Abstract
The c-Myb and GATA-3 transcription factors play important roles in T-cell development. We recently reported that c-Myb, GATA-3, and Menin form a core transcription complex that regulates GATA-3 expression and ultimately Th2 cell development in human peripheral blood T-cells. However, c-Myb roles for Th2 cytokine expression were not demonstrated. Here, we report that c-Myb and GATA-3 cooperatively play an essential role in IL-13 expression though direct binding to a conserved GATA-3 response element (CGRE), an enhancer for IL-13 expression. c-Myb and GATA-3 were shown to activate the CGRE-IL-13 promoter by ~160 fold, and mutation of the canonical Myb binding site completely abrogated CGRE enhancer activity. In contrast, mutation of the GATA binding site partially decreased CGRE enhancer activity. GATA-3 did not bind to CGRE when c-myb expression was silenced. c-Myb, GATA-3, Menin and MLL bound to CGRE in human primary CD4+ effector/memory cells. Moreover, c-myb silencing significantly decreased both methylation of histone H3K4 and acetylation of histone H3K9 at the IL-13 locus in CD4+ effector/memory cells. Therefore, in addition to the strong enhancer effect for the transcription of IL-13, the c-Myb/GATA-3 complex recruits MLL to the CGRE for histone modification of the IL-13 locus during the differentiation of memory Th2 cells.
Introduction
T helper cells are critical in regulating immune responses to specific pathogens by controlling the function of other immune cells (1). The three distinct subsets of Th cells, Th1, Th2, and Th17, have different cytokine expression and effector functions. Th1 cells help promote the clearance of intracellular bacteria and viruses via production of IFN-γ, TNF-β, and IL-2. Th17 cells express IL-17 and play a critical function in protection against extracellular bacteria and fungi (2). Th2 cells express IL-4, IL-5, and IL-13. Coordinate regulation of IL-4, IL-5, and IL-13 are important for typical immune responses observed during infection by extracellular pathogens, helminthic parasites, and in atopic or allergic response such as asthma (3–5). The genes encoding IL-4, IL-5, and IL-13 are closely linked over a 150-kb genomic region of human chromosome 5 called the “Th2 cytokine gene” (6). Of these, the IL-4 and IL-13 genes reside only 13 kb apart. Given the close proximity of the IL-4, IL-5, and IL-13 genes and their coordinate expression after T cell activation, their expression is regulated similar to a single transcriptional locus.
The c-myb proto-oncogene encodes a transcription factor, c-Myb, which regulates genes important for early myeloid, B, and T lymphoid cell development (7, 8). c-Myb has an important role at multiple points during early T cell development (9–11). While the function of c-Myb in developing cells has been intensively studied, there is a paucity of investigations focused on c-Myb function in peripheral blood T cells, despite the very high level of c-Myb expression in primary peripheral T cells after activation.
GATA-3 is a member of the zinc finger transcription factor family (12, 13). In addition to its crucial role in regulating Th2 cell differentiation, GATA-3 is also essential for thymic T-cell development (14, 15). In peripheral T cells activation of STAT6 through IL-4 signaling induces GATA-3 mRNA expression, which finally results in Th2 cell differentiation. During Th2 cell development, GATA-3 works as a transcription factor for Th2 cytokine genes, particularly for IL-5 and IL-13 (12, 16, 17). However, the molecular mechanism by which GATA-3 regulates the Th2 cytokine genes is not well understood.
While c-Myb and GATA-3 were demonstrated to have a crucial role in early T cell development (14), a recent report first showed that c-Myb plays a direct role in the regulation of GATA-3 expression in early T-cell development (18). We have also recently demonstrated that c-Myb forms a complex with GATA-3 and binds a conserved Myb binding site in the GATA-3 promoter, which upregulates GATA-3 expression, and that this plays an important role in human Th2 cell development (19). We observed that silencing c-myb in primary human peripheral blood CD4+ T-cells stimulated under Th2 cell promoting conditions results in decreased expression of Th2 cytokines, IL-4, IL-5 and IL-13 (19). The decreased Th2 cytokine expression observed may have been a direct effect by c-Myb or an indirect effect by the decrease of GATA-3, or may have been a consequence of affecting the c-Myb/GATA-3 complex. Here we address c-Myb roles in regulating Th2 cytokine gene expression, and demonstrate that c-Myb critically regulates IL-13 expression via direct binding to the conserved GATA-3 response element (CGRE).
Materials and Methods
Lymphocyte preparation and cell culture
Normal human peripheral blood CD4+ T lymphocytes were obtained from consenting donors via the Human Immunology Core Facility at the University of Pennsylvania School of Medicine. Naive and memory/effector cells were sorted by CD45RO or CD45RA MicroBeads (Miltenyi Biotec), or PE-anti-human CD45RA antibody (BD Biosciences). Freshly isolated cells were cultured in RPMI-1640 medium supplemented with 10% FCS. Naive CD4+ T-cells were cultured with either IL-12 (10 ng/ml), IL-2 (20 ng/ml) and anti-IL-4 antibody (10μg/ml) for Th1 cell development, or with IL-4 (25–40 ng/ml), IL-2 (20ng/ml) and anti-IL-12 antibody (5–10 μg/ml) to promote Th2 cell formation in 1–10 days. CD3/CD28 antibodies (Dynabeads CD3/CD28 T cell expander, Invitrogen) were used for primary T cell stimulation. Jurkat and 293T cells were cultured in RPMI-1640 or DMEM medium, respectively and supplemented with 10% FCS. PMA (200ng/mL) and ionomycin (300ng/mL) were used for the stimulation of Jurkat cells.
RNAi for c-myb silencing
To generate lentivirus constructs for c-myb RNAi, c-myb-1(tgttattgccaagcactta), c-myb-3 (ctgcctggacgaactgata) or control c-myb-1 scramble (ctttatacgtagtcataag) siRNA sequence was inserted into the H1UG lentivirus vector (a gift of Dr. E.J. Brown, University of Pennsylvania, Philadelphia, PA). The mixed lentiviruses with c-myb-1 and c-myb-3 shRNA were transduced into human primary CD4+ T cells as described previously (19). Forty-eight hours post-transduction, GFP positive cells were sorted using a FACSAria™ cell sorter (BD Biosciences). Jurkat cells were infected with lentivirus expressing GFP and c-myb-3 shRNA. GFP positive cells were sorted and used to establish stable cell lines expressing c-myb or control shRNA. cmyb-1 and -3 siRNA were also used for transient c-myb silencing in human T cells using Amaxa nucleofection system (Lonza).
Dual-Luciferase reporter assay
Seven reporter constructs were prepared by inserting the human IL-13 and IL-4 promoter with or without the regulatory element(s) which includes CGRE, CNS-1, IE and HSV into pGL3-basic vector (Promega). The constructs contain the following regions. IL13P: −254 to +60 bp, IL4P: −766 to +60 bp, CGRE: a 65 bp fragment from −1740 to −1676 bp upstream of the IL-13 transcription start site, CNS-1: a 371 bp fragment from −9652 to −9282 bp upstream of the IL-4 transcription start site, IE: a 127 bp fragment from +1485 to +1611 bp downstream of the IL-4 transcription start site and HSV: a 364 bp fragment from +15089 to +15452 bp downstream of the IL-4 transcription start site. CGRE-IL-13P has the region from −1740 to +60. The pcDNA3 vectors containing full length c-myb (1 μg) and/or full length GATA-3 (1 μg) were cotransfected with phRL (0.01 μg) and appropriate promoter and the regulatory element up/downstream of luciferase in pGL3 (0.5 μg) into human primary T-cells using Human T cell Nucleofector kit (Lonza) or into Jurkat cells using Cell line Nucleofector kit (Lonza) or into 293T cells using Lipofectamine 2000 (Invitrogen). Firefly and Renilla luciferase activities were measured from the pGL3 and phRL reporter constructs, respectively, according to instructions in the Dual-Luciferase reporter assay kit (Promega) using a luminometer (Orion Microplate Luminometer, Berthold Detection Systems).
Chromatin immunoprecipitation (ChIP) assay
ChIP assays were completed using ChIP Assay kit (Millipore) according to the instruction manual. CD4+ naive or effector/memory T cells were cultured under Th2 conditions for 3–5 days except where noted (see below). The cells were re-stimulated with IL-4 and IL-2 at 1–6 hours before crosslinking with formaldehyde. Immunoprecipitation was performed with anti-c-Myb (a mixture of clone 1-1; Millipore and H-141; Santa Cruz), anti-GATA-3 (a mixture of H-48; Santa Cruz and HG3-31; Santa Cruz), anti-MLL1 (Bethyl), or anti-Menin (Bethyl). The precipitated DNA fraction was then amplified by quantitative real time PCR using standard protocols with Sybr green and specific primers for each regulatory element (CGRE, forward: 5'-CCCCATCTCCCGTTACATAAG-3'; reverse: 5'-CCCCATGGCCACCAAAAG-3' and forward: 5'-CCTCCGCAGCTTCCAGACT-3'; reverse: 5'-GGTGGCCTTATGTAACGGGA-3'; CNS-1, forward: 5'-ACTATGTTCCAGACAGGACAGCCT-3'; reverse: 5'-TTGTTGCAAACCCTTGCCCTCTTG-3'; IE, forward: 5'-CTCTCTGCCTCCTGTGCTG-3'; reverse: 5'-CTTTGGGGCTTTCATTCTCA-3'; HSV, forward: 5'-TGCTGCTTTTAGGTTGGGTTA-3'; reverse: 5'-CAGATATTCCTCTCCACCTGCT-3'). In ChIP assays for histone modification, CD4+ naive and effector/memory T cells were stimulated under Th2 promoting conditions for 7 and 4 days, respectively. Antibodies against di- and tri-methylated H3K4 (ab6000), acetyl K9 (ab4441) and di-methyl K9 (ab1220) (Abcam, Cambridge, MA) were utilized for the ChIP assays probing the IL-13 and IL-4 locus in primary CD4+ T cells. The 22 primer-pairs are listed in supplemental table 1.
Enzyme-linked immunosorbent assay (ELISA)
ELISA was performed using human IL-4, IL-5 and IL-13 ELISA Ready-SET-Go! kit (eBioscience). Human primary T cells or naïve CD4+ T cells were stimulated with Th2 promoting conditions to measure IL-5 and IL-13, or stimulated with IL-2 and CD3/CD28 antibodies to measure IL-4 by the ELISA kits. All procedures are done according to the instruction manual of the kit.
Electrophoretic Mobility Shift Assay (EMSA)
These experiments used 5 μl of in vitro translated c-Myb or GATA-3 proteins bound to 50 ng double-stranded DNA oligonucleotide. The c-Myb and GATA-3 proteins were in vitro translated from their respective pcDNA expression vector using TNT Quick Coupled Transcription/Translation Systems (Promega). The binding reactions proceeded for 20 min in a buffer containing 50 μg/ml of poly(dI-dC), 10 mM HEPES, pH 7.9, 1 mM EDTA, 5 mM MgCl2, 30 mM KCl, 10 % glycerol and protease inhibitors. Samples were separated by electrophoresis on a native 5 % polyacrylamide gel (0.5 × Tris-boric acid-EDTA buffer and 2 % glycerol) for 2 h at 200 V. Gels were exposed to X-ray film. For competition assays, a 10-fold excess of a cold competitor was included. The sequences of the two strands of the oligonucleotides used were as follows (consensus c-Myb and GATA3 binding site sequences in underlined and italic, respectively): CGRE, 5'-CTTATCGGGCCCATCTCCCGTTACATAAGGCCACCCCCCTATCTCCGCGGGCCATCGC CG-3'. The sequences of the two strands of the oligonucleotides with the mutated consensus c-Myb binding sites were as follows (mutated nucleotides in bold): CGRE mut Myb, 5'-CTTATCGGGCCCATCTCCCGTCACATAAGGCCACCCCCCTATCTCCGCGGGCCATCG CCG-3'.
Statistical analyses
Statistical comparisons of the data were completed using the two-tailed Student's t test. The level of significance was set at P values of < 0.05 in all cases.
Results
c-Myb expression levels are concordant with the production of Th2 cytokine
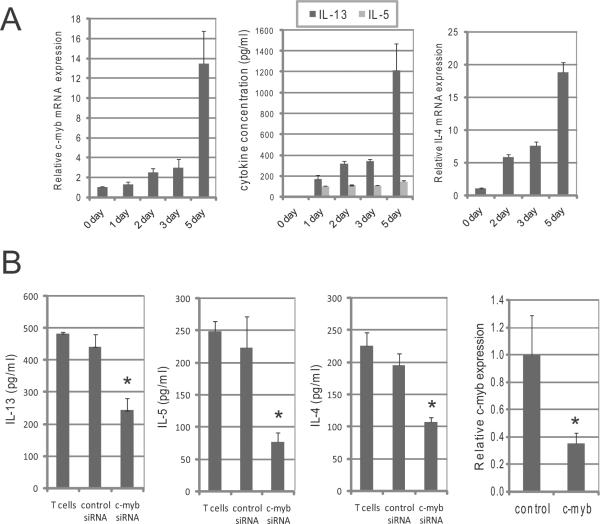
To initially test whether c-Myb expression levels affect the production of Th2 cytokines in human primary T cells, we first measured the amounts of c-myb and IL-4 mRNA and concentration of IL-13 and IL-5 using quantitative real time PCR (QRT-PCR) and enzyme-linked immunosorbent assay (ELISA) in primary human CD4+ CD45RA+ (naïve) T cells that were stimulated under Th2 cell promoting conditions. The expression of c-myb and IL-4 and the secretion of IL-13 were the greatest after 5 days of stimulation, indicating that the expression levels were concordant (Figure 1A). We next silenced c-myb expression by siRNA in human primary T cells, and measured the secretion of Th2 cytokines using ELISA after stimulation. Silencing c-myb in the cells significantly decreased the concentration of IL-5, IL-13 and IL-4 in the cell culture media (Figure 1B), suggesting that the expression level of c-Myb directly affects the production of Th2 cytokines.
Figure 1. c-Myb expression levels are concordant with the production of Th2 cytokine.
(A) Naive (CD45RA+) CD4+ T cells were cultured with Th2 cell promoting stimulatory conditions, and the cells and supernatants were collected before stimulation (0 day) and 1,2, 3 and 5 days after stimulation. Left graph shows the amount of c-myb mRNA, as measured by QRT-PCR. Middle graph shows the secreted IL-13 and IL-5 in each supernatant. Right graph shows the IL-4 mRNA expression in the cells. (B) Human primary T-cells were stimulated with IL-2 and CD3/CD28 antibodies for 3 days, and then the siRNA against c-myb or control siRNA were transfected into the cells. The transfected cells were stimulated with IL-4, IL-2 and CD3/CD28 antibodies or with IL-2 and CD3/CD28 antibodies for 3 days to measure IL-13 and IL-5 or IL-4, respectively, and then the concentration of IL-13, IL-5 and IL-4 in supernatants were measured by ELISA. Gene expression of c-myb was measured by QRT-PCR in the same cells. T cells = no transfection with siRNA.* Statistically significant (P < 0.05) compared control siRNA.
c-Myb regulates IL-13 and IL-4 expression through regulatory elements in the Th2 cytokine gene in cooperation with GATA-3
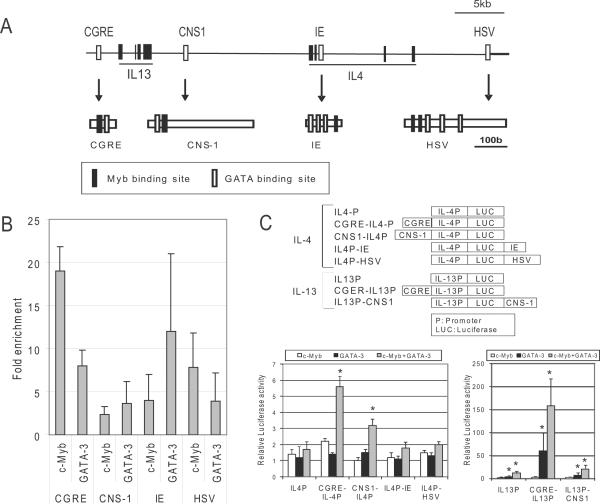
Because we previously reported that c-myb silencing decreases GATA-3 expression in Th2 cells (19), we next examined whether the observed decrease in Th2 cytokine expression is a direct effect by c-Myb or an indirect effect by the decrease of GATA-3. The Th2 cytokine locus has numerous DNase I hypersensitive sites (DHSs) which are known to act as regulatory elements for Th2 cytokine gene expression. Among the DHSs known as regulatory elements activated by GATA-3 directly in the Th2 cytokine gene locus are the conserved GATA-3 response element (CGRE) (20, 21), the conserved noncoding sequence 1 (CNS-1) (22) (23), the intronic enhancer (IE) (22) and the hypersensitive site V (HSV) (22) (24) (Figure 2A). Sequence comparison revealed that all four regulatory elements are highly conserved between human and mouse, and further sequence analysis revealed that all four elements contain strong GATA binding sites and surprisingly contain c-Myb binding sites in close proximity to the identified GATA binding sites within all four regulatory elements (Figure 2A). Due to this rare arrangement of c-Myb and GATA binding sites in the four regulatory elements, we hypothesized that c-Myb might play an important role with GATA-3 in the function of the regulatory elements.
Figure 2. c-Myb regulates IL-13 and IL-4 expression through regulatory elements in the Th2 cytokine gene in cooperation with GATA-3.
(A) Schematic representation of the IL-13 and IL-4 gene locus. Exons (Black rectangles) and regulatory elements (blank rectangles) of IL-13 and IL-4 are depicted in the locus. The schemas below each arrow show the Myb (Black rectangles) and GATA binding sites (blank rectangles) in the regulatory elements. CGRE: Conserved GATA-3 Response Element, CNS-1: Conserved Noncoding Sequence 1, IE: Intronic Enhancer, HSV: Hypersensitive Site V. (B) ChIP assays were performed with anti-c-Myb and anti-GATA-3 antibodies using primary human CD4+ T cells after stimulation with IL-4, IL-2 and antibodies against CD3/CD28. Fold enrichments were measured by QRT-PCR with specific primers for CGRE, CNS-1, IE and HSV regions. All results shown (all grey bars) are statistically significant (P < 0.05) compared to control IgG. (C) Upper schema shows reporter constructs in the pGL3 vector for promoter analysis. The order of promoter and regulatory elements in the constructs is the same as the arrangement in the human gene as shown in the upper schema of Figure 1A. Graphs show the results of reporter assays for IL-13 (right) and IL-4 (left). Reporter assays were carried out in 293T cells 24 hours after transfection with each construct in the presence of c-myb and/or GATA-3 expression constructs, or control empty vectors. * Statistically significant (P < 0.05) compared to the activity of the promoter alone.
To explore the function of c-Myb and the relationship between c-Myb and GATA-3 in these four elements, ChIP assays were performed to test if c-Myb and GATA-3 bound to the four elements in primary human CD4+ T cells with stimulation under Th2 promoting conditions for 3-5 days. ChIP assays revealed that in the stimulated CD4+ T cells, both c-Myb and GATA-3 bound to all four of the elements (Figure 2B). Strikingly, c-Myb binding to the CGRE locus was increased ~18-fold compared to control IgG.
To develop an understanding of the role of c-Myb and GATA-3 in Th2 cytokine gene expression we carried out Dual-Luciferase reporter assays using IL-4 and IL-13 promoter constructs that include the well-conserved promoter region, with or without the four regulatory elements in 293T cells that express neither c-Myb nor GATA-3 (Figure 2C, upper schema). Using the IL-4 promoter construct either with or without the regulatory elements, the effects of c-Myb or GATA-3 expression alone were minimal (Figure 2C, left graph). However, the combined expression of c-Myb and GATA-3 significantly increased luciferase activity for the CGRE-IL-4 promoter and CNS1-IL4 promoter 5.6- and 3.2-fold, respectively (Figure 2C, left graph). When we performed the luciferase reporter assays using IL-13 promoter constructs without/with regulatory elements, we, surprisingly, observed that combined c-Myb and GATA-3 expression increased CGRE-IL-13 promoter activity by ~160-fold and increased IL-13 promoter-CNS-1 element activity by~22-fold (Figure 2C, right graph). These results suggest that CGRE is an enhancer for IL-13 and that the enhancer is regulated by c-Myb and GATA-3.
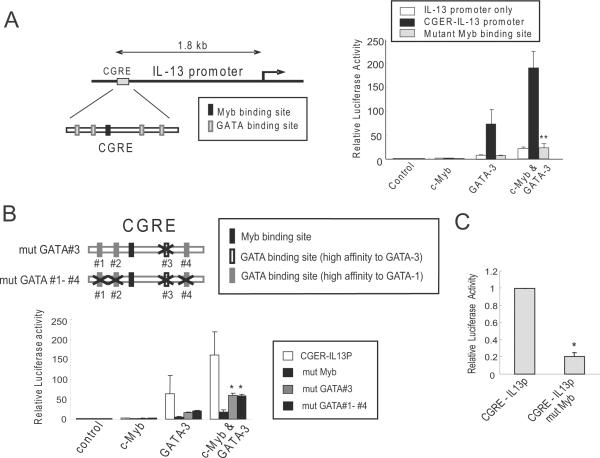
The canonical Myb binding site in the CGRE is essential for the strong enhancer activity of the CGRE for IL-13 expression by c-Myb and GATA-3
Having determined that the CGRE markedly enhanced IL-13 promoter activity by c-Myb and GATA-3 (Figure 2C, right graph), we next performed mutational analysis of the Myb and GATA binding sites within the CGRE locus to more precisely define regulation of IL-13 gene expression by c-Myb and GATA-3. The CGRE locus has one canonical Myb binding site which was mutated (TAACGG to TGACGG) in the CGRE-IL13 promoter construct and was tested in dual-luciferase reporter assays in 293T cells. The IL-13 promoter construct containing the CGRE enhancer was activated ~70-fold by GATA-3 alone and ~190-fold by the addition of c-Myb (Figure 3A), with similar results shown in Figure 1C. Surprisingly, this strong enhancer effect on IL-13 expression by c-Myb and GATA-3 was completely lost when the Myb binding site in the CGRE was mutated (Figure 3A). The levels of luciferase activity of the CGRE-IL-13 promoter containing a mutated Myb binding site was similar to the activity of the minimal IL-3 promoter (25) with expression of c-Myb and GATA-3 or GATA-3 alone. These results suggest that c-Myb and GATA-3 cooperatively regulate IL-13 expression through CGRE directly. The Myb binding site is essential for the enhancer activity in CGRE.
Figure 3. The canonical Myb binding site in the CGRE is essential for the strong enhancer activity of the CGRE for IL-13 expression by c-Myb and GATA-3.
(A) Left schema shows CGRE and IL-13 promoter locus and Myb and GATA binding sites in CGRE. Dual-luciferase reporter assays were performed in 293T cells 24 hours after cotransfection with IL-13 promoter (IL13P), wild-type CGRE-IL13 promoter (CGRE-IL13P) and CGRE-IL13 promoter containing a mutated canonical Myb binding site within the CGRE (mut Myb). (B) Upper schema shows GATA binding sites and the generated mutation sites in CGRE. Reporter assays carried out in 293T cells 24 hours after co-transfection with CGRE-IL13P, CGRE-IL13 promoter containing a mutated canonical Myb binding site within the CGRE (mut Myb), CGRE-IL13P containing a mutated (shown with X in upper schema) high affinity GATA-3 binding site (mut GATA#3), or CGRE-IL13P containing four mutated GATA binding sites (mut GATA#1-#4). (C) Dual-luciferase reporter assays were performed with Jurkat cells with CGRE-IL13P, or CGRE-IL13P containing a mutated Myb binding site (CGRE-IL13P mut Myb). Cells were stimulated 24 hours post-transfection with PMA (200ng/mL) and ionomycin (300ng/mL) for 24 hours.
Next, GATA binding sites in CGRE were mutated and tested in the dual-luciferase reporter assay. Mutation of a consensus GATA binding site, AGATAG, with high affinity for GATA-3 (26), within the CGRE locus decreased IL-13 promoter activity by approximately 60% (Figure 3B). Mutation of this site still resulted in higher enhancer activity compared to the IL-13 minimal promoter alone. It was possible that the three remaining GATA binding sites contributed to the enhancer activity. Based on sequence analysis the three remaining GATA binding sites should have high affinity for GATA-1 (26). Dual-luciferase reporter assays in the absence and presence of c-Myb and/or GATA-3 in 293T cells showed that mutation of all four GATA binding sites in CGRE (mut GATA #1-#4) resulted in the same level of enhancer activity as mutation of the consensus GATA-3 binding site (mut GATA#3) (Figure 3B).
The IL-13 promoter reporter constructs with or without a mutated c-Myb binding site within the CGRE locus were tested in Jurkat cells which are a human T leukemic cell line that expresses IL-13 upon stimulation with phorbol 12-myristate 13-acetate (PMA) and ionomycin (27). Mutation of the c-Myb binding site resulted in loss of the CGRE enhancer effect in Jurkat cells (Figure 3C). This confirms the observations in 293T cells.
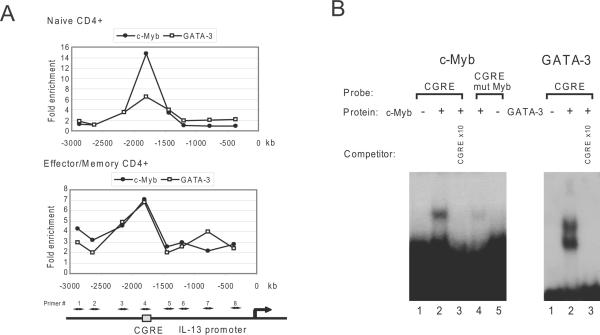
c-Myb and GATA-3 bind to CGRE directly
CGRE is positioned at a locus approximately 1.7Kb downstream from the IL-13 transcription start site. Three potential GATA-3 binding sites and four possible sites for Myb binding lie between human CGRE and the transcription start site of IL-13. ChIP was used to detect c-Myb binding to the locus using eight primers approximately −360 to −2900 bp from the transcription start site of the IL-13 gene. ChIP assays were performed with c-Myb, GATA-3 and control antibodies in human primary CD4+ naive and effector/memory T cells with stimulation under Th2 promoting conditions. The peak fold enrichment by c-Myb and GATA-3 antibodies for the locus was shown with the #4 primers that amplify the region including the Myb binding site in CGRE in both stimulated CD4+ naive and effector/memory T cells (Figure 4A). This result strongly demonstrated both c-Myb and GATA-3 bound to Myb and GATA binding sites in the CGRE locus in human primary CD4+ T-cells.
Figure 4. c-Myb and GATA-3 bind to CGRE directly.
(A) ChIP assays were performed with anti-c-Myb and anti-GATA-3 antibodies using primary human CD4+ naive and effector/memory T cells after stimulation with IL-4, IL-2 and anti-CD3/CD28 antibodies for 3–4 days. The fold enrichments were measured by QRT-PCR with 8 specific primers upstream of the IL-3 locus including CGRE as shown in the bottom schema. The transcription start site is indicated. (B) Electrophoretic Mobility Shift Assay for binding of c-Myb and GATA-3 to CGRE. Proteins were in vitro translated from their respective pcDNA expression vector using TNT Quick Coupled Transcription/Translation Systems (Promega) and subjected to EMSA with a radiolabeled CGRE oligonucleotide (CGRE) and CGRE oligonucleotide containing Myb's mutant binding sequence (CGRE mut Myb). Oligonucleotide competition was carried out with 10-fold excess of unlabeled CGRE oligonucleotide (CGRE).
To confirm binding to CGRE and to test for direct binding an electrophoretic mobility shift assay (EMSA) was used. A specific c-Myb and CGRE complex was detected using the radiolabeled CGRE oligonucleotide probe (CGRE) and the complex formation was inhibited by an excess of the oligonucleotide containing the consensus binding site for c-Myb (Figure 4B). The specific complex was not detected using the CGRE oligonucleotide containing Myb's mutant binding sequence (CGRE mut Myb) as probe. A specific GATA-3 and CGRE complex was detected and the complex formation was inhibited by an excess of the oligonucleotide containing the consensus binding site for GATA-3 (Figure 4B). These results indicate that c-Myb and GATA-3 directly bind the CGRE locus.
GATA-3 associates with c-Myb to bind CGRE
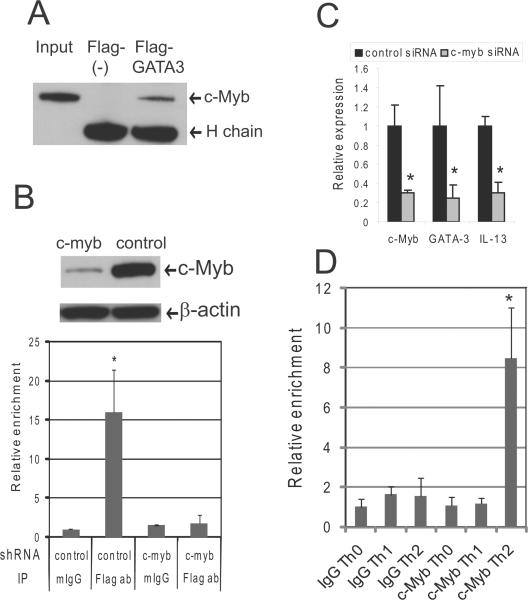
We next determined the importance of c-Myb associating with GATA-3 within CGRE and affecting IL-13 expression. To identify a GATA-3/c-Myb complex in Jurkat cells, we used Flag-tagged GATA-3 expression vectors. Immunoprecipitation assays with the cells showed that Flag-GATA-3 proteins bound to c-Myb (Figure 5A). We next silenced c-myb expression in Jurkat cells using a lentivirus c-myb shRNA expressing system as we previously reported (19). Forty-eight hours after transfection of the Flag-GATA-3 vector into Jurkat cells, ChIP assays performed with anti-Flag antibodies revealed a dramatic enrichment the CGRE upstream of IL-13 compared to when the ChIP assays were completed with mIgG. When Jurkat cells, with silenced c-myb expression, were transfected with Flag-GATA-3, we observed a marked decrease in the enrichment of CGRE by ChIP assays compared to when Jurkat cells express c-Myb (Figure 5B), suggesting that GATA-3 requires the association with c-Myb to bind to the CGRE. To determine the importance of the c-Myb/GATA-3 complex on GATA-3 and IL-13 expression, we treated Jurkat cells with control or c-myb siRNA and used QRT-PCR to determine gene expression. The c-myb siRNA suppressed c-myb gene expression 70% and decreased GATA-3 and IL-13 gene expression by approximately 75% and 70%, respectively, in Jurkat cells after stimulation with PMA and ionomycin compared to control siRNA treated cells (Figure 5C).
Figure 5. GATA-3 associates with c-Myb to bind CGRE.
(A) Immunoprecipitation (IP) was performed with Jurkat cells 48 hrs post-transfection of pcDNA GATA-3-Flag tag expression vector or control pcDNA Flag-tag vector using anti-Flag tag antibodies. The Western blot was performed with anti-c-Myb antibody. H chain = the immunoglobulin heavy chain (B) The upper panel shows c-Myb and β-actin protein expression as determined by Western blot in the lentivirus c-myb shRNA expressing or control Jurkat cells that were used for ChIP assays. The graph in the lower panel shows the relative enrichment of CGRE gene fragments by ChIP assays that were carried out with anti-Flag tag antibody (Flag ab) or normal mouse IgG (mIgG). The precipitated DNA fractions were analyzed by QRT-PCR with primers specific to the CGRE region. (C) Jurkat cells transfected with c-myb siRNA or control scrambled siRNA (19) were cultured with PMA and ionomycin for 24 hours, and QRT-PCR was then performed with primers specific to measure c-myb, GATA-3, and IL-13 gene expression. (D) Purified nucleoprotein complex was obtained from CD4+CD45RA+ cells cultured under Th1 or Th2 promoting conditions and then utilized for ChIP assays using either anti-c-Myb (c-Myb) or mouse IgG (IgG). The precipitated DNA fractions were analyzed by QRT-PCR with primers specific to the CGRE. *P<0.05
We also examined whether c-Myb binds to CGRE differentially in Th0, Th1 or Th2 promoting conditions using ChIP assays. CD45RA+CD4 (naïve) T cells were cultured under Th1 or Th2 promoting conditions for 4 days, and, then the cells were re-stimulated with IL-12 and IL-2 or IL-4 and IL-2, respectively, for one hour before crosslinking with 1% formaldehyde. The ChIP assays showed that c-Myb bound to CGRE under Th2 promoting conditions but did not bind to CGRE under Th1 promoting conditions or in Th0 cells (Figure 5D).
MLL and Menin bind to CGRE in CD4+ effector/memory T cells while MLL does not bind to CGRE in CD4+ naive T cells under Th2 cell promoting conditions
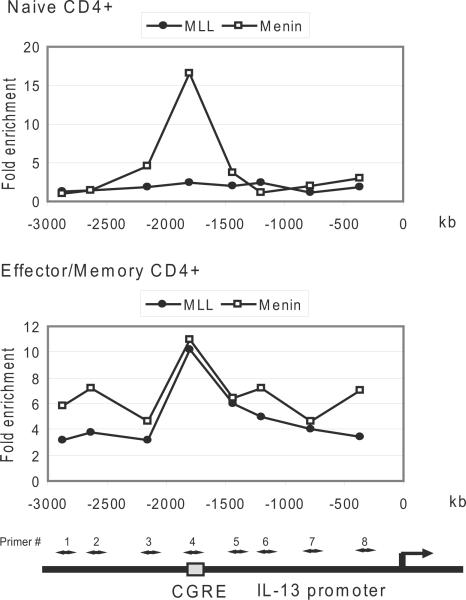
Our recent work showed that c-Myb bound to GATA-3 via an adaptor protein, Menin and cooperatively regulated GATA-3 expression (19). Additionally we demonstrated that c-Myb bound mixed lineage leukemia (MLL) though Menin in human leukemic cells and primary T cells (28). c-Myb silencing decreases methylation of histone H3K4 and acetylation of histone H3K9 at the GATA-3 locus in primary human CD4+ effector/memory T cells stimulated under Th2 promoting conditions (19). Therefore we tested our hypothesis that c-Myb, GATA-3 and Menin form a transcriptional complex which recruits MLL for histone modification of the Th2 cytokine gene locus. ChIP was used to test if MLL and Menin bind to the CGRE locus in human primary CD4+ T cells. Under Th2 promoting conditions Menin strongly bound to CGRE in CD4+ naïve T cells stimulated for 3– 4 days while MLL binding was not detected (Figure 6, upper graph). However, both MLL and Menin bound to CGRE in CD4+ effector/memory T cells under the same stimulation conditions (Figure 6, lower graph).
Figure 6. MLL and Menin bind to CGRE in CD4+ effector/memory T cells while MLL does not bind to CGRE in CD4+ naive T cells under Th2 cell promoting conditions.
ChIP assays were performed with anti-MLL and anti-Menin antibodies using primary human CD4+ naïve (upper panel) and effector/memory (lower panel) T cells after stimulation with IL-4, IL-2 and anti-CD3/CD28 antibodies for 3–4 days as detailed in materials and methods.
c-Myb silencing decreases methylation of histone H3K4 and acetylation of histone H3K9 at the IL-13 locus in stimulated primary human CD4+ effector/memory T cells
Histone modification of the IL-13 locus was examined and extended to the IL-4 locus due to the potential for coordinate regulation. Antibodies directed to Histone H3 di- and tri- methylated K4, Histone H3 acetyl K9, and Histone H3 di-methylated K9 were used in ChIP assays utilizing at least 22 primer sets from within the ~35kb Th2 cytokine gene locus including the IL-13 and IL-4 loci.
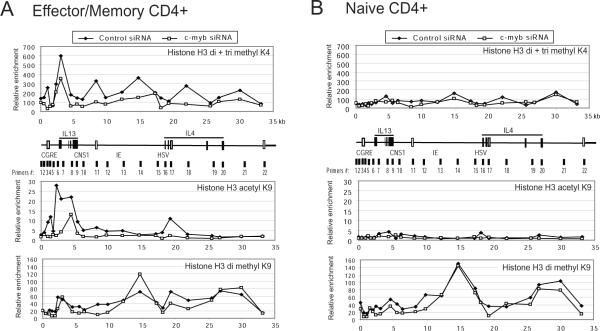
Because MLL bound CGRE only in CD4+ effector/memory T cells, we tested histone modification in CD4+ effector/memory T cells with and without silenced c-Myb. Compared to control siRNA treated cells, silencing c-Myb led to considerably decreased levels of H3K4 di- and tri-methylation, as well as decreased H3K9 acetylation in the loci in the cells (Figure 7A). Although the levels of H3K4 di- and tri-methylation and H3K9 acetylation were very low in CD4+ naive T cells stimulated under Th2 promoting conditions compared to CD4+ effector/memory T cells, the levels of histone methylation and acetylation were decreased overall following c-Myb silencing in the naive T cells (Figure 7B). Interestingly, silencing c-Myb did not affect the level of histone H3K9 di-methylation in both naïve and CD4+ effctor/memory T cells. Moreover, the methylation status of the IL13 and IL-4 loci in naive and effector/memory T cells are similar, suggesting that histone H3K9 di-methylation is not important for regulation of Th2 cytokine gene locus (Figure 7A, 7B bottom graphs). These results demonstrate that c-Myb significantly contributes to chromatin re-modeling at the Th2 cytokine gene locus in primary human memory Th2 cells. The data suggest that c-Myb forms a complex with GATA-3 and/or Menin, and recruits MLL in the transition from CD4+ effector to memory T cells and/or maintenance of memory Th2 cells.
Figure 7. c-Myb silencing decreases methylation of histone H3K4 and acetylation of histone H3K9 at the IL-13 locus in stimulated primary human CD4+ effector/memory T cells.
Representative results of ChIP assays with a series of primer pairs covering the IL-13 and IL-4 gene locus in primary human CD4+ effector/memory T cells (A), and naïve T cells (B) stimulated under Th2 promoting conditions, expressed with c-myb shRNA or control scramble shRNA using the lentivirus systems. The human CD4+CD45RA+ T-cells (naïve) were cultured for 7 days under Th2 promoting conditions and CD4+CD45RA- T-cells (effector/memory) were cultured for 4 days with stimulation prior to ChIP assay. ChIP assays were carried out with antibodies directed against di- and tri- methylated HistoneH3K4 (upper graph in each panel), acetyl H3K9 (middle graph in each panel), and di-methyl H3K9 (bottom graph in each panel) and analyzed by QRT-PCR. Line graphs show the relative specific antibody mediated enrichment in comparison to control mouse, or rabbit IgG. The schematic below the base line of each top panel illustrates ~35 kb of the Th2 gene locus including the IL-13 and IL-4 genes. Four regulatory elements are shown with blank rectangles.
Discussion
Despite the high level of c-Myb expression in primary peripheral T cells following activation, little is known regarding the function of c-Myb in this population of cells. It is reported here that c-Myb has a direct role in regulating Th2 cytokine expression in primary CD4+ T cells. The importance of the c-Myb binding site found within the CGRE for enhanced transcription of IL-13 was shown (Figure 3). In primary human naive and effector/memory CD4+ T cells under Th2 promoting conditions, both c-Myb and GATA-3 bound to CGRE (Figure 4). GATA-3 required c-Myb to bind CGRE (Figure 5). Moreover, MLL bound to CGRE in primary CD4+ effector/memory T cells (Figure 6), and silencing c-Myb considerably decreased levels of H3K4 di- and tri-methylation, as well as H3K9 acetylation in the IL-13 and IL-4 loci in the cells (Figure 7).
DNase I hypersensitivity is postulated to reflect a localized “open” chromatin configuration where the DNA is accessible to binding by many factors (29). It has been shown that regulatory elements like tissue-specific enhancers and locus control regions are located in DHSs (30) and many DHSs develop in the Th2 cytokine gene during Th2 lineage commitment. Among DHSs in the Th2 cytokine gene, CGRE is located 1.7 kb upstream of IL-13 and was described as a highly conserved Th2-specific hypersensitive site (20, 21). We clearly demonstrated that c-Myb and GATA-3 cooperatively increased the activity of CGRE for the IL-13 promoter while no obvious regulation to IE and HSV were observed and a small effect for CNS-1 was shown (Figure 2C). Important studies suggested that CNS-1, a DHS located between the IL-13 and IL-4 loci, is a coordinate regulator of Th2 cytokine genes. Deletion of human CNS-1 resulted in a reduction of cells producing IL-4, IL-5 and IL-13 (31). Subsequent studies in transgenic reporter mice demonstrated that CNS-1 has strong enhancer activity for the IL-4 promoter (22), whereas other studies revealed CNS-1 to be responsive to GATA-3 enhancer activity as well as chromatin remodeling which is consistent with the identification of potential GATA-3 sites by gel shift analyses (22, 23). The enhancer activity of CNS-1 with c-Myb and GATA-3 was strong for IL-13 but it was limited for the IL-4 promoter, while c-Myb and GATA-3 bound CNS-1. Many factors have roles for the IL-4 locus (12, 16, 17), and GATA-3 and c-Myb may require additional molecules or binding sites to activate CNS-1 for IL-4 expression in 293T cells. This concern also should apply to IE and HSV. Using transgenic reporter assays, IE was determined to enhance IL-4 promoter activity, and when IE was combined with CNS-1, it conferred GATA-3-dependent enhancement of IL-4 promoter activity (22). In transient transfection and transgenic reporter assays, the combination of HSV and HSVa had strong enhancer activity (22) (24), when NFAT1 and GATA-3 bound to HSVa. HSV/CNS-2+HSVa knockout mice showed reduced expression of IL-4, IL-5, and IL-13 in Th2 cells (32). Because both c-Myb and GATA-3 bound to IE and HSV in stimulated human CD4+ cells in ChIP assays (Figure 2B), c-Myb would have a role for regulation of those elements. However, significant activation of IE and HSV by c-Myb and GATA-3 was not observed (Figure 2C). Because the combination of IE and HSV regulatory elements showed enhancer activity for IL-4 promoter (22) (24), we hypothesize that a combination of regulatory elements might be required for the activation of IL-4 promoter by c-Myb.
c-Myb augmented the enhancer activity of GATA-3 for the IL-13 promoter. The mechanism by which c-Myb and GATA-3 regulate IL-13 expression was determined. Mutation of the canonical Myb binding site within the CGRE abrogated the strong enhancer activity of the CGRE, however the enhancer activity was only partially abrogated when the GATA binding site was mutated (Figure 3). GATA-3 activated the CGRE without c-Myb while this activity was suppressed by the mutation of the Myb binding site in the CGRE. It is possible that B-Myb might contribute to this complex regulation because B-Myb and c-Myb bind to the same Myb binding site (33) and could partially compensate for their respective functions when c-Myb or B-Myb expression is individually silenced by RNAi (34). B-Myb could bind to the CGRE in 293T cells and weakly assist GATA-3's function. However, exogenous expression of B-Myb did not change CGRE-IL-13 promoter activity in our reporter assays (data not shown). Endogenously high B-Myb expression would suppress the affect of the promoter activity by the exogenous expression of B-Myb. Neither c-Myb nor B-Myb can bind to the mutated Myb binding site, and the mutation completely removed the activity of CGRE. However, the mutated GATA binding sites partially decreased CGRE activity. We previously showed that c-Myb bound to GATA-3 through Menin or when c-Myb was in an activated truncated form (19). GATA-3 formed a complex with c-Myb to bind to CGRE and the complex might partially bind to the mutated GATA binding sites with the assistance of c-Myb or another protein. GATA binding sites in the IL-13 promoter might compensate for the mutated binding site in CGRE for the activity.
MLL and Menin bound to the CGRE locus in stimulated CD4+ effector/memory T cells (Figure 6). MLL is known to have intrinsic histone methyl transferase activity for histone H3-K4 (35) (36), and the methylation of histone H3-K4 is important for the maintenance of acetylation of histone H3-K9. Since we have already shown that c-Myb can bind MLL through Menin (28), it is possible that histone modification of the Th2 cytokine locus occurs through c-Myb and MLL. A notable decrease in the methylation of histone H3-K4 at the CGRE gene locus was observed after c-Myb silencing in human primary CD4+ effector/memory T cells (Figure 7). c-Myb silencing also decreased methylation of histone H3-K4 in a broad range of the Th2 cytokine locus including the IL-4 gene. Thus, it is likely that a complex of c-Myb, MLL and Menin with or without GATA-3 regulates the Th2 cell phenotype through maintenance of histone methyl transferase activity. Although c-Myb mainly regulates CGRE IL-13 expression, our results suggest that c-Myb plays a role in other regions including CNS-1, IE and HSV for maintenance of histone methyl transferase activity.
We conclude that c-Myb directly contributes to the regulation of IL-13 expression by two distinct mechanisms. First, c-Myb augments IL-13 promoter activity together with GATA-3. In this activation process, MLL would have no significant function because MLL does not bind to CGRE in naïve CD4+ T cells under Th2 promoting conditions. The second, c-Myb contributes to induce an open chromatin status in the Th2 cytokine gene locus, especially IL-13 locus, during the process of generating memory Th2 cells and maintenance of memory Th2 cells. In this process, MLL that is recruited by c-Myb complex has a critical role for the histone modification in the locus. This work further implicates c-Myb as a critical factor for Th2 cell differentiation.
Supplementary Material
Acknowledgements
We thank Dr. Hui Hu and Dr. Martin Carroll for critical reading of the manuscript and helpful editorial suggestions.
This work was supported in part by grants to AMG from the NIH (RO1-CA101859), the Leukemia and Lymphoma Society, and from the State of Pennsylvania Dept of Health Tobacco Settlement Grant via the Abramson Cancer Center at the University of Pennsylvania.
Abbreviations
- CGRE
conserved GATA-3 response element
- CNS-1
conserved noncoding sequence 1
- IE
intronic enhancer
- HSV
hypersensitive site V
- MLL
mixed lineage leukemia
References
- 1.Reiner SL. Development in motion: helper T cells at work. Cell. 2007;129:33. doi: 10.1016/j.cell.2007.03.019. [DOI] [PubMed] [Google Scholar]
- 2.Zhu J, Paul WE. CD4 T cells: fates, functions, and faults. Blood. 2008;112:1557. doi: 10.1182/blood-2008-05-078154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O'Garra A. Cytokines induce the development of functionally heterogeneous T helper cell subsets. Immunity. 1998;8:275. doi: 10.1016/s1074-7613(00)80533-6. [DOI] [PubMed] [Google Scholar]
- 4.Murphy KM, Ouyang W, Farrar JD, Yang J, Ranganath S, Asnagli H, Afkarian M, Murphy TL. Signaling and transcription in T helper development. Annu Rev Immunol. 2000;18:451. doi: 10.1146/annurev.immunol.18.1.451. [DOI] [PubMed] [Google Scholar]
- 5.Glimcher LH, Murphy KM. Lineage commitment in the immune system: the T helper lymphocyte grows up. Genes Dev. 2000;14:1693. [PubMed] [Google Scholar]
- 6.Frazer KA, Ueda Y, Zhu Y, Gifford VR, Garofalo MR, Mohandas N, Martin CH, Palazzolo MJ, Cheng JF, Rubin EM. Computational and biological analysis of 680 kb of DNA sequence from the human 5q31 cytokine gene cluster region. Genome Res. 1997;7:495. doi: 10.1101/gr.7.5.495. [DOI] [PubMed] [Google Scholar]
- 7.Greig KT, Carotta S, Nutt SL. Critical roles for c-Myb in hematopoietic progenitor cells. Semin Immunol. 2008 doi: 10.1016/j.smim.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 8.Ramsay RG, Gonda TJ. MYB function in normal and cancer cells. Nat Rev Cancer. 2008;8:523. doi: 10.1038/nrc2439. [DOI] [PubMed] [Google Scholar]
- 9.Emambokus N, Vegiopoulos A, Harman B, Jenkinson E, Anderson G, Frampton J. Progression through key stages of haemopoiesis is dependent on distinct threshold levels of c-Myb. Embo J. 2003;22:4478. doi: 10.1093/emboj/cdg434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bender TP, Kremer CS, Kraus M, Buch T, Rajewsky K. Critical functions for c-Myb at three checkpoints during thymocyte development. Nat Immunol. 2004;5:721. doi: 10.1038/ni1085. [DOI] [PubMed] [Google Scholar]
- 11.Lieu YK, Kumar A, Pajerowski AG, Rogers TJ, Reddy EP. Requirement of c-myb in T cell development and in mature T cell function. Proc Natl Acad Sci U S A. 2004;101:14853. doi: 10.1073/pnas.0405338101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou M, Ouyang W. The function role of GATA-3 in Th1 and Th2 differentiation. Immunol Res. 2003;28:25. doi: 10.1385/IR:28:1:25. [DOI] [PubMed] [Google Scholar]
- 13.Chou J, Provot S, Werb Z. GATA3 in development and cancer differentiation: cells GATA have it! J Cell Physiol. 222:42. doi: 10.1002/jcp.21943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rothenberg EV, Moore JE, Yui MA. Launching the T-cell-lineage developmental programme. Nat Rev Immunol. 2008;8:9. doi: 10.1038/nri2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ho IC, Tai TS, Pai SY. GATA3 and the T-cell lineage: essential functions before and after T-helper-2-cell differentiation. Nat Rev Immunol. 2009;9:125. doi: 10.1038/nri2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ansel KM, Djuretic I, Tanasa B, Rao A. Regulation of Th2 differentiation and Il4 locus accessibility. Annu Rev Immunol. 2006;24:607. doi: 10.1146/annurev.immunol.23.021704.115821. [DOI] [PubMed] [Google Scholar]
- 17.Nakayama T, Yamashita M. Initiation and maintenance of Th2 cell identity. Curr Opin Immunol. 2008;20:265. doi: 10.1016/j.coi.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 18.Maurice D, Hooper J, Lang G, Weston K. c-Myb regulates lineage choice in developing thymocytes via its target gene Gata3. Embo J. 2007;26:3629. doi: 10.1038/sj.emboj.7601801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakata Y, Brignier AC, Jin S, Shen Y, Rudnick SI, Sugita M, Gewirtz AM. c-Myb, Menin, GATA-3, and MLL form a dynamic transcription complex that plays a pivotal role in human T helper type 2 cell development. Blood. 2010;116:1280. doi: 10.1182/blood-2009-05-223255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamashita M, Ukai-Tadenuma M, Kimura M, Omori M, Inami M, Taniguchi M, Nakayama T. Identification of a conserved GATA3 response element upstream proximal from the interleukin-13 gene locus. J Biol Chem. 2002;277:42399. doi: 10.1074/jbc.M205876200. [DOI] [PubMed] [Google Scholar]
- 21.Agarwal S, Rao A. Modulation of chromatin structure regulates cytokine gene expression during T cell differentiation. Immunity. 1998;9:765. doi: 10.1016/s1074-7613(00)80642-1. [DOI] [PubMed] [Google Scholar]
- 22.Lee GR, Fields PE, Flavell RA. Regulation of IL-4 gene expression by distal regulatory elements and GATA-3 at the chromatin level. Immunity. 2001;14:447. doi: 10.1016/s1074-7613(01)00125-x. [DOI] [PubMed] [Google Scholar]
- 23.Takemoto N, Kamogawa Y, Jun Lee H, Kurata H, Arai KI, O'Garra A, Arai N, Miyatake S. Cutting edge: chromatin remodeling at the IL-4/IL-13 intergenic regulatory region for Th2-specific cytokine gene cluster. J Immunol. 2000;165:6687. doi: 10.4049/jimmunol.165.12.6687. [DOI] [PubMed] [Google Scholar]
- 24.Agarwal S, Avni O, Rao A. Cell-type-restricted binding of the transcription factor NFAT to a distal IL-4 enhancer in vivo. Immunity. 2000;12:643. doi: 10.1016/s1074-7613(00)80215-0. [DOI] [PubMed] [Google Scholar]
- 25.Kishikawa H, Sun J, Choi A, Miaw SC, Ho IC. The cell type-specific expression of the murine IL-13 gene is regulated by GATA-3. J Immunol. 2001;167:4414. doi: 10.4049/jimmunol.167.8.4414. [DOI] [PubMed] [Google Scholar]
- 26.Merika M, Orkin SH. DNA-binding specificity of GATA family transcription factors. Mol Cell Biol. 1993;13:3999. doi: 10.1128/mcb.13.7.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Silbermann K, Schneider G, Grassmann R. Stimulation of interleukin-13 expression by human T-cell leukemia virus type 1 oncoprotein Tax via a dually active promoter element responsive to NF-kappaB and NFAT. J Gen Virol. 2008;89:2788. doi: 10.1099/vir.0.2008/003699-0. [DOI] [PubMed] [Google Scholar]
- 28.Jin S, Zhao H, Yi Y, Nakata Y, Kalota A, Gewirtz AM. c-Myb binds MLL through menin in human leukemia cells and is an important driver of MLL-associated leukemogenesis. J Clin Invest. 2010;120:593. doi: 10.1172/JCI38030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gross DS, Garrard WT. Nuclease hypersensitive sites in chromatin. Annu Rev Biochem. 1988;57:159. doi: 10.1146/annurev.bi.57.070188.001111. [DOI] [PubMed] [Google Scholar]
- 30.Stamatoyannopoulos JA, Goodwin A, Joyce T, Lowrey CH. NF-E2 and GATA binding motifs are required for the formation of DNase I hypersensitive site 4 of the human beta-globin locus control region. Embo J. 1995;14:106. doi: 10.1002/j.1460-2075.1995.tb06980.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loots GG, Locksley RM, Blankespoor CM, Wang ZE, Miller W, Rubin EM, Frazer KA. Identification of a coordinate regulator of interleukins 4, 13, and 5 by cross-species sequence comparisons. Science. 2000;288:136. doi: 10.1126/science.288.5463.136. [DOI] [PubMed] [Google Scholar]
- 32.Solymar DC, Agarwal S, Bassing CH, Alt FW, Rao A. A 3' enhancer in the IL-4 gene regulates cytokine production by Th2 cells and mast cells. Immunity. 2002;17:41. doi: 10.1016/s1074-7613(02)00334-5. [DOI] [PubMed] [Google Scholar]
- 33.Oh IH, Reddy EP. The myb gene family in cell growth, differentiation and apoptosis. Oncogene. 1999;18:3017. doi: 10.1038/sj.onc.1202839. [DOI] [PubMed] [Google Scholar]
- 34.Nakata Y, Shetzline S, Sakashita C, Kalota A, Rallapalli R, Rudnick SI, Zhang Y, Emerson SG, Gewirtz AM. c-Myb contributes to G2/M cell cycle transition in human hematopoietic cells by direct regulation of cyclin B1 expression. Mol Cell Biol. 2007;27:2048. doi: 10.1128/MCB.01100-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, Allis CD, Hess JL. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell. 2002;10:1107. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- 36.Nakamura T, Mori T, Tada S, Krajewski W, Rozovskaia T, Wassell R, Dubois G, Mazo A, Croce CM, Canaani E. ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol Cell. 2002;10:1119. doi: 10.1016/s1097-2765(02)00740-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.