Abstract
Microsomal cytochrome P450s (CYPs) are anchored to the endoplasmic reticulum membrane by the N-terminal signal-anchor sequence which is predicted to insert into the membrane as a type 1 transmembrane helix with a luminally located N-terminus. We have mapped amino acids of the CYP2C1 signal-anchor, fused to Cys-free glutathione S-transferase, within the membrane by Cys-specific labeling with membrane-impermeant maleimide polyethylene glycol. At the C-terminal end of the signal-anchor, Trp-20 was mapped to the membrane-cytosol interface and Leu-19 was within the membrane. Unexpectedly, at the N-terminal end, Glu-2 and Pro-3 were mapped to the cytoplasmic side of the membrane rather than the luminal side as expected of a type 1 transmembrane helix. Similar results were observed for the N-terminal amino acids of the signal-anchor sequences of CYP3A4 and CYP2E1. These observations indicate that contrary to the current model of the signal-anchor of CYPs as a type 1 transmembrane helix, CYP2C1, CYP2E1, and CYP3A4 are monotopic membrane proteins with N-terminal signal-anchors that have a hairpin or wedge orientation in the membrane.
Keywords: cytochrome P450, endoplasmic reticulum, membrane protein
INTRODUCTION
Microsomal cytochromes P450 (CYPs)1 are integral membrane proteins localized in the ER. Their membrane topology has been studied extensively leading to a model in which the hydrophobic N-terminal signal-anchor peptide is the single transmembrane helix and functions as a membrane anchor and ER retention signal whereas the remaining catalytic domain resides on the cytosolic side of the ER partially imbedded in the membrane (reviewed in [1–3]). The localization of the catalytic domain of the CYPs on the cytosolic side of the ER membrane has been well documented, but evidence for the orientation of the N-terminal signal-anchor in the membrane has been conflicting. The most generally accepted model is that the N-termini of CYPs are located in the lumen of the ER and the hydrophobic region of the signal sequence spans the membrane bilayer with a following hydrophilic linker region and the catalytic domain in the cytosol. Evidence supporting this model includes glycosylation of N-terminal peptide sequences [4,5] and the reversal of topology induced by changing the charge balance of the regions flanking the signal-anchor of CYPs [6–8]. Studies employing methods that label the N-terminus with membrane-impermeant reagents resulted in conflicting conclusions that either agree with the model above [2,9] or to the contrary, indicate that the N-terminus is on the cytoplasmic side of the membrane [10,11].
The N-terminal signal-anchors of microsomal CYPs are very heterogeneous in length and amino acid sequence, but they all contain a hydrophobic core with an average length of 15–18 amino acids (http://drnelson.uthsc.edu/p450bpub237.html), but are as short as 12–14 amino acids in some CYPs such as CYP21A1 [12], CYP52C2 [13] and CYP3A11 [14]. Since the mean length of eukaryotic ER transmembrane domains is about 20 residues [15], in most cases the hydrophobic core of membrane insertion sequences of CYPs is either too short or of borderline length to span a membrane in a α-helix conformation as predicted by the current model.
We initially undertook these studies to determine whether hydrophobic changes in the signal anchor sequence of CYP2C1 that affected ER retention [16] also altered the position of the signal-anchor sequence relative to the membrane-cytosol interface. The position of the signal-anchor sequence was mapped by modification of Cys with a membrane-impermeable reagent MAL-PEG [17]. Surprisingly, the results indicated that the CYP2C1 signal anchor sequence inserts into the cytosolic leaflet of the bilayer as a hairpin, with both N- and C-terminal ends of the signal-anchor sequence at the cytosolic side of the ER membrane.
MATERIALS AND METHODS
Materials
Cell culture materials were purchased from Invitrogen. Antibodies were from Santa Cruz Biotechnology, except that HRP-conjugated anti-FLAG antibody was from Sigma-Aldrich and Cy-5 conjugated secondary antibody was from Jackson ImmunoResearch Laboratories. The chemiluminescence western blotting detection kit was from Pierce Chemical Co. and MAL-PEG was from Fluka Chemicals.
Plasmid construction
Plasmid pGEXC4S-6P-1 containing Cys-free glutathione S-transferase (GST) was obtained from Dr. Nikolay Tzvetkov, Department of Biophysical Chemistry, School of Medicine Hannover, Germany [18]. The GST coding region was amplified by PCR with a 5′ primer containing a HindIII site and 3′ primer with an XhoI site and the product was digested with HindIII and XhoI. The CYP2C1 DNA fragment coding for the 29 N-terminal amino acids was amplified from the plasmid C1(1–29)/GFP [19] with a 5′ primer containing a KpnI site and a 3′ primer with a HindIII site and the product was digested with these enzymes. Both fragments were cloned into the CYP2C2/Ad-Track plasmid [20,21], digested with KpnI and XhoI to delete the CYP2C2 insert, to produce 2C1/GST in which the resulting CYP2C1/GST coding sequence was in frame with the C-terminal FLAG tag. To construct the Cys-free chimera ALA/GST, QuickChange mutagenesis with 2C1/GST as a template was used to replace Cys at positions 10 and 13 with Ala. This plasmid was subsequently used as a template for constructing mutants with selected Cys substitutions using the QuickChange site-directed mutagenesis kit (Agilent Technologies) following the manufacturer’s protocol. To construct 2E1/GST and 3A4/GST, plasmids 2E1/GFP [22] and CYP3A4/GFP [3], respectively, were used as templates for amplification of the N-terminal coding regions with oligonucleotide primers containing KpnI and HindIII sites in the 5′ and 3′ primers, respectively. DNA fragments encoding amino acids 1–31 and 1–33 for CYP2E1 and CYP3A4, respectively, were amplified for cloning. PCR products were digested with KpnI and HindIII and ligated to plasmid C1/GST digested with the same enzymes. Mutants substituting Cys for Ala-2 in 3A4/GST and for Ser-2 in 2E1/GST were constructed using the QuickChange mutagenesis kit and confirmed by sequencing.
Cell culture and Transfection
Cell culture of HEK293 and COS1 cells and transfection with Lipofectamine 2000 reagent was done as described [23].
Preparation of Microsomes
To isolate microsomes, HEK293 or COS1 cells were grown and transfected in 100 mm plates. Two days after transfection, cells were washed, suspended in phosphate-buffered saline and pelleted by low speed centrifugation. The cell pellet was resuspended in 1.5 ml of microsomal buffer (50 mM Hepes-KOH, pH 7.2, 250 mM sorbitol, 70 mM potassium acetate, 5 mM EGTA, 1.5 mM magnesium acetate, and protease inhibitors) and homogenized with 35 strokes (pestle B) in a Dounce homogenizer. The homogenate was centrifuged for 8 min at 3,000×g and the supernatant was again centrifuged at 105,000×g for 45 min to pellet the membranes. The microsomal pellet was resuspended in 100 μl of microsomal buffer and aliquots were stored at −80°C.
Cys modification assay
For modification with maleimide PEG 5 kDa (MAL-PEG), reactions were performed in microsomal buffer and contained 6–10 μl of microsomes and 2 mM MAL-PEG (diluted from freshly prepared 50 mM stock in DMSO) with or without 1% Triton X-100 in a final volume of 20 μl. Incubations were carried out for 5 min at room temperature and then for 30 min at 4°C. To stop the reactions, DTT was added to a final concentration of 100 mM, and after incubation for additional 10 min at 4°C, an equal volume of 2× SDS PAGE loading buffer was added and samples were incubated for 5 min at 95°C before separation of proteins by SDS/PAGE and western analysis.
Immunoblot analysis
Proteins were separated by SDS/PAGE and electrophoretically transferred to nitrocellulose. The Flag containing peptides were detected by western analysis with anti-FLAG HRP-conjugated antibody (1:10,000). To detect calnexin, the blots were stripped and re-probed with anti-calnexin antibody (1:1,000) followed by the incubation with the secondary HRP-anti rabbit antibody (1:10,000). The blots were analyzed using a chemiluminescence detection kit from Pierce Chemical Co.
Immunofluorescence
For confocal fluorescent microscopy studies, HEK293 cells were grown and transfected on coverslips in 6-well plates, and 24–36 h after transfection, cells were fixed in 3.7% paraformaldehyde, permeabilized with 0.1% Triton X-100, and incubated with the mouse anti-FLAG antibody (1:100), followed by Cy5 conjugated anti-mouse antibody (1:100). Fixed cells were imaged with a Zeiss LSM510 confocal microscope as described [24].
RESULTS
Mapping the membrane protected region of the CYP2C1 signal-anchor by Cys scanning using MAL-PEG modification
Because of its specificity and usefulness for labeling proteins in natural membranes, MAL-PEG has been used successfully to map the membrane topology of other proteins [17,25]. MAL-PEG is a membrane-impermeant conjugate of maleimide and 5 kDa polyethylene glycol, which can react with a Cys SH group either exposed outside of the membrane or located at the interface and to all Cys after membranes are solubilized with detergents. Attachment of this reagent to a single Cys adds about 10 kDa to the apparent molecular weight of a protein after PAGE, so that the protein modification and the number of modified Cys residues can be easily detected by the change in mobility [17]. For the required Cys-free reporter protein, we used a Cys-free version of GST [18] which contained a FLAG sequence at its C-terminus and the CYP N-terminal sequences fused at its N-terminus.
Similar to other microsomal CYPs, the N-terminal signal of CYP2C1 consists of the membrane targeting hydrophobic sequence flanked on its N-terminal side by a negatively charged amino acid and positively charged amino acids on its C-terminal side, which is followed by a linker region (amino acids 21–29, also called stop-transfer signal) (Fig 1A). The chimeric protein, 2C1/GST localized to the ER membranes in HEK293 cells ( Fig. 1B), as expected from our previous studies in other cell lines with other reporter proteins [16,22,26] which showed that the 29-amino acid N-terminal signal-anchor is sufficient to mediate ER targeting and retention. Two Cys (10 and 13) are present in the hydrophobic region of the CYP2C1 signal-anchor (Fig. 1A). A Cys-free chimera (ALA/GST) that was constructed by substituting Ala for Cys-10 and Cys-13 also localized normally to the ER (Fig. 1C). ALA/GST was subsequently used as a template to prepare mutants containing single Cys substitutions. As analyzed by confocal microscopy, none of these mutations affected ER localization with the W20C mutation shown as a representative example (Fig. 1D).
Figure 1.
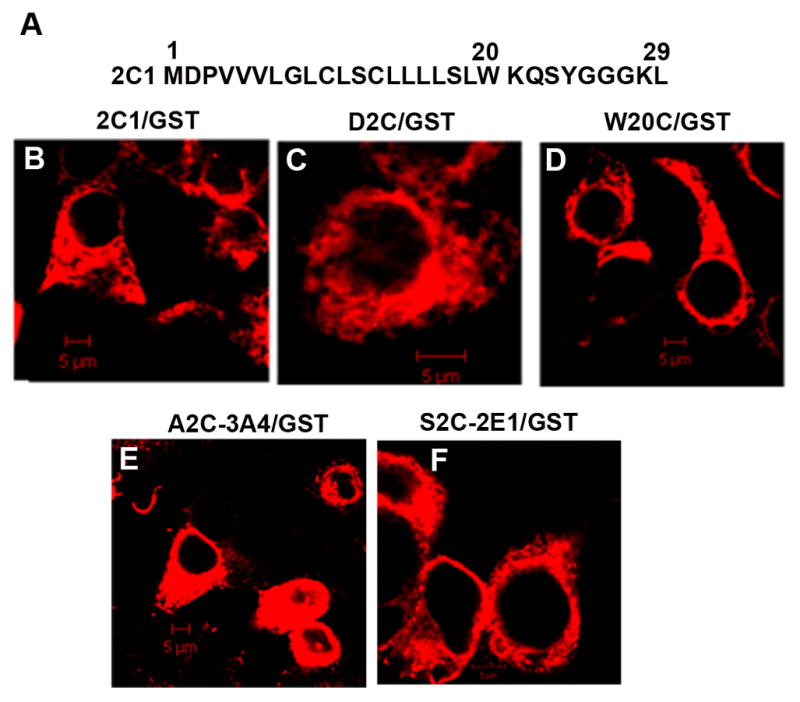
Subcellular localization of wt and Cys-mutants of chimera 2C1/GST. A. Sequence of the N-terminal 29 amino acids of CYP2C1. B–F. HEK293 cells were transfected with expression vectors for 2C1/GST (B) or the 2C1 mutants, D2C/GST (C) and W20C/GST (D) or for A2C-3A4/GST (E) or S2C-2E1/GST (F). After 24 h, fixed cells were permeabilized and subjected to immunostaining with anti-FLAG antibody followed by Cy5-conjugated secondary antibody. Cells were analyzed by confocal microscopy. Scale bars, 5 μm.
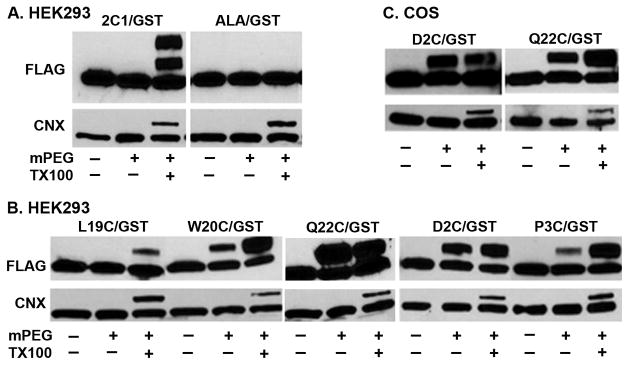
The mobility of 2C1/GST expressed in HEK293 cells was not changed after treatment of intact microsomes with MAL-PEG, but after solubilization of the membranes with Triton X-100, two slower migrating bands were observed (Fig. 2A). This result is consistent with the location of Cys-10 and Cys-13 in the membrane so that one or both residues in individual 2C1/GST molecules are modified only after solubilization of the membranes. In contrast, mobility of the Cys-free chimera ALA/GST was not altered in the presence or absence of detergent (Fig 2A), demonstrating the specificity of MAL-PEG for Cys. The control protein calnexin, which is an integral ER membrane protein and contains Cys only in its large luminal domain, was labeled with MAL-PEG only after solubilization of the membranes indicating that the ER membranes were intact and not inverted (Fig. 2A). For both 2C1/GST and calnexin, only a fraction of the proteins react with MAL-PEG, presumably because the reaction is incomplete and the extent is affected by the sequence context and conformation of the protein at the Cys residue. Thus, the Triton X-100 control is critical, and the labeling in the absence of Triton X-100 relative to labeling in its presence indicates the extent of protection of the Cys residue by the membrane.
Figure 2.
Membrane topology of CYP2C1 signal-anchor analyzed by MAL-PEG labeling. A–B. HEK293 cells were transfected with expression vectors for 2C1/GST or Cys-free mutant ALA/GST (A) or for mutants of 2C1/GST containing a single Cys substituted for Leu-19, Trp-20, Gln-22, Asp-2, or Pro-3 (B). Forty h after transfection, microsomes were prepared and used for MAL-PEG labeling assay, as described in Materials and Methods. Equal aliquots of microsomes were incubated with or without 2 mM MAL-PEG (mPEG) in the presence or absence of 1% Triton X-100 (TX100), and samples were analyzed by Western blot with anti-FLAG antibody (upper panel). The lower panel shows Western blots of the same samples probed with an anti-calnexin antibody. C. COS1 cells were transfected with expression vectors for mutants of 2C1/GST containing single Cys substituted for Asp-2 or Gln-22. The cells were analyzed as described in parts A and B.
To identify the C-terminal end of the CYP2C1 signal-anchor that is protected from MAL-PEG modification by the membrane, Cys was substituted for Leu-19, Trp-20, or Gln-22 (Fig. 2B). As expected, the Q22C mutant was modified in both the intact and solubilized membranes consistent with its expected location outside the membrane on the cytosolic side (Fig. 2B). Trp is often located at the membrane interface of transmembrane helices [27], and in CYP2C1, Trp-20 is followed by the positively charged Lys-21 at the beginning of the linker region located in the cytosol (Fig. 1A). The W20C mutant was modified by MAL-PEG in microsomes, but not as efficiently as after membrane solubilization (Fig. 2B) suggesting it was partially protected by the membrane which is consistent with the location of Trp-20 at the membrane-cytosol interface. Consistent with this idea, the L19C mutant was completely protected by the membranes from modification by MAL-PEG (Fig 2B), as expected if the hydrophobic Leu-19 is positioned within the membrane. These studies indicate that Leu-19 is the C-terminal most amino acid of the CYP2C1 signal-anchor within the membrane and Trp-20 is at the interface. Gln-22 and, presumably, the positively charged Lys-21 are completely exposed on the cytosolic side of the ER membrane.
To map the N-terminal amino acids of the CYP2C1 signal-anchor that are protected by the membrane, Cys was substituted for Glu-2 or Pro-3. Based on the accepted model placing the N-terminus of the signal-anchor on the luminal side of the membrane, modification of these residues by MAL-PEG should have required solubilization of the membranes. Surprisingly, in intact microsomes, both D2C and P3C mutants were modified with MAL-PEG (Fig. 2B). The P3C mutant was only partially protected by the membrane from modification by MAL-PEG suggesting that this position is at the membrane-cytosol interface. Analysis of the control ER protein calnexin by reprobing the same blot showed that the microsomal membranes were intact vesicles and not inverted (Fig. 2B). Similar results were observed when the proteins were expressed in COS1 cells (Fig. 2C), which have been used most often in previous studies on CYP membrane topology and targeting. These observations indicate that, contrary to the generally accepted model, the N-terminus of CYP2C1 is on cytosolic side of the ER membrane. These mapping results indicate that the sequence of the CYP2C1 signal-anchor inserted in the membrane extends from Val-4 to Leu-19 and that these 16 residues are inserted in a hairpin like configuration and penetrate only partially through the membrane in the cytosolic leaflet of the ER membrane.
Mapping the localization of the N-terminus of CYP2E1 and CYP3A4
To determine whether a similar hairpin-like structure of the N-terminal signal-anchor is present in other CYPs, particularly with longer hydrophobic sequences, we examined human CYP2E1 and human CYP3A4. The N-terminal signal-anchor of CYP2E1 contains 20 amino acids between Ser-2 and Trp-23 and CYP3A4 contains 21 amino acids between Asp-6 and His-28 (Fig. 3A) so in contrast to CYP2C1, both hydrophobic regions could span the membrane. CYP2E1 and CYP3A4 have no Cys residues in their N-terminal signal-anchors. Cys was substituted for Ser-2 of CYP2E1 and Ala-2 of CYP3A4 in the signal-anchor sequences fused to Cys-free GST. These mutants still exhibited normal ER localization (Fig. 1E, F).
Figure 3.
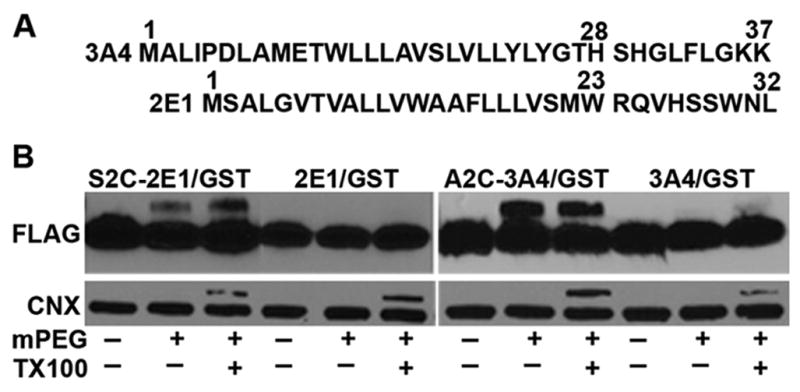
Subcellular localization and N-terminal labeling of chimeras 2E1/GST and 3A4/GST. A. Sequences of the N-terminal sequences of CYP3A4 and CYP2E1. B. HEK293 cells were transfected with the expression vectors for wt and single Cys containing mutants of 2E1/GST and 3A4/GST and after 40 h microsomes were prepared from the cells. Labeling with 2 mM MAL-PEG and analysis were performed as described in the legend to Fig. 2.
Treatment of microsomes from HEK293cells expressing these GST chimeras with MAL-PEG did not affect the mobility of the Cys-free parent CYP2E1 and CYP3A4 signal sequence-GST chimeric proteins as expected (Fig 3B). In contrast, the Cys at position 2 of both proteins were modified by MAL-PEG with or without Triton X-100 treatment (Fig. 3B) indicating that the N-termini of both proteins were on the cytoplasmic side. Although the level of modification was lower than that observed for the CYP2C1 D2C substitution, solubilization of the microsomes with Triton X-100 only modestly increased the modification. The decreased labeling, thus, most likely represents sequence-specific effects on the efficiency of the reaction. These results are consistent with the localization of the N-terminus of CYP2E1 and CYP3A4 signal-anchors on the cytosolic side of the ER membrane, as was observed with CYP2C1.
DISCUSSION
These studies unexpectedly demonstrate that the N-terminal amino acids of the microsomal CYPs, CYP2C1, CYP2E1, and CYP3A4 are exposed on the cytoplasmic side of the ER membrane, contrary to the generally accepted model of CYP membrane topology. Similar levels of labeling of D2C and Q22C with and without Triton X-100 indicate that these positions are totally exposed on the cytoplasmic side while partial protection of P3C and W20C indicate that these residues are present at the membrane-cytosol interface. An important control is the labeling of calnexin, which occurs only after solubilization of membranes with Triton X-100, providing evidence that the microsomal membranes are intact vesicles and not inverted. These results, therefore, provide strong evidence that the N-termini of the CYPs that were studied are on the cytoplasmic side and, thus that the hydrophobic core of the signal-anchor inserts into the membrane as a hairpin or wedge structure. In support of this model, NMR studies on the signal-anchor of a prostaglandin I synthase, which is structurally similar to CYPs, indicated that residues 1–20 formed a helix-turn-helix structure in the membrane which did not span the membrane [28]. While in prostaglandin I synthase, the uncharged N-terminus was proposed to be in the membrane core, we would propose that the charged Asp-2 results in the cytosolic orientation of the N-terminus in CYP2C1.
In earlier studies, labeling of CYPs in microsomes with the membrane impermeant label, FITC, was observed and a cytosolic orientation for the N-terminus was proposed [10] as we observe, but the opposite conclusion was reached in similar studies showing that the N-terminal Met of CYP2B1 in liposomes or natural membranes was protected from labeling with FITC, but was labeled after Triton X-100 treatment [9,29]. Support for the luminal orientation of the N-terminus also came from studies showing that CYP2C1 with a glycosylation signal fused at the N-terminus [4] and CYP19, which has a natural glycosylation signal at the N-terminal end [5], were glycosylated, which only occurs on the luminal side of the membrane. The earlier CYP2C1 experiments may have been misleading since the glycosylation was inefficient and the glycosylation could have “locked” the N-terminus in a luminal orientation that may normally occur transiently during the insertion into the membrane [30]. The reasons for the differences in this study with other studies supporting a luminal orientation for the N-terminus are not clear, but possibilities include CYP-specific differences in the orientation of the N-terminus or the different experimental conditions used. In this study of the membrane position of the signal anchor, the use of exogenously expressed recombinant and mutated proteins in cultured cells is required for the analysis which possibly could alter the topology the signal anchor. The present studies cannot exclude this possibility. The appropriate cellular localization of the mutated proteins, however, suggests that the signal anchor sequence is functioning normally and has a normal membrane topology.
It has been shown that the mean length of the hydrophobic transmembrane segment found in different ER membrane proteins in vertebrates is 20.3 residues, which is sufficient to span the hydrocarbon core of about 30 Å of a typical membrane, which is flanked on both sides by 15 Å long interfacial regions [15]. The N-terminal hydrophobic cores of microsomal CYPs average 15–18 amino acids, but may be as short as 12–14 amino acids and, thus may not be long enough to extend across the membrane. The insertion of the signal-anchor sequence as a hairpin eliminates the need for 20 hydrophobic amino acids and is a structure that can accommodate the varying lengths of hydrophobic cores in CYPs.
The impetus for these studies was to determine if mutations in the signal-anchor sequence that altered its hydrophobicity and eliminated ER retention changed the position of the amino acids at the C-terminus that bordered the membrane-cytosol interface [16]. Studies of the position of these mutated signal anchors in the membrane (not shown) revealed only minor changes in the membrane position and no consistent correlation of position with ER retention. The position within the membrane, therefore, does not appear to be the determining factor for the ER retention function. The results suggest that the hairpin structure, rather than specific amino acids sequences may be important for ER retention. Further support for this idea comes from earlier studies in which we have shown that the CYP2C1 signal-anchor positioned at the C-terminal end or in the middle of a type 1 chimeric membrane protein did not function as an ER retention signal [23]. At that time the type 1 transmembrane helix model was accepted for the N-terminal signal anchor so we hypothesized that the ER retention function was lost because of the sequences fused at the N-terminal side of the signal-anchor. Based on our current results, a more likely explanation is that the hairpin structure of the signal-anchor is required for ER retention and when the signal-anchor is in the middle or at the C-terminal end of a protein, it inserts into the membrane as a typical type1 transmembrane helix.
The hairpin structure may increase the affinity of CYP2C1 for the curved membranes in the smooth ER where CYPs are located. Interestingly, reticulon 4 has been recently shown to be a part of the multicomponent complex formed by CYP2C2 in a mouse liver [20,21]. Reticulons are ER membrane structural proteins which induce curvature in the smooth ER by insertion of two strongly conserved long hydrophobic sequences into the membrane as two wedges which expands the surface of the cytosolic leaflet [31]. If the CYP2C1signal anchor is a hairpin sequence mainly in the cytosolic leaflet, the signal anchor may have higher affinity for the expanded cytosolic leaflet in curved smooth ER, thus, accounting for its localization in smooth ER. Alternatively, or in addition, direct interactions with reticulons may contribute to the ER localization of CYP2C1.
In conclusion, these studies have shown that the N-terminal signal-anchor sequences of 3 microsomal CYPs are inserted in hairpin-like structures, contrary to the commonly accepted model of a transmembrane α-helix structure. This hairpin-like structure may be able to accommodate heterogeneity in sequence and length of signal-anchors that is observed in CYPs, although this structure may not be characteristic of all CYPs. The hairpin structure of the signal-anchor may contribute to its ER retention properties.
Highlights.
N-terminal cytochromes P450 sequences are thought to be transmembrane helices.
N-terminal ends of three cytochromes P450 were cytoplasmically accessible.
These N-terminal sequences are inserted partially into the membrane as a hairpin.
The hairpin structure may play a role in cellular localization of cytochromes P450.
Acknowledgments
This work was supported in part by a grant from the National Institute of Health: Grant GM35897. We thank Dr. Nikolay Tzvetkov, Department of Biophysical Chemistry, School of Medicine, Hannover, Germany, for providing the plasmid, pGEXC4S-6P-1.
Footnotes
Abbreviations: CYP, cytochrome P450; ER, endoplasmic reticulum; GST, glutathione S-transferase; MAL-PEG, maleimide polyethylene glycol.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Neve EP, Ingelman-Sundberg M. Intracellular transport and localization of microsomal cytochrome P450. Anal Bioanal Chem. 2008;392:1075–1084. doi: 10.1007/s00216-008-2200-z. [DOI] [PubMed] [Google Scholar]
- 2.Vergeres G, Winterhalter KH, Richter C. Microsomal cytochrome P-450: substrate binding, membrane interactions, and topology. Mutat Res. 1989;213:83–90. doi: 10.1016/0027-5107(89)90034-1. [DOI] [PubMed] [Google Scholar]
- 3.Szczesna-Skorupa E, Kemper B. Influence of protein-protein interactions on the cellular localization of cytochrome P450. Expert Opin Drug Metab Toxicol. 2008;4:123–136. doi: 10.1517/17425255.4.2.123. [DOI] [PubMed] [Google Scholar]
- 4.Szczesna-Skorupa E, Kemper B. An N-terminal glycosylation signal on cytochrome P450 is restricted to the endoplasmic reticulum in a luminal orientation. J Biol Chem. 1993;268:1757–1762. [PubMed] [Google Scholar]
- 5.Shimozawa O, Sakaguchi M, Ogawa H, Harada N, Mihara D, Omura T. Core glycosylation of cytochrome P-450(arom). Evidence for localization of N terminus of microsomal cytochrome P-450 in the lumen. J Biol Chem. 1993;268:21339–21402. [PubMed] [Google Scholar]
- 6.Szczesna-Skorupa E, Browne N, Mead D, Kemper B. Positive charges at the NH2 terminus convert the membrane-anchor signal peptide of cytochrome P-450 to a secretory signal peptide. Proc Natl Acad Sci USA. 1988;859:738–742. doi: 10.1073/pnas.85.3.738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sato T, Sakaguchi M, Mihara K, Omura T. The amino-terminal structures that determine topological orientation of cytochrome P-450 in microsomal membrane. EMBO J. 1990;9:2391–2397. doi: 10.1002/j.1460-2075.1990.tb07414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neve EP, Ingelman-Sundberg M. Molecular basis for the transport of cytochrome P450 2E1 to the plasma membrane. J Biol Chem. 2000;275:17130–17135. doi: 10.1074/jbc.M000957200. [DOI] [PubMed] [Google Scholar]
- 9.Vergeres G, Winterhalter KH, Richter C. Localization of the N-terminal methionine of rat liver cytochrome P-450 in the lumen of the endoplasmic reticulum. Biochim Biophys Acta. 1991;1063:235–241. doi: 10.1016/0005-2736(91)90376-j. [DOI] [PubMed] [Google Scholar]
- 10.Bernhardt R, Kraft R, Ruckpaul K. A simple determination of the sideness of the NH2-terminus in the membrane bound cytochrome P-450 LM2. Biochem Int. 1988;17:1143–1150. [PubMed] [Google Scholar]
- 11.Bernhardt R, Ngoc Dao NT, Stiel H, Schwarze W, Friedrich J, Janig GR, Ruckpaul K. Modification of cytochrome P-450 with fluorescein isothiocyanate. Biochim Biophys Acta. 1983;745:140–148. doi: 10.1016/0167-4838(83)90042-0. [DOI] [PubMed] [Google Scholar]
- 12.Zhou MY, del Carmen Vila M, Gomez-Sanchez EP, Gomez-Sanchez CE. Cloning of two alternatively spliced 21-hydroxylase CDNAs from rat adrenal. J Steroid Biochem Mol Biol. 1997;62:277–286. doi: 10.1016/s0960-0760(97)00040-x. [DOI] [PubMed] [Google Scholar]
- 13.Ohkuma M, Muraoka S, Tanimoto T, Fujii M, Ohta A, Takagi M. CYP52 (cytochrome P450alk) multigene family in Candida maltosa: identification and characterization of eight members. DNA Cell Biol. 1995;14:163–173. doi: 10.1089/dna.1995.14.163. [DOI] [PubMed] [Google Scholar]
- 14.Yanagimoto T, Itoh S, Muller-Enoch D, Kamataki T. Mouse liver cytochrome P-450 (P-450IIIAM1): its cDNA cloning and inducibility by dexamethasone. Biochim Biophys Acta. 1992;1130:329–332. doi: 10.1016/0167-4781(92)90447-8. [DOI] [PubMed] [Google Scholar]
- 15.Sharpe HJ, Stevens TJ, Munro S. A comprehensive comparison of transmembrane domains reveals organelle-specific properties. Cell. 2010;142:158–169. doi: 10.1016/j.cell.2010.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szczesna-Skorupa E, Kemper B. Endoplasmic reticulum retention determinants in the transmembrane and linker domains of cytochrome P450 2C1. J Biol Chem. 2000;275:19409–19415. doi: 10.1074/jbc.M002394200. [DOI] [PubMed] [Google Scholar]
- 17.Lu J, Deutsch C. Pegylation: a method for assessing topological accessibilities in Kv1.3. Biochemistry. 2001;40:13288–13301. doi: 10.1021/bi0107647. [DOI] [PubMed] [Google Scholar]
- 18.Tzvetkov N, Breuer P, Boteva R. Cysteine-free glutathione-S-tranferase as a tool for thiol-specific labeling of proteins. BioTechniques. 2006;40:145–146. doi: 10.2144/000112113. [DOI] [PubMed] [Google Scholar]
- 19.Szczesna-Skorupa E, Chen C, Rogers S, Kemper B. Mobility of cytochrome P450 in the endoplasmic reticulum membrane. Proc Natl Acad Sci USA. 1998;95:14793–14798. doi: 10.1073/pnas.95.25.14793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dong MS, Bell LC, Guo Z, Phillips DR, Blair IA, Guengerich FP. Identification of retained N-formylmethionine in bacterial recombinant mammalian cytochrome P450 proteins with the N-terminal sequence MALLLAVFL…: Roles of residues 3–5 in retention and membrane topology. Biochemistry. 1966;35:10031–10040. doi: 10.1021/bi960873z. [DOI] [PubMed] [Google Scholar]
- 21.Li B, Yau P, Kemper B. Identification of cytochrome P450 2C2 protein complexes in mouse liver. Proteomics. 2011;11:3359–3368. doi: 10.1002/pmic.201100001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szczesna-Skorupa E, Chen C, Kemper B. Cytochromes P450 2C1/2 and P450 2E1 are retained in the endoplasmic reticulum membrane by different mechanisms. Arch Biochem Biophys. 2000;374:128–136. doi: 10.1006/abbi.1999.1628. [DOI] [PubMed] [Google Scholar]
- 23.Szczesna-Skorupa E, Kemper B. The juxtamembrane sequence of cytochrome P-450 2C1 contains an endoplasmic reticulum retention signal. J Biol Chem. 2001;276:45009–45014. doi: 10.1074/jbc.M104676200. [DOI] [PubMed] [Google Scholar]
- 24.Szczesna-Skorupa E, Kemper B. BAP31 is involved in the retention of cytochrome P450 2C2 in the endoplasmic reticulum. J Biol Chem. 2006;281:4142–4148. doi: 10.1074/jbc.M509522200. [DOI] [PubMed] [Google Scholar]
- 25.Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 2006;124:573–586. doi: 10.1016/j.cell.2005.11.047. [DOI] [PubMed] [Google Scholar]
- 26.Szczesna-Skorupa E, Ahn K, Chen C, Doray B, Kemper B. The cytoplasmic and N-terminal transmembrane domains of cytochrome P450 contain independant signals for retention in the endoplasmic reticulum. J Biol Chem. 1995;270:24327–24333. doi: 10.1074/jbc.270.41.24327. [DOI] [PubMed] [Google Scholar]
- 27.Ridder AN, Morein S, Stam JG, Kuhn A, de Kruijff B, Killian JA. Analysis of the role of interfacial tryptophan residues in controlling the topology of membrane proteins. Biochemistry. 2000;39:6521–6528. doi: 10.1021/bi000073v. [DOI] [PubMed] [Google Scholar]
- 28.Ruan KH, So SP, Zheng W, Wu J, Li D, Kung J. Solution structure and topology of the N-terminal membrane anchor domain of a microsomal cytochrome P450: prostaglandin I2 synthase. Biochem J. 2002;368:721–728. doi: 10.1042/BJ20021001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vergeres G, Winterhalter KH, Richter C. Identification of the membrane anchor of microsomal rat liver cytochrome P-450. Biochemistry. 1989;28:3650–3655. doi: 10.1021/bi00435a005. [DOI] [PubMed] [Google Scholar]
- 30.von Heijne G. Membrane-protein topology. Nat Rev Mol Cell Biol. 2006;7:909–918. doi: 10.1038/nrm2063. [DOI] [PubMed] [Google Scholar]
- 31.Zurek N, Sparks L, Voeltz G. Reticulon short hairpin transmembrane domains are used to shape ER tubules. Traffic. 2011;12:28–41. doi: 10.1111/j.1600-0854.2010.01134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]