Abstract
Synthesis of amphiphilic oligosaccharides is problematic because traditional methods for separating and purifying oligosaccharides, including sulfated oligosaccharides, are generally not applicable to working with amphiphilic sugars. We report here RPIP-LC and LC-MS methods that enable the synthesis, separation, and characterization of amphiphilic N-arylacyl O-sulfonated aminoglycosides, which are being pursued as small-molecule glycosaminoglycan mimics. The methods described in this work for separating and characterizing these amphiphilic saccharides are further applied to a number of uses: monitoring the progression of sulfonation reactions with analytical RP-HPLC, characterizing sulfate content for individual molecules with ESI-MS, determining the degree of sulfation for products having mixed degrees of sulfation with HPLC and LC-MS, and purifying products with benchtop C18 column chromatography. We believe the methods described here will be broadly applicable to enabling the synthesis, separation and characterization of amphiphilic, sulfated and phosphorylated oligosaccharides and other types of molecules substituted to varying degrees with both anionic and hydrophobic groups.
Keywords: Biomimetic synthesis, Aliphatic oligosaccharide, Mass spectrometry, Reversed-phase ion-pairing, Sulfation, Electrospray ionization
1. Introduction
1.1 Therapeutic potential and limitations of GAG mimics
Heparan sulfate (HS) and other glycosaminoglycans (GAGs) are sulfated, polyanionic polysaccharides located in the extracellular matrix or on the surface of mammalian cells.1–3 As extracellular carbohydrates, HS and other GAGs are bound by many endogenous and exogenous proteins, often termed heparin-binding or HS-binding proteins.4–10 These carbohydrate-protein interactions can be inhibited by GAG-mimicking bind-and-block antagonists for potential therapeutic applications such as: slowing angiogenesis, preventing metastasis, inhibiting inflammation and blocking numerous pathogens from binding to and invading host cells.5, 6, 11
To date, the development of molecules that bind HS-binding proteins and block interaction with HS or other GAGs has primarily focused on preparing persulfated oligosaccharides and polyanionic polysaccharides.4, 5, 12 Therapeutic application of these highly anionic structures has been limited by poor pharmacodynamic and pharmacokinetic properties and by their nonspecific binding to many proteins.13–15 Working to overcome these limitations we recently demonstrated that heparin derivatives substituted with aromatic residues, in place of anionic groups to reduce anionic content, bind with high affinity and selectivity to select HS-binding proteins.12, 16, 17 The next challenge is to exploit this discovery and synthesize structurally similar but smaller, oligosaccharide-size heparin mimics that would be more drug-like and would be more selective in binding individual proteins.18
1.2 Preparing small amphiphilic oligosaccharides as GAG mimics
Aminoglycoside antibiotics are small, naturally occurring glycoconjugates with multiple hydroxyl and amine groups. The orthogonal nature of the amine and hydroxyl groups provide synthetically useful handles for chemical modification and the introduction of structural diversity while the different aminoglycoside core structures orient these substituent groups in different 3-dimensional space. Thus we envisioned the aminoglycosides to be readily available scaffolds for making libraries of relatively small oligosaccharide-size heparin mimics, where the amine groups would be substituted with structurally diverse N-arylacyl moieties and hydroxyl groups would be sulfonated.
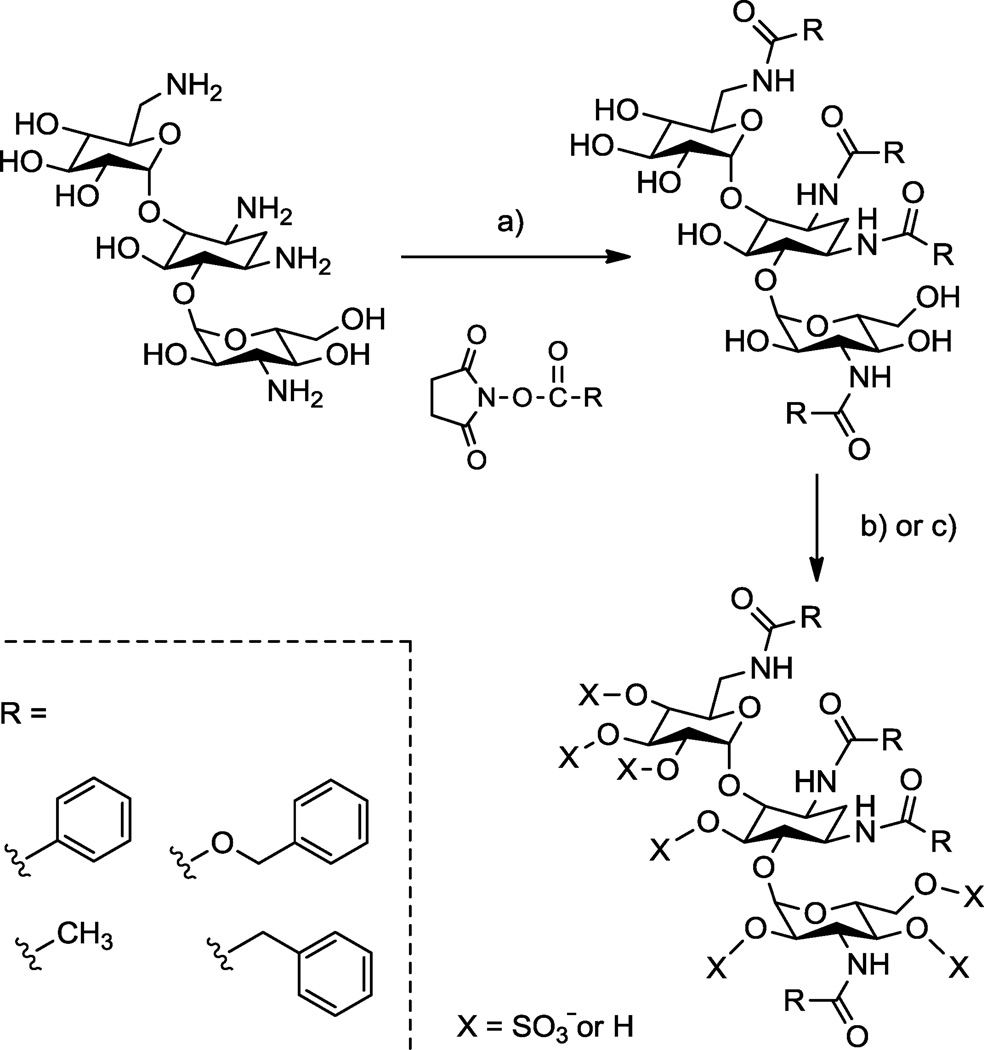
Toward this end, we set out to use aminoglycosides as scaffolds for preparing structurally defined oligosaccharides substituted with both aromatic rings and sulfate groups (see Scheme 1). Although modification of the aminoglycosides in this manner was expected to be straightforward, initial efforts in our laboratory to prepare these amphiphilic, N-arylacyl O-sulfonated oligosaccharides was fraught with problems. First, standard thin-layer chromatography and HPLC methods were insufficient for monitoring sulfonation reactions to establish complete, persulfation. Second, approaches previously used to separate and purify sulfated oligosaccharides from reaction byproducts after sulfonation failed with these amphiphilic structures. Third, incomplete sulfonation reactions produce product mixtures that vary by the degree and position of sulfate groups; HPLC and HPLC-MS methods previously used to separate sugars based on the number of sulfate groups and to characterize the degree of sulfation, or to demonstrate persulfation, did not work due to the amphiphilic nature of these molecules.
Scheme 1.
Representative synthesis showing concept for making N-aryl O-sulfonated aminoglycosides; shown is kanamycin. a) R-NHS, NaHCO3, H2O, 23 °C, 12–16 hr, 30–85%; b) i: Pyr·SO3, DMF, anhydrous pyr, 66 °C, 5–7 hr ii: H2O, 10 mM NaOH, 4 °C, 45 65%; c) i: ClSO3H, pyr, 57 °C, 4–6 hr, ii: H2O, NaHCO3, 35–95%.
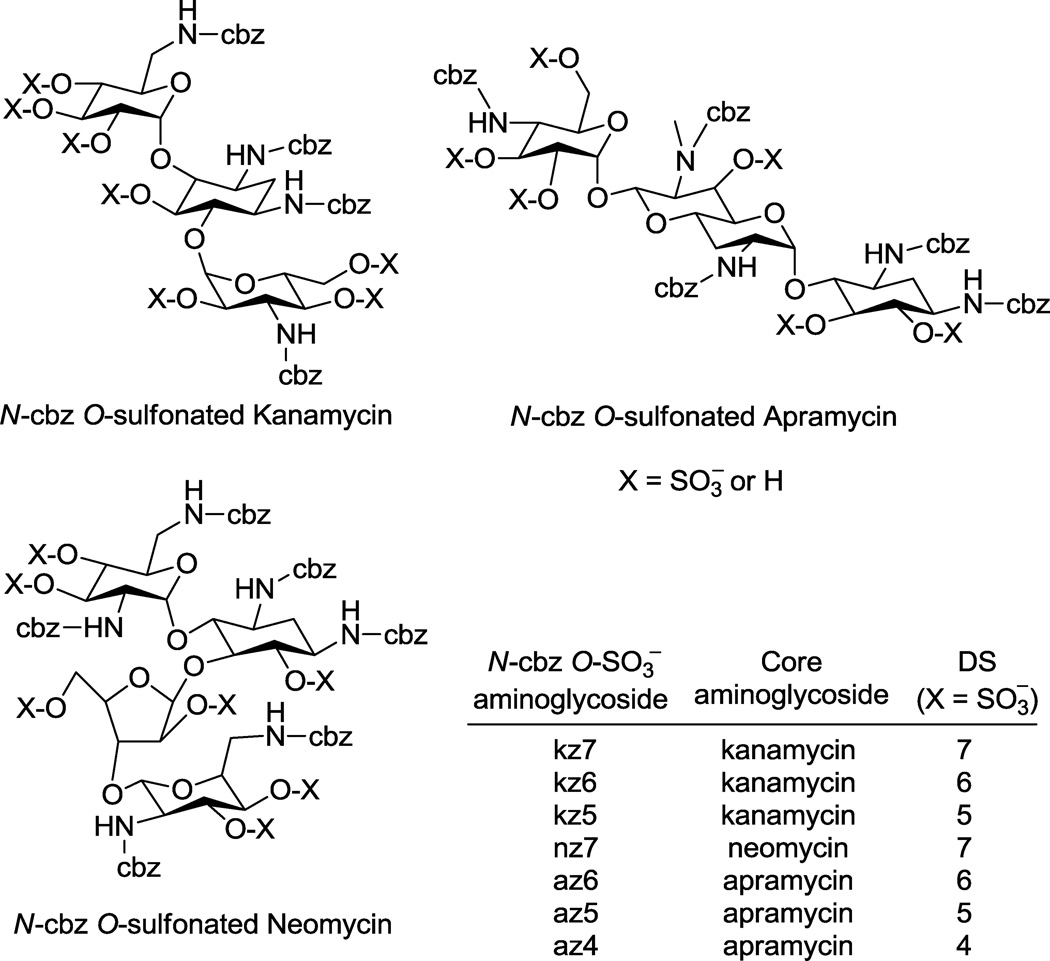
In this report we outline the limitations of current approaches for reversed phase ion pairing liquid chromatography (RPIP-LC) and LC-mass spectrometry (MS) of various anionic saccharides. We then describe adaptation of these approaches to afford LC and LC-MS methods that facilitated the synthesis and enabled the characterization of N-arylacyl O-sulfonated aminoglycoside derivatives. Utility of these methods are exemplified in this work using aminoglycosides where the amine groups were functionalized to N-benzyloxycarbonyl (N-cbz) groups and the hydroxyl groups were sulfonated to produce N-cbz O-sulfonated apramycin, kanamycin and neomycin (Figure 1). These methods provide a means to monitor sulfonation of aryl-substituted carbohydrates, to separate complex mixtures of the sugars based on degree of aromatic and/or sulfate groups, and to characterize the structure of these novel oligosaccharides using LC-MS. The procedures described in this work are expected to be useful for separating and characterizing many additional types of amphiphilic oligosaccharides and other molecules substituted to varying degrees with anionic and hydrophobic groups.
Figure 1.
Sulfonation of N-cbz aminoglycosides (kanamycin, apramycin, and neomycin) with Pyr·SO3 or ClSO3H resulted in products with varied degrees of sulfation (DS). RPIP-HPLC methods developed here were used to separate reaction products based on degree and position of sulfate groups. Sulfonation of N-cbz neomycin was complete, giving persulfated product, nz7. Sulfonation of N-cbz kanamycin and N-cbz apramycin produced per sulfated derivatives kz7 and az6, respectively, as well as under-sulfated kanamycin derivatives (kz5, kz6) and under-sulfated apramycin derivatives (az4, az5), where the number indicates the number of sulfate groups attached.
1.3 RPIP-LC-MS analysis of sulfated oligosaccharides
The separation of sulfated carbohydrates and nucleic acids has been described by various methods that employ RPIP-LC.19–21 RPIP-chromatography relies on alkyl ammonium salts to separate charged species on reversed phase resin and is particularly useful for the separation and analysis of sulfated oligosaccharides because this method is readily compatible with LC-MS.22–25 For sulfated oligosaccharides, a common problem in MS ionization is sulfate fragmentation; this complicates structure identification and renders it difficult to determine the number of sulfate groups on a saccharide.23, 26, 27 It has been shown, however, that RPIP with quaternary ammonium salts decreases sulfate fragmentation during electrospray ionization.27 For example, tetraethyl and tetrabutylammonium salts have been used to separate a wide range of sulfated saccharides and phosphorylated nucleotide sugars.23, 27–33 Nevertheless, a limiting factor with these methods is that the quaternary ammonium ions are non-volatile salts and are thus difficult to rapidly clear from the mass spectrometer.22
Sulfated oligosaccharides have also been separated using RPIP methods with volatile salts, including mono-, di- and trialkylammonium salts. The tertiary amines N-hexylaminedibutylamine, tributylamine, and tripropylamine were shown to be useful for separating sulfated oligosaccharides based on degree of sulfation, stereoisomers of sulfate groups, or both.22–24, 29, 34, 35 Unfortunately, application of these methods using tributylammonium salts did not provide chromatographic resolution of our N-arylacyl O-sulfonated oligosaccharides, which have a considerable hydrophobic component and thus likely prone to aggregation. To accommodate the unique differences in polarity between other sulfated oligosaccharides and our amphiphilic N-arylacyl modified sulfated oligosaccharides, we looked to methods designed for separating oligonucleotides. Oligonucleotides have phosphate groups and hydrophobic bases, and were thus anticipated to have physical and chromatographic properties more similar to our novel O-sulfonated N-aryl aminoglycosides.
1.4 RPIP-LC separation of oligonucleotides
Like oligosaccharides, oligonucleotides have been separated and characterized with RPIP-HPLC and LC-MS methods. Separation of oligonucleotides with 1,1,1,3,3,3-hexafluoro-2-propanol (HFIP) is very successful when used on conjunction with triethylamine (TEA).36–39 The HFIP method for separating oligonucleotides was recently adapted to the separation of sulfated oligosaccharides.25, 34 Regardless, HFIP-based methods require column heating, an ultraperformance LC system, and in some instances high concentrations of ion pairing reagents. For our analytical and preparative needs, we desired a single RPIP-separation method that could be used with HPLC, benchtop C18 chromatography and LC-MS. Single-use application hampers many methods for RPIP-HPLC-MS of oligonucleotides and sulfated oligosaccharides, such as those that require a second eluent spray or pre or post-column derivitization prior to LC-ESI-MS.23, 24, 27, 29, 30, 40–42 Moreover, other methods require special columns such as amino columns, columns designed for hydrophilic compounds, pre-loaded microcolumns or capillary HPLC.22, 43–46 These multiple derivitization steps or specialty columns are cost, scale, and time-prohibitive.
In this report, we adapt an oligodeoxyribonucleotide RPIP method to the HPLC and benchtop chromatographic separation, and the LCMS characterization, of amphiphilic N-arylacyl O-sulfonated aminoglycosides.47 Specifically reported here are data and results obtained when developing these methods with N-cbz modified aminoglycosides. This method is well suited for LC-MS, because it relies on low salt concentration with common reagents (10 mM ammonium acetate and <0.1 mM TEA).47 Furthermore, since separation is achieved with C18 resin and without a column oven or an ultraperformance LC system, the analytical methods are shown to be scalable for preparative-scale desalting and benchtop product purification.
2. Results and Discussion
2.1 Preparing N-cbz O-sulfonated aminoglycosides having varied degrees of sulfate
The synthesis of O-sulfonated N-arylacyl aminoglycoside derivatives, as exemplified with kanamycin in Scheme 1, is accomplished in two steps: 1) selective acylation of the amine group on the aminoglycoside and 2) the N-arylacyl aminoglycoside is sulfonated to convert hydroxyl groups to sulfate groups. Using this methodology, the aminoglycosides apramycin, kanamycin, and neomycin were converted to their N-benzyloxycarbonyl (N-cbz) derivatives by coupling with the N-(Benzyloxycarbonyloxy)succinimide under alkaline conditions. Subsequently, the N-cbz aminoglycosides were sulfated with either pyridine sulfur trioxide complex (Pyr·SO3) or chlorosulfonic acid (ClSO3H) (Figure 1).
By using the two different sulfonation reagents under different reaction conditions we obtained varied results in attempts to generate per-sulfated products. For example, when Pyr·SO3 was used to O-sulfonate N-cbz kanamycin, the resulting product was a mixture of the per-sulfated derivative (with seven hydroxyls sulfonated, kz7) and under-sulfated derivatives: six hydroxyls sulfonated (kz6) and five hydroxyls sulfonated (kz5). When ClSO3H was used to O-sulfonate N-cbz neomycin, the reaction produced N-cbz per-O-sulfonated neomycin (nz7, Figure 1). In two separate reactions, both Pyr·SO3 and ClSO3H were used to O-sulfonate N-cbz apramycin. Sulfonation with Pyr·SO3 produced a mixture of products containing N-cbz per-O-sulfonated apramycin (az6) as well as under-sulfated products with either five sulfates (az5) or four sulfates (az4) per molecule. Sulfonation of N-cbz apramycin with ClSO3H produced per Ncbz O-sulfonated apramycin (az6, Figure 1). Thus by using the LC and LC-MS methods developed in this work to rigorously follow these sulfonation reactions over time and to characterize product mixtures (methods discussed in section 2.4), it is shown here that ClSO3H is the superior reagent to achieve complete, per-sulfation of these glycoconjugates.
2.2 Optimizing separation of N-cbz O-sulfonated aminoglycosides with ion pair-RP-HPLC
Since the O-sulfonation of N-cbz aminoglycosides often resulted in products with varied degrees of sulfation, product characterization required a method that would: 1) separate components in the product mixture based on the degree of sulfation 2) show robust ability for separating and characterizing different N-cbz O-sulfonated aminoglycosides and 3) minimize sulfate fragmentation on LC-MS without losing signal intensity so that LC-based separation could be coupled with MS to characterize the sulfated products.
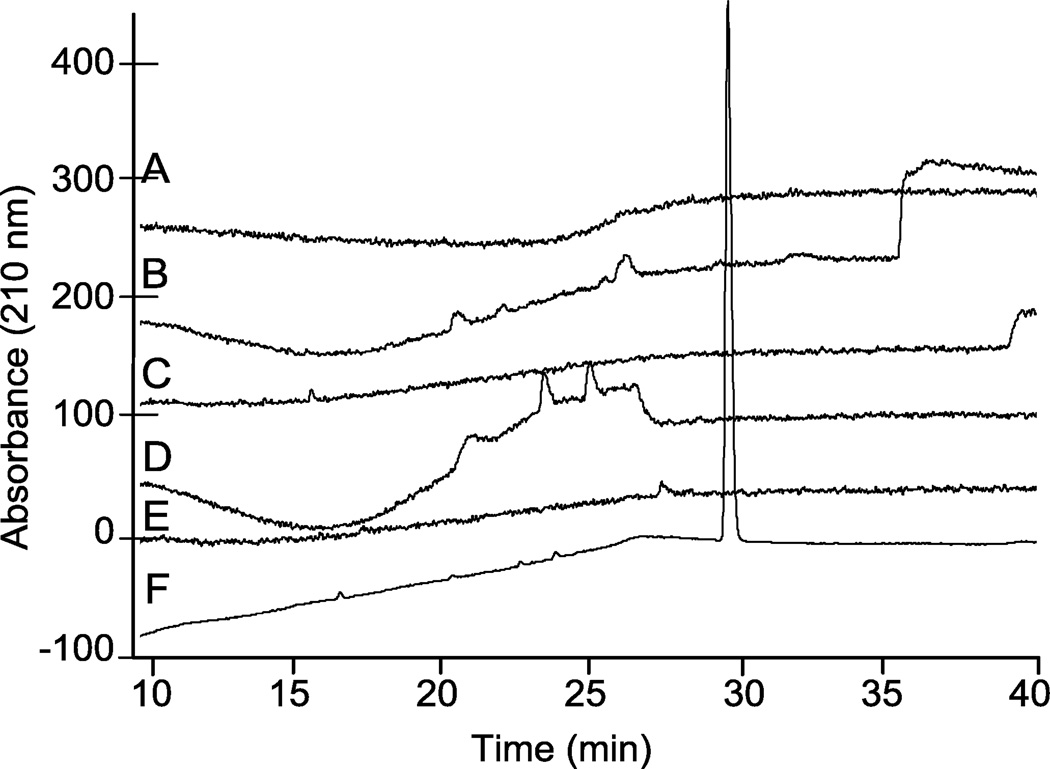
Initially, different mobile phase compositions that varied in alkyl ammonium salts, pH, and acetonitrile gradient were evaluated to identify a method that would elute N-cbz per-O-sulfonated neomycin as a single, separable peak. Representative results for a number of different methods tested are shown (Figure 2). In each chromatogram shown, the mobile phase contained 10 mM ammonium acetate. In chromatograms A–D, the counter ion was a quaternary ammonium salt, tetramethyl ammonium iodide (TMAI): 10 mM with pH = 7.3 (Figure 2, A), 0.8 mM with pH adjusted to 5.0 (Figure 2, B), 0.8 mM with pH = 7.4 (Figure 2, C) and 0.8 mM with pH adjusted to 8.0 (Figure 2, D). For each of these analyses, A–D, the run gradient was 10–80% ACN in water over 20 min then isocratic elution at 80% ACN until the end of run time. Another quaternary ammonium salt, tetrabutylammonium iodide (TBAI), was evaluated at 10 mM, pH = 7.3, with gradient elution of 10–70% ACN for 20 minutes followed by isocratic elution at 70% ACN for the remaining run time (Figure 2, E). In each of these chromatograms (Figure 2, A–E), N-cbz O-sulfated neomycin eluted as a broad or undefined peak. However, using a trialkyl ammonium salt, triethylamine, as the alkyl ammonium ion pairing agent afforded elution of N-cbz O-sulfated N-cbz neomycin as a single sharp peak (Figure 2, F).
Figure 2.
Representative comparison of analytical HPLC chromatograms from various RPIP conditions eluting persulfated N-cbz neomycin. Each chromatogram was generated from a 20 min. gradient increase in percent ACN, and the ending percent ACN was then maintained isocratic for an additional 22 min. (A) 10–80% ACN; 10 mM ammonium acetate, 10 mM TMAI, pH = 7.3. (B) 10–80% ACN; 10 mM ammonium acetate, 0.8 mM TMAI, pH = 5.0. (C) 10–80% ACN; 10 mM ammonium acetate, 0.8 mM TMAI, pH = 7.4. (D) 10–80% ACN; 10 mM ammonium acetate, 0.8 mM TMAI, pH = 8.0. (E) 10–70% ACN; 10 mM ammonium acetate, 10 mM TBAI, pH = 7.3. (F) 10–30% ACN; 10 mM ammonium acetate, 0.8 mM TEA, pH = 8.3.
As shown, optimal separation on HPLC was achieved with adjusting the mobile phase to pH 8.3 with triethylamine (TEA, ~0.8 mM) and using a gradient elution of 10–30% ACN over 20 min, followed by isocratic elution at 30% ACN until the end of the run. Moreover, the triethylammonium salt is better suited for LC-MS analyses than the quaternary ammonium salts, because ternary salts are more quickly cleared from the instrument thus allowing for more frequent injections. Other studies have described the use of triethylammonium acetate (TEAA) ion pairing for the separation of oligonucleotides.37, 48 In these reported methods, TEA is used at concentrations two orders of magnitude greater than the concentration of TEA in the optimized method for amphiphilic saccharides described here. When ion pairing with TEAA was used to separate and characterize N-cbz O-sulfonated aminoglycosides, it produced sharp peaks and similar elution profiles on HPLC, but often resulted in decreased ionization on LC-MS. This finding agrees with the previous observation that increasing TEA in the elution buffer decreases signal intensity in MS detection.47
2.3 LC-MS detection of N-cbz O-sulfonated aminoglycosides
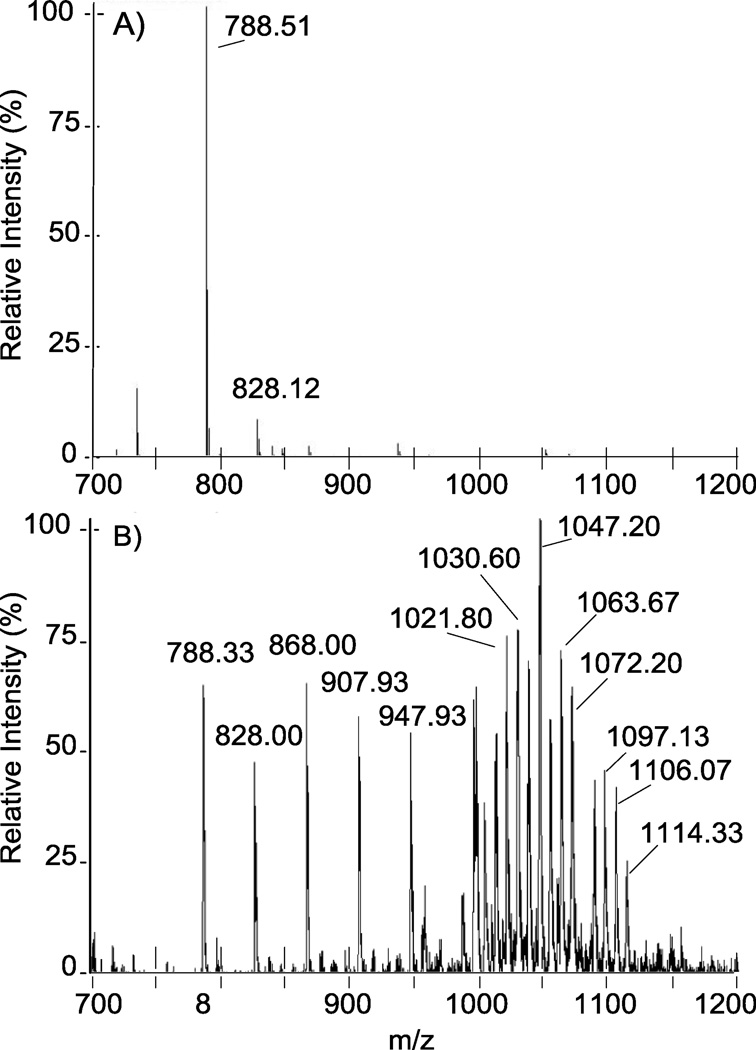
Initial analysis of N-cbz O-sulfonated aminoglycosides on LC-MS with the optimized TEA-containing ammonium acetate buffer (section 2.2) detected primarily di-sulfate, doublycharged products for all peaks observed in the total ion chromatogram (TIC). As shown for nz7 in Figure 3, A, initially, only m/z = 788.51 [M-5SO3 + 5H]−2 and m/z = 828.12 [M-4SO3 + 4H]2 were observed. However, decreasing both capillary temperature and auxiliary gas flow and increasing the tube lens offset captured persulfated species (M = nz7) in high abundance (Figure 3, B). As shown, the persulfated material is not concentrated in a single ionic species but rather distributed across a wide range of distinct ions. These per-sulfated ions of nz7 were detected in association with hydrogen, triethylammonium, ammonium, or combinations thereof (Table 1). The various ion states of the persulfated product and the high propensity for sulfate fragmentation in ESI likely account for the low abundance initially observed for persulfated material. Furthermore, even after tuning the instrument to decrease fragmentation, loss of sulfate during ionization still occurred, and ions were observed for the various fragmented states (Table 1 and Figure 3).
Figure 3.
Tuning of the ESI-ion trap mass spectrometer is critical for decreasing sulfate fragmentation and detecting the many ionic states of highly sulfated products. . Panel A: Before tuning, direct inject ESI of persulfated N-cbz neomycin, nz7, detects only desulfated ions m/z = 788.51 [M-5SO3 + 5H]−2 and m/z = 828.12 [M-4SO3 + 4H]2. Panel B: After tuning, higher molecular weight ions are more abundant and persulfated nz7 is observed in various adducts with ion pairing reagents (denoted in Table 1).
Table 1.
N-cbz per O-sulfonated neomycin (nz7) adducts and in source sulfate fragmentation observed in ESI direct inject spectra (Figure 3, B). nz7 forms adducts with cations: H+, TEA[a], NH4+ All ions are observed in negative mode and are doubly-charged. M = C71H75N6O46S77− Mr = 1972.83
| m/z theoretical | m/z observed | Interpretation | ΔMr |
|---|---|---|---|
| 788.22 | 788.33 | [M − 5SO3 + 5H]−2 | 0.11 |
| 828.19 | 828.00 | [M − 4SO3 + 4H]2 | −0.19 |
| 868.68 | 868.00 | [M − 3SO3 + 3H]−2 | −0.68 |
| 908.15 | 907.93 | [M − 2SO3 + 2H]−2 | −0.22 |
| 948.13 | 947.93 | [M − SO3 + H]−2 | −0.20 |
| 988.11 | 988.53 | [M + 5H]−2 | 0.42 |
| 996.62 | 996.20 | [M + 4H+NH4]−2 | −0.42 |
| 1005.13 | 1005.13 | [M + 3H + 2NH4]−2 | 0.00 |
| 1013.64 | 1014.13 | [M + 2H + 3NH4]−2 | 0.49 |
| 1022.15 | 1021.80 | [M + H + 4NH4]−2 | −0.35 |
| 1030.67 | 1030.60 | [M + 5NH4]−2 | −0.07 |
| 1038.67 | 1038.27 | [M + 4H + TEA]−2 | −0.40 |
| 1047.18 | 1047.20 | [M + 3H + TEA + NH4]−2 | 0.02 |
| 1055.69 | 1055.13 | [M + 2H + TEA + 2NH4]−2 | −0.56 |
| 1064.20 | 1063.67 | [M + H + TEA + 3NH4]−2 | −0.53 |
| 1072.71 | 1072.20 | [M + TEA + 4NH4]−2 | −0.51 |
| 1089.23 | 1089.87 | [M + 3H + 2TEA]−2 | 0.64 |
| 1097.74 | 1097.13 | [M + 2H + 2TEA + NH4]−2 | −0.61 |
| 1106.25 | 1106.07 | [M + H + 2TEA + 2NH4]−2 | −0.18 |
| 1114.76 | 1114.33 | [M + 2TEA + 3NH4]−2 | −0.43 |
TEA = (CH3CH2)3NH+
2.4 Characterizing N-cbz O-sulfonated aminoglycoside mixtures using LC-MS
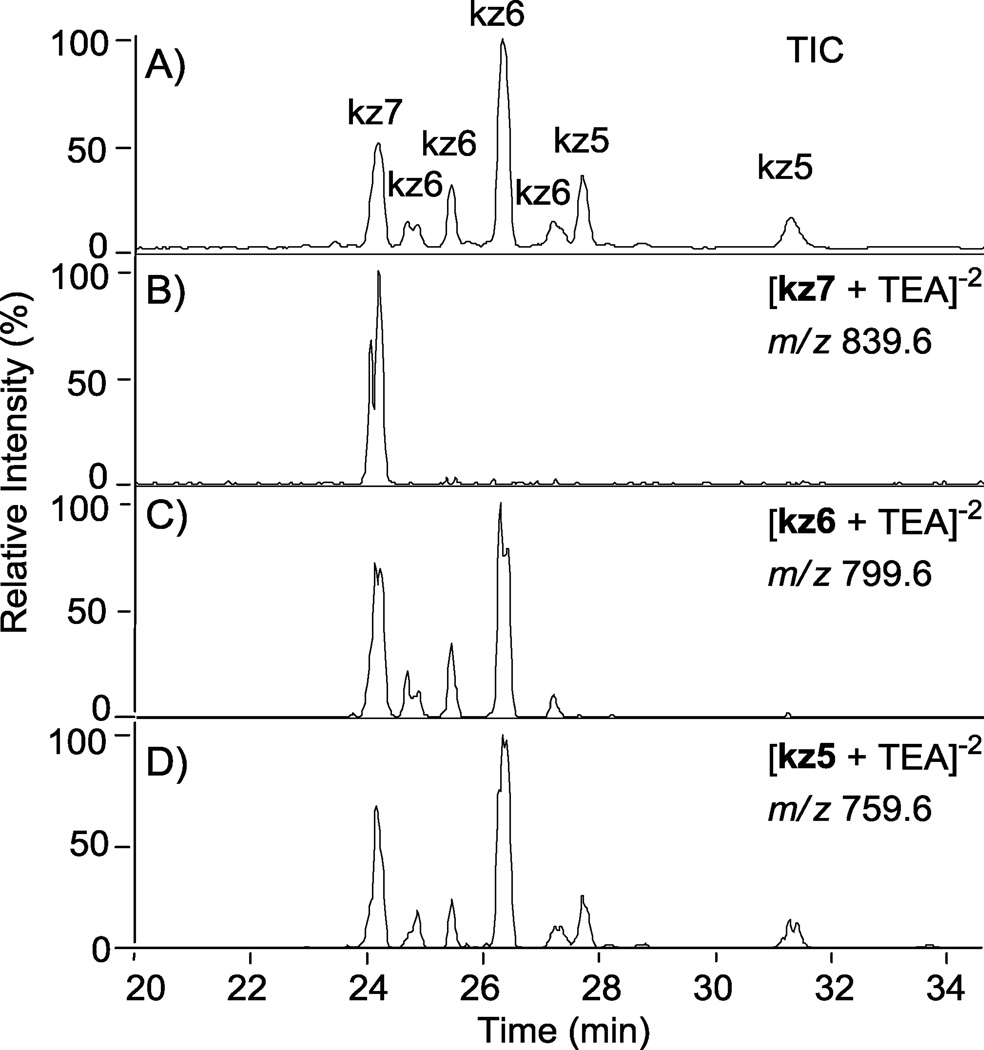
Tuning the MS to nz7 as outlined in section 2.3 allowed for the first time characterization of these sulfonation reaction products. The O-sulfonation of N-cbz kanamycin with Pyr·SO3 resulted in a variety of under-sulfated products that were only now possible to separate based on both degree of sulfation and position of sulfate groups using the RPIP LC-MS (Figure 4). All peaks detected by the ion trap mass analyzer are shown in the total ion chromatogram (TIC, Figure 4, A). From the TIC, ion chromatograms were extracted by selecting m/z ratios calculated for various ions corresponding to specific degrees of sulfation (Figure 4, B–D); an example of how m/z ratios were calculated for the various ions is shown in Table 1. Ions corresponding to persulfated compound, kz7, (m/z = 839.6 ± 0.5) correlate to the peak eluting at 24.10 min in the TIC (Figure 4, B). Likewise, ions that have a mass to charge ratio of kz6 (m/z = 799.6 ± 0.5) are found within peaks eluting at 24.58, 25.34, 26.22, and 27.11 minutes (Figure 5, C). Ions with a mass to charge ratio of kz5 (m/z = 759.6 ± 0.5) are found within peaks eluting at 27.58, 28.03, 28.72, and 31.18 minutes (Figure 4, D). Additionally, there is evidence of in-source sulfate fragmentation: The persulfated product peak at 24.10 min also appears with loss of one or two sulfates in chromatograms C and D, corresponding to kz6 and kz5, respectively. Peaks within each TIC were assigned a degree of sulfation based on extracted ion chromatograms; the ion representing the maximum degree of sulfation detected within a particular peak was assigned as the degree of sulfation.
Figure 4.
Separation of N-cbz O-sulfonated kanamycin derivatives that differ by degree of sulfation and the position of sulfate groups using RPIP-HPLC interfaced with an ESI- ion trap mass analyzer. Peaks for all sulfonation products are shown in the total ion chromatogram (TIC, A). The extracted ion chromatograms (B–D) were extracted based on m/z ratios that fall within the range predicted for a degree of sulfation (5, 6, or 7 sulfate groups); see Table 1 for an example of predicted m/z ratios. Ion chromatograms were extracted from the TIC for the indicated degrees of sulfation (DS): (B) persulfated, DS = 7, kz7; (C) DS = 6, kz6; (D) DS =5, kz5.
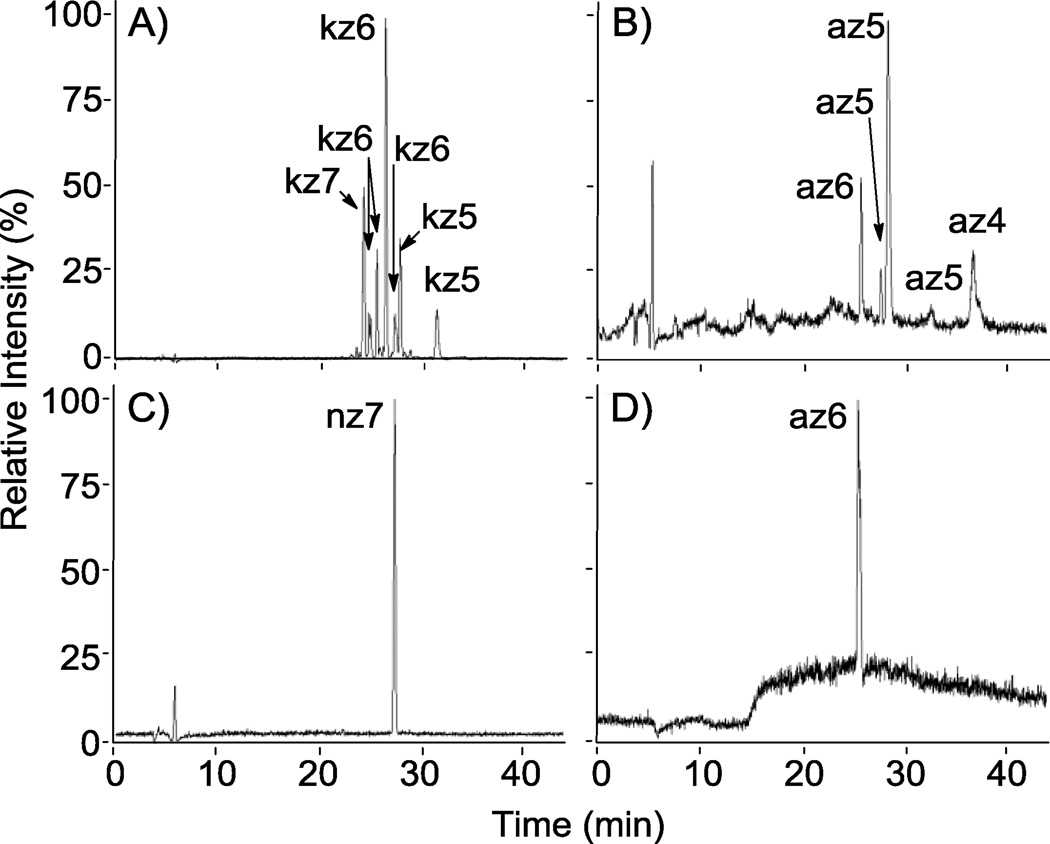
Figure 5.
RPIP-HPLC-MS total ion chromatograms (TICs) of reaction products from the sulfonation of N-cbz aminoglycosides. Shown are product mixtures from the sulfonation of kanamycin and apramycin with Pyr·SO3 (A and B, respectively) and the sulfonation of neomycin and apramycin with ClSO3H (C and D, respectively). A and B demonstrate separation of different sulfated states of N-cbz kanamycin (DS = 5–7) and N-cbz O-sulfonated apramycin (DS = 4–6), respectively. O-sulfonation with ClSO3H afforded persulfated products: (C) nz7 and (D) az6.
2.5 Determining degree of sulfation
The methods employed for O-sulfonation with Pyr·SO3 resulted in mixtures of products having varied degrees of sulfation for both N-cbz kanamycin (Figure 5, A) and N-cbz apramycin (Figure 5, B). For each of these sulfated compounds, MS analysis was used to assign a degree of sulfation for each peak within the product mixtures (Figures 4, 5). Next, the relative concentration of each sulfated compound was calculated by integrating peak area with UV detection at each compound’s λ max between 254–258 nm. Peak areas for compounds having the same number of sulfate groups were added together to then calculate average degree of sulfation for the entire product mixture (Table 2). The average degree of sulfation was notably lower in sulfonation reactions with Pyr·SO3 than wit ClSO3H (Table 2). This is most evident in the O-sulfonation of N-cbz apramycin: O-sulfonation with ClSO3H produced persulfated product (az6) that eluted as a single peak on LC-MS (Figure 5, D) while O-sulfonation with Pyr·SO3 provided a mixture of sulfated compounds that varied in both positioning and degree of sulfation, as ascertained by LC-MS (Figure 5, B).
Table 2.
Calculated degree of sulfation (DS) for reaction product mixtures afforded by O-sulfonation of N-cbz kanamycin (A), N-cbz apramycin (B and D), and N-cbz neomycin (C). Two different sulfating reagents were employed, Py·SO3 and ClSO3H. The percent at each sulfated state is expressed as %DS and the number of sulfates per molecule
| Rxn | Starting Material | Sulf Method | avg DS |
% DS 7SO4 |
% DS 6SO4 |
% DS 5SO4 |
% DS 4SO4 |
|---|---|---|---|---|---|---|---|
| A | N-cbz kanamycin | PySO3 | 6.0 | 17.8 | 66.0 | 16.2 | -- |
| B | N-cbz apramycin | PySO3 | 4.9 | NA | 18.4 | 55.5 | 26.1 |
| C | N-cbz neomycin | ClSO3H | 7 | 100 | -- | -- | -- |
| D | N-cbz apramycin | ClSO3H | 6 | NA | 100 | -- | -- |
NA, not applicable
--, below limit of detection
2.6 Monitoring O-sulfonation reactions and product purification using RP-IP-HPLC
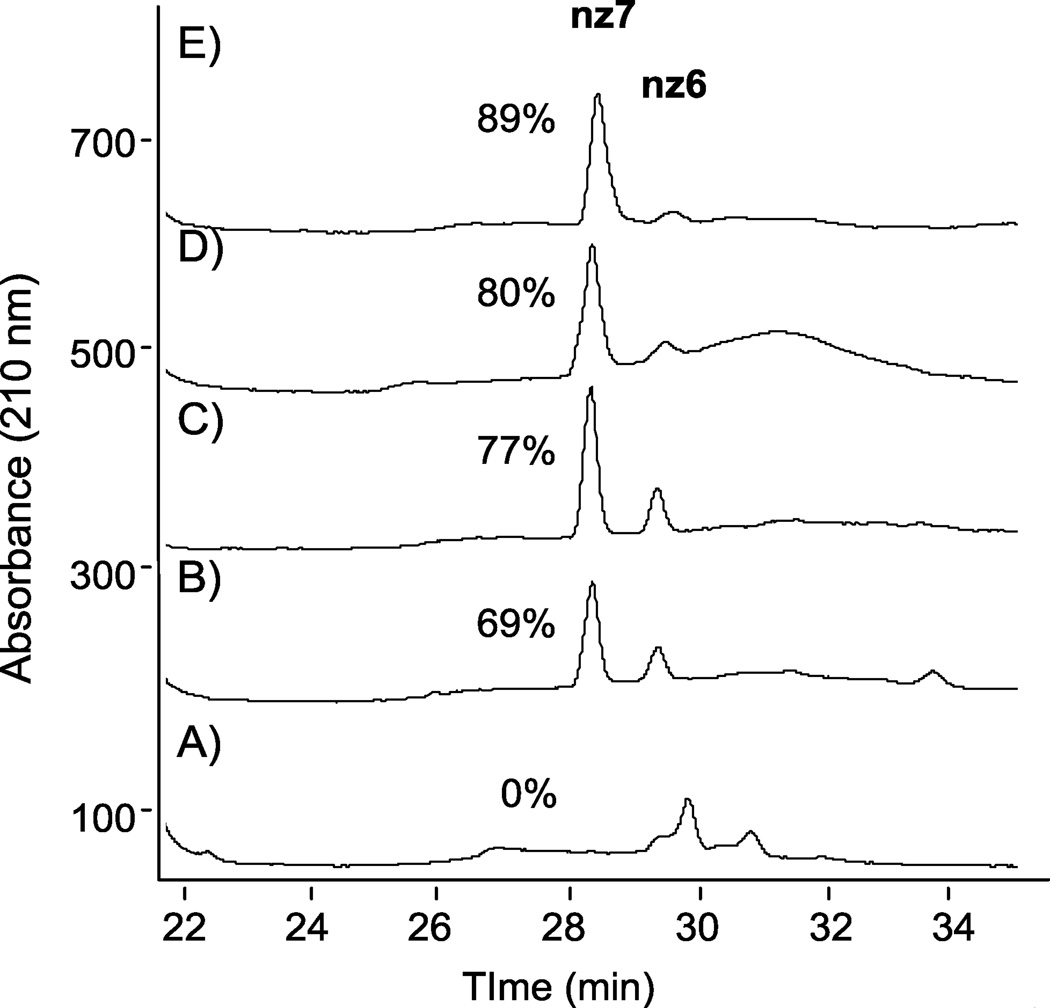
Once methods had been established to characterize reaction products using LC-MS, subsequent O-sulfonation reactions to be monitored over time to drive synthesis to the persulfated product. The utility of these methods for monitoring sulfonation reactions is exemplified in Figure 6, which shows the RPIP HPLC analysis of reaction aliquots at various times during the sulfonation of N-cbz neomycin with ClSO3H. In these chromatograms, N-cbz per-O-sulfonated neomycin correlates to the peak at 28 minutes. Within 5 minutes of adding ClSO3H to N-cbz neomycin, a number of partially sulfated products appear, including a small amount of hexasulfated product at 29 minutes (Figure 6, A). Fifteen minutes after adding ClSO3H, the reaction shows nearly 70% of the product to be persulfated (Figure 6, B). After 20 minutes, 77% of the N-cbz neomycin is persulfated (Figure 6, C) and by 275 minutes about 90% of the saccharide is persulfated (Figure 6, E).
Figure 6.
RPIP-HPLC to follow reaction progression over time for O-sulfonation of N-cbz neomycin. Following addition of ClSO3H, reaction aliquots were analyzed at: (A) 5 min, (B) 15 min, (C) 20 min, (D) 50 min, (E) 275 min. Percent of reaction product that is persulfated product (nz7) is indicated next to the nz7 peak at 28 minutes.
2.7 Purifying and desalting of N-cbz O-sulfonated aminoglycosides with RPIP benchtop chromatography
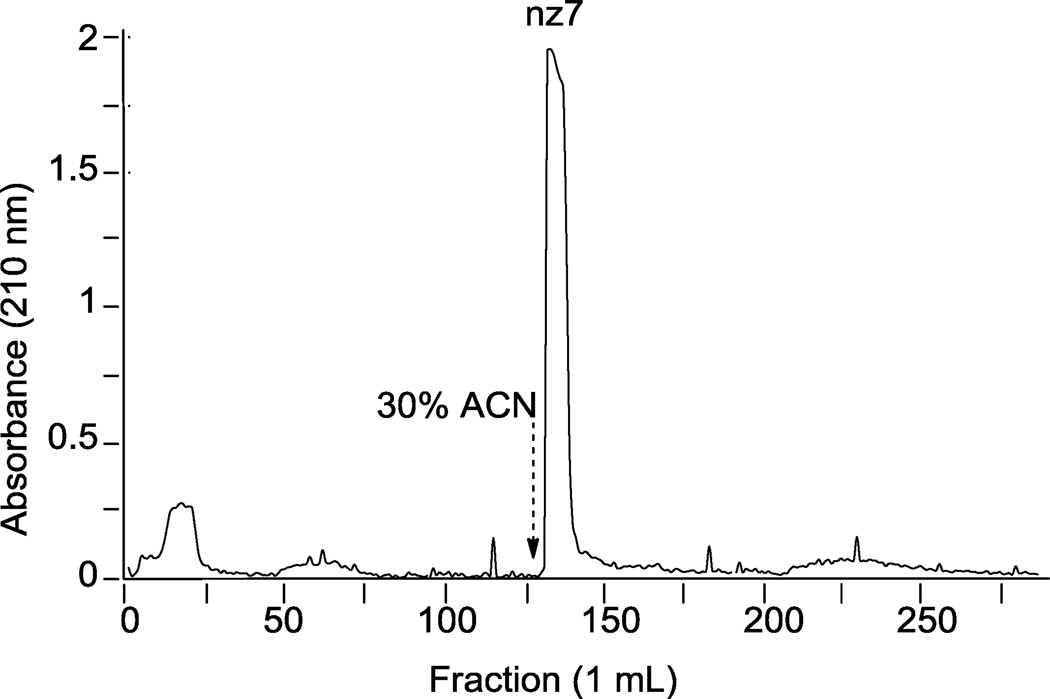
After aqueous quenching of the sulfonation reactions, the desired N-cbz O-sulfonated aminoglycoside products are in an aqueous-organic mixture with reaction by-products including sodium sulfate, trace amounts of pyridine, and at times under-sulfated N-cbz aminoglycosides. To remove pyridine and sodium sulfate, sulfated N-aryl aminoglycosides were separated using a benchtop C18 employing the RPIP buffer conditions developed here for LC-MS separation (Figure 7). In this example, the column was loaded with the aqueous reaction mixture from the O-sulfonation of N-cbz neomycin with ClSO3H. After elution with ~20 column volumes of the LC buffer (sans organic solvent), 30% ACN was added to the mobile phase and elution continued (Figure 7, dashed arrow indicates ACN in elution buffer). UV-absorbing product was eluted off the column in one peak; absorbing fractions were pooled, dried, and confirmed on LC-MS to be persulfated N-cbz neomycin. The product was analyzed for presence of sodium sulfate by CE using indirect detection; no sodium sulfate was detected in product purified with the optimized RPIP separation (data not shown).
Figure 7.
Adaptation of RPIP-HPLC method developed here to separate sulfonated products from reaction mixtures by gravity flow benchtop C18 resin. N-cbz O-sulfonated neomycin was eluted in 10 mM ammonium acetate, initially in 100% water, and then with addition of 30% ACN as indicated by the dashed arrow. 1 mL fractions were collected and analyzed for UV absorbance at 258 nm. UV-absorbing fractions were pooled, freeze-dried and injected on analytical HPLC, detected at 258 nm.
3. Conclusions
Described here is the optimization and exploitation of a highly useful methodology for separating and characterizing amphiphilic, sulfated N-aryl substituted oligosaccharides. Compared with other RPIP-HPLC methods for the separation of sulfated carbohydrates, the sulfated N-cbz aminoglycosides presented here were best resolved by adaptation of a method originally developed for oligonucleotide separation. The aryl substituents on the amphiphilic sugars likely enhance interaction with the C18 resin; thus lower concentrations of ion pairing reagents were sufficient, even required, to achieve separation of various sulfated states compared with RPIP conditions used for sulfated oligosaccharides having no aryl substituents.
This separation method is shown to be applicable to LC-MS characterization of single compounds in complex product mixtures, useful in monitoring sulfation reaction progression, adaptable to using UV detection combined with LC-MS for calculating degree of sulfation, and useful for purifying amphiphilic saccharides by benchtop chromatography. We believe the methods described here will be useful for the separation of other sulfated or phosphorylated amphiphilic small molecules. We are currently employing this methodology in the preparation, purification, and characterization of structurally diverse O-sulfonated N-aryl aminoglycosides for further evaluation as bind-and-block antagonists of heparin-binding proteins.
4. Experimental Section
4.1 General
Neomycin sulfate, apramycin sulfate, and pyridine sulfur trioxide complex were from Sigma Aldrich (St. Louis, MO, USA); kanamycin sulfate was from Bristol Laboratories Inc. (Syracuse, NY, USA). N-(Benzyloxycarbonyloxy)succinimide and chlorosulfonic acid were purchased from Acros Organics (Morris Plains, NJ, USA). HPLC-grade acetonitrile (ACN) and triethylamine (TEA) were from Fisher Scientific (Hampton, NH, USA). Filtered, deionized water used with all buffers was from a Barnstead Nanopure Diamond system, Thermo Fisher Scientific (Hampton, NH, USA). Ammonium acetate (99.999% metal basis) was obtained from Sigma Aldrich. All buffers were filtered and degassed prior to use. A Fisher Accumet AB15 pH meter was used for all pH determinations.
Analytical HPLC used a Phenomenex Luna C18 100Å LC column (4.6 mm × 250 mm) (Torrance, CA, USA) with a Shimadzu chromatography system consisting of an LC-10AT VP pump and a SPD-M10A VP photodiode array detector (Kyoto, Japan). Semi-preparative HPLC used a Phenomenex Luna 10 µm C18(2) 100 Å LC Column (250 × 21.2 mm) with a Shimadzu HPLC system: dual LC-10AT VP pumps and SPD-M10A VP diode array detector. Centrifugation was done in a Fisher Marathon 21000R, Fisher Scientific. Cation exchange chromatography used Amberlite IR 120 resin, Sigma Aldrich. Benchtop column C18 chromatography used silica gel 60 RP-18 resin, EMD Chemicals (Gibbstown, NJ, USA). Product collected from benchtop chromatography was identified by UV absorbance in 96-well plates using a BioTek Synergy 2 Multimode plate reader (Winooski, VT, USA). Capillary electrophoresis (CE) was employed to determine presence/absence of inorganic sulfate using inverse detection with a UV detector at 214 nm on a Beckman Coulter P/ACE MDQ CE system (Brea, CA, USA). For LC-MS characterization of compounds, LC separation was performed with either a Thermo Fisher Scientific Surveyor LC system or a Dionex UltiMate 3000 (Sunnyvale, CA, USA) coupled to a Thermo Fisher Scientific LCQ Deca ESI-quadrupole- ion trap mass spectrometer.
4.2 Synthesis of N-cbz aminoglycosides
Synthesis and characterization of per N-cbz derivatives of neomycin, kanamycin, and apramycin have been reported.49, 50 Synthesis of the per-N-cbz derivatives of these aminoglycosides used N-(Benzyloxycarbonyloxy)succinimide (NHS-cbz) as previously reported for introducing cbz groups onto free amine groups of N-desulfonated heparin.17 Briefly, aminoglycoside (0.2–0.5 mmol) was dissolved in aqueous NaHCO3 and stirred at room temperature. Aliquots of NHS-cbz (1.2 equivalents per amine moiety) dissolved in 4.5 mL DMF were then added to the above solution fraction-wise (0.9 mL) at 60–90 min intervals. During the reaction, a white solid precipitate formed. The reaction was monitored using analytical HPLC: 20 µL of the reaction mixture and was diluted in 80 µL ACN and 40 µL water; 100 µL of this solution was injected and eluted with a gradient of 10–95% ACN in water (0.1% TFA) over 40 min at 1 mL/min. After complete acylation of amine groups, ~5–7 hours, the reaction was diluted with water (4 mL) to give additional white precipitate. The precipitate was collected by centrifugation (20 min, 4°C, 3500 rpm) and decanting followed by washing the solid with cold water (10 mL), centrifugation and decanting. This wash procedure was repeated seven times followed by lyophilization of a final suspension to give dry white solid. In reactions where the N-acylation was incomplete, as detected by analytical HPLC, under-reacted product was removed by dissolving the product in ACN:water (3:1) and passing the solution through a column of amberlite cation (H+) exchange resin (4 mL bed volume).
In some reactions, over-acylated product (having at least one O-cbz) was detected by ESI-MS; the desired product was then purified using semi-preparative HPLC. To this end, the product mixture was dissolved in 3:1 ACN:water and replicate aliquots injected and separated. The per-N-cbz product was collected after about 20 minutes, depending on the parent aminoglycoside, when eluted with 67% ACN in water with 0.1% TFA, 7 mL/min. Product purified with cation exchange or semi-prep chromatography was rotary evaporated to remove organic solvent and the resulting aqueous suspension lyophilized to give white solid.
4.3 RPIP-HPLC for monitoring sulfonation reaction progression
Many sulfonation procedures as well as TLC and HPLC methods were employed throughout the early stages of this work in efforts to find a way to efficiently follow the sulfonation reactions and to separate final products. The method to monitor sulfonation reactions described below is the method used once we developed the RPIP-LC-MS system described in this report. Thus analytical HPLC was used to monitor O-sulfonation of the N-cbz aminoglycosides as follows: As reactions progressed, 10 µL aliquots of a reaction mixture were diluted with 20 µL aq. NaHCO3, 20 µL ACN and 60 µL H2O and mixed, followed by loading 100 µL of this solution onto the analytical column. The mobile phase for elution consisted of two buffers: Buffer A: 10 mM ammonium acetate, 10% ACN, pH adjusted to 8.3 with addition of TEA; Buffer B: 10 mM ammonium acetate, 30% ACN, pH adjusted to 8.3 with addition of TEA. The mobile phase gradient was: 0–20 min, 0–100% B; 20–38 min, 100% B; 38–40 min, 100-0% B; 40–45 min, 100% A. The flow rate was held at 0.5 mL/min. Each N-cbz O-sulfated aminoglycoside was detected at 210 and 254 nm.
4.4 Sulfonation of N-cbz aminoglycosides
All glassware was oven-dried; all reactions were performed under argon gas. Two of the sulfonation methods were compared in the results here. In one method, aminoglycoside (100 mg) was dissolved in anhydrous DMF (1 mL) and stirred at 66 °C. To this solution was added Pyr·SO3 (3 equivalents per aminoglycoside hydroxyl) dissolved in DMF (1 mL) with anhydrous pyridine (1 mol equivalent).51 The reaction was stirred at 66 °C for 12–16 hours and monitored by analytical HPLC as outlined in section 4.3. The completed reaction was cooled to 4°C with an ice bath. Residual Pyr·SO3 was quenched by the addition of water (2 mL), and the resulting aqueous solution made alkaline with cold 10 M NaOH (added in 10 µL increments). Alternatively, ClSO3H was used to sulfonate the N-cbz aminoglycosides.52, 53 With this method, ClSO3H (8 equivalents per hydroxyl) was added drop-wise to anhydrous pyridine (3 mL) stirring at room temperature in a round bottom flask and the mixture then heated to 57 °C. Separately, the N-cbz aminoglycoside (50 mg) was dissolved in anhydrous pyridine and evaporated for azeotropic removal of residual water; this procedure was repeated 2–3 times before dissolving the N-cbz aminoglycoside in anhydrous pyridine (2 mL) and cannulating the resulting solution into the stirring ClSO3H/pyridine solution. Heating was maintained at 57 °C and reaction progress monitored by analytical HPLC as described in section 4.3. Upon reaction completion, about 4–6 hr, the mixture was cooled in an ice bath and adjusted to pH = 8 with addition of aqueous NaHCO3 (7.5 mL). The resulting aqueous solution was transferred to a separatory funnel and extracted with CH2Cl2 (7–10 × 15 mL) to remove pyridine. The water layer was then condensed on a rotary evaporator to remove residual organic solvent. Desalting and product separation was accomplished by loading the aqueous product on to a benchtop C18 column as described in section 4.5.
4.5 RPIP-Benchtop C18 chromatography
The aqueous mixture of sulfonation reaction product(s) was loaded onto a benchtop column packed with C18 silica resin (1 × 5.8 cm, 6.5 mL bed volume). Non-volatile salts were first eluted with the ammonium acetate buffer (10 mM ammonium acetate adjusted to pH = 8.3 with TEA) under gravity flow. Eluent was collected in fractions (1 mL) and analyzed at 210 and 258 nm to detect elution of sodium sulfate and sugar, respectively. As determined with 210 nm detection, the sodium sulfate eluted within the first 100 mL; this was verified with CE (see section 4.6). Then, sulfated product was eluted by addition of 10% ACN. As determined with 258 nm detection, fractions containing sulfated N-Cbz aminoglycoside were pooled, and ACN was removed en vacuo. The remaining water and volatile salts were removed by lyophilization to give product as a white solid. Products were analyzed by CE to show complete removal of sulfate and LC-MS to determine product purity or composition (methods described in section 4.6).
4.6 Capillary electrophoresis to demonstrate removal of sulfate
N-Cbz O-sulfonated aminoglycoside was dissolved in DI water to give a stock solution (1 mg/mL). Similarly, a stock solution of Na2SO4 was prepared and serially diluted to generate a standard curve of sulfate concentrations between 0–25 mM Na2SO4. An autosampler was used to inject samples over 10 sec at 0.5 psi. Separation was performed at 20.0 kV (25 °C, 50 cm capillary, 5 µm linear diameter, 4 Hz) using a buffer composed of 10 mM 5-sulfosalicyic acid pH 3.0.54, 55 Sodium sulfate was detected by indirect absorbance at 214 nm. Sodium sulfate concentration was calculated from the standard curve, and products desalted until sulfate was undetectable.
4.7 RPIP-HPLC coupled to ESI-MS
HPLC separations prior to mass spectra analysis used the Phenomenex analytical column on either a Surveyor LC system or a Dionex UltiMate 3000. Column temperature was not controlled; sample tray was maintained at 4°C. An autosampler was used to inject N-cbz O-sulfated aminoglycoside (20 µg); all samples were dissolved in water. (Pre-dissolving the samples in Buffer A made no noticeable difference in ion intensity or mass spectra.) Separation was achieved as described section 4.3 for monitoring sulfonation reactions with HPLC. The mass spectra was acquired on a Thermo LCQ Deca mass spectrometer with ESI ionization and quadrupole ion trap mass analyzer collecting centroid data in the negative mode. Mass range for samples with neomycin or apramycin cores was set from 600–2000 amu, and 500–2000 amu for kanamycin-core samples.
4.8 Tuning the ESI-MS for sulfated N-cbz aminoglycosides
To tune the MS, 2 mg/mL N-cbz O-sulfonated neomycin in water was injected via T-flow with buffer A. The instrument was focused on the mass corresponding to the doubly charged persulfated ion [M − 2H + TEA]−2 (m/z = 1038.7). Optimal ion detection was achieved by setting the spray voltage to 3.00 kV, capillary temp at 203 °C, capillary voltage to −46 V, increasing the tube lens offset to 25 V and decreasing the sheath flow rate to 60 psi.
4.9 Calculating degree of sulfation
Degree of sulfation was determined by peak integration of UV absorbance chromatograms. HPLC separations to determine UV absorbance were performed on a Shimadzu LC10AT VP series HPLC pump with PDA detector. Peak area was calculated by measuring absorbance at the wavelength of maximum absorbance at 254–258 nm for each compound. Each peak was assigned a sulfated state based on molecular mass of the corresponding peak in the total ion chromatogram (TIC). Percent area was calculated for each degree of sulfation in the UV chromatogram and used to assign an average degree of sulfation for the sample.
Acknowledgements
We are grateful to Dr. Lynn Teesch, the University of Iowa High Resolution Mass Spectrometry Facility, for her insightful advice in developing and troubleshooting ESI-MS and LC-MS methods in this work. We are also grateful to Professor Lei Geng and Claudiu Brumaru, the University of Iowa Department of Chemistry, for advice and assistance with CE.
Role of funding source
Funding was provided by in part by an NIH Predoctoral Training Grant in the Pharmacological Sciences to AF (T32 GM067795), an ACS Division of Medicinal Chemistry Predoctoral Fellowship sponsored by Bristol-Myers Squibb to AF, and a Predoctoral Fellowship from the American Foundation for Pharmaceutical Education to AF. Study design, data collection and analysis, writing the report and decision to publish the work were done independently of study sponsors.
Abbreviations
- ACN
acetonitrile
- cbz
benzyloxycarbonyl
- CE
capillary electrophoresis
- ClSO3H
chlorosulfonic acid
- DS
degree of sulfation
- ESI
electrospray ionization
- GAG
glycosaminoglycan
- HFIP
1,1,1,3,3,3-hexafluoro-2-propanol
- HS
heparan sulfate
- LC-MS
liquid chromatography mass spectrometry
- MeOH
methanol
- NHS
N-hydroxysuccinamide
- Pyr·SO3
pyridine sulfur trioxide complex
- RPIP
reversed-phase ion pairing
- TBAI
tetrabutylammonium iodide
- TEA
triethylamine
- TEAA
triethylammonium acetate
- TIC
total ion chromatogram
- TMAI
tetramethylammonium iodide
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dreyfuss JL, Regatieri CV, Jarrouge TR, Cavalheiro RP, Sampaio LO, Nader HB. An. Acad. Bras. Cienc. 2009;81:409–429. doi: 10.1590/s0001-37652009000300007. [DOI] [PubMed] [Google Scholar]
- 2.Hacker U, Nybakken K, Perrimon N. Nat. Rev. Mol. Cell Biol. 2005;6:530–541. doi: 10.1038/nrm1681. [DOI] [PubMed] [Google Scholar]
- 3.Zhang L. In: Glycosaminoglycan (GAG) Biosynthesis and GAG-Binding Proteins. Lijuan Z, editor. Vol. Volume 93. San Diego: Academic Press; 2010. pp. 1–17. [DOI] [PubMed] [Google Scholar]
- 4.Capila I, Linhardt RJ. Angew. Chem. Int. Ed. Engl. 2002;41:391–412. doi: 10.1002/1521-3773(20020201)41:3<390::aid-anie390>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 5.Powell AK, Yates EA, Fernig DG, Turnbull JE. Glycobiology. 2004;14:17R–30R. doi: 10.1093/glycob/cwh051. [DOI] [PubMed] [Google Scholar]
- 6.Casu B, Vlodavsky I, Sanderson RD. Pathophysiol. Haemost. Thromb. 2008;36:195–203. doi: 10.1159/000175157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laremore TN, Zhang F, Dordick JS, Liu J, Linhardt RJ. Curr. Opin. Chem. Biol. 2009;13:633–640. doi: 10.1016/j.cbpa.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernfield M, Gotte M, Park PW, Reizes O, Fitzgerald ML, Lincecum J, Zako M. Annu. Rev. Biochem. 1999;68:729–777. doi: 10.1146/annurev.biochem.68.1.729. [DOI] [PubMed] [Google Scholar]
- 9.Tumova S, Woods A, Couchman JR. Int. J. Biochem. Cell Biol. 2000;32:269–288. doi: 10.1016/s1357-2725(99)00116-8. [DOI] [PubMed] [Google Scholar]
- 10.Kirn-Safran C, Farach-Carson MC, Carson DD. Cell. Mol. Life Sci. 2009;66:3421–3434. doi: 10.1007/s00018-009-0096-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang L, Fuster M, Sriramarao P, Esko JD. Nat. Immunol. 2005;6:902–910. doi: 10.1038/ni1233. [DOI] [PubMed] [Google Scholar]
- 12.Fernandez C, Hattan CM, Kerns RJ. Carbohydr. Res. 2006;341:1253–1265. doi: 10.1016/j.carres.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 13.Lander AD. Chem. Biol. 1994;1:73–78. doi: 10.1016/1074-5521(94)90043-4. [DOI] [PubMed] [Google Scholar]
- 14.Lever R, Page CP. Nat. Rev. Drug Discov. 2002;1:140–148. doi: 10.1038/nrd724. [DOI] [PubMed] [Google Scholar]
- 15.Luscher-Mattli M. Antivir. Chem. Chemother. 2000;11:249–259. doi: 10.1177/095632020001100401. [DOI] [PubMed] [Google Scholar]
- 16.Huang L, Fernandez C, Kerns RJ. Bioorg. Med. Chem. Lett. 2007;17:419–423. doi: 10.1016/j.bmcl.2006.10.026. [DOI] [PubMed] [Google Scholar]
- 17.Huang L, Kerns RJ. Bioorg. Med. Chem. 2006;14:2300–2313. doi: 10.1016/j.bmc.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 18.Petitou M, Herault JP, Bernat A, Driguez PA, Duchaussoy P, Lormeau JC, Herbert JM. Nature. 1999;398:417–422. doi: 10.1038/18877. [DOI] [PubMed] [Google Scholar]
- 19.Huber CG, Oberacher H. Mass Spectrom. Rev. 2001;20:310–343. doi: 10.1002/mas.10011. [DOI] [PubMed] [Google Scholar]
- 20.Korir AK, Larive CK. Anal. Bioanal. Chem. 2009;393:155–169. doi: 10.1007/s00216-008-2412-2. [DOI] [PubMed] [Google Scholar]
- 21.Yang B, Solakyildirim K, Chang Y, Linhardt RJ. Anal. Bioanal. Chem. 2011;399:541–557. doi: 10.1007/s00216-010-4117-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuberan B, Lech M, Zhang LJ, Wu ZLL, Beeler DL, Rosenberg RD. J. Am. Chem. Soc. 2002;124:8707–8718. doi: 10.1021/ja0178867. [DOI] [PubMed] [Google Scholar]
- 23.Thanawiroon C, Rice KG, Toida T, Linhardt RJ. J. Biol. Chem. 2004;279:2608–2615. doi: 10.1074/jbc.M304772200. [DOI] [PubMed] [Google Scholar]
- 24.Henriksen J, Roepstorff P, Ringborg LH. Carbohydr. Res. 2006;341:382–387. doi: 10.1016/j.carres.2005.11.030. [DOI] [PubMed] [Google Scholar]
- 25.Doneanu CE, Chen W, Gebler JC. Anal. Chem. 2009;81:3485–3499. doi: 10.1021/ac802770r. [DOI] [PubMed] [Google Scholar]
- 26.Chai W, Luo J, Lim CK, Lawson AM. Anal. Chem. 1998;70:2060–2066. doi: 10.1021/ac9712761. [DOI] [PubMed] [Google Scholar]
- 27.Gunay NS, Tadano-Aritomi K, Toida T, Ishizuka I, Linhardt RJ. Anal. Chem. 2003;75:3226–3231. doi: 10.1021/ac034053l. [DOI] [PubMed] [Google Scholar]
- 28.Thanawiroon C, Linhardt RJ. J. Chromatogr. A. 2003;1014:215–223. doi: 10.1016/s0021-9673(03)00779-9. [DOI] [PubMed] [Google Scholar]
- 29.Zhang ZQ, Xie J, Liu HY, Liu J, Linhardt RJ. Anal. Chem. 2009;81:4349–4355. doi: 10.1021/ac9001707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meyer A, Raba C, Fischer K. Anal. Chem. 2001;73:2377–2382. doi: 10.1021/ac001402s. [DOI] [PubMed] [Google Scholar]
- 31.Karamanos NK, Vanky P, Tzanakakis GN, Tsegenidis T, Hjerpe A. J. Chromatogr. A. 1997;765:169–179. doi: 10.1016/s0021-9673(96)00930-2. [DOI] [PubMed] [Google Scholar]
- 32.Nakajima K, Kitazume S, Angata T, Fujinawa R, Ohtsubo K, Miyoshi E, Taniguchi N. Glycobiology. 2010;20:865–871. doi: 10.1093/glycob/cwq044. [DOI] [PubMed] [Google Scholar]
- 33.Grondahl F, Tveit H, Akslen-Hoel LK, Prydz K. Carbohydr. Res. 2011;346:50–57. doi: 10.1016/j.carres.2010.10.025. [DOI] [PubMed] [Google Scholar]
- 34.Solakyildirim K, Zhang ZQ, Linhardt RJ. Anal. Biochem. 2010;397:24–28. doi: 10.1016/j.ab.2009.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones CJ, Membreno N, Larive CK. J. Chromatogr. A. 2010;1217:479–488. doi: 10.1016/j.chroma.2009.11.064. [DOI] [PubMed] [Google Scholar]
- 36.Apffel A, Chakel JA, Fischer S, Lichtenwalter K, Hancock WS. J. Chromatogr. A. 1997;777:3–21. doi: 10.1021/ac960916h. [DOI] [PubMed] [Google Scholar]
- 37.Apffel A, Chakel JA, Fischer S, Lichtenwalter K, Hancock WS. Anal. Chem. 1997;69:1320–1325. doi: 10.1021/ac960916h. [DOI] [PubMed] [Google Scholar]
- 38.Irungu J, Dalpathado DS, Go EP, Jiang H, Ha HV, Bousfield GR, Desaire H. Anal. Chem. 2006;78:1181–1190. doi: 10.1021/ac051554t. [DOI] [PubMed] [Google Scholar]
- 39.Zhang G, Lin J, Srinivasan K, Kavetskaia O, Duncan JN. Anal. Chem. 2007;79:3416–3424. doi: 10.1021/ac0618674. [DOI] [PubMed] [Google Scholar]
- 40.Nordstrom A, Tarkowski P, Tarkowska D, Dolezal K, Astot C, Sandberg G, Moritz T. Anal. Chem. 2004;76:2869–2877. doi: 10.1021/ac0499017. [DOI] [PubMed] [Google Scholar]
- 41.Nielsen TC, Rozek T, Hopwood JJ, Fuller M. Anal. Biochem. 2010;402:113–120. doi: 10.1016/j.ab.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 42.Mason KE, Meikle PJ, Hopwood JJ, Fuller M. Anal. Chem. 2006;78:4534–4542. doi: 10.1021/ac052083d. [DOI] [PubMed] [Google Scholar]
- 43.Rogatsky E, Jayatillake H, Goswami G, Tomuta V, Stein D. J. Am. Soc. Mass Spectrom. 2005;16:1805–1811. doi: 10.1016/j.jasms.2005.07.017. [DOI] [PubMed] [Google Scholar]
- 44.Thomsson KA, Karlsson H, Hansson GC. Anal. Chem. 2000;72:4543–4549. doi: 10.1021/ac000631b. [DOI] [PubMed] [Google Scholar]
- 45.Djordjevic NM, Houdiere F, Fowler P, Natt F. Anal. Chem. 1998;70:1921–1925. doi: 10.1021/ac9711697. [DOI] [PubMed] [Google Scholar]
- 46.Huber CG, Buchmeiser MR. Anal. Chem. 1998;70:5288–5295. doi: 10.1021/ac980791b. [DOI] [PubMed] [Google Scholar]
- 47.Bleicher K, Bayer E. Chromatographia. 1994;39:405–408. [Google Scholar]
- 48.Gilar M, Fountain KJ, Budman Y, Holyoke JL, Davoudi H, Gebler JC. Oligonucleotides. 2003;13:229–243. doi: 10.1089/154545703322460612. [DOI] [PubMed] [Google Scholar]
- 49.Igarashi K, Honma T. In: Aprosamine derivatives, Vol. 81–206352; 80–150926 Eds.: Shionogi and Co, L. J.), Index IPC: C07; A61., Patent Application Country: Application: BE; Patent Country: BE; Priority Application Country: JP. Shionogi, Co LJ, editors. 1982. p. 26. [Google Scholar]
- 50.Chen G, Pan P, Yao Y, Chen Y, Meng X, Li Z. Tetrahedron. 2008;64:9078–9087. [Google Scholar]
- 51.Vogl H, Paper DH, Franz G. Carbohyd. Polym. 2000;41:185–190. [Google Scholar]
- 52.Zhang LY, Huang W, Tanimura A, Morita T, Harihar S, DeWald DB, Prestwich GD. Chemmedchem. 2007;2:1281–1289. doi: 10.1002/cmdc.200700071. [DOI] [PubMed] [Google Scholar]
- 53.Wang X, Zhang L. Carbohydr. Res. 2009;344:2209–2216. doi: 10.1016/j.carres.2009.04.033. [DOI] [PubMed] [Google Scholar]
- 54.Johnstone KD, Karoli T, Liu L, Dredge K, Copeman E, Li CP, Davis K, Hammond E, Bytheway I, Kostewicz E, Chiu FC, Shackleford DM, Charman SA, Charman WN, Harenberg J, Gonda TJ, Ferro V. J. Med. Chem. 2010;53:1686–1699. doi: 10.1021/jm901449m. [DOI] [PubMed] [Google Scholar]
- 55.Yu G, Gunay NS, Linhardt RJ, Toida T, Fareed J, Hoppensteadt DA, Shadid H, Ferro V, Li C, Fewings K, Palermo MC, Podger D. Eur. J. Med. Chem. 2002;37:783–791. doi: 10.1016/s0223-5234(02)01347-8. [DOI] [PubMed] [Google Scholar]