Abstract
Purpose
To describe a new semi-automated method for segmenting and measuring the volume of the muscle, bone and adipose (subcutaneous and inter-muscular) tissue in calf muscle compartments using magnetic resonance (MR) images and determine the intra- rater and inter-rater reproducibility of the measures.
Materials and Methods
Proton-density weighted MR images were acquired from the right calf for 21 subjects. Three raters segmented and measured the volumes of bones, adipose tissue, and 5 individual muscle compartments. Two raters repeated the segmentations. The intra- and inter-rater reproducibility of the measures (intra-class correlation coefficients; ICC) were determined using generalizability theory.
Results
All ICC values were greater than 0.96. The average SEM of all measures was 1.21 cm3 and none were greater than 2.3 cm3. Essentially all variation (≥ 97% for all measures) was due to subject differences indicating low error in the measurements.
Conclusion
The volumetric measurements for the bones, adipose tissue, and muscle in each of the compartments using MR imaging were highly reproducible. MR imaging can provide quantitative, reproducible volumetric measures of bone, adipose tissue, and individual muscles in the calf. We believe these methods can be used to quantify specific muscle or adipose volumetric measures for other clinical or research purposes.
Keywords: MRI, Fat, Muscle, Reproducibility, Diabetes Mellitus, Peripheral Neuropathy
INTRODUCTION
Clinicians and researchers often seek to investigate the relationship between muscle structure, composition, and performance. Magnetic resonance imaging (MRI) is a non-radiographic, non-invasive method to visualize and quantify muscle cross-sectional areas and volumes. These areas and volumes can be segmented (i.e. separated) into bone, lean tissue (muscle), and fat tissue according to the intensity (brightness) of the individual voxels. Fat-based tissues such as adipose tissue and bone marrow have much higher relative signal intensity (brighter) than water based tissues such as muscle in proton-density weighted images without fat saturation. Furthermore, MRI can help characterize fat as subcutaneous adipose tissue (SAT) or inter-muscular adipose tissue (IMAT) (1,2).
SAT is the fat below the skin and above the muscle fascial plane and considered to be fairly benign (1). IMAT is defined as the visible adipose tissue beneath the muscle fascia, between muscles, and even within the muscle, but outside the actual muscle cell (3,4). Considerable IMAT in a muscle gives it a “marbled” appearance and has been associated with insulin resistance(1,2), decreased muscle performance and decreased physical function (4), and can occur after physical injury (5,6). IMAT has been compared to visceral adipose tissue in its risk for metabolic impairments (1–3).
Previous methods using MRI have quantified volumes in the entire calf (4,7,8) and some have segmented muscles into plantar flexor and dorsi flexor muscle groups (9,10,11). Segmenting individual muscles can be challenging because a clear demarcation between adjacent muscles can be difficult to identify. Manual segmentation is time consuming and the results can be dependent upon individual interpretation of muscle boundaries. Automated methods to segment neurological tissues or visceral adipose tissues have been widely developed, but automated methods to segment individual muscles or muscle groups in the calf are not well documented. Boettcher et al.(2) developed methods using a semi-automatic segmentation program (Matlab) for measuring the SAT and IMAT in the calf using MR images. They measured the IMAT in all of the calf muscles combined, but not in individual muscle compartments. Likewise, Broderick et al. (8) recently described fully automated methods to segment the whole calf muscle. Many researchers have used MR images to measure the IMAT in the body (4,7,8,9,11–14), some of which have looked at IMAT in the calf (7–11). To our knowledge, no one has reported research on semi-automated methods to segment muscle and adipose tissue in individual calf muscle compartments.
The purposes of our study were to: 1) describe a new semi-automated method for segmenting and measuring the volume of the calf into 5 muscle compartments to obtain volumes of muscle, bone and adipose (subcutaneous and intermuscular) tissue using MR images, and 2) determine the intra-rater and inter-rater reproducibility for all measures. We also report mean muscle and adipose tissue volumes for the 5 muscle compartments. Our previous attempts at segmenting calf muscle structures indicated that reproducibility of repeated measuring of the segmented total calf muscle was good (i.e., less than 3 percent error), but manual segmentation of individual calf muscles was considerably more challenging and resulted in large errors (11–37% differences in repeated measures) which were not acceptable. Therefore, we developed these automated methods that would make segmenting decisions quantitatively rather than by sight alone. Although our own research interests are to investigate the calf muscle structure and function of people with diabetes mellitus (DM), peripheral neuropathy and/or obesity, the methods and results of this study could be generalized to others who wish to quantify specific muscle or adipose tissue volumetric measures for other clinical or research purposes.
MATERIALS AND METHODS
Subjects
MR images from twenty one older adult subjects were utilized for the study [11 men, 10 women, 60.7(11.7) yrs]. Seven of the subjects had type 2 DM [3 men, 4 women, 52.4 (7.9) yrs, body mass index (BMI) = 35.7(5.6) kg/m2], seven subjects had type 2 DM and peripheral neuropathy without foot deformity or history of foot ulcer [5 men, 2 women, 66.3(12.5) yrs, BMI = 32.6(5.2) kg/m2], and seven of the subjects (controls) were weight matched, but did not have DM or peripheral neuropathy [3 men, 4 women, 63.4(10.7) yrs, BMI = 32.3(5.2) kg/m2]. The detailed demographics for the three groups of subjects are shown in Table 1. The subjects were recruited from diabetes clinics, advertisements and local volunteers for health-related studies in the CITY and surrounding areas. The UNIVERSITY NAME Institutional Review Board approved our study protocol. We obtained informed consent from each subject prior to testing. Peripheral neuropathy was assessed in each foot using previously described, reliable methods for all subjects (15). Briefly, peripheral neuropathy was determined by an inability to sense a single-thickness (5.07/10 gram) Semmes-Weinstein monofilament on at least one point on the plantar surface of each foot and based on a vibration perception threshold of greater than 25 volts using a biothesiometer.
Table 1.
Subject Demographics
| DM | n | DM+PN | n | Control | n | |
|---|---|---|---|---|---|---|
| Age (yrs) | ||||||
| Female | 51.8 (9.3) | 4 | 60.5 (6.4) | 2 | 58.5 (12.3) | 4 |
| Male | 53.3 (7.4) | 3 | 68.6 (14.2) | 5 | 70.0 (2.0) | 3 |
| DM Duration (yrs) | ||||||
| Female | 6.0 (3.3) | 4 | 5.0 (0.0) | 2 | ||
| Male | 4.0 (5.2) | 3 | 7.2 (4.4) | 5 | ||
| Weight (kg) | ||||||
| Female | 99.1 (16.5) | 4 | 108.0 (12.8) | 2 | 83.7 (15.1) | 4 |
| Male | 103.3 (15.7) | 3 | 95.6 (10.8) | 5 | 112.6 (12.1) | 3 |
| Height (cm) | ||||||
| Female | 163.2 (2.4) | 4 | 168.9 (5.4) | 2 | 166.4 (6.7) | 4 |
| Male | 176.1 (6.4) | 3 | 177.3 (2.8) | 5 | 179.5 (7.8) | 3 |
| BMI (kg/m2) | ||||||
| Female | 37.5 (6.3) | 4 | 38.0 (6.9) | 2 | 30.4 (6.4) | 4 |
| Male | 33.3 (4.4) | 3 | 30.5 (2.9) | 5 | 34.9 (1.5) | 3 |
Data are mean ± SD
Abbreviations: DM, diabetes mellitus; DM+PN, diabetes mellitus and peripheral neuropathy; yrs, years; kg, kilograms; cm, centimeters; m, meters; SD, standard deviation;
Image Acquisition
The Siemens Magnetom Trio 3T MRI scanner was used to acquire the images of the right calf (Siemens Medical Systems, Inc., Malvern, PA, USA) of each subject using previously established methods (4). The subjects were placed in a supine position with a Siemens circularly polarizing (CP) no-tune transmit/receive extremity coil placed over the right calf muscle. The CP coil is suited for obtaining the highest resolution with excellent signal strength/noise ratio. The following MR parameters were used to acquire proton-density weighted MR images: spin echo pulse sequence, TR/TE = 1,500/12 msec, field of view = 180 mm, bandwidth = 130 Hz/pixel, 30 slices, transverse orientation, signal averages = 1, flip angle = 90°, matrix = 256×256, echo train length = 1, acquisition time ~ 4.5 minutes, and a pixel size of 0.703 mm. The MR images without fat saturation were collected beginning at the tibiofemoral joint space and proceeding distally with each slice having a thickness of 7 mm and no inter-slice gap for a total of 21 cm. The fat saturated images were acquired subsequently using the spectral fat suppression technique (i.e. Siemens “strong” mode) covering the same anatomical location. Automatic 3D local shimming was used to minimize B1 non uniformity.
Calf, Bone, Muscle, SAT, Individual Muscle, and IMAT Segmentation Using Semi-automatic Methods
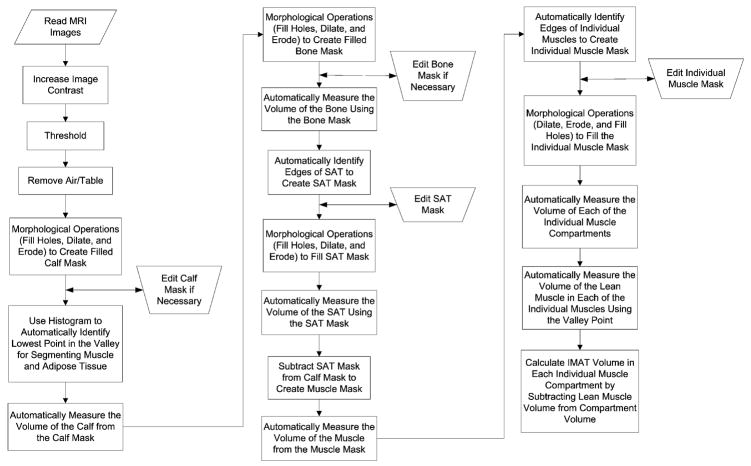
Images of the calf were loaded into Analyze (Biomedical Imaging Resource, Mayo Clinic, Rochester, MN) (16,17) in DICOM format and were converted to two AnalyzeImage 7.5 formatted volumes, one for each image acquisition (with and without fat saturation). The voxel size was 0.70 mm by 0.70 mm by 7 mm. The original voxel size was retained for each subject’s images to eliminate errors due to isotropic voxel interpolation. Such interpolation errors could affect the precision of the volume measurements. A program was developed using Matlab 7.4.0 (R2007a) software (Mathworks, Inc., Matick, MA) to semi-automatically segment and measure the calf compartments (Figure 1). A graphical user interface was developed to allow easy usage of the program and reading the Analyze formatted volumes.
Figure 1.
A flowchart of the Matlab program shows the steps of execution to measure the calf.
Nine consecutive slices (slice numbers 11–19 out of the 30 slices) were selected from the mid portion of the image volume to calculate the 16 volume measures listed in Tables 2–5. SAT was defined as the volume of adipose tissue below the skin and above the muscle fascial plane (1). IMAT was defined as the visible adipose tissue beneath the muscle fascia, between muscles, and within the muscle. The lean muscle and IMAT volume was determined for the following five muscle compartments: 1) Anterior compartment containing the tibialis anterior, the extensor digitorum longus and the extensor hallucis; 2) Lateral compartment containing the peroneus longus and brevis; 3) Deep compartment containing the tibialis posterior, the flexor digitorum longus and the flexor hallucis longus; 4) Soleus, and 5) Gastrocnemius (both heads). The “total muscle volume” was a sum of all lean muscle volume across the 5 muscle compartments. “Total IMAT volume” was the sum of the IMAT volumes across the 5 muscle compartments. “Bone” was the segmented volume for the tibia and fibula. “Whole leg” was the sum of the volumes from all component parts within the 9 slices.
Table 2.
Mean Volumes and Stand Error of the Measure (SEM) Values
| Measure | Mean volume (cm3) | SD | SEM (cm3) | SEM/Volume | IMAT/muscle Ratio |
|---|---|---|---|---|---|
| Whole Leg | 761.7 | 109.7 | 0.05 | <0.001 | |
| Bone | 61 | 13.8 | 0.05 | 0.004 | |
| Subcutaneous Fat | 211.4 | 76.5 | 2.31 | 0.030 | |
| Total Muscle | 489.4 | 74.5 | 2.3 | 0.031 | |
| Anterior Muscle | 69.6 | 11 | 0.62 | 0.056 | |
| Lateral Muscle | 36.8 | 9.5 | 0.41 | 0.043 | |
| Deep Muscle | 56.8 | 14.4 | 1.12 | 0.078 | |
| Soleus Muscle | 127.7 | 27.1 | 1.24 | 0.046 | |
| Gastroc Muscle | 139.4 | 32.1 | 1.08 | 0.034 | |
| Total Muscle Vol | 430.2 | 73.3 | 1.61 | 0.022 | |
| Anterior IMAT | 6.5 | 3.3 | 0.17 | 0.052 | 0.093 |
| Lateral IMAT | 5.5 | 4 | 0.44 | 0.110 | 0.149 |
| Deep IMAT | 8.1 | 4.7 | 0.43 | 0.091 | 0.143 |
| Soleus IMAT | 12.5 | 10.3 | 2.07 | 0.201 | 0.098 |
| Gastroc IMAT | 28.3 | 20.5 | 1.74 | 0.085 | 0.203 |
| Total IMAT Vol | 60.8 | 40.2 | 2.31 | 0.057 | 0.141 |
Abbreviations: IMAT, intramuscular adipose tissue; Vol, volume; cm, centimeters;
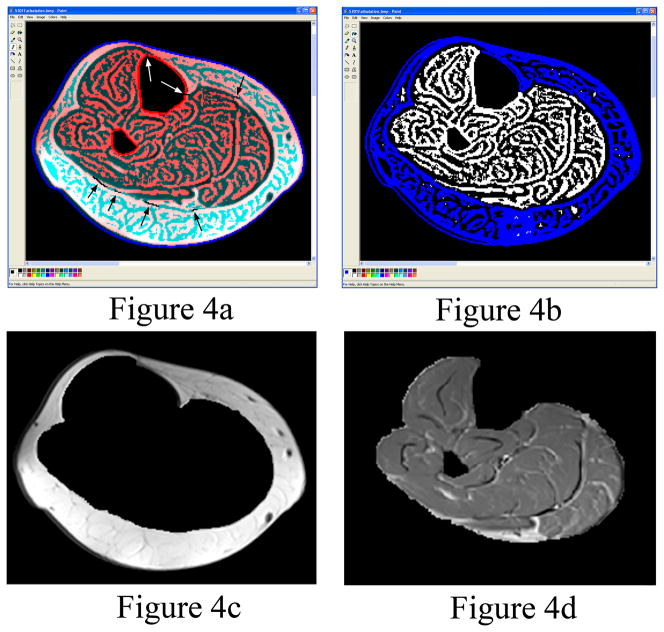
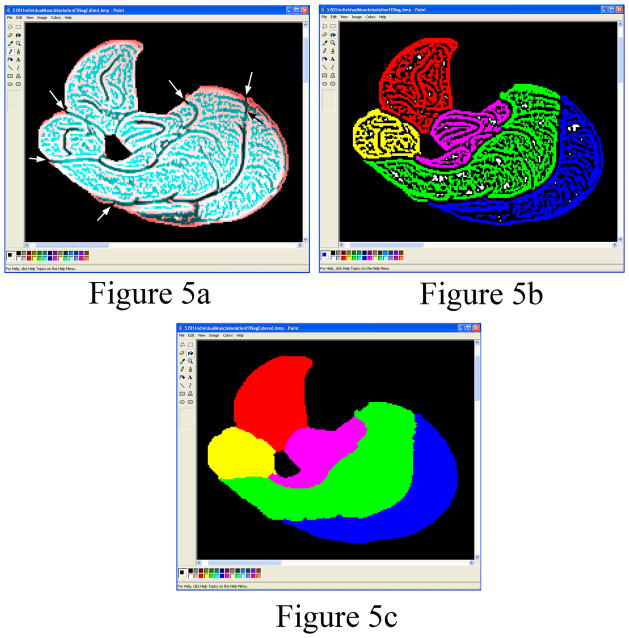
After importing the AnalyzeImage 7.5 formatted images into the Matlab program, the image contrast was improved by contrast stretching which maps the intensity values in the grayscale image to new values so that 1% of the data is saturated at low intensities and 1% at high intensities. The program uses a threshold to automatically segment the non-fat saturated images of the calf from the air and surrounding materials including morphological operations (fill holes, dilate, and erode) to create a filled calf mask. The filled calf mask was used to remove the air surrounding the calf in the fat saturated MR images (Figure 2a). In the rare case where MR artifacts existed in the images, Microsoft Paint (Microsoft Corporation, Redmond, WA) was used to edit the perimeter of the calf (i.e. a few subjects were edited). The volume of the calf is automatically calculated. A grayscale histogram from the fat saturated images of the segmented calf is displayed for the user to allow confirmation of the automatically identified threshold located at the lowest point in the valley between the adipose tissue peak and the muscle peak (Figure 3). The histogram from the fat saturated images produced better shaped peaks and fewer peaks compared to the non-fat saturated images. The histogram is smoothed using moving averages. The two peaks are automatically identified using a maximum function and the minimum between the two maximum peaks is identified using a minimum function. In the rare case where one or both peaks do not exist, the user can select the threshold from the curve and change the threshold setting. The threshold is utilized by the program to calculate lean muscle and IMAT volumes. The bones are automatically segmented from the calf using an intensity threshold (Figure 2b), except in rare cases where the cortical shell is thin which may require some manual editing of the bone perimeter. The fat saturated images were used to segment the bones because the contrast between the cortical shell of the fibula was greatest in the fat saturated images compared to the non-fat saturated images resulting in better automated threshold segmentations. The volume of the bone is automatically calculated. In the non-fat saturated MR images, the perimeter of the SAT is found automatically utilizing the second derivative of brightness to find the edges in the images (18). After edge detection, the edges of the subcutaneous fat were easily seen in the non-fat saturated images as compared to the edge detection in the fat saturated images. In most cases, the SAT has some small connections to the IMAT. The program creates a SAT binary mask containing the edges found in the images. The user is required to break the small connections in the binary mask between the SAT and the IMAT in the images using a pencil tool (Figure 4a). After breaking the connections, colored muscle, SAT and IMAT image is subtracted from the image leaving the edited binary mask. The SAT portion of the binary mask is colored blue using the ”Fill With Color” tool in Microsoft Paint to distinguish it from IMAT and lean muscle tissue (Figure 4b). Matlab uses the blue region to automatically segment the SAT from the calf. The Matlab program automatically fills the holes in the SAT binary mask by applying morphological operations of filling, dilation and erosion (18). The program displays the SAT and lean muscle tissue with the IMAT for the user to verify the images look correct (Figure 4c and 4d). The edges of the muscle compartments in the fat saturated MR images are automatically identified utilizing the same edge detection methods (Figure 5a). The fat saturated images were utilized to segment the individual muscles, because the fascia between the individual muscles can be more clearly identified visually in the fat saturated image than in the non-fat saturated images (i.e. the fascia is darker in intensity relative to the bright adjacent muscles in the fat saturated images). The user is required to break small connections in the binary mask between the individual muscle compartments using the Paint program’s pencil tool for each slice to separate the compartments. The edges of the binary mask that define the boundary of individual muscle compartments are colored with the Paint program’s “Fill With Color” tool (Figure 5b). The Matlab program automatically fills the holes in each individual muscle compartment (color) by applying morphological operation of dilation, erosion, and filling (Figure 5c). The volume of each individual muscle and IMAT within each muscle is automatically calculated based on the threshold that was automatically chosen by the program and displayed in the graphical user interface. All calculated volumes are stored into a text file as the user makes the measurements. The program shows a binary representation of the IMAT and the lean muscle tissue compared to the grayscale fat saturated image.
Figure 2.
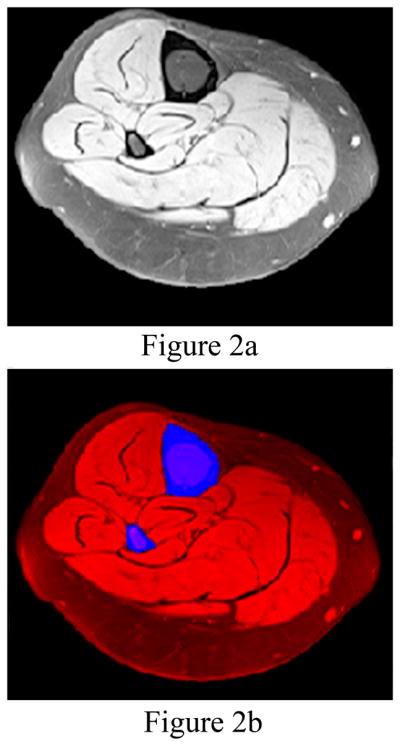
(a) The leg is automatically segmented from the MR image (left). (b) The bones (tibia and fibula colored blue) are automatically identified for segmentation from the leg (colored red in the right image). The fat saturated images make segmenting the bone from the muscle and fat automatic in most cases.
Figure 3.
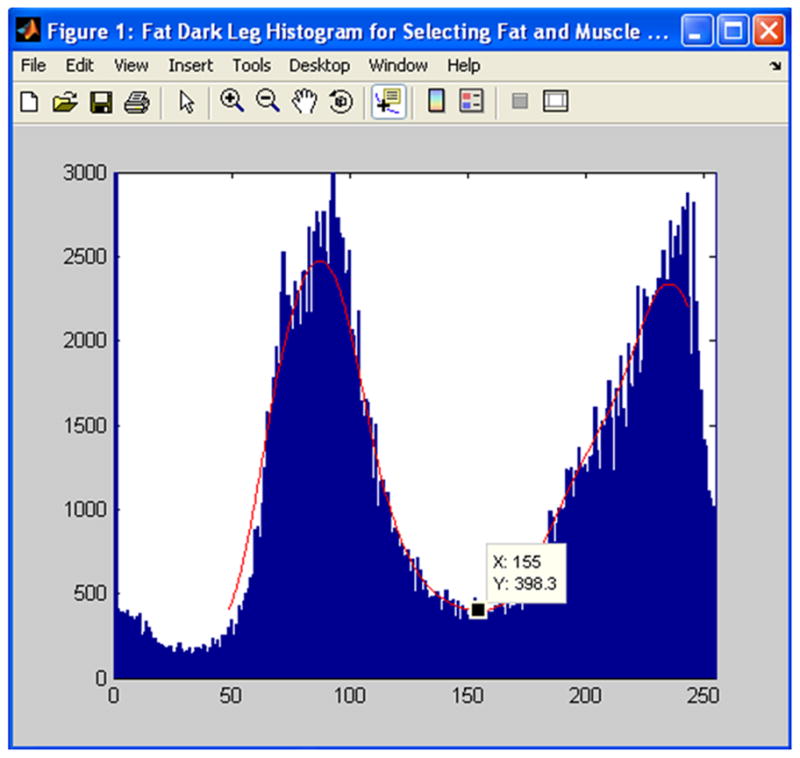
A histogram of calf bone, adipose tissue and muscle is shown. The lowest point in the valley between the two peaks is automatically identified (black square, x=155). The program used this lowest point to find the individual muscle and IMAT volumes.
Figure 4.
(a) The internal edge of the subcutaneous adipose tissue is semi-automatically identified, but the user is required to break small connections (arrows) between the subcutaneous adipose tissue and IMAT using a Paint program. (b) A “Fill With Color” tool within Paint is used to automatically fill the region denoted as subcutaneous fat with the color blue. (c) Representative subcutaneous fat segmentation. (d) Representative muscle and IMAT segmentation. Figures 4c and 4d are automatically created using the filled blue colored region shown in figure 4b.
Figure 5.
(a) Edges of the individual muscles are automatically identified by the program. The user is required to break (arrows) the few small connections between the individual muscles using the Paint program’s black pencil tool. (b) Each of the individual muscles is filled using the Paint program’s ”Fill With Color” tool when the user selects a muscle. The anterior compartment is colored red, lateral yellow, deep pink, soleus green and gastroc blue. (c) After using the ”Fill With Color” tool, the muscle mask for each compartment is automatically filled by the program to identify each individual muscle compartment.
Intra-rater and Inter-rater Measurements
Two physical therapists who also are engaged in full time clinical research and one engineer in radiology served as raters. One physical therapist (LJT, rater 1) had approximately one year of experience segmenting the SAT from the muscle and several months of experience segmenting individual muscles using B-splines in Analyze. The second physical therapist (MKH, rater 2) had no prior experience using the Analyze or Matlab segmentation tools. The engineer (PKC, rater 3) who developed the Matlab program had some experience with SAT and IMAT segmentation while developing the program. Each rater received approximately 8 hours of training from the experienced rater (LJT) to determine how SAT and each muscle compartment were to be defined. The training was necessary to improve the intra- and inter-rater reproducibility when identifying the SAT and IMAT boundary connections (four lower black arrows in Figure 4a) that need to be broken. A user’s manual was created in an effort to enhance consistency between raters. Each rater then learned how to utilize the Matlab program to identify and measure the following volumes using a few training cases that were not included in the 21 subjects: calf, bones, SAT, whole muscle, and individual muscles. All three raters measured the five volumes for the 21 subjects once. The two physical therapists repeated volume measurements with five or more days between measurements. Each rater recorded the time needed to complete all measurements.
Statistical Analysis
Generalizability theory (GT) procedures were used to estimate the reproducibility of the measures (bone, muscle, adipose and lean tissue volumes) (19,20). Analyses producing the generalizability theory results were done in SPSS Version 19. GT partitions measurement error into multiple sources such as from raters or time and allows estimating reproducibility under a variety of expected applications (e.g., a single rater collecting measures on one occasion). GT can furthermore be used to estimate reproducibility that reflects simple consistency in measurement or it can take rater mean differences into account. We were most interested in estimating the absolute error of the measure (rather than a rank ordering), so we report the more conservative estimates of absolute reproducibility in which mean differences among raters are considered a source of error. We report separate reproducibility indices for the two raters who provided measurements at two time points (simple intra-rater reproducibility) in order to illustrate how similarly and well they were able to use the procedure. We report the reproducibility for all three raters based on a single assessment (inter-rater reproducibility) to illustrate how well raters with different kinds of background and training are able to use the procedure. More importantly, we report reproducibility estimates that account for error due to rater differences and different measurement times simultaneously. These estimates provide the best indication of reproducibility by taking all relevant error sources into account. The intra-class correlation coefficients (ICC) we report provide a ratio of true variance (subject variance) to the total variance in each measure. We also report the standard error of the measure (SEM) which is expressed in cubic centimeters and provides an indication of measurement error in the metric of the original variables. As an additional indicator of reproducibility and to illustrate the individual differences between repeated measures, we provide representative Bland-Altman plots for selected measures. Finally, we report descriptive volumetric measures for all variables (mean and standard deviation).
RESULTS
There were no group differences in any of the volume measures using an ANOVA (p>0.05), so all subjects (n=21) were analyzed as one group in the repeated measures analysis to determine indices of reproducibility. The mean and standard deviation for each measure for all subjects along with the ratio of IMAT to lean muscle tissue are contained in Table 2 and included for descriptive purposes.
Reproducibility estimates are contained in Table 3. The ICC values indicating intra-rater reproducibility were greater than 0.99 for all measures except for the measure of IMAT in the soleus muscle compartment for rater 2 which was equal to 0.987. The ICC values indicating inter-rater reproducibility were greater than 0.99 for all measures except for measures of IMAT in the Soleus muscle compartment (0.974). The combined reproducibility estimates take into account error due to both multiple raters and multiple measurement times. These provide the most conservative and appropriate estimates for reproducibility in future applications because raters are unlikely to provide measurements at similar times. These reproducibility estimates all exceeded 0.99, with the exception of measures of IMAT in the lateral compartment (0.989) and for IMAT in the Soleus muscle compartment (0.967).
Table 3.
Results: Intra-class Correlation Coefficient values for Testers 1 and 2 (Intra-rater reproducibility), all testers at one time (Inter-rater reproducibility), and combined reproducibility accounting for tester and time error sources.
| Measure | Intra-rater | Intra-rater | Inter-rater | Combined |
|---|---|---|---|---|
| Reproducibility | Reproducibility | Reproducibility | Reproducibility | |
| Tester 1 | Tester 2 | 3 Testers 1 Time | 2 Testers 2 Times | |
| Whole Leg | 1.000 | 1.000 | 1.000 | 1.000 |
| Bone | 1.000 | 1.000 | 1.000 | 1.000 |
| Subcutaneous Fat | 1.000 | 1.000 | 0.999 | 0.999 |
| Total Muscle | 1.000 | 1.000 | 0.999 | 0.999 |
| Anterior Muscle | 1.000 | 0.996 | 0.997 | 0.997 |
| Lateral Muscle | 1.000 | 0.996 | 0.998 | 0.998 |
| Deep Muscle | 1.000 | 0.996 | 0.996 | 0.994 |
| Soleus Muscle | 1.000 | 0.998 | 0.999 | 0.998 |
| Gastroc Muscle | 1.000 | 0.999 | 0.999 | 0.999 |
| Total Muscle Vol | 1.000 | 1.000 | 0.999 | 1.000 |
| Anterior IMAT | 1.000 | 0.999 | 0.997 | 0.997 |
| Lateral IMAT | 0.998 | 0.992 | 0.990 | 0.989 |
| Deep IMAT | 0.997 | 0.997 | 0.993 | 0.993 |
| Soleus IMAT | 0.999 | 0.987 | 0.974 | 0.967 |
| Gastroc IMAT | 1.000 | 0.992 | 0.990 | 0.993 |
| Total IMAT Vol | 1.000 | 0.999 | 0.997 | 0.997 |
Abbreviations: IMAT, intramuscular adipose tissue; Vol, volume
The SEMs for each measurement based on the combined reproducibility analysis are contained in Table 2 and ranged from 0.48 to 2.31 cm3. The SEMs provide an indication of measurement error in the metric of the original variables for individual measures. The ratio of SEM to mean total volume of the segment also is contained in Table 2 to provide a context to the SEM value.
The proportions of variance for all measures attributed to different sources are listed in Table 4. Nearly all of the variance in measures was related to the (true score) ”Subject” source, with very low proportions of variance due to any of the error sources. Of the error sources, the highest proportion was associated with Subject × Raters, indicating that different raters had slightly different rank orderings of subjects. But, even for this source, the highest proportion was only 0.017, for the measure of IMAT in the Soleus muscle compartment.
Table 4.
Sources of variance for all measures and all factors. All variance is related to “Subject” with very low variance (error) associated with Subject × Raters indicating excellent reproducibility of measures.
| Source of Variance | |||||||
|---|---|---|---|---|---|---|---|
| Measure | Subject | Rater | Time | Subject × Rater | Rater × Time | Subject × Time | Error |
| Whole Leg | 1.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Bone | 1.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Subcutaneous Fat | 0.999 | 0.000 | 0.000 | 0.001 | 0.000 | 0.000 | 0.000 |
| Total Muscle | 0.999 | 0.000 | 0.000 | 0.001 | 0.000 | 0.000 | 0.000 |
| Anterior Muscle | 0.997 | 0.000 | 0.000 | 0.001 | 0.000 | 0.000 | 0.002 |
| Lateral Muscle | 0.998 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 |
| Deep Muscle | 0.994 | 0.000 | 0.000 | 0.004 | 0.000 | 0.001 | 0.002 |
| Soleus Muscle | 0.998 | 0.000 | 0.000 | 0.001 | 0.000 | 0.000 | 0.001 |
| Gastroc Muscle | 0.999 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Total Muscle Vol | 1.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Anterior IMAT | 0.997 | 0.000 | 0.000 | 0.001 | 0.000 | 0.000 | 0.001 |
| Lateral IMAT | 0.989 | 0.000 | 0.000 | 0.003 | 0.001 | 0.000 | 0.007 |
| Deep IMAT | 0.993 | 0.000 | 0.000 | 0.002 | 0.000 | 0.000 | 0.005 |
| Soleus IMAT | 0.967 | 0.006 | 0.000 | 0.017 | 0.003 | 0.000 | 0.007 |
| Gastroc IMAT | 0.993 | 0.000 | 0.000 | 0.002 | 0.000 | 0.000 | 0.004 |
| Total IMAT Vol | 0.997 | 0.000 | 0.000 | 0.003 | 0.000 | 0.000 | 0.000 |
Abbreviations: Vol, volume; IMAT, intramuscular adipose tissue
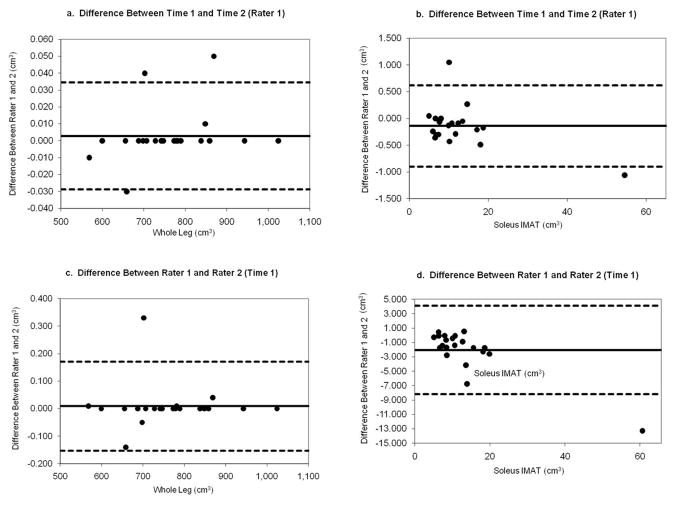
Representative Bland-Altman plots are provided representing measures with the highest reproducibility (Whole leg volume) and for the lowest reproducibility (Soleus IMAT volume) in Figure 6a–d. Plots are provided for the differences between Time 1 and Time 2 for rater 1 (intra rater reproducibility) for measures of Whole Leg volume (Figure 6a) and for Soleus IMAT Volume (Figure 6b). Next, plots are provided for the difference between measures obtained from Rater 1 and Rater 2 at Time 1 (inter rater reproducibility) for Whole Leg volume (Figure 6c) and for Soleus IMAT Volume (Figure 6d). Note the wider 95% confidence interval when a different rater repeats the measure compared to when the same rater repeats the measure for Soleus IMAT volume (Figure 6d vs 6b).
Figure 6.
Representative Bland-Altman plots for Intra rater (6a and 6b) and Inter rater reproducibility (6c and 6d) of measures with best (Whole Leg volume, 6a and 6c) and worst reproducibility values (Soleus IMAT volume, 6b and 6d).
DISCUSSION
These results indicate excellent reproducibility for all volumetric measures of bone, adipose tissue, or muscle tissues in the calf in individual and group muscle compartments using MRI. All ICC values were greater than 0.96 and most were greater than 0.99. The semi-automated methods used quantitative approaches to make decisions about adipose tissue and muscle boundaries and required few decisions from the individual raters. Essentially all variance was related to “Subject” with little variance attributed to other sources (Table 4). These results indicate that essentially all variance was related to actual differences in the measures between subjects rather than the methods individual raters used to quantify the volumes or changes in the use of the measurement procedure over time.
IMAT in the Soleus muscle compartment had the greatest Subject × Rater error, yet this error was still only 1.7% (SEM= 2.07 cm3). The fascial border around the soleus muscle occasionally was difficult to identify, and seemed to contribute to this error. Discussion between the raters prior to testing helped to establish consistent decisions about placement of fascial borders. For repeated testing over time and/or following various interventions, reproducibility will be optimal if the same rater repeats the Soleus IMAT volume measure compared to having different raters take the measure (Figure 6d compared to 6b).
In addition to being highly reproducible, the semi-automated methods also reduced data analysis time compared to previous manual segmenting methods. Manual segmenting methods to estimate the values for the 16 variables reported in this study required a minimum of 60 minutes per subject. Using the semi-automated methods, the raters reported that the same 16 variables were measured for the nine slices and recorded in a text document in an average time of approximately 30 minutes with a range of 20–50 minutes. The saving in time can be attributed to the automatic identification of the majority of SAT and muscle boundaries using the Matlab program (Figures 4a and 5a) compared to the manual selection of the boundary using B-splines in Analyze. Therefore, in addition to enhancing reproducibility, the methods also reduced analysis time by at least 50% compared to manual methods.
These methods provide quantitative volumetric measures of muscle and adipose tissue in the calf that can be used to better understand muscle changes as a result of disease, injury, or intervention. Previous research has shown that IMAT can be related to impaired glucose tolerance (1,2), impaired muscle and physical performance (4), and lower levels of walking (i.e, steps per day) (21). In this sample of obese subjects, there appears to be high variability in the amount of IMAT volume between various subjects and even between individual muscle compartments (Table 2). Some compartments appear to have greater percentages of IMAT than other compartments, but additional research is needed to determine the muscle specific changes due to obesity, diabetes, and peripheral neuropathy. The causal relationship between IMAT, disease, and function still is not clear. Given the rising incidence of diabetes and obesity, and the physical dysfunction associated with these groups, additional research is needed to determine specifically what types of exercise are optimal to improve the muscle composition, and more importantly, to improve muscle function and overall physical performance. Furthermore, fat infiltration into muscle has been noted in numerous types of musculoskeletal injury including injury to the back muscles (6), and rotator cuff muscles (5). Muscle and fat volumes, such as those described in this study, may help to understand mechanisms of disability and be important outcome measures in clinical studies where changes in body composition are desired. For example, Marcus et al. (22) used MR volume images of the thigh to help demonstrate that an intensive aerobic and eccentric resistance exercise program can help patients with type 2 DM improve long-term glycemic control, thigh composition, and physical performance.
Despite the high reproducibility of repeated measures reported in this study, several limitations are acknowledged. One of the purposes of this study was to assess raters’ ability to quantify muscle and adipose tissue volume reproducibly from nine slices located approximately in the middle of the calf from a single MR series of images. We did not test if repeated MR image acquisitions or variation in MR sequence parameters or varying the number of slices measured would introduce additional errors. Changing the number of slices measured would affect the total volume measured for each subject, but we expect only a minimal change in measurement error if the image quality remains constant. For a volumetric study, we recommend the number of slices to remain fixed for the entire study or the number of slices measured be based on the length of the tibia. Because the Matlab thresholding methods automatically identify a new threshold to distinguish lean individual muscle tissue from IMAT for each subject’s scan, we believe our methods would produce consistent results (independent of rater) even with a slight variation in repeated MR image acquisitions. For clinical musculoskeletal imaging, high spatial resolution, high contrast and a good signal to noise ratio (SNR) are very important to distinguish tissues such as muscle, adipose tissue, cartilage and bone (23). Proton-density weighted images with long TRs above 1000 msec provide high SNR and contrast for depicting normal structures. By increasing the TR, the SNR improves for tissues such as muscle with a long T1. Utilizing the MR sequence parameters we chose for this study along with contrast stretching resulted in a large separation in the muscle and fat histogram peaks (Figure 3). The large separation allowed the program to easily and automatically identify a unique threshold repeatedly in the valley between the two peaks for each subject. Small variations in the MR sequence parameters may cause a slight change in the actual volume of fat, bone, muscle and IMAT measured.
Furthermore, B1 non uniformity should have little effect on the subcutaneous fat and individual muscle segmentations since we did not utilize a global threshold based method for segmenting these two tissues, but instead used an edge-detection method to determine the edges between the muscle and subcutaneous fat, and between individual muscles. Edge detection does not rely on a single threshold for the entire image, but only a local intensity change between tissues. The B1 non uniformity may have an effect on the IMAT measure since a threshold for the entire volume was used to determine the IMAT content in the individual muscles, but our Siemens MR scanner uses a standard 3D local shimming adjustment technique which adjusts for every sequence unless the field of view does not change between sequences which was true in our case.
For optimal results, all testing should be conducted on the same magnet with identical testing parameters. Boettcher et al.(2) determined reproducibility of their semiautomatic segmentation methods by 3 repeated measurements in five volunteers after they were repositioned on a MR scanner. The coefficient of variation (SD/mean*100) was low for these repeat MR scans (2.8±0.8% for IMAT and 2.2±1.5% for SAT). In addition to the previous limitations noted, we only measured a portion of the calf muscle (9 slices equal to 6.3 cm) and not the entire length of the calf. Imaging the entire calf would be optimal for studying complete muscle changes, but would require a longer coil to be placed over the extremity and a longer scanning time than used in this study. Because the slice thickness was 7 mm, volume averaging could compromise our ability to measure small quantities of IMAT when the IMAT thickness is less than the 0.7 mm in-plane resolution within the 7 mm slice. Thinner slices and smaller in-plane resolution would allow us to measure smaller quantities of IMAT. This study did not assess the accuracy of measures against a true gold standard; i.e. we did not determine validity of measures. However, visual appraisal of the segmented muscle did have good face validity by visual assessment; i.e. the segmented muscle looked like the given muscle as documented in traditional anatomy textbooks. Future studies should determine the accuracy of the muscle and IMAT volume measures against a true measure of these volumes. Finally, we measured only 5 muscle compartments and not every individual muscle in the calf. Additional individual muscles can be added in the future as the MR images become more precise and automated methods for edge detection improve. Although this study investigated only calf muscles, we hope these semi-automated methods can be applied to other specific muscles or muscle groups (i.e, thigh or arm).
In conclusion, the volumetric measurements for the bones, adipose tissue, and muscle in each of the calf compartments using MR imaging were highly reproducible. MR imaging can provide quantitative, reproducible volumetric measures of bone, adipose tissue, and individual muscle in the calf. These measures can be used to help understand underlying pathology and determine treatment effectiveness of exercise and other medical interventions.
Acknowledgments
We wish to thank Dr. Jie Zheng for his MR imaging expertise. We wish to thank Dr. John Kotyk for his MR guidance and support.
This work was supported by grants R21 HD058938-01A1 and T32 HD0073434 (NIH, NICHHD, NCMRR); NSMRC R24HD650837, UL1-RR024992 CTSA, Center for Clinical Imaging Research (CCIR), and Recruitment Cores; and Promotion of Doctoral Studies Awards from the Foundation for Physical Therapy (Tuttle).
Footnotes
The project was reviewed and approved by the Washington University School of Medicine Human Research Protection Office.
References
- 1.Goodpaster BH, Thaete FL, Kelley DE. Thigh adipose tissue distribution is associated with insulin resistance in obesity and in type 2 diabetes mellitus. Am J Clin Nutr. 2000;71:885–892. doi: 10.1093/ajcn/71.4.885. [DOI] [PubMed] [Google Scholar]
- 2.Boettcher M, Machann J, Stefan N, et al. Intermuscular Adipose Tissue (IMAT): Association With Other Adipose Tissue Compartments and Insulin Sensitivity. J Magnetic Resonance Imaging. 2009;29:1340–1345. doi: 10.1002/jmri.21754. [DOI] [PubMed] [Google Scholar]
- 3.Gallagher D, Kuznia P, Heshka S, et al. Adipose tissue in muscle: a novel depot similar in size to visceral adipose tissue. Am J Clin Nutr. 2005;81:903–910. doi: 10.1093/ajcn/81.4.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hilton TN, Tuttle LJ, Bohnert KL, Mueller MJ, Sinacore DR. Excessive adipose tissue infiltration in skeletal muscle in individuals with obesity, diabetes mellitus, and peripheral neuropathy: association with performance and function. Phys Ther. 2008;88:1336–1344. doi: 10.2522/ptj.20080079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doro LC, Ladd B, Hughes RE, Chenevert TL. Validation of an adapted MRI pulse sequence for quantification of fatty infiltration in muscle. Magn Reson Imaging. 2009;27(6):823–827. doi: 10.1016/j.mri.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 6.Kjaer P, Bendix T, Sorensen JS, Korsholm L, Leboeuf-Yde C. Are MRI-defined fat infiltrations in the multifidus muscles associated with low back pain? BMC Med. 2007 Jan 25;5:2. doi: 10.1186/1741-7015-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gadeberg P, Andersen H, Jakobsen J. Volume of ankle dorsiflexors and plantar flexors determined with stereological techniques. Journal of Applied Physiology. 1999;86:1670–1675. doi: 10.1152/jappl.1999.86.5.1670. [DOI] [PubMed] [Google Scholar]
- 8.Broderick BJ, Dessus S, Grace PA, Olaighin G. Technique for the computation of lower leg muscle bulk from magnetic resonance images. Med Eng Phys. 2010;32(8):926–933. doi: 10.1016/j.medengphy.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 9.Andersen H, Gadeberg PC, Brock B, Jakobsen J. Muscular atrophy in diabetic neuropathy: a stereological magnetic resonance imaging study. Diabetologia. 1997;40(9):1062–1069. doi: 10.1007/s001250050788. [DOI] [PubMed] [Google Scholar]
- 10.Mathur S, Lott DJ, Senesac C, Germain SA, Vohra RS, Sweeney HL, Walter GA, Vandenborne K. Age-related differences in lower-limb muscle cross-sectional area and torque production in boys with Duchenne muscular dystrophy. Arch Phys Med Rehabil. 2010;91(7):1051–1058. doi: 10.1016/j.apmr.2010.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stevens JE, Pathare NC, Tillman SM, Scarborough MT, Gibbs CP, Shah P, Jayaraman A, Walter GA, Vandenborne K. Relative contributions of muscle activation and muscle size to plantarflexor torque during rehabilitation after immobilization. J Orthop Res. 2006;24(8):1729–36. doi: 10.1002/jor.20153. [DOI] [PubMed] [Google Scholar]
- 12.Ruan XY, Gallagher D, Harris T, et al. Estimating whole body intermuscular adipose tissue from single cross-sectional magnetic resonance images. J Appl Physiol. 2007;102:748–754. doi: 10.1152/japplphysiol.00304.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song MY, Ruts E, Kim J, Janumala I, Heymsfield S, Gallagher D. Sarcopenia and increased adipose tissue infiltration of muscle in elderly African American women. Am J Clin Nutr. 2004;79:874–880. doi: 10.1093/ajcn/79.5.874. [DOI] [PubMed] [Google Scholar]
- 14.Janssen I, Fortier A, Hudson R, Ross R. Effects of an energy restrictive diet with or without exercise on abdominal fat, intermuscular fat, and metabolic risk factors in obese women. Diabetes Care. 2002;25:431–438. doi: 10.2337/diacare.25.3.431. [DOI] [PubMed] [Google Scholar]
- 15.Diamond JE, Mueller MJ, Delitto A, Sinacore DR. Reliability of a diabetic foot evaluation. Phys Ther. 1989;69:797–802. doi: 10.1093/ptj/69.10.797. [DOI] [PubMed] [Google Scholar]
- 16.Robb RA. Three Dimensional Biomedical Imaging: Principles and Practice. New York: VCH Publishers, Inc; 1995. [Google Scholar]
- 17.Robb RA, Hanson DP, Karwoski RA, Larson AG, Workman EL, Stacy MC. Analyze: a comprehensive, operator-interactive software package for multidimensional medical image display and analysis. Comput Med Imaging Graph. 1989;13:433–454. doi: 10.1016/0895-6111(89)90285-1. [DOI] [PubMed] [Google Scholar]
- 18.Russ JC. The Image Processing Handbook. 2. Boca Raton, FL: CRC Press, Inc; 1995. [Google Scholar]
- 19.Brennan RL. Generalizability Theory. New York: Springer-Verlag; 2001. [Google Scholar]
- 20.Shavelson RJ, Webb NM. Generalizability theory: A primer. Newbury Park, CA: Sage; 1991. [Google Scholar]
- 21.Tuttle LJ, Sinacore DR, Cade WT, Mueller MJ. Lower physical activity is associated with higher intermuscular adipose tissue in people with type 2 diabetes and peripheral neuropathy. Phys Ther. 2011;91:923–930. doi: 10.2522/ptj.20100329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marcus RL, Smith S, Morrell G, et al. Comparison of combined aerobic and high-force eccentric resistance exercise with aerobic exercise only for people with type 2 diabetes mellitus. Phys Ther. 2008;88(11):1345–1354. doi: 10.2522/ptj.20080124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitchell DG, Cohen MS. MRI Principles. 2. Philadelphia, PA: Saunders – Elsevier Inc; 2004. [Google Scholar]