4.309 | β-Man-(1→ [β-Man-(1→2)]n2)-β-Man-(1→2)-α-Man- (1→PO4 | = | ⇓ | ⇔ | ⇓ | ⇓ | ⇓ |
| s | 4.904 | 4.293 | β-Man-(1→ [β-Man-(1→2)]n2)-β-Man-(1→2)-α-Man- (1→PO4 | = | ⇔ | ⇔ | ⇓ | ⇓ | ⇓ |
| t | 4.866 | 4.276 | β-Man-(1→2)-β-Man-(1→2)-α-Man-(1→ | = | ⇑ | ⇓ | ⇓ | ⇓ | ⇓ |
| t | 4.862 | 4.168 | β-Man-(1→2)-β-Man-(1→2)-α-Man-(1→PO4 | = | ⇑ | ⇓ | ⇓ | ⇓ | ⇓ |
| t | 4.857 | 4.168 | β-Man-(1→2)-β-Man-(1→2)-α-Man-(1→2(3) β-Man-(1→2)-β-Man-(1→2)-α-Man-(1→PO4 | = | ⇑ | ⇓ | ⇓ | ⇓ | ⇓ |
| u | 4.840 | 4.088 | β-Man-(1→2)-α-Man-(1→PO4 | ⇓ | ⇔ | ⇓ | ⇓ | ⇓ | ⇓ |
| v | 4.791 | 4.056 | β-Man-(1→2)-α-Man-(1→ | ⇓ | ⇑ | ⇓ | ⇓ | ⇓ | ⇓ |
Growth temperatures are shown as follows: growth temperature during three passages, slash separator, then final growth temperature.
Relative to presence of the structural feature in mannan grown on the stated medium at 30°C/30°C with resonance n for →6α-Man-(1→ at 5.067 ppm set to the same height for both spectra.
Symbols: =: no difference; ⇔: very slight difference; ⇑: increase; ⇓: decrease
Repeat unit along the backbone with different side chains attached.
COSY crosspeak not observed.
Chemical shift for the anomeric proton of Man next to terminal β–Man-(1→
See text for discussion.
Figure 2.
2D NOESY mannan 950 MHz NMR spectrum expanded to show detailed correlations between the anomeric proton spectral region and the rest of the carbohydrate spectral region. Specific COSY (blue) and NOESY (red) 2D correlations observed between and within monomers in the β-Man-(1→2)-α-Man-(1→PO4 acid-labile structural motif are indicated in the structure and in the spectrum. In addition generalized assignment of the anomeric proton resonances to specific mannan structural motifs are indicated relative to the 1D spectrum. Resonance labels are defined in Tables 1 and 2.
Table 3.
Tabulation of structural motif assignments for mannan grown on YPD under 37 °C/37 °C conditions based upon the chemical shifts (in ppm) for the anomeric protons of the mannosyl repeat units.
| Label | Chemical Shifts (ppm) H-1 | Structural Feature |
|---|---|---|
| a | 5.568 | β-Man-(1→2)-α-Man-(1→PO4 |
| b | 5.556 | β-Man-(1→ [β-Man-(1→2)]n2)-β-Man-(1→2)-α-Man-(1→PO4 |
| c | 5.474 | α-Man-(1→PO4 |
| d | 5.385 | α-Man-(1→2)-α-Man-(1→3)-α-Man-(1→ |
| a′ | 5.375 | α-Man-(1→2)-α-Man-(1→3)-α-Man-(1→2) |
| b′ | 5.373 | α-Man-(1→2)-α-Man-(1→3)-α-Man-(1→2) |
| e | 5.308 | α-Man-(1→3)-[? →]-α-Man-(1→2)-α-Man-(1→3)[α-Man-(1→6)]-α-Man-(1→2)- |
| f | 5.291 | α-Man-(1→3)-α-Man-(1→6)-α-Man-(1→2)-α-Man-(1→3)][α-Man-(1→6)]-α-Man-(1→2) |
| g | 5.282 | α-Man-(1→2)-α-Man-(1→2) |
| h | 5.276 | α-Man-(1→2)-α-Man-(1→2) |
| h | 5.260 | α-Man-(1→2)-α-Man-(1→2) |
| c′ | 5.257 | α-Man-(1→2)-α-Man-(1→2)-α-Man-(1→2) |
| d′ | 5.247 | α-Man-(1→2)-α-Man-(1→2) |
| e′ | 5.226 | α-Man-(1→3)-α-Man-(1→6)-α-Man-(1→2)-α-Man-(1→2) |
| i | 5.183 | β-Man-(1→2)-α-Man-(1→2) |
| i | 5.179 | β-Man-(1→2)-α-Man-(1→ |
| j | 5.163 | β-Man-(1→2)-α-Man-(1→2) |
| f′ | 5.157 | α-Man-(1→3) ? |
| g′ | 5.131 | α-Man-(1→3) |
| k | 5.121 | →6)-α-Man-(1→ a |
| l | 5.110 | →6)-α-Man-(1→ a |
| m | 5.095 | →6)-α-Man-(1→ a |
| h′ | 5.072 | α-Man-(1→6)-[α-Man-(1→2)]-α-Man-(1→6)-α-Man-(1→6) |
| n | 5.064 | α-Man-(1→2; →6)-α-Man-[α-Man-(1→2)-α-Man]-(1→ |
| i′ | 5.053 | α-Man-(1→6(→2)α-Man-(1→6(α-Man-(1→2)-α-Man-(1→6(→2)-α-Man-(1→6 |
| p | 4.939 | β-Man-(1→2)-[β-Man-(1→2)]n-α-Man-(1→b |
| q | 4.928 | α-Man-(1→6) |
| r | 4.915 | β-Man-(1→ [β-Man-(1→2]n2)-β-Man-(1→2)-α-Man-(1→PO4 |
| s | 4.902 | β-Man-(1→ [β-Man-(1→2]n2)-β-Man-(1→2)-α-Man-(1→PO4 |
| j′ | 4.890 | β-Man-(1→ [β-Man-(1→2]n2)-β-Man-(1→2)-α-Man-(1→PO4 |
| t | 4.855 | β-Man-(1→2)-β-Man-(1→2)-α-Man-(1→2(3) |
| t | 4.853 | β-Man-(1→2)-β-Man-(1→2)-α-Man-(1→PO4 |
| u | 4.832 | β-Man-(1→2)-α-Man-(1→PO4 |
| v | 4.784 | β-Man-(1→2)-α-Man-(1→ |
Repeat unit along the backbone with different side chains attached.
Chemical shift for the anomeric proton of Man next to terminal β–Man-(1→
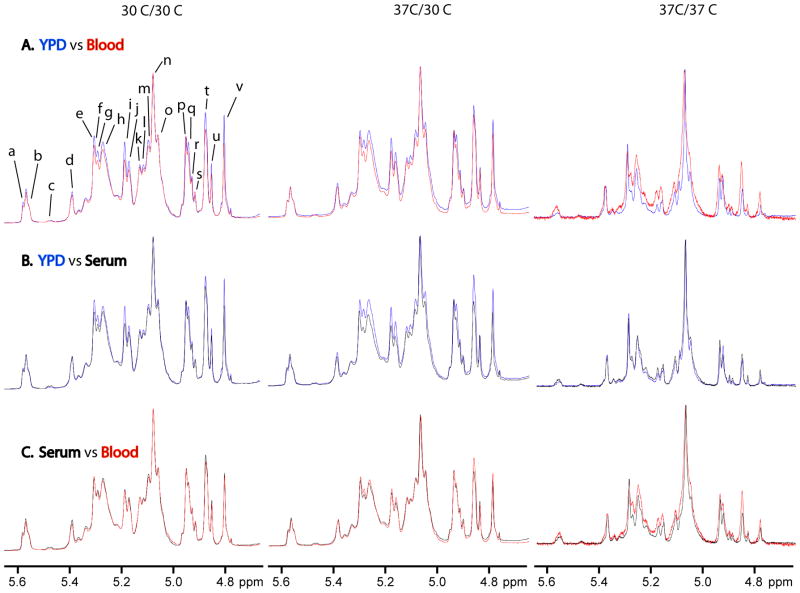
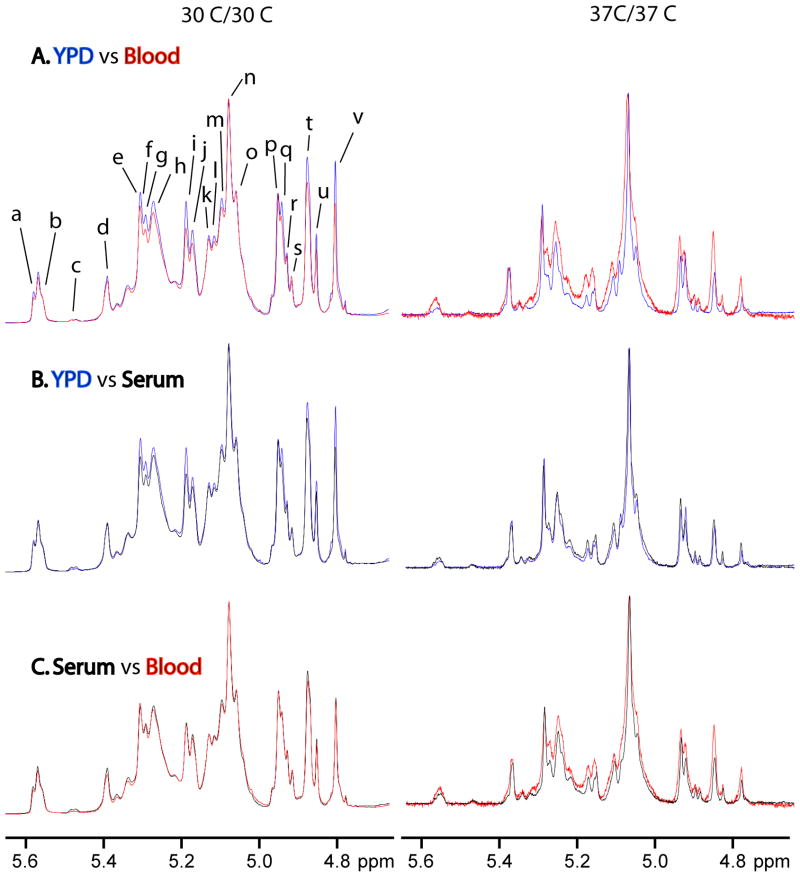
Characterization of these structural motifs enables rapid structural comparison of isolated mannans grown under different conditions of medium and temperature and allows us to define the resulting structural changes (Figs. 3–4). Under 30 °C/30 °C growth conditions, the acid-stable side chains show structural changes when grown in blood (Fig. 3A, left) and serum (Fig. 3B, left) relative to growth in YPD while the acid-labile side chains change only when grown in blood relative to YPD (Table 1). Mannan structures resulting from growth in serum and blood basically have the same structures (Fig. 3C, left). Structural changes for the acid-stable side chains grown in blood and serum relative to YPD show fewer terminal β-(1→2)-D-linked Man chain motifs and α-(1→2)-D-linked Man-containing side chains. The acid-labile side chains show a reduction in β-Man-(1→2)-α-Man-(1→PO4 side chains when grown in blood and serum, longer side chains remain constant when grown in either medium, and α-Man-(1→PO4 side chains remain constant when grown in blood but increase when grown in serum relative to growth in YPD.
Figure 3.
Comparison of the 600 MHz proton NMR spectra of mannans isolated from yeast grown on YPD, blood or serum (30 °C/30 °C (left), 37 °C/30 °C (middle), and 37 °C/37 °C (right)) for the following media: (A) YPD versus blood, (B) YPD versus serum, and (C) serum versus blood. Resonance labels are defined in Tables 1 and 2.
Figure 4.
Comparison of the 600 MHz proton NMR spectra of mannans isolated from yeast grown on YPD, blood or serum (37 °C/30 °C (left) and 37 °C/37 °C (right)) relative to conditions at 30 °C/30 °C for the following media: (A) YPD, (B) Blood, and (C) Serum. Resonance labels are defined in Tables 1 and 2.
Under 37 °C/30 °C growth conditions, the acid stable portion changes for both blood (Fig. 3A, middle) and serum (Fig. 3B, middle) relative to growth in YPD. The acid-labile portion remains nearly unchanged for both media except that α-Man-(1→PO4 side chains increase when grown on blood and β-Man-(1→2)-α-Man-(1→PO4 side chains increase when grown in serum relative to YPD (Table 1). Mannan structures resulting from growth on serum and blood exhibit only minor differences in the acid-stable side chains (Fig.3C, middle). Structural changes for the acid-stable side chains grown on blood and serum involved the same structural motifs as seen at 30 °C/30 °C, but with a greater loss in these side chains.
Under 37 °C/37 °C growth conditions, significant structural differences are observed for mannan isolated from yeast grown on blood relative to growth on either YPD (Fig. 3A, right) or serum (Fig. 3C, right). Mannan structures resulting from growth on serum and YPD have comparable levels of structural motifs with minor variations in their levels (Fig. 3B, right). The differences in the levels of structural motifs are not as dramatic as when either is compared to growth on blood. Side chains containing →2)-α-Man-(1→3)-α-Man-(1→ structural motifs have comparable levels when grown on serum and lower levels when grown on blood but higher levels of all other structural motifs in the acid-stable and acid-labile side chains compared to growth on YPD. Growth on serum compared to blood has slightly higher levels of α-Man-(1→ but lower levels of all other identified structural motifs. Also, the content of the acid-labile side chains is reduced in mannan isolated from yeast grown on YPD and serum relative to growth on blood.
Mannans isolated from yeast grown on YPD under 30 °C/30 °C and 37 °C/30 °C conditions exhibit similar structures with minor variation in the levels of structural motifs (Fig.4A, left; Table 2) while growth under 37 °C/37 °C conditions results in a mannan with a significantly different structure (Fig. 4A, right; Table 2) irrespective of the growth media. These structural comparisons for growth under 37°C/30 °C and 37 °C/37 °C conditions relative to 30 °C/30 °C conditions hold true as well for mannan isolated from yeast grown on either blood (Fig. 4B) or serum (Fig. 4C). For mannan isolated from yeast grown under the highest constant temperature conditions (37 °C/37 °C) on any of the three media, their structures contain reduced content of all structural motifs in the acid-stable and acid-labile side chains except →6)-α-Man-(1→ in the acid-stable side chains and side chains containing α-(1→3)-Man motifs. Structural motif assignments for mannan grown at 37 °C/37 °C on YPD, for example, are shown in Table 3. In addition to most of the structural motifs a to v shown in Tables 1 and 2, there are several new structural motifs a′ to j′ (Table 3) present, supporting the new structural differences observed in mannans grown under 37 °C/37 °C conditions. Some of the structural motifs in Table 3 exhibit chemical shifts similar to, but slightly different from, those shown in Tables 1 and 2 along with new structural motifs not observed in Tables 1 and 2. These slight differences are most likely caused by the presence or absence of neighboring structural motifs and differences in solution conformation of the mannans. Koyama and coworkers18 reported a reduction of β-(1→2)-linked D-mannosyl motifs in both acid-stable and the acid-labile side chains of mannan when C. albicans serotype A strain was grown at 37 °C relative to 30 °C on Sabouraud medium. Okawa and coworkers also reported a reduction in both β-(1→2)-D-linked mannosyl motifs and acid-labile side chains through loss of phosphate groups as well as an increase in α-Man-(1→3)- structural motifs when C. albicans NIH A-207 strain was grown at 37 °C33,34 and 40 °C33 relative to 27 °C on galactose-added yeast nitrogen base medium34 and Sabouraud medium.33
Our data indicate that the C. albicans cell wall is a dynamic and adaptive organelle, which alters its phenotype in response to growth in host-derived media at physiological temperature. Specifically, our data indicate that C. albicans mannan undergo significant structural changes as a function of growth conditions including temperature (30 °C versus 37 °C) and media (YPD, blood, and serum). The structural differences impact mannan acid-stable and acid-labile side chains. Additionally, we have demonstrated the ease of following these changes in mannan structural motifs using 950 MHz NMR and published literature to develop an understanding of these resonance assignments. Using the resonance assignments, we have employed 600 MHz NMR to characterize motifs of mannans isolated from fungi grown under different conditions. This approach to structural elucidation of mannans is fast and does not require extensive sample degradation and isolation of individual mannan side chains. Recognition of structural changes as a function of growth conditions in fungal cell wall carbohydrates, such as mannans, is critical to our ability to define the structure of the fungal cell wall. These data demonstrate that the response of C. albicans cell wall mannan to changing environmental conditions is dynamic and dramatic.
When considered as a whole our data indicate that when C. albicans is grown on media containing host-derived components, such as blood or serum, there is a reduction in the overall structural complexity of cell wall mannan. The structural complexity of mannan is further reduced when the cells are cultured at 37 °C. These are intriguing observations, which may have significant implications for how the fungal pathogen is recognized by and interacts with its environment. In a separate, but parallel study, we have reported that the amount of mannosylated proteins in the C. albicans cell wall are increased when cells are grown on blood or serum at physiological temperature.35 In contrast, cultivation on blood or serum at 37 °C results in decreased amounts of mannan in the cell wall.35 We speculate that by reducing the structural complexity of cell wall mannan the fungal pathogen may alter its recognition by pattern recognition receptors in the host innate immune system. However, additional studies will be required to fully elucidate how cell wall structural changes benefit C. albicans.
3. Experimental
3.1 Strains and media
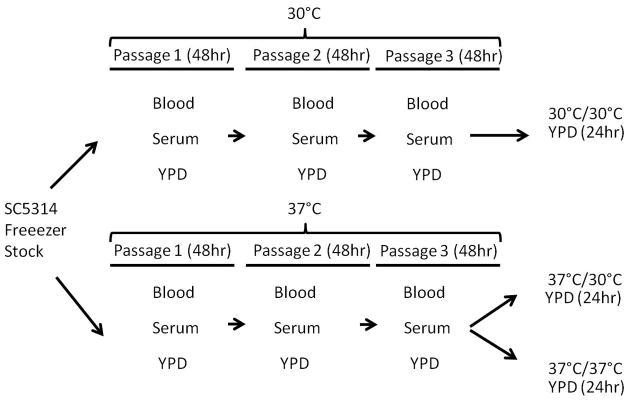
C. albicans SC5314 strain was taken directly from frozen stock and passaged on YPD (1% yeast extract, 2% peptone, 2% dextrose, 2% agar), blood (5% sheep’s blood, 4% Tryptic soy broth, 2% agar), and serum (5% serum, 2% agar) plate media at 30 °C and 37 °C. Cells were passaged at 48 h intervals three times (Figure 1). For mannan extraction, cells were inoculated from the third passage of 30 °C and 37 °C plates into 2L of YPD for growth at 30 °C for 18 h. In addition, cells that were cultivated strictly at 37 °C were also grown in 2L of YPD at 37 °C for 18 h. Overnight growth in 2L of YPD was required to extract enough mannan for analysis. For shorthand notation, 30 °C/37 °C indicates that cells were grown for three passages at 30 °C, then cultivated at 37 °C. Similar notation is used for the other temperature growth condition combinations.
Figure 1.
Schematic presentation of the growth conditions employed in this work. Yeast was grown in three passages over blood, serum, or YPD at 30°C and 37°C, and then grown at 30 °C or 37 °C on YPD.
3.2 Mannan extraction
Mannan was isolated from cells using a modified extraction procedure reported previously.21 Briefly, cells were delipidated with acetone, and the pellet disrupted by bead beating in dH2O. The extracted cells were then autoclaved for 3 h, and the extract centrifuged. The supernatant was saved and subjected to Fehling’s reagent for precipitation. The resulting precipitate was used for NMR analysis after removal of the Fehling’s reagent. From our cell preparations we generally recover 100–250 mg of material from 10–15 g cell wet weight.
3.3 NMR
Proton NMR spectra for mannan were collected on Bruker Avance III 600 and 950 NMR spectrometers using a CH cryoprobe and a TCI inverse cryoprobe, respectively, operating at 333 °K (60° C) in 5-mm NMR tubes. Mannan (variable sample sizes ranging from 7 to 23 mg) was dissolved in ca. 600 μL D2O (Cambridge Isotope Laboratories, 99.8+% deuterated). Proton 1D and 2D NMR spectra including COSY36,37 and NOESY38,39 were obtained in this study. Chemical shift referencing was accomplished relative to TMSP at 0.0 ppm. NMR spectra at 950 MHz were collected and processed as follows: for 1D NMR, 16 90° scans, 65,536 points, 20.5 ppm sweep width centered at 5.0 ppm, exponential apodization with 0.3 Hz broadening, and 15 sec pulse delay; for 2D COSY and NOESY NMR, 2048 × 256 point matrix was zero-filled to 1024 points in f1, 8 (COSY) and 24 (NOESY) scans per row, 3 ppm sweep width centered at 4.7 ppm, SINE apodization in both dimensions, NOESY mixing time 150 msec, and 2 sec pulse delay. NMR spectra at 600 MHz were collected and processed as follows: for 1D NMR, 256 30° scans, 65,536 points, 20.5 ppm sweep width centered at 6.175 ppm, exponential apodization with 0.3 Hz broadening, and 1 sec pulse delay. Mannan NMR spectra were processed using wxMacNUTS (2nd Generation NMR Utility Transform Software, Version 1.0.1, Acorn NMR, Inc.) on a Macintosh MacBook Pro running OSX version 10.5.8. Spectral comparisons in pairs are used to detect structural changes as indicated by changes in assigned peak intensities. For each set of comparisons, the spectra are height normalized to the largest peaks in each spectrum. The tallest peak in each spectrum at 5.067 ppm is assigned to the anomeric proton of α-D-(1→2)-linked mannosyl repeat units.
Supplementary Material
Acknowledgments
This work was supported, in part, by NIH GM53552 to D.L. Williams, and faculty startup funding to M. Kruppa.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wenzel RP. Clin Infect Dis. 1995;20:1531–1534. doi: 10.1093/clinids/20.6.1531. [DOI] [PubMed] [Google Scholar]
- 2.Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. Clin Infect Dis. 2004;39:309–317. doi: 10.1086/421946. [DOI] [PubMed] [Google Scholar]
- 3.Braun BR, Johnson AD. Genetics. 2000;155:57–67. doi: 10.1093/genetics/155.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fu Y, Ibrahim AS, Sheppard DC, Chen YC, French SW, Cutler JE, Filler SG, Edwards JE., Jr Mol Microbiol. 2002;44:61–72. doi: 10.1046/j.1365-2958.2002.02873.x. [DOI] [PubMed] [Google Scholar]
- 5.Monod M, Borg-von ZM. Biol Chem. 2002;383:1087–1093. doi: 10.1515/BC.2002.117. [DOI] [PubMed] [Google Scholar]
- 6.Brown GD, Herre J, Williams DL, Willment JA, Marshall ASJ, Gordon S. J Exp Med. 2003;197:1119–1124. doi: 10.1084/jem.20021890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williams DL, Lowman DW, Ensley HE. Toxicology of 1->3-beta-glucans. In: Young SH, Castranova V, editors. Glucans as a Marker for Fungal Exposure. 1. Taylor & Francis; New York: 2004. pp. 1–34. [Google Scholar]
- 8.Chauhan N, Li D, Singh P, Calderone R, Kruppa M. In: Candida and Candidiasis. 1. Calderone RA, editor. ASM Press; Washington, DC: 2002. pp. 159–175. [Google Scholar]
- 9.Netea MG, Brown GD, Kullberg BJ, Gow NA. Nat Rev Microbiol. 2008;6:67–78. doi: 10.1038/nrmicro1815. [DOI] [PubMed] [Google Scholar]
- 10.Brown GD, Gordon S. Immunity. 2003;19:311–315. doi: 10.1016/s1074-7613(03)00233-4. [DOI] [PubMed] [Google Scholar]
- 11.Brown GD, Williams DL. In: Chemistry, Biochemistry and Biology of (1,3)-Beta Glucans and Related Polysaccharides. Bacic A, Fincher GB, Stone BA, editors. Academic Press, Elsevier Inc; San Diego, USA: 2009. pp. 579–620. [Google Scholar]
- 12.Klippel N, Cui S, Groebe L, Bilitewski U. Microbiology. 2010;156:3432–3444. doi: 10.1099/mic.0.040006-0. [DOI] [PubMed] [Google Scholar]
- 13.Kobayashi H, Shibata N, Mitobe H, Ohkubo Y, Suzuki S. Arch Biochem Biophys. 1989;272:364–375. doi: 10.1016/0003-9861(89)90230-0. [DOI] [PubMed] [Google Scholar]
- 14.Kobayashi H, Shibata N, Nakada M, Chaki S, Mizugami K, Ohkubo Y, Suzuki S. Arch Biochem Biophys. 1990;278:195–204. doi: 10.1016/0003-9861(90)90248-w. [DOI] [PubMed] [Google Scholar]
- 15.Kobayashi H, Giummelly P, Takahashi S, Ishida M, Sato J, Takaku M, Nishidate Y, Shibata N, Okawa Y, Suzuki S. Biochem Biophys Res Comm. 1991;175:1003–1009. doi: 10.1016/0006-291x(91)91664-x. [DOI] [PubMed] [Google Scholar]
- 16.Kobayashi H, Shibata N, Sukuki S. Infec Immun. 1992;60:2106–2109. doi: 10.1128/iai.60.5.2106-2109.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kobayashi H, Takahashi S, Shibata N, Miyauchi M, Ishida M, Sato J, Maeda K, Suzuki S. Infect Immun. 1994;62:968–973. doi: 10.1128/iai.62.3.968-973.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koyama T, Makita M, Shibata N, Okawa Y. Carbohyd Res. 2009;344:2195–2200. doi: 10.1016/j.carres.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 19.Kruppa M, Goins T, Cutler JE, Lowman D, Williams D, Chauhan N, Menon V, Singh P, Li D, Calderone R. FEMS Yeast Res. 2003;3:289–299. doi: 10.1111/j.1567-1364.2003.tb00170.x. [DOI] [PubMed] [Google Scholar]
- 20.Kruppa M, Jabra-Rizk MA, Meiller TF, Calderone R. FEMS Yeast Res. 2004;4:409–416. doi: 10.1016/S1567-1356(03)00201-0. [DOI] [PubMed] [Google Scholar]
- 21.Li D, Williams D, Lowman D, Monteiro MA, Tan X, Kruppa M, Fonzi W, Roman E, Pla J, Calderone R. Fungal Genet Biol. 2009;46:731–741. doi: 10.1016/j.fgb.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ikuta K, Shibata N, Kanehiko H, Kobayashi H, Suzuki S, Okawa Y. FEBS Letters. 1997;414:338–342. doi: 10.1016/s0014-5793(97)01028-4. [DOI] [PubMed] [Google Scholar]
- 23.Kobayashi H, Watanabe M, Komido M, Matsuda K, Ikeda-Hasebe T, Suzuki M, Shibata N, Hisamichi K, Suzuki S. Carbohyd Res. 1995;267:299–306. [Google Scholar]
- 24.Kobayashi H, Oyamada H, Matsuda K, Shibata N, Suzuki S. Arch Microbiol. 2003;180:76–80. doi: 10.1007/s00203-003-0550-7. [DOI] [PubMed] [Google Scholar]
- 25.Lizicarova I, Maatulova M, Capek P, Machova E. Carbohyd Poly. 2007;70:89–100. [Google Scholar]
- 26.Shibata N, Ikuta K, Imai T, Satoh Y, Satoh R, Suzuki A, Kojima C, Kobayashi H, Hisamichi K, Suzuki S. J Biol Chem. 1995;270:1113–1122. doi: 10.1074/jbc.270.3.1113. [DOI] [PubMed] [Google Scholar]
- 27.Shibata N, Kobayashi H, Okawa Y, Suzuki S. Eur J Biochem. 2003;270:2565–2575. doi: 10.1046/j.1432-1033.2003.03622.x. [DOI] [PubMed] [Google Scholar]
- 28.Shibata N, Suzuki A, Kobayashi H, Okawa Y. Biochem J. 2007;404:365–372. doi: 10.1042/BJ20070081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tada R, Magi-Miura N, Yoshiyuki A, Ohno N. Microbial Pathogenesis. 2008;44:379–388. doi: 10.1016/j.micpath.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 30.Vinogradov E, Petersen B, Bock K. Carbohyd Res. 1998;307:177–183. doi: 10.1016/s0008-6215(98)00042-1. [DOI] [PubMed] [Google Scholar]
- 31.Maes E, Mille C, Trivelle X, Janbon G, Poulain D, Guerardel Y. J BIochem. 2009;145:413–419. doi: 10.1093/jb/mvp008. [DOI] [PubMed] [Google Scholar]
- 32.Kath F, Kulicke WM. Angew Makromolek Chem. 1999;268:69–80. [Google Scholar]
- 33.Okawa Y, Takahata T, Kawamata M, Miyauchi M, Shibata N, Suzuki A, Kobayashi H, Suzuki S. FEBS Letters. 1994;345:167–171. doi: 10.1016/0014-5793(94)00434-x. [DOI] [PubMed] [Google Scholar]
- 34.Okawa Y, Miyauchi M, Goto K, Giummelly P. Biol Pharm Bull. 2005;28:391–393. doi: 10.1248/bpb.28.391. [DOI] [PubMed] [Google Scholar]
- 35.Kruppa M, Greene RR, Lowman DW, Williams DL. Glycobiology. 2011;21:1173–1180. doi: 10.1093/glycob/cwr051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shaw AA, Salaun C, Dauphin JF, Ancian B. J Magn Reson. 1996;A 120:110–115. [Google Scholar]
- 37.Ancian B, Bourgeois I, Dauphin JF, Shaw AA. J Magn Reson. 1997;125:348–354. [Google Scholar]
- 38.Jeener J, Meier BH, Bachmann P, Ernst RR. J Chem Phys. 1979;71:4546–4553. [Google Scholar]
- 39.Wagner R, Berger S. J Magn Reson A. 1996;123:119–121. doi: 10.1006/jmra.1996.0222. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.