Abstract
The frequency of a splice variant of mucin 1 (MUC1), MUC1/A was lower in dry eye disease patients compared to normal controls, suggesting a link between the absence of MUC1/A and the development of dry eye disease which is characterized by chronic inflammation. The objectives of the present study were to clone and characterize the phenotype of cells expressing solely MUC1/A versus MUC1/B or a variant lacking the extracellular domain (ΔEX) and to determine whether MUC1/A and MUC1/B differentially modulate inflammatory responses in transfected cells. The additional 27 bp and SNP present in the N-terminus of MUC1/A were cloned into a FLAG-MUC1/B expression vector. Transient transfection of MUC1/A and MUC1/B plasmids into MUC1-null COS-7 cells resulted in similar protein expression and plasma membrane localization. MUC1/B and MUC1/A differed in their ability to modulate tumor necrosis α (TNFα)-induced transcription of IL-1β and IL-8. MUC1/B and MUC1/A inhibited IL-8 induction by TNFα at 4 h. However with 24 h TNFα, MUC1/A increased IL-1β and IL-8 whereas MUC1/B had no effect on cytokine expression. MUC1/B inhibited TNFα-induced luciferase activity from an NF-κB reporter whereas MUC1/A either inhibited or increased this luciferase activity depending on the time of TNFα treatment. MUC1/A, but not MUC1/B, increased the basal TGFβ expression. Both MUC1/B and MUC1/A blocked TNFα-induced miR-21 expression. These data demonstrate that MUC1/A and MUC1/B have different inflammatory activities and support the hypothesis that MUC1 genotypic differences may affect susceptibility to ocular surface damage in dry eye disease.
Keywords: dry eye disease, IL-1β, IL-8, TNFα, TGFβ, miR-21
Introduction
Mucin 1 (MUC1) is a membrane-bound glycoprotein expressed at the apical border of epithelial cells (Kufe et al., 1984). MUC1 undergoes autoproteolysis generating two subunits: 1) the >250 kDa N-terminal extracellular domain (MUC1-N) and 2) the ~25 kDa C-terminal cytoplasmic domain (MUC1-CD) (Levitin et al., 2005; Macao et al., 2006). The MUC1-N subunit contains the plasma membrane localization signal sequence and a variable number of 20 aa tandem repeats (VNTR) that are extensively O-glycosylated (Gendler et al., 1990). The MUC1-CD includes a 58 aa extracellular domain, 28 aa transmembrane (TM) domain, and 72 aa cytoplasmic tail (MUC1.CT). The two subunits of MUC1 form a stable non-covalent heterodimer localized to the plasma membrane. The fully glycosylated MUC1-N extends above the glycocalyx forming a physical barrier protecting cells from damage induced by changes in pH, reactive oxygen species (ROS), pathogens, physical interactions and stresses (Hollingsworth and Swanson, 2004). MUC1-N is further cleaved and released as a soluble protein (the ectodomain) into the surrounding fluid, leaving the MUC1-CD free to interact with putative partners in the plasma membrane or to move to different cellular compartments, e.g., cytoplasm, nucleus, and mitochondria (Abe and Kufe, 1989). The highly conserved MUC1.CT contains tyrosine, threonine and serine residues that are potential phosphorylation sites located within Src homology-2 and non-Src homology docking sites (Park et al., 1996; Wang et al., 2003; Zrihan-Licht et al., 1994). Depending on the phosphorylation pattern, MUC1.CT interacts with cytoplasmic and nuclear proteins including transcription factors altering their activities (Carson, 2008; Gendler, 2001; Hollingsworth and Swanson, 2004).
At least twelve splice variants of MUC1 have been described (Williams et al., 1999). We reported that MUC1/A was reduced in the non-Sjögrens aqueous deficient dry eye and concluded that the lack of expression of MUC1/A is one factor involved in susceptibility to dry eye disease (Imbert et al., 2006). Likewise, differential expression of MUC1/A and MUC1/B have been demonstrated in prostate, ovarian and breast cancer (Obermair et al., 2002; Schmid et al., 2003; Strawbridge et al., 2008). However, the mechanism by which MUC1/A and MUC1/B may contribute to the pathogenesis of dry eye disease and cancer is unknown.
The MUC1/A splice variant differs from the MUC1/B form by having an additional 9 aa within the MUC1-N domain (Figure 1). This additional sequence is predicted to change the signal peptidase cleavage site, thereby altering the localization signal within the amino terminus (Ligtenberg et al., 1990; Wreschner et al., 1990), and potentially changing its intracellular trafficking and/or subsequent processing. Although much is known about how the MUC1 CT regulates signal transduction (Jonckheere and Van Seuningen, 2010) the contribution of MUC1-N in the cell signaling properties of MUC1 is poorly understood. The impact of the 9 additional aa in the MUC1-N in MUC1/A versus MUC1/B on the intracellular localization and signaling functions of MUC1 has not been examined.
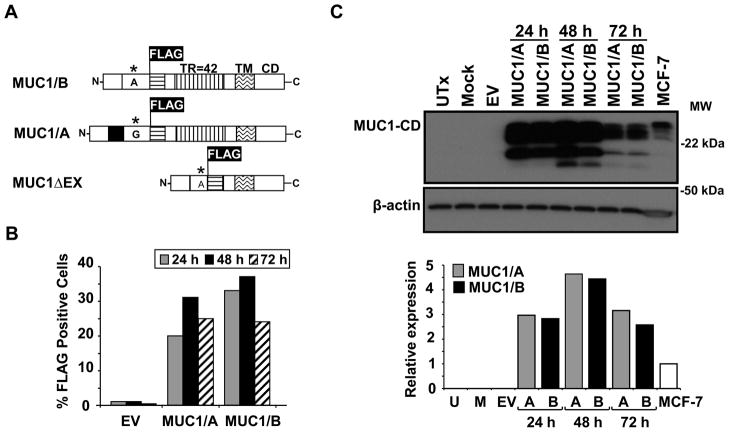
Figure 1. Expression of MUC1/A and MUC1/B in COS-7 cells.
(A) Schematic diagram of MUC1/A, MUC1/B and MUC1ΔEX constructs used for transfection of COS-7 cells. The insertion of the FLAG tag within the N-terminal domain (NTD) is indicated. There are 42 tandem repeats (TR) in both MUC1/A and MUC1/B whereas MUC1ΔEX lacks the tandem repeat. The transmembrane (TM) and C-terminal domains (CD) are indicated. * indicates the SNP and nucleotide change between MUC1/B and MUC1/A. The black box indicates the additional 27 bp in MUC1/A. (B) Flow cytometry analysis of FLAG expression in COS-7 cells 24, 48 and 72 h post-transfection using M2 anti-FLAG antibody and PE-conjugated secondary antibody. Values are from one representative FACS analysis. (C) Western analysis of MUC1 protein expression in WCE from COS-7 cells either untransfected (UTx), transfected with the empty vector (EV), a mock control (no DNA), or expression vectors for MUC1/A or MUC1/B for 24, 48 and 72 h. 6 μg of WCE from MCF-7 cells was loaded into the far right lane as a positive control for MUC1. The blot was probed with CT2 anti-MUC1 antibody and β-actin which was used as a loading control. The bar graph shows the quantitation of MUC1/A and MUC1/B relative to β-actin expression relative to MCF-7 that was set to one for comparison.
The goal of this study was to characterize for the first time the phenotype of cells expressing MUC1/A versus MUC1/B and to elucidate the roles of these splice variants in cellular responses to tumor necrosis factor alpha (TNFα). The importance of examining MUC1 splice variants in the context of an inflammatory signal is two-fold: first, MUC1 regulates inflammatory responses via modulation of the NF-κB pathway in vitro and in vivo (Ahmad et al., 2007; Lu et al., 2006; Ueno et al., 2008). Second, dry eye disease is an inflammatory disorder affecting the ocular surface (Pflugfelder, 2004). Thus, determining if MUC1/A and MUC1/B splice variants differentially regulate inflammatory mediators may provide insight into the pathogenesis of dry eye disease. In this report, we demonstrate that MUC1/B and MUC1/A splice variants differ in their ability to modulate certain TNFα-induced inflammatory responses.
2. Materials and methods
2.1. Cell Culture
COS-7 cells were purchased from ATCC (Manassas, VA, USA). COS-7 cells were maintained in DMEM supplemented with 10% fetal bovine serum (FBS) Invitrogen, Carlsbad, CA, USA).
2.2. Plasmids and MUC1/A cloning
The MUC1/B and MUC1ΔEX in pCMV-DNA3 constructs were a kind gift from J.A. Schroeder, Arizona Cancer Center. MUC1/B cDNA was originally cloned by M.A. Hollingsworth at the University of Nebraska Medical Center (Burdick et al., 1997). MUC1/B cDNA was subcloned into HindIII/EcoRI sites of Litmus38i vector (New England Biolabs (NEB), Beverly, MA, USA). MUC1/B in Litmus38i was digested with XmnI and BsmI (NEB). The additional 27 bp present in the MUC1/A variant was cloned into XmnI and BsmI sites of MUC1/B using In-Fusion cloning (Clontech, Mountain View, C, USA). Briefly, a 262 bp fragment containing the 27 bp corresponding to the MUC1/A splice variant (Imbert et al., 2006; Imbert et al., 2009) was generated by PCR using cDNA from a MUC1/A homozygous donor and primers: 5’-CCT GCC TGA ATC TGT TCT GC-3’ (forward) and 5’-CTG GAG AGT ACG CTG CTG GT-3’ (reverse) that flank the XmnI and BsmI sites in exons 1 and 2 of the MUC1 gene, respectively. The PCR product was ligated into pCR®-Blunt II-TOPO® vector (Invitrogen). In-Fusion PCR was performed to generate a 252 bp product using pCR®-Blunt II-TOPO® vector as a template and the following primers: 5’-CAG CGC CTG CCT GAA TCT GTT CTG CCC CCT CCC CA-3’ (forward) and 5’-TCA TCC TTG TAA TCA GCA TTC TTC TCA GTA GAG CT-3’ (reverse). In-Fusion Dry Down reaction (Clontech) was used to ligate MUC1/A into the MUC1/B Litmus vector. Plasmid DNA from clones was sequenced to confirm the full length MUC1/A sequence in pCDNA3.
2.3. Transfection of COS-7 cells
Cells seeded in six-well plates were transfected with 1μg of MUC1/A, MUC1/B, MUC1ΔEX or pCDNA3 (empty vector, EV) using FuGENE 6 (Roche, Indianapolis, IN, USA). FACS — Transfected COS-7 cells were harvested at 24, 48 and 72 h and stained using M2 anti-FLAG antibody (M2, Sigma, St. Louis, MO, USA) diluted (1:1000) in PBS containing 5 % FBS and sodium azide. Stained cells were analyzed using a FACS calibur (Becton Dickinson). Cell Quest Software 1.2.2 was used for acquisition and analysis of each cell sample (n = 10,000).
2.4. WCE and western blotting
WCE were prepared by incubating the cells for 30 min in lysis buffer (20 mM Tris pH 7.5, 150 mM NaCl, 1 % NP40 and 5 mM EDTA pH 8.0) containing protease inhibitors (Roche). Protein concentrations were determined by a BioRad assay (BioRad, Hercules, CA, USA). Proteins were separated on 14 % Tris-Glycine gels (Invitrogen) and transferred to PVDF membranes (Millipore, Billerica, MA, USA). The membranes were immunoblotted with the following antibodies: Armenian hamster against MUC1-CD (CT2, Thermo Scientific), β-actin (Sigma), TGFβ (Cell Signaling, Beverly, MA), DF3 against MUC1-N (Signet, Berkeley, CA, USA). HyGlo chemiluminescent HRP antibody detection reagent (Denville Scientific, Metuchen, NJ, USA) was used to detect bands on Kodak BioMax ML film (Eastman Kodak, Rochester, NY, USA). Resulting immunoblots were scanned using a Microtek ScanMaker i800 scanner. Un-Scan-It 6.1 for Windows (Silk Scientific, Orem, UT, USA) was used to quantitate the integrated optical densities (IOD) for each band and analyze the relative immunoreactive proteins as described (Dougherty et al., 2006).
2.5. Immunofluorescence
Cells were seeded onto glass coverslips and transfected with MUC1/A, MUC1/B, MUC1ΔEX or pCDNA3 (empty vector, EV) as above. Cells were washed once with PBS and chilled on ice for 10 min. The cells were then incubated in cold 5 % MEM with M2 anti-FLAG antibody (Sigma) at a dilution of 1:1000 for 1 h at 4 °C. After incubation with the primary antibody, the cells were incubated with goat anti-mouse DyLight 488 conjugated (Invitrogen) and wheat germ agglutinin (WGA) Alexa Fluor 633 conjugated (Invitrogen), both diluted 1:200 in cold 5 % MEM. The cells were then fixed on ice for 15 min in 4% paraformaldehyde and mounted with ProLong Anti-fade (Invitrogen). Images were captured using an Olympus FV1000 confocal microscope.
2.6. RNA isolation, RT-PCR, and quantitative real-time RT-PCR (QRT-PCR)
Total RNA was extracted using RNeasy micro columns (QIAGEN, Valencia, CA, USA). Reverse transcription was performed by using random hexamers and the High-Capacity cDNA Archive Kit (Applied Biosystems, Foster City, CA, USA). QRT-PCR was performed with the ABI PRISM 7900 SDS 2.1 by relative quantification with standard thermal cycler conditions. TaqMan primers and probes for IL-1β, IL-8, TGFβ, PDCD4 and control gene 18S rRNA were purchased as Assays-on-DemandTM Gene Expression Products (Applied Biosystems). Expression of each gene was determined in triplicate and normalized to 18S rRNA. Analysis and fold differences were determined using the comparative CT method. Unless otherwise indicated, fold change was calculated from the ΔΔCT values with the formula 2−ΔΔCT. Experiments were repeated 3–4 times and values averaged.
2.7. ELISA
COS-7 cells were transfected with EV, MUC1/A, MUC1/B or MUC1ΔEX expression vectors. After 48 h incubation, cells were treated with 10 ng/ml TNFα (Prepotech, Rocky Hill, NJ, USA) for 24 h and media was collected. The media was concentrated using Amicon ultra 10K centrifugation columns (Millipore). IL-8 quantification was performed using a kit from BD Biosciences c
2.8. NF-κB Luciferase reporter assay
COS-7 cells were transfected in 24-well plates as described above. Each well received 250 ng of a NF-κB luciferase reporter (Promega, Fitchburg, WI, USA), 5 ng of a Renilla luciferase reporter (pRL-tk, Promega) and 500 ng plasmid DNA for EV, MUC1/A, MUC1/B or MUC1ΔEX. 48 h after transfection, triplicate wells were left untreated or treated with 1 or 10 ng/ml TNFα. The cells were harvested 4 and 24 h after treatment using Promega’s Passive Lysis buffer. Luciferase and Renilla luciferase activities were determined using Promega’s Dual Luciferase assay (Dougherty et al., 2006).
2.9. miRNA isolation and QRT-PCR
COS-7 cells were transfected as described above. 48 h after transfection, cells were treated with 10 ng/ml TNFα (PrepoTech) for 24 h. RNA was extracted using the miRCURY miRNA isolation kit (Exiqon, Copenhagen, Denmark). QPCR performed using the miRCURY LNATM SYBR Green master mix using the primer set for miR-21 (Exiqon). 5S RNA was used for normalization of miRNA expression. Analysis and fold change was determined using the comparative threshold cycle (Ct) method.
2.10. MUC1 ectodomain release
Conditioned media was collected from transfected COS-7 grown in 6-well plates and treated with 1 or 10 ng/ml TNFα for 24 h. 500 μl of media was concentrated to 15 μl using Amicon ultra 10K columns (Millipore). 13 μl of conditioned media was separated on 3–8% Tris-Acetate gels (Invitrogen) and immunoblotted with DF3 Mab (Signet) to MUC1-N. Coomassie stain (Sigma) was performed following standard protocol.
2.11. Statistical analyses
Results are expressed as mean ± S.E.M of 3–5 independent experiments, each treatment was done in triplicate. Student's t test was performed when indicated (Graph Pad Prism). p < 0.05 was considered significant.
3. Results
3.1. Expression of MUC1/A and MUC1/B splice variants in COS-7 cells
To determine the functional consequence of the extra 9 aa present at the MUC1-N domain of MUC1/A, MUC1/A was cloned (data not shown, Fig. 1A) and its cellular distribution compared with MUC1/B in transfected COS-7 (MUC1-null) cells The transfection efficiency and MUC1/A and MUC1/B protein expression was determined by flow cytometry (Fig. 1B) and western blot using an antibody specific for the MUC1-CD (Fig. 1C). Although MUC1/B was reported to be localized to the plasma membrane (PM) in transfected cells (Schroeder et al., 2001), this is the first evaluation of the cellular localization of MUC1/A. Cell surface MUC1/A and MUC1/B expression was detected in ~ 20–35 % of the cells 24–72 h after transfection (Fig. 1B). These results indicate that MUC1/A and MUC1/B localized to the PM, as seen for native MUC1/B in epithelial cells (Argueso et al., 2009). MUC1/A and MUC1/B protein expression in COS-7 cells 24, 48 and 72 h post-transfection was similar (Fig. 1C), confirming the FACS data. Endogenous MUC1 in MCF-7 human breast cancer cells, which have high MUC1 (Wei et al., 2006), is shown for comparison. The highest expression of MUC1/A and MUC1/B was achieved 48 h after transfection. Thus, this time of transfection was used for further experiments. Overexpression of MUC1/A and MUC-1B had no effect on COS-7 viability (Supplementary Fig. 1).
Immunocytochemistry was performed on nonpermeabilized COS-7 cells using an anti-FLAG antibody and Alexa-Flour-conjugated wheat germ agglutinin (WGA) as a PM marker. The PM localization of the MUC1ΔEX, lacking most of the MUC1-N but containing the transmembrane domain (Fig. 1A), was evaluated in parallel as a control to examine how lack of MUC1-N affected PM localization. This is the first evaluation of the cellular localization of MUC1ΔEX. MUC1/A, MUC1/B and MUC1ΔEX, all localize to the PM of COS-7 cells, as indicated by the overlap in signal (yellow) with WGA (Figure 2). The empty vector (EV, negative control), panel demonstrates the specificity of the FLAG antibody.
Figure 2. Surface expression of MUC1/A and MUC1/B in COS-7 cells.
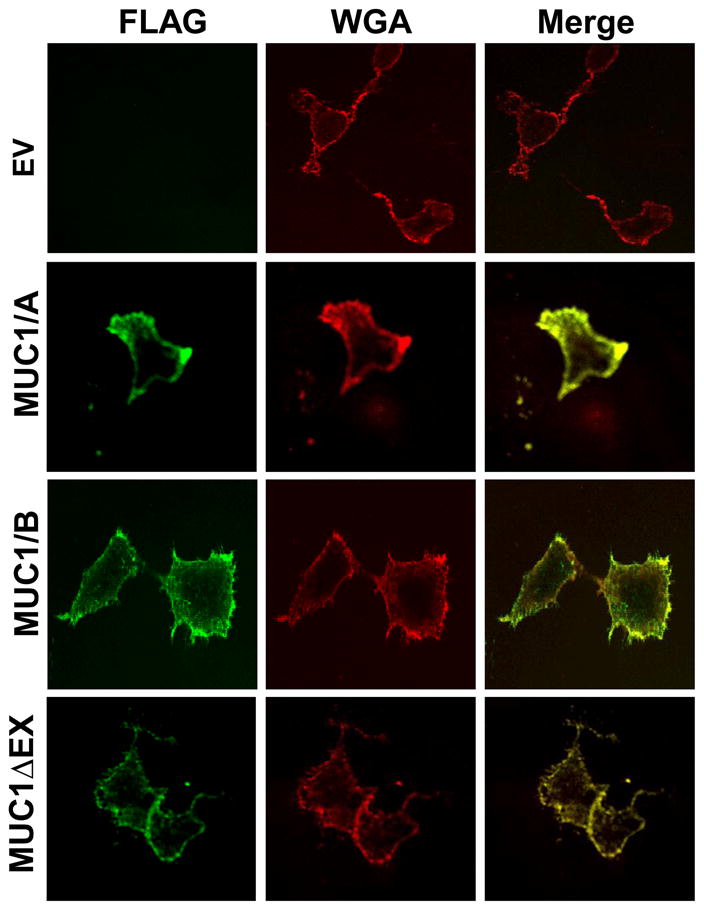
COS-7 cells were transfected with the empty vector (EV, negative control), MUC1/A, MUC1/B, or MUC1ΔEX as described in Experimental Procedures. Forty-eight hours after transfection, the cells were stained with the M2 anti-FLAG antibody (green) and Wheat Germ Agglutinin (WGA, red) as a plasma membrane marker. Images were captured using an Olympus FV1000 confocal microscope. The colocalization of FLAG and WGA is indicated in yellow in the merged image.
3.2. Regulation of the inflammatory response by MUC1/A and MUC1/B
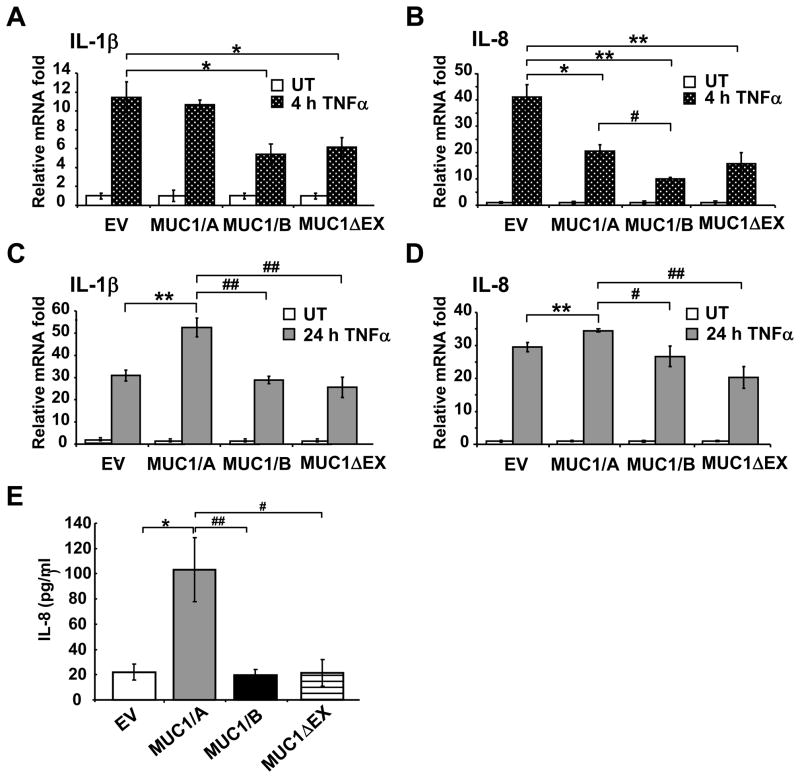
MUC1/B has both anti- and pro- inflammatory properties (Ahmad et al., 2007; Guang et al., 2010; Ueno et al., 2008). To determine if MUC1/A and MUC1/B differentially modulate inflammatory responses, COS-7 cells were transfected with EV (negative control), MUC1/A, MUC1/B or MUC1ΔEX, testing for lack of MUC1-N, for 48 h and treated with 10 ng/ml TNFα for 4 and 24 h to initiate the inflammatory response (Sonoda et al., 2006; Thathiah et al., 2004). QRT-PCR was used to determine the mRNA levels of pro-inflammatory cytokines known to be induced by TNFα: IL-1β (Schroder and Tschopp, 2010) and IL-8 (Lane et al., 2002). Because IL-8 and TNFα are increased in the tears of patients with dry eye disease (Pflugfelder et al., 1999), IL-8 regulation by MUC1 splice variants is relevant to this study. TNFα induced a 12- and 40-fold increase in IL-1β and IL-8 mRNA at 4 h, respectively, in EV-transfected cells (Fig. 3A and B). Transfection with either MUC1/B or MUC1ΔEX inhibited the 4 h TNFα-induced IL-1β and IL-8 expression compared to the EV control. In contrast, MUC1/A-expressing cells did not attenuate the induced expression of IL-1β after 4 hrs of TNFα (Fig. 3A). At the 4 h time point, the TNFα-induced expression of IL-8 was also inhibited in MUC1/A expressing cells compared to EV (Fig. 3B). After 24 h of TNFα, COS-7 cells expressing MUC1/A exhibited an increase in IL-1β and IL-8 compared to EV-transfected cells (Fig. 3C and D). Interestingly, MUC1/B or MUC1ΔEX no longer inhibited the induction of IL-1β and IL-8 mRNA after 24 h TNFα treatment.
Figure 3. MUC1/A and MUC1/B differentially regulate IL-1β and IL-8 expression.
COS-7 cells were transfected with empty vector (EV), MUC1/A, MUC1/B, or MUC1ΔEX for 48 h. Cells were left untreated (UT) or treated with 10 ng/ml TNFα for 4 and 24 h. QRT-PCR analysis of IL-1β (A and C) or IL-8 (B and D) mRNA was performed. TNFα-induced expression in EV-transfected cells at 4 h was set to one for comparison. (E) COS-7 cells were treated for 24 h with 10 ng/ml TNFα. IL-8 protein levels were measured in the cultured media by ELISA. Values in panels A-D are the mean ± SEM of 3–5 experiments. *, p<0.05 compared to EV. **, p<0.001 compared to EV. #, p<0.05 compared to MUC1/A. ##, p<0.001 compared to MUC1/A.
The amount of IL-8 protein released into the media of transfected COS-7 cells was examined 24 h after TNFα treatment (Fig. 3E). Cells transfected with MUC1/A released 5-fold more IL-8 protein compared to cells transfected with the EV control. No change in IL-8 protein secretion was observed in cells expressing MUC1/B or MUC1ΔEX compared to EV. These data agree with the 24 h mRNA data for IL-8 shown in Fig. 3D.
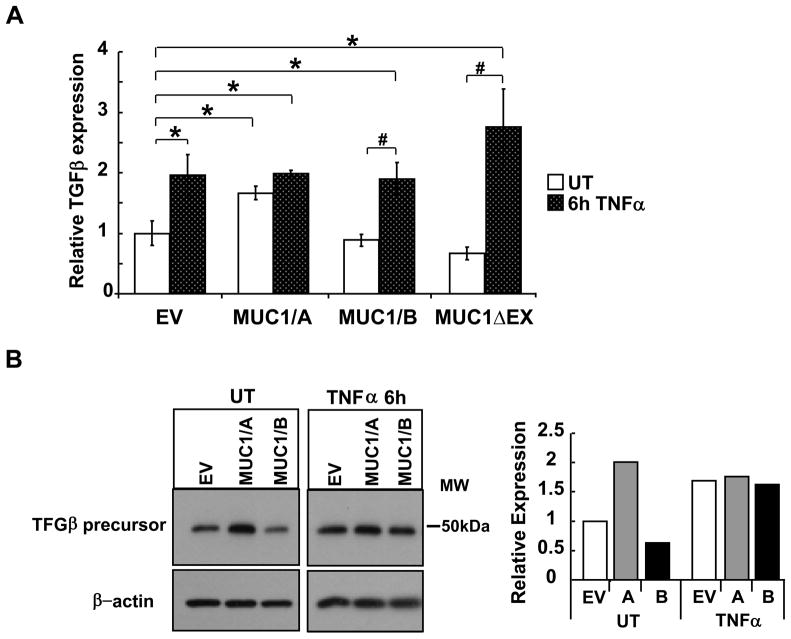
3.3. MUC1/A increases basal TGFβ mRNA and protein expression
TGFβ has anti-inflammatory activity (Hong et al., 2010) as well as stimulating differentiation and survival of Th-17 cells (De Paiva et al., 2009). MUC1/A overexpression in COS-7 cells significantly increased basal TGFβ mRNA levels (Fig. 4A). In contrast, no change in basal TGFβ mRNA was detected in cells transfected with MUC1/B or MUC1ΔEX. TNFα increased TGFβ mRNA in EV, MUC1/B or MUC1ΔEX-transfected cells, but not in MUC1/A-transfected cells. TNFα-induced levels of TGFβ mRNA were comparable in EV, MUC1/B or MUC1ΔEX transfected cells. Western blot analysis of TGFβ revealed an increase in basal TGFβ precursor protein in the MUC1/A-tranfected COS-7 cells compared to EV- and MUC1/B- transfected cells (Fig. 4B). These data demonstrate that differences in the MUC1-N in MUC1/A versus MUC1/B result in differential regulation of basal and TNFα-induced TGFβ expression.
Figure 4. MUC1/A stimulates TGFβ expression.
COS-7 cells were transfected with EV control, MUC1/A, MUC1/B, or MUC1ΔEX for 48 h prior to treatment with 10 ng/ml TNFα for 6 h, UT= untreated. (A) QRT-PCR analysis of TGFβ mRNA was performed. TGFβ expression in EV-transfected/UT cells was set to one for comparison. Values are the mean ± SEM of 3 experiments. *, p<0.05 compared to EV-UT. #, p<0.05 compared to MUC1/B or MUC1ΔEX UT. (B) Western blot analysis of TGFβ precursor expression in WCE (6 μg). The blot was probed with TGFβ antibody and then stripped and reprobed for β-actin as a loading control. This blot is representative of three blots that showed the same results. The bar graph shows the quantitation of TGFβ relative to β-actin expression relative to EV that was set to one for comparison.
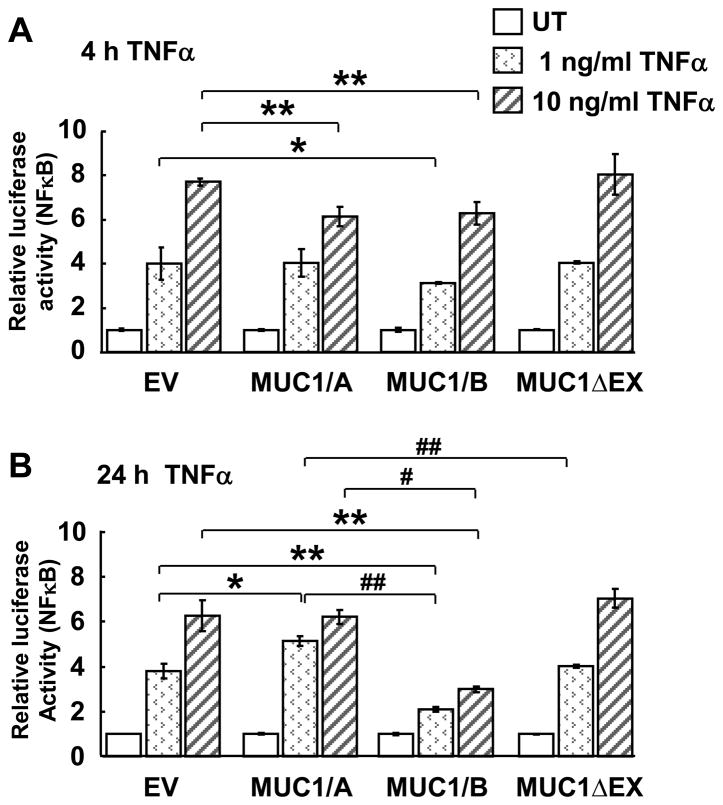
3.4. Regulation of NF-κB-Luciferase reporter by MUC1/A and MUC1/B
TNFα activation of NF-κB induces IL-1β and IL-8 transcription (Lane et al., 2002; Schroder and Tschopp, 2010). To determine if MUC1/A and MUC1/B affect NF-κB activation, a NF-κB luciferase reporter assay was performed in COS-7 cells transfected with EV, MUC1/A, MUC1/B or MUC1ΔEX (Fig. 5). As expected, TNFα increased NF-κB luciferase activity in a concentration-dependent manner. MUC1/B overexpressing cells showed reduced TNFα-induced reporter activity when treated with 1 ng/ml TNFα for 4 h compared to the EV control (Fig. 5A). With 10 ng/ml TNFα, both MUC1/A and MUC1/B overexpressing cells showed inhibition of the NF-κB reporter activity compared to EV at 4 h. Interestingly, MUC1ΔEX overexpression did not inhibit the NF-κB luciferase response, suggesting that MUC1-N is required to inhibit the NF-κB response. When COS-7 cells were incubated with 1 or 10 ng/ml TNFα for 24 h, similar responses to the 4 h were observed (Fig. 5B). MUC1/B significantly inhibited 1 and 10 ng/ml TNFα-activated NF-κB luciferase activity, whereas MUC1/A increased the NF-κB luciferase activity at 1 ng/ml but had no effect at 10 ng/ml.
Figure 5. Effect of MUC1/A and MUC1/B overexpression in TNFα-induced NF-κB activation.
COS-7 cells were transfected with NF-κB luciferase reporter, pRL-tk and 500 ng plasmid DNA for EV, MUC1/A or MUC1/B as described in Experimental Procedures. 48 h after transfection, wells were left untreated (UT) or treated with 1 or 10 ng/ml TNFα and WCE were harvested 4 and 24 h after treatment. Luciferase and Renilla luciferase activities were determined. Values were calculated as described in Experimental Procedures and are the mean ± SEM of 3 experiments. *, p<0.05 compared to EV. **, p<0.001 compared to EV. #, p<0.05 compared to MUC1/A. ##, p<0.001 compared to MUC1/A.
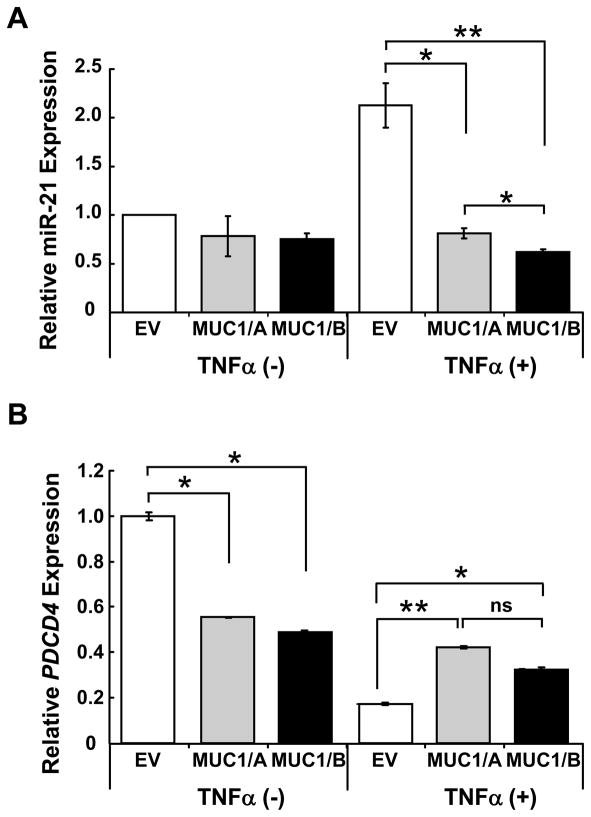
3.5. Regulation of microRNA miR-21 by MUC1/A and MUC1/B
A recent report showing that IL-1β and TNFα induced miR-21 expression via NF-κB activation in pancreatic cancer cells (Roggli et al., 2010) prompted us to examine if MUC1/A or MUC1/B would affect TNFα-induced miR-21 expression. Because MUC1/B inhibits TNFα-induced IL-1β expression at 4 h (Fig. 3B) and 24 h TNFα-activation of NFκB-reporter activity (Fig. 5B), we hypothesized that MUC1/B would abrogate miR-21 induction by TNFα. The results agreed with the predicted outcome: MUC1/B inhibited TNFα-induced miR-21 expression at 24 h (Fig. 6A). MUC1/A also inhibited the TNFα-induced miR-21 up-regulation, but to a lesser extent than MUC1/B. This was unexpected because MUC1/A did not inhibit TNFα-induced IL-1β transcription (Fig. 3A and C) or NF-κB-luciferase activity (Fig. 5B).
Figure 6. MUC1/A and MUC1/B inhibit TNFα-induced miR-21 expression.
COS-7 cells transfected with EV, MUC1/A or MUC1/B were treated with 10 ng/ml TNFα for 24 h. (A) QRT-PCR for mature miR-21was quantified using 5S rRNA for normalization. (B) QRT-PCR for PDCD4 mRNA was normalized to 18S rRNA. Values are the avg. +/− SEM from triplicate determinations in one experiment.
We next examined if the suppression of TNFα-induced miR-21 by MUC1/A and MUC1/B leads to de-repression of a gene downstream of miR-21. Programmed cell death 4 (PDCD4) is an established target of miR-21 (Wickramasinghe et al., 2009). MUC1/A and MUC1/B reduced basal PDCD4 expression and blocked the suppression of PDCD4 detected in TNFα-treated, EV-transfected cells (Fig. 6B). The reduction in PDCD4 is commensurate with the increase in miR-21 seen in TNFα-treated-EV-transfected cells (Fig. 6A). Together, these data indicate that both MUC1/A and MUC1/B inhibit TNFα-induced miR-21 expression and thus ameliorate the blockade of PDCD4 expression.
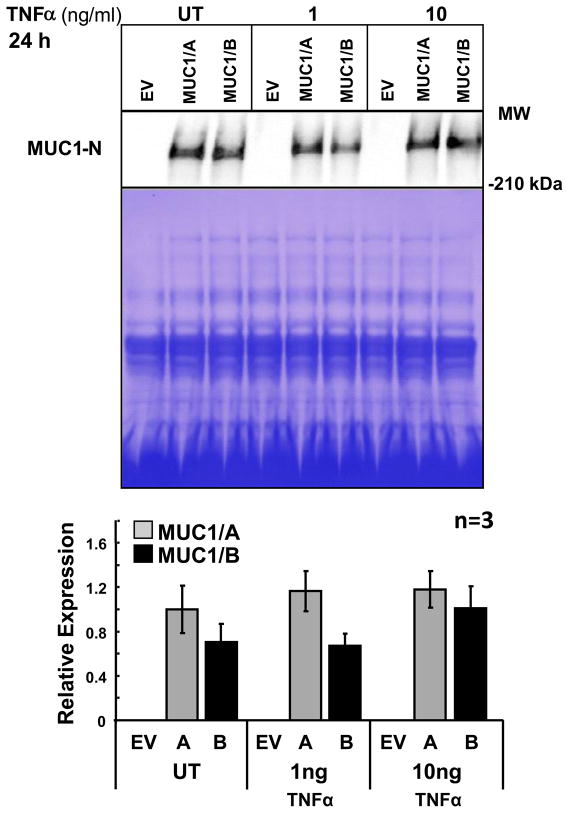
3.6. MUC1 ectodomain release
MUC1-N, the ectodomain, is shed into the media from human corneal limbal epithelial cell line and human uterine epithelial cells under normal conditions as well as in response to TNFα (Albertsmeyer et al., 2010; Thathiah et al., 2004). MUC1/A and MUC1/B ectodomain release was examined. TNFα increased MUC1-N release from cells expressing MUC1/B (Fig. 7). In contrast, MUC1/A-expressing cells did not increase MUC1-N release in response to TNFα. There was no significant difference in MUC1-N shed between cells transfected with MUC1/A or MUC1/B; however, there was a trend for higher MUC1/A ectodomain release in cells untreated or treated with 1 ng/ml TNFα.
Figure 7. MUC1 ectodomain release in COS-7 cells transfected with MUC1/A or MUC1/B and treated with TNFα.
Conditioned media was collected COS-7 transfected with EV, MUC1/A (A), or MUC1/B (B) grown in 6-well plates and treated with the indicated concentrations of TNFα for 24 h. The media was concentrated and identical amounts were separated on 3–8 % Tris-acetate gels. The immunoblots were probed with an antibody (DF3) to the extracellular domain of MUC1. To control for loading, an identical gel was stained with Coomassie blue. The graph represents the quantification of the MUC1-N bands normalized to the Coomassie net pixel intensity of the entire lane. Values are the average of 3 separate experiments ± SEM.
4. Discussion
This report demonstrates for the first time that MUC1/A and MUC1/B, previously demonstrated to be differentially expressed in conjunctival tissue from non-Sjögren’s and evaporative dry eye patients compared to normal controls (Imbert et al., 2006), differ in their ability to regulate the TNFα-induced inflammatory responses. The lack of expression of MUC1/A in samples from patients with non-Sjögren’s dry eye disease indicates a possible protective role of MUC1/A in preventing the symptoms accompanying dry eye disease. Whereas MUC1/B was reported to have anti-inflammatory activity (Lu et al., 2006; Ueno et al., 2008), this is the first report to demonstrate that MUC1/A regulates cellular inflammation and that there are specific differences in the anti-inflammatory activities of MUC1/A and MUC1/B. MUC1/B has greater efficacy in inhibiting pro-inflammatory responses induced by TNFα compared to MUC1/A. Specifically, depending on the time of TNFα-treatment, MUC1/B inhibits the increase in IL-1β and IL-8 stimulated by TNFα whereas MUC1/A increases the expression of these two pro-inflammatory cytokines. Not all cellular responses are different between MUC1/A and MUC1/B, e.g., both inhibit the induction of miR-21 by TNFα and inhibit basal PDCD4 expression. In addition, MUC1/A, and not MUC1/B, increases basal TGFβ expression. This is the first report showing that MUC1 regulates TGFβ expression.
In patients with dry eye disease, the ocular surface is exposed to pro-inflammatory cytokines produced by infiltrating lymphocytes (Barabino and Dana, 2007). Here, MUC1/B blocked the TNFα-mediated increase in the transcription of IL-1β and IL-8. These findings are consistent with a report showing that MUC1/B overexpression inhibits activation of Toll-like receptors in cultured HEK-293T cells (Ueno et al., 2008). Our data indicate that MUC1/A has less anti-inflammatory activity than MUC1/B. Although these data do not explain the correlation between the lower frequency of MUC1/A expression in patients with non-Sjögrens aqueous deficient and evaporative dry eye disease, they demonstrate that MUC1/A and MUC1/B mediate different cytokine responses to TNFα. Interestingly, the TNFα-stimulated cytokine response in MUC1ΔEX-expressing cells was similar to MUC1/B- expressing cells. This result was unexpected because MUC1ΔEX lacks most of the extracellular domain which is the part of MUC1 that is different between MUC1/A and MUC1/B. Thus, the pro-inflammatory phenotype of MUC1/A indicates that the extra 9 aa in the N-terminus affects its signaling properties.
The precise mechanisms accounting for the differential role of MUC1/A and MUC1/B in regulating TNFα-induced IL-1β and IL-8 transcription remain to be fully elucidated. However, we postulate that differential regulation of the NF-κB pathway by MUC1/A and MUC1/B plays a key role in the concomitant differences in cytokine expression since MUC1/B, but not MUC1/A, inhibited TNFα-induced NF-κB driven luciferase activity. However, in agreement with our data showing that MUC1/A inhibited TNFα-induced IL-8 mRNA only at the 4 h time point, MUC1/A inhibited TNFα activation of the NF-κB luciferase reporter activity only at 10 ng/ml TNFα at 4 h. These findings indicate different time-dependent effects of MUC1/A versus MUC1/B in response to TNFα. Surprisingly, MUC1ΔEX did not inhibit TNFα-induced activation of the NF-κB driven luciferase reporter. These results were unexpected because the MUC1-CD is thought to mediate the intracellular activities of MUC1 (Gendler, 2001) and also because MUC1ΔEX blocked TNFα-induced induction of IL-1β and IL-8 at 4 h. This intriguing finding further supports the idea that MUC1-N and MUC1-CD subunits work together to modulate signal transduction events.
MUC1/A and MUC1/B showed similar PM localization; however, there may be subtle differences in their localization that were not detected. It is also possible that MUC1/A and MUC1/B interact with a different repertoire of regulatory proteins in the PM and cytoplasm or other intracellular compartments that differentially mitigate the TNFα-induced inflammatory response. Another possibility is that the MUC1-CD of MUC1/A and MUC1/B are differently phosphorylated which would impact downstream signaling.
Although COS-7 cells were used because they are readily transfected and MUC1 null, these cells are not a good model for the polarized epithelial cells that constitute the ocular surface. Nevertheless, the results presented here clearly indicate functional differences between MUC1/A and MUC1/B.
TGFβ is a potent anti-inflammatory cytokine that plays an important role in maintaining lacrimal gland homeostasis (Rocha et al., 1998). TGFβ ablation in mice leads to “crusty eyes” due to lacrimal gland inflammation (McCartney-Francis et al., 1997). Desiccating stress, a mimetic of dry eye, increased TGF-β1 mRNA and protein in conjunctiva from mouse eyes which, in turn, enhanced differentiation and survival of Th-17 cells (De Paiva et al., 2009). Th-17-committed cells produce pro-inflammatory cytokines, e.g., IL-17A, that increase TNFα, IL-6, IL-8 and matrix metalloproteinases (De Paiva et al., 2009; Zheng et al., 2010). Thus, while the ability of MUC1/A to induce TGFβ, a new finding, suggests that one mechanism by which MUC1/A confers protection to the ocular surface; whether TGFβ ultimately acts as a a pro- or anti- inflammatory cytokine is likely cell-type dependent.
Our data indicate that both MUC1/A and MUC1/B inhibit TNFα-induced miR-21 expression and thus de-repress PDCD4 expression. PDCD4−/− mice show reduced inflammatory responses (Hilliard et al., 2006). TNFα-downregulation of PDCD4 in pro-inflammatory environments decreased inflammation (Hilliard et al., 2006; Yasuda et al., 2009). To our knowledge, this is the first investigation of MUC1 regulation of miR-21 expression and activity. Further investigation of MUC1 regulation of miRNAs in dry eye disease is of clear interest.
In conclusion, our findings indicate that MUC1/A and MUC1/B differ in their anti-inflammatory properties as reflected in cytokine activation and NF-κB regulation, but similar activities in regulating miR-21 expression. The difference in the function of MUC1/A and MUC1/B detected in transfected COS-7 cells is the first step in understanding how their expression in conjunctival tissue is involved in dry eye disease.
Supplementary Material
Highlights.
Cloned MUC1/A splice variant
MUC1/B inhibits TNFα-induced IL-1β and IL-8
MUC1/A stimulates TNFα-induced IL-1β and IL-8
MUC1/A and MUC1/B inhibit TNFα-induced miR-21
MUC1/B, but not MUC1/A, inhibited TNFα-induced NF-κB luciferase reporter activity
MUC1/A and MUC1/B have different inflammatory activities
Acknowledgments
We thank Dr. J.A. Schroeder for providing the MUC1/B and MUC1ΔEX expression vectors. We thank Dr. Ronald G. Gregg for advice on MUC1/A cloning. Y. Imbert-Fernandez was supported by a pre-doctoral fellowship: NIH F31 EY017275. This work was supported in part by NIH R01 CA138410 to C.M. Klinge. The DNA sequencing core is supported by NIEHS P30ES01443.
Abbreviations
- EV
empty vector
- FBS
fetal bovine serum
- IL
interleukin
- miRNA
micro RNA
- MUC1
mucin 1
- TNFα
tumor necrosis factor α
- MUC1-N
MUC-1N-terminal extracellular domain
- MUC1-CD
MUC-1C-terminal cytoplasmic domain
- MUC1.CT
MUC-1 cytoplasmic tail
- PM
plasma membrane
- PDCD4
Programmed cell death 4
- ROS
reactive oxygen species
- TM
transmembrane
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abe M, Kufe D. Structural analysis of the DF3 human breast carcinoma-associated protein. Cancer Res. 1989;49:2834–2839. [PubMed] [Google Scholar]
- Ahmad R, Raina D, Trivedi V, Ren J, Rajabi H, Kharbanda S, Kufe D. MUC1 oncoprotein activates the I[kappa]B kinase [beta] complex and constitutive NF-[kappa]B signalling. Nat Cell Biol. 2007;9:1419–1427. doi: 10.1038/ncb1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertsmeyer AC, Kakkassery V, Spurr-Michaud S, Beeks O, Gipson IK. Effect of pro-inflammatory mediators on membrane-associated mucins expressed by human ocular surface epithelial cells. Experimental Eye Research. 2010;90:444–451. doi: 10.1016/j.exer.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argueso P, Guzman-Aranguez A, Mantelli F, Cao Z, Ricciuto J, Panjwani N. Association of Cell Surface Mucins with Galectin-3 Contributes to the Ocular Surface Epithelial Barrier. J Biol Chem. 2009;284:23037–23045. doi: 10.1074/jbc.M109.033332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabino S, Dana MR. Dry eye syndromes. Chem Immunol Allergy. 2007;92:176–184. doi: 10.1159/000099268. [DOI] [PubMed] [Google Scholar]
- Burdick MD, Harris A, Reid CJ, Iwamura T, Hollingsworth MA. Oligosaccharides Expressed on MUC1 Produced by Pancreatic and Colon Tumor Cell Lines. Journal of Biological Chemistry. 1997;272:24198–24202. doi: 10.1074/jbc.272.39.24198. [DOI] [PubMed] [Google Scholar]
- Carson DD. The cytoplasmic tail of MUC1: a very busy place. Sci Signal 1. 2008:pe35. doi: 10.1126/scisignal.127pe35. [DOI] [PubMed] [Google Scholar]
- De Paiva CS, Chotikavanich S, Pangelinan SB, Pitcher JD, III, Fang B, Zheng X, Ma P, Farley WJ, Siemasko KF, Niederkorn JY, Stern ME, Li DQ, Pflugfelder SC. IL-17 disrupts corneal barrier following desiccating stress. Mucosal Immunol. 2009;2:243–253. doi: 10.1038/mi.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty SM, Mazhawidza W, Bohn AR, Robinson KA, Mattingly KA, Blankenship KA, Huff MO, McGregor WG, Klinge CM. Gender difference in the activity but not expression of estrogen receptors alpha and beta in human lung adenocarcinoma cells. Endocr-Relat Cancer. 2006;13:113–134. doi: 10.1677/erc.1.01118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendler SJ. MUC1, the renaissance molecule. J Mammary Gland Biol Neoplasia. 2001;6:339–353. doi: 10.1023/a:1011379725811. [DOI] [PubMed] [Google Scholar]
- Gendler SJ, Lancaster CA, Taylor-Papadimitriou J, Duhig T, Peat N, Burchell J, Pemberton L, Lalani EN, Wilson D. Molecular cloning and expression of human tumor-associated polymorphic epithelial mucin. Journal of Biological Chemistry. 1990;265:15286–15293. [PubMed] [Google Scholar]
- Guang W, Ding H, Czinn SJ, Kim KC, Blanchard TC, Lillehoj EP. MUC1 cell surface mucin attenuates epithelial inflammation in response to a common mucosal pathogen. Journal of Biological Chemistry. 2010 doi: 10.1074/jbc.M110.121319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliard A, Hilliard B, Zheng SJ, Sun H, Miwa T, Song W, Goke R, Chen YH. Translational Regulation of Autoimmune Inflammation and Lymphoma Genesis by Programmed Cell Death 4. J Immunol. 2006;177:8095–8102. doi: 10.4049/jimmunol.177.11.8095. [DOI] [PubMed] [Google Scholar]
- Hollingsworth MA, Swanson BJ. Mucins in cancer: protection and control of the cell surface. Nat Rev Cancer. 2004;4:45–60. doi: 10.1038/nrc1251. [DOI] [PubMed] [Google Scholar]
- Hong S, Lee HJ, Kim SJ, Hahm KB. Connection between inflammation and carcinogenesis in gastrointestinal tract: focus on TGF-beta signaling. World J Gastroenterol. 2010;16:2080–2093. doi: 10.3748/wjg.v16.i17.2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbert Y, Darling DS, Jumblatt MM, Foulks GN, Couzin EG, Steele PS, Young JWW. MUC1 splice variants in human ocular surface tissues: Possible differences between dry eye patients and normal controls. Experimental Eye Research. 2006;83:493–501. doi: 10.1016/j.exer.2006.01.031. [DOI] [PubMed] [Google Scholar]
- Imbert Y, Foulks GN, Brennan MD, Jumblatt MM, John G, Shah HA, Newton C, Pouranfar F, Young WW., Jr MUC1 and estrogen receptor [alpha] gene polymorphisms in dry eye patients. Experimental Eye Research. 2009;88:334–338. doi: 10.1016/j.exer.2008.05.019. [DOI] [PubMed] [Google Scholar]
- Jonckheere N, Van Seuningen I. The membrane-bound mucins: From cell signalling to transcriptional regulation and expression in epithelial cancers. Biochimie. 2010;92:1–11. doi: 10.1016/j.biochi.2009.09.018. [DOI] [PubMed] [Google Scholar]
- Kufe D, Inghirami G, Abe M, Hayes D, Justi-Wheeler H, Schlom J. Differential reactivity of a novel monoclonal antibody (DF3) with human malignant versus benign breast tumors. Hybridoma. 1984;3:223–232. doi: 10.1089/hyb.1984.3.223. [DOI] [PubMed] [Google Scholar]
- Lane BR, Liu J, Bock PJ, Schols D, Coffey MJ, Strieter RM, Polverini PJ, Markovitz DM. Interleukin-8 and Growth-Regulated Oncogene Alpha Mediate Angiogenesis in Kaposi's Sarcoma. J Virol. 2002;76:11570–11583. doi: 10.1128/JVI.76.22.11570-11583.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitin F, Stern O, Weiss M, Gil-Henn C, Ziv R, Prokocimer Z, Smorodinsky NI, Rubinstein DB, Wreschner DH. The MUC1 SEA Module Is a Self-cleaving Domain. Journal of Biological Chemistry. 2005;280:33374–33386. doi: 10.1074/jbc.M506047200. [DOI] [PubMed] [Google Scholar]
- Ligtenberg MJ, Vos HL, Gennissen AM, Hilkens J. Episialin, a carcinoma-associated mucin, is generated by a polymorphic gene encoding splice variants with alternative amino termini. Journal of Biological Chemistry. 1990;265:5573–5578. [PubMed] [Google Scholar]
- Lu W, Hisatsune A, Koga T, Kato K, Kuwahara I, Lillehoj EP, Chen W, Cross AS, Gendler SJ, Gewirtz AT, Kim KC. Cutting Edge: Enhanced Pulmonary Clearance of Pseudomonas aeruginosa by Muc1 Knockout Mice. The Journal of Immunology. 2006;176:3890–3894. doi: 10.4049/jimmunol.176.7.3890. [DOI] [PubMed] [Google Scholar]
- Macao B, Johansson DG, Hansson GC, Hard T. Autoproteolysis coupled to protein folding in the SEA domain of the membrane-bound MUC1 mucin. Nat Struct Mol Biol. 2006;13:71–76. doi: 10.1038/nsmb1035. [DOI] [PubMed] [Google Scholar]
- McCartney-Francis NL, Mizel DE, Frazier-Jessen M, Kulkarni AB, McCarthy JB, Wahl SM. Lacrimal gland inflammation is responsible for ocular pathology in TGF-beta 1 null mice. Am J Pathol. 1997;151:1281–1288. [PMC free article] [PubMed] [Google Scholar]
- Obermair A, Schmid BC, Packer LM, Leodolter S, Birner P, Ward BG, Crandon AJ, McGuckin MA, Zeillinger R. Expression of MUC1 splice variants in benign and malignant ovarian tumours. International Journal of Cancer. 2002;100:166–171. doi: 10.1002/ijc.10456. [DOI] [PubMed] [Google Scholar]
- Park H, Hyun SW, Kim KC. Expression of MUC1 mucin gene by hamster tracheal surface epithelial cells in primary culture. Am J Respir Cell Mol Biol. 1996;15:237–244. doi: 10.1165/ajrcmb.15.2.8703480. [DOI] [PubMed] [Google Scholar]
- Pflugfelder SC. Antiinflammatory therapy for dry eye. American Journal of Ophthalmology. 2004;137:337–342. doi: 10.1016/j.ajo.2003.10.036. [DOI] [PubMed] [Google Scholar]
- Pflugfelder SC, Jones D, Ji Z, Afonso A, Monroy D. Altered cytokine balance in the tear fluid and conjunctiva of patients with Sjogren's syndrome keratoconjunctivitis sicca. Curr Eye Res. 1999;19:201–211. doi: 10.1076/ceyr.19.3.201.5309. [DOI] [PubMed] [Google Scholar]
- Rocha EM, Wickham LA, Huang Z, Toda I, Gao J, da Silveira LA, Sullivan DA. Presence and testosterone influence on the levels of anti- and pro-inflammatory cytokines in lacrimal tissues of a mouse model of Sjogren's syndrome. Adv Exp Med Biol. 1998;438:485–491. doi: 10.1007/978-1-4615-5359-5_67. [DOI] [PubMed] [Google Scholar]
- Roggli E, Britan A, Gattesco S, Lin-Marq N, Abderrahmani A, Meda P, Regazzi R. Involvement of MicroRNAs in the Cytotoxic Effects Exerted by Proinflammatory Cytokines on Pancreatic β-Cells. Diabetes. 2010;59:978–986. doi: 10.2337/db09-0881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid BC, Rudas M, Fabjani G, Speiser P, Kaserer K, Leodolter S, Zeillinger R. Evaluation of MUC1 splice variants as prognostic markers in patients with ductal carcinoma in situ of the breast. Oncol Rep. 2003;10:1981–1985. [PubMed] [Google Scholar]
- Schroder K, Tschopp J. The Inflammasomes. Cell. 2010;140:821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- Schroeder JA, Thompson MC, Gardner MM, Gendler SJ. Transgenic MUC1 Interacts with Epidermal Growth Factor Receptor and Correlates with Mitogen-activated Protein Kinase Activation in the Mouse Mammary Gland. Journal of Biological Chemistry. 2001;276:13057–13064. doi: 10.1074/jbc.M011248200. [DOI] [PubMed] [Google Scholar]
- Sonoda S, Uchino E, Nakao K, Sakamoto T. Inflammatory cytokine of basal and reflex tears analysed by multicytokine assay. Br J Ophthalmol. 2006;90:120–122. doi: 10.1136/bjo.2005.076737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strawbridge RJ, Nister M, Brismar K, Gronberg H, Li C. MUC1 as a Putative Prognostic Marker for Prostate Cancer. Biomark Insights. 2008;3:303–315. doi: 10.4137/bmi.s666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thathiah A, Brayman M, Dharmaraj N, Julian JJ, Lagow EL, Carson DD. Tumor Necrosis Factor {alpha} Stimulates MUC1 Synthesis and Ectodomain Release in a Human Uterine Epithelial Cell Line. Endocrinology. 2004;145:4192–4203. doi: 10.1210/en.2004-0399. [DOI] [PubMed] [Google Scholar]
- Ueno K, Koga T, Kato K, Golenbock DT, Gendler SJ, Kai H, Kim KC. MUC1 Mucin Is a Negative Regulator of Toll-Like Receptor Signaling. Am J Respir Cell Mol Biol. 2008;38:263–268. doi: 10.1165/rcmb.2007-0336RC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Lillehoj EP, Kim KC. Identification of four sites of stimulated tyrosine phosphorylation in the MUC1 cytoplasmic tail. Biochemical and Biophysical Research Communications. 2003;310:341–346. doi: 10.1016/j.bbrc.2003.09.030. [DOI] [PubMed] [Google Scholar]
- Wei X, Xu H, Kufe D. MUC1 Oncoprotein Stabilizes and Activates Estrogen Receptor [alpha] Molecular Cell. 2006;21:295–305. doi: 10.1016/j.molcel.2005.11.030. [DOI] [PubMed] [Google Scholar]
- Wickramasinghe N, Manavalan T, Dougherty S, Riggs K, Li Y, Klinge C. Estradiol downregulates miR-21 expression and increases miR-21 target gene expression in MCF-7 breast cancer cells. Nucleic Acids Res. 2009;37:2584–2595. doi: 10.1093/nar/gkp117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SJ, McGuckin MA, Gotley DC, Eyre HJ, Sutherland GR, Antalis TM. Two novel mucin genes down-regulated in colorectal cancer identified by differential display. Cancer Res. 1999;59:4083–4089. [PubMed] [Google Scholar]
- Wreschner DH, Hareuveni M, Tsarfaty I, Smorodinsky N, Horev J, Zaretsky J, Kotkes P, Weiss M, Lathe R, Dion A, et al. Human epithelial tumor antigen cDNA sequences. Differential splicing may generate multiple protein forms. Eur J Biochem. 1990;189:463–473. doi: 10.1111/j.1432-1033.1990.tb15511.x. [DOI] [PubMed] [Google Scholar]
- Yasuda M, Nishizawa T, Ohigashi H, Tanaka T, Hou DX, Colburn NH, Murakami A. Linoleic acid metabolite suppresses skin inflammation and tumor promotion in mice: possible roles of programmed cell death 4 induction. Carcinogenesis. 2009;30:1209–1216. doi: 10.1093/carcin/bgp106. [DOI] [PubMed] [Google Scholar]
- Zheng X, Bian F, Ma P, De Paiva CS, Stern M, Pflugfelder SC, Li DQ. Induction of Th17 differentiation by corneal epithelial-derived cytokines. Journal of Cellular Physiology. 2010;222:95–102. doi: 10.1002/jcp.21926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zrihan-Licht S, Baruch A, Elroy-Stein O, Keydar I, Wreschner DH. Tyrosine phosphorylation of the MUC1 breast cancer membrane proteins Cytokine receptor-like molecules. FEBS Letters. 1994;356:130–136. doi: 10.1016/0014-5793(94)01251-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.