Abstract
Signaling by innate immune receptors initiates and orchestrates the overall immune responses to infection. Macrophage receptors recognizing pathogens can be broadly grouped into surface receptors and receptors restricted to intracellular compartments, such as phagosomes and the cytoplasm. There is an expectation that ingestion and degradation of microorganisms by phagocytes contributes to activation of intracellular innate receptors, although direct demonstrations of this are rare and many model ligands are studied in soluble form, outside of their microbial context. By comparing a wild-type strain of Staphylococcus aureus and a lysozyme-sensitive mutant, we have been able to directly address the role of degradation of live bacteria by mouse macrophages in determining the overall innate cellular inflammatory response. Our investigations revealed a biphasic response to S. aureus that consisted of an initial signal resulting from the engagement of surface TLR2, followed by a later, second wave on inflammatory gene induction. This second wave of inflammatory signaling was dependent on and correlated with the timing of bacterial degradation in phagosomes. We found that TLR2 signaling followed by TLR2/TLR9 signaling enhanced sensitivity to small numbers of bacteria. We further found that treating wild-type bacteria with the peptidoglycan synthesis-inhibiting antibiotic vancomycin made S. aureus more susceptible to degradation and resulted in increased inflammatory responses, similar to those observed for mutant degradation-sensitive bacteria.
Introduction
Phagocytosis is the process by which myeloid cells such as macrophages and dendritic cells internalize and kill microorganisms (1, 2). The process is intimately linked to the activation of inflammatory signals that help to coordinate broader tissue and systemic immune responses to potential pathogens. Since many soluble microbial components can stimulate strong inflammatory responses, it is clear that such responses do not always require phagocytosis. However, recent studies are beginning to define the processes by which phagosomes influence the nature and magnitude of an inflammatory response.
Toll-like receptors (TLRs) are key innate immune receptors that detect microorganisms and are responsible for a large fraction of the inflammatory response elicited from macrophages during bacterial, fungal, and viral infection. Some TLRs including TLR3, TLR7, and TLR9 are found exclusively in intracellular compartments where they detect nucleic acids (3). As such, the expectation that ingestion and degradation of microorganisms should contribute to activation of these receptors is implicit in their biology, although direct demonstrations of this are rare and many model ligands are studied in soluble form. Other TLRs including TLR1, TLR2, TLR4, and TLR6 are expressed at the cell surface and can detect astonishingly small amounts of soluble ligands including lipopolysaccharides and lipoproteins shed from or imbedded in the bacterial cell walls (4). In these cases, signaling occurs primarily at the cell surface, and no intracellular processing of the ligands is necessary.
Nevertheless, these surface TLRs are recruited to phagosomes during ingestion of microbes, although the significance of their recruitment to phagosomes has remained uncertain (5).
While TLRs sense stimuli that are outside of the cell or in the lumen of intracellular compartments, another class of innate immune receptors, the Nod-like receptors, is poised to detect microbial components in the cell cytoplasm (6). Nod1 and Nod2 are specifically important for detecting fragments of peptidoglycan (PGN) from bacterial cell walls (7, 8), and are implicated in host defense against a variety of intracellular and extracellular pathogens. In the case of pathogenic bacteria that penetrate the cytoplasm, Nod1 and Nod2 would have direct access to peptidoglycan and potential ligands. However, in the case of extracellular pathogens, the mechanism by which peptidoglycans are detected is not clear. It can be hypothesized that degradation of peptidoglycans within phagosomes releases fragments that are transported into the cytosol for detection by Nods. Indeed, Marina-Garcia et al. has demonstrated that soluble muramyl dipeptide (a ligand sensed through Nod2) must be endocytosed in order to signal effectively (9).
Staphylococcus aureus is a Gram-positive bacterium that is both a commensal organism colonizing human mucosal surfaces and a serious pathogen that can cause life-threatening mucosal and systemic infections. Mouse studies have demonstrated that both TLR2 (10) and Nod2 (11, 12) are important for controlling infection by S. aureus. Recently Ip et al. have demonstrated that phagocytosis of S. aureus by macrophages contributes to the activation of MyD88-dependent signaling (13). In this manuscript, we have made use of a variant of S. aureus that is especially sensitive to degradation by lysozyme inside macrophage phagosomes to explore the role of phagocytosis and phagosomal degradation in the activation of inflammatory responses through TLRs and Nod2. We recently made use of these bacteria to document an absolute requirement for phagosomal degradation of peptidoglycan in activation of the inflammasome and IL-1β secretion by macrophages (14). In the present study, we have determined that phagosomal degradation can be additionally important for determining the sensitivity of macrophages to live bacteria through TLRs. In contrast, Nod2 plays little or no role in detection of infection with live bacteria. Our results suggest that S. aureus cell walls contain TLR2 ligands that are directly accessible to macrophage surface receptors. At reasonably low multiplicities of infection, this accessible pool is insufficient to fully activate cytokine production by macrophages. S. aureus mutants that are susceptible to phagosomal degradation allow for additional activation of TLR9 that enhances and prolongs inflammatory responses to even small numbers of organisms.
Materials and Methods
Bacterial Strains
All experiments were done using wild-type S. aureus SA113 (WT-SA) and oatA mutant (ΔoatA-SA) kindly provided by Fiedrich Götz (15). S. aureus was grown in LB or THB at 37°C to stationary phase. Bacteria were diluted to OD600 =0.4, determined to be 1×108cfu/ml.
Mice and Cell culture
TLR2−/−, NOD2−/−, and MyD88−/− mice were bred and housed under SPF conditions in the Cedar-Sinai Medical Center animal facilit y. C57BL/6 mice were purchased from Jackson Laboratories (Bar Harbor, Maine). TLR9−/− bone marrow macrophages were derived from bones kindly provided by Dr. Miao (Institute for Systems Biology). Bone marrow derived macrophages (BMDM) were generated from femurs and tibias by culturing for 6-7 days in complete RPMI 1640 (10% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, 2 mM L-glutamine; Mediatech) suppemented with 10% L-cell conditioned media. BMDM were plated the night before infection. Infections were done by adding indicated MOIs of bacteria onto cells and spinning at 450xg for 5 min. The infection was allowed to progress for 30 min, the cells were washed and put in new media containing 100 μg/ml gentamycin to kill extracellular bacteria. Lysates were harvested at designated times, and cytokine levels were measured by ELISA (Biolegend and BD Biosciences).
Lysozyme digestion and treatment
Bacteria were grown overnight to stationary phase as described above. Bacteria were washed 1x, resuspended in PBS, and diluted to yield a OD595 =0.3 in a 200 μl volume in 96-well plate reader. WT-SA and ΔoatA-SA cultures were split, and 300 μg/ml fresh hen egg lysozyme (Sigma) was added to one culture of each. Cultures + lysozyme were incubated at 37°C with shaking, and a 200 μl aliquot was removed each hour to measure the OD595. Serial dilutions of the starting cultures and the 4 hour time point were plated to determine the cfu in the cultures. Percent survival was determined to be the percentage of cfu in the lysozyme-treated compared to untreated cultures at 4 hours. Each culture was passed through a 0.2 μm syringe filter to remove whole bacteria. BMDM were treated with 5, 10, 25, 50 μl of filtered lysate for 8-12 hrs. DNA was removed by incubating a portion of the lysate with RNase-free DNase I (Qiagen) at 2.5U/ml for 1 hour at 37°C.
Antibiotic Treatment
Wild-type SA113 bacteria were grown in Bacto-Todd Hewitt Broth (THB) for 18 hours at 37°C with shaking to reach stationary phase. The overnight culture was diluted in THB media to obtain 108CFU/ml by measuring 0.4 at OD600. The antibiotics, Vancomycin Hydrochloride (Acros Organics) or Linezolid (Pfizer), were prepared in ddH2O, filtered, and added to the bacteria at concentrations of 32, 4 and 0.5 μg/ml in RPMI 1640 + 10% FBS media for 2 hours at 37°C with shaking (Minimum inhibitory concentration for both antibiotics = 4 μg/ml). After treatment, the OD was re-adjusted, and bacteria were plated on THB-agar plates to measure bacterial concentration. Dilutions of treated bacteria were made to obtain multiplicity of infections (MOI) of 6, 12, 25 and 50 in RPMI 1640 + 10% FBS media, and added to 1×105 BMDM in 100 μl/well. Media containing Vancomycin or Linezolid, or media containing bacteria only were used as controls. The plates were centrifuged to settle bacteria at 500g for 5 min at room temperature, and incubated for 30 minutes at 37°C + 5% CO2. After infection, cells were washed four times in 150 μl prewarmed RPMI (no additions) to remove non-adherent extra-cellular bacteria, and 200 μl/well of freshly made Gentamycin (200 μg/ml) was added. BMDMs and intracellular bacteria were incubated at 37°C + 5% CO2 for a total of 24 hours. After specific time points, the plates were centrifuged at 500g for 5 min at room temperature to remove cellular debris, and supernatants were collected for analysis by ELISA. The cells were lysed using autoclaved distilled water, and intracellular survival was calculated by counting CFU on THB plates. Degradation sensitivity was assessed by centrifuging the antibiotic treated bacteria and resuspending in sterile PBS with or without 250 ng/ml lysostaphin (Sigma). Bacteria were incubated at 37°C and the OD600 was measured at different time points.
Internal Bacterial pH Determination
Bacteria were grown as above and 1 ml of overnight culture was washed 1x with sterile PBS. Bacteria were resuspended in 5ml of sterile PBS containing 1 μM CFDA-SE (CFSE) (Invitrogen) and incubated at 37°C for 15 minutes (17). Bacteria were washed with PBS+ 20% FCS 1x followed by PBS 1x. Bacteria were resuspended in 1ml PBS containing 20 μg/ml tetramethylrhodamine B isothiocyanate (TRITC) (Fluka) and incubated at 37°C for 15 minutes. Labeled bacteria were washed 7x with TBS + 10% FCS. The toxicity of the labeling was checked by determining the cfu with and without labeling which we found to have a negligible effect. The bacteria were then diluted to OD600 = 0.4 and infected at an MOI=25 as described above. At different times post-infection several TRITC and CFSE images were taken of both WT-SA and ΔoatA-SA infected cells under identical magnification and exposure settings using a Leica SP5 confocal microscope. A standard curve of the ratio of CFSE/TRITC vs. pH was established for each strain of bacteria by incubating each strain in buffers at different pHs containing 10 μM Nigericin (Calbiochem) to permeablize the bacteria. These images were collected with the same settings as the cell images. The fluorescence intensity of CFSE and TRITC were determined using the ImageJ software particle analysis tool and converted to pH using the standard curves.
Quantitative PCR
BMDM were plated and infected with each strain of bacteria as described above. At designated time points cells were lysed and RNA was isolated using Qiagen RNeasy kit following the manufactures protocol. cDNA synthesis and Taq-man real-time PCR reactions were done as previously described (18).
Results
The lysozyme-resistance of S. aureus peptidoglycan results in significantly reduced inflammatory responses by macrophages
In our previous investigations into the mechanisms by which S. aureus activates the NLRP3 inflammasome in macrophages and dendritic cells we utilized a bacterial strain deficient for the enzyme necessary for O-acetylation of PGN, a modification that normally renders S. aureus PGN highly resistant to lysozyme (15). We observed that live S. aureus and S. aureus PGN are poor activators of the NLRP3 inflammasome even at very high doses and that the lysozyme-resistance conferred by PGN O-acetylation was a key feature of S. aureus’s reduced inflammasome activation (14).
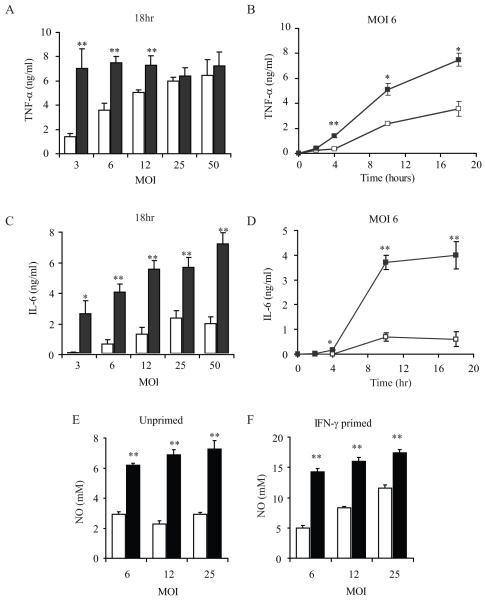
We realized that directly comparing degradation-resistant or degradation-sensitive bacterial strains could provide a unique way to define the role of phagosomal degradation in contributing to general innate inflammatory signaling. Specifically, we hypothesized that degradation could release ligands that activate macrophages and that as a result, macrophages would be more sensitive to bacteria that are degradable. We infected bone marrow-derived macrophages (BMDM) with various doses of wild-type (WT-SA) or O-acetyl transferase (oatA) deficient S. aureus (ΔoatA-SA) and measured TNF-α production. Consistent with our previous report, infection with high doses (MOI>25) of wild-type or mutant S. aureus stimulated similar amounts of TNF-α production (Fig. 1A). However, low dose infection with ΔoatA S. aureus (MOI<12) produced significantly more TNF-α than cells infected with the same dose of wild-type bacteria (Fig. 1A). The enhanced sensitivity of macrophages to ΔoatA-SA was apparent within 4 hours of infection and persisted for at least 18 hours (Fig. 1B).
Figure 1. The lysozyme resistance of S. aureus peptidoglycan reduces inflammatory responses by macrophages.
BMDM were infected with different MOI of WT-SA (open bars and squares) or ΔoatA-SA (black bars and squares) bacteria and supernatants were harvested at different time points to assess cytokine production in by ELISA. All experiments represent + SD of triplicates. After 18 hours of infection the ΔoatA-SA bacteria induced significantly higher amounts of TNF-α (A) and IL-6 (C) in an MOI dependent manner. The time course of TNF-α (B) and IL-6 (D) induction at a low MOI=6 shows similar early induction by WT-SA and ΔoatA-SA that diverges after 4 hours. BMDM, unprimed (E) or primed with IFN-γ for 24 hrs (F) were infected with WT-SA or ΔoatA-SA for 18 hr. Nitric oxide (NO) was measured by Griess assay. Statistics were done by student t-test * p<0.05, ** p<0.001.
We next analyzed whether other common inflammatory responses to S. aureus exhibited similarly enhanced sensitivity to degradable bacteria. Like TNF-α, IL-6 was induced more potently by the mutant bacteria, and this difference was greatest at lower levels of infection, although unlike TNF-α there was still a significant (2-3 fold) difference at high levels of infection (Fig. 1C). Like TNF-α, the induction of IL-6 by the wild-type and mutant bacteria was similar (and very low) at early time points and diverged strongly after 4 hours of infection (Fig. 1D). In addition to cytokines, we also looked at the induction of nitric oxide (NO) which has been shown to play a role in killing of S. aureus in mice (19, 20). Previous work has indicated that for optimal NO production BMDM need to be primed with IFN-γ (21), so we assessed NO in BMDM that were either unprimed or primed with IFN-γ for 24 hours. In both primed and unprimed BMDM the ΔoatA-SA bacteria induced on average 2-3 fold more NO than WT-SA (Fig. 1E and F). The combination of these results demonstrates that the lysozyme-resistance of S. aureus PGN significantly influences the level of proinflammatory responses mounted against the microbe and that this effect is most prominent at lower levels of microbial exposure.
TLR2 and TLR9 are both involved in sensing internalized S. aureus
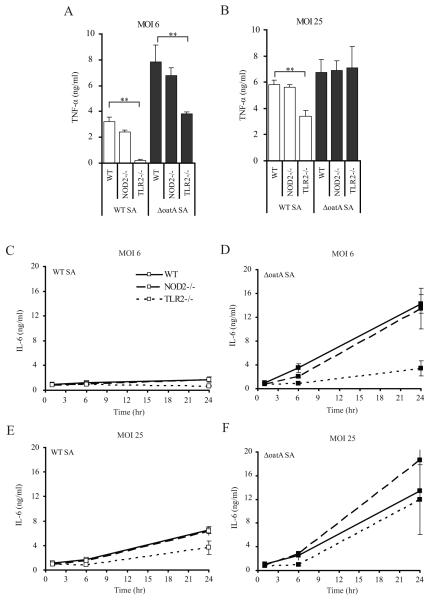
S. aureus has been shown to a possess TLR2 activating lipoproteins and lipoteichoic acid, and S. aureus PGN’s degradation products are able to activate responses through the cytoplasmic pattern recognition receptor Nod2. Therefore we compared the proinflammatory cytokine responses of Nod2- and TLR2-deficient BMDM to determine the relative contributions of each receptor to the proinflammatory responses induced by the WT-SA compared to the ΔoatA-SA. Considering the ΔoatA-SA mutation results in degradation-sensitive PGN, we expected the increased inflammation would be mediated through the detection of PGN degradation products by Nod2. However, we observed little requirement for Nod2 in macrophage responses to infection with live S. aureus (Fig. 2). When we examined TNF-α induction at 18 hours post infections, we found that the BMDM response to low MOI=6 of WT-SA was largely dependent on TLR2 (85%), while the enhanced response to ΔoatA-SA was only 50% dependent on TLR2 (Fig. 2A). This suggests that the ΔoatA-SA release a component that stimulates an additional inflammatory response through a mechanism other than TLR2. This additional component was unlikely to be MDP derived from PGN degradation, since the absence of Nod2 had little or no effect on TNF-α production in response to infection with wild-type or mutant bacteria. When cells were exposed to a high MOI of ΔoatA-SA bacteria (>25), TNF-α production was no longer dependent on TLR2 (Fig. 2B). This suggests that at high multiplicities of infection, the other signal is sufficient to induce the cytokine maximally, while at lower multiplicities of infection, both TLR2 and the other signal are required to drive maximal signal.
Figure 2. The contribution of TLR2 to the induction of TNF-α and IL-6 following S. aureus infection of BMDMs is dose-dependent, while Nod2 is not required.
WT-SA BMDM were infected with either a low dose (A) or a high dose (B) of WT-SA (open bars) or ΔoatA-SA (black bars), and TNF-α production was measured by ELISA after 24 hr. The kinetics of IL-6 production was also measured. BMDM from Nod2−/− (dashed) and TLR2−/− (dotted) mice were compared to wild-type C57BL/6 (black line) during low dose (MOI=6) (C and D) and high dose (MOI=25) (E and F) infection with either WT-SA (open squares) (C and E) or ΔoatA-SA (filled squares)(D and F). At different times post infection triplicate supernatants were analyzed by ELISA for IL-6 + SD. Statistics were done by student t-test * p<0.05, ** p<0.001.
Similar results were obtained when examining IL-6 production. During low dose infection (MOI=6) 76% of the IL-6 induced by ΔoatA-SA bacteria was TLR2-dependent (Fig. 2D). While the induction of IL-6 by WT-SA at an MOI=6 was dramatically lower than ΔoatA-SA, it was similarly TLR2-dependent (61%) (Fig. 2C). However, when the dose of ΔoatA-SA bacteria was increased to an MOI=25 there was no longer any dependence on TLR2 (Fig. 2F) while WT-SA induction of IL-6 was still 44% TLR2-dependent (Fig. 2E). Nod2 was not required for these responses.
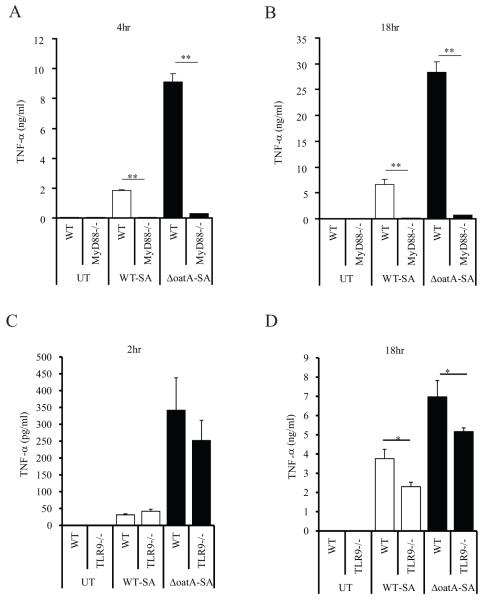
In order to identify principle mediators of this non-TLR2-dependent response, we infected MyD88-deficient cells with bacteria. TNF-α production in response to wild-type and mutant bacteria was entirely MyD88-dependent at an MOI=6 (Fig. 3A, B). Even at a high MOI=25, the response was still largely MyD88 dependent (data not shown). The lack of response in the MyD88−/− macrophages also supports our previous observation that Nod2 contributes very little to the response to S. aureus in macrophages (Fig. 2A, B). We therefore examined whether the additional response could come from TLR9 activated by DNA. TLR9-deficient cells respond similarly to wild-type and mutant bacteria at an early time point (Fig. 3C). However, later during infection optimal TNF-α production is dependent on TLR9, with TLR9-deficiency reducing cytokine production induced by ΔoatA-SA to levels identical to that induced by wild-type bacteria (Fig. 3D).
Figure 3. The role of MyD88 and TLR9 in detecting live S. aureus.
(A and B) BMDM from WT C57BL/6 mice and MyD88−/− mice were infected at an MOI=6 and TNF-α was assessed in the supernatant at 4 (A) and 18 (B) hours. All responses were MyD88-dependent. (C and D) BMDM from C57BL/6 and TLR9−/− mice were infected at and MOI=25. After 2 (C) and 18 (D) hours, TNF-α levels in the supernatant were measured by ELISA. TLR9 contributes to later responses induced by ΔoatA-SA. All data represent triplicates + SD. Statistics were done by student t-test * p<0.05, ** p<0.001.
Phagocytosis and phagosomal maturation are essential for the induction of maximal inflammatory responses to S. aureus
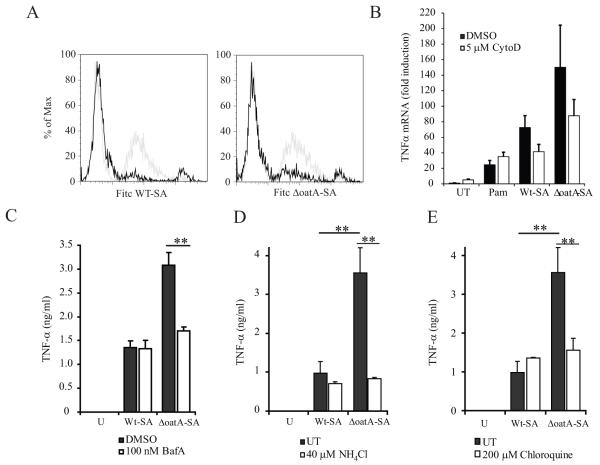
Taken together, the above data suggest that low dose infection of macrophages with ΔoatA-SA induces more cytokine because, unlike wild-type S. aureus, it is more susceptible to degradation and release of DNA within phagosomes. However, it is equally possible that ΔoatA-SA bacteria are simply more fragile than wild-type bacteria and that preparations of these bacteria contain more free DNA. To address this possibility, we first examined the requirement for phagocytosis and phagosome maturation. Macrophage internalization of wild-type and ΔoatA S. aureus was blocked by cytochalasin D, an inhibitor of the actin remodeling necessary for phagocytosis (Fig. 4A). Cytochalasin D strongly inhibited both basal TNF-α gene expression induced in response to low dose infection with wild-type bacteria and the enhanced response to mutant bacteria (Fig. 4B). Blockade of phagosome maturation with inhibitors of acidification including bafilomycin A, ammonium chloride, or chloroquine had no effect on TNF-α production in response to infection with wild-type bacteria, but completely inhibited the enhanced response to mutant bacteria reducing them to levels identical to wild-type (Fig. 4C-E). These data are consistent with internalization being important for both the initial TLR2-dependent signal (through efficient binding and capture of bacteria and concentration of TLR2 to new phagosomes) as well as a later phagosome maturation-dependent TLR9-dependent enhanced response to degradable mutant bacteria.
Figure 4. Phagocytosis is required for optimal detection of live S. aureus, while phagosome maturation is specifically required for enhanced response to mutant bacteria.
(A) BMDM were pretreated for 30 min with 5 μM cytoclasin D and infected with FITC-labeled WT-SA and ΔoatA-SA at MOI=25 for 30 minutes. Phagocytosis was assessed by flow cytometry. Light grey histogram represents phagocytosis by untreated BMDM and black histogram is in the presence of cytoclasin D. (B) BMDM were pretreated for 30 min with 5 μM cytoclasin D (ctyoD) or DMSO control and infected with WT-SA or ΔoatA-SA at an MOI=6 for 4hr. TNF-α gene expression was measured by QPCR. (C-E) BMDM were pre-treated for 1 hr with either 100 nM Bafilomycin A, 40 μM NH4Cl, 200 μM chloroquine to block phagasome acidification. Control samples were treated with DMSO for the case of Bafilomycin A or left untreated for treatment with the aqueous soluble NH4Cl and chloroquine. Cells were then infected with WT-SA or ΔoatA-SA at an MOI=6 for 4hrs. TNF-α was assayed by ELISA. All data represent measurements done in triplicate + SD and statistics were done by student t-test * p<0.05, ** p<0.001.
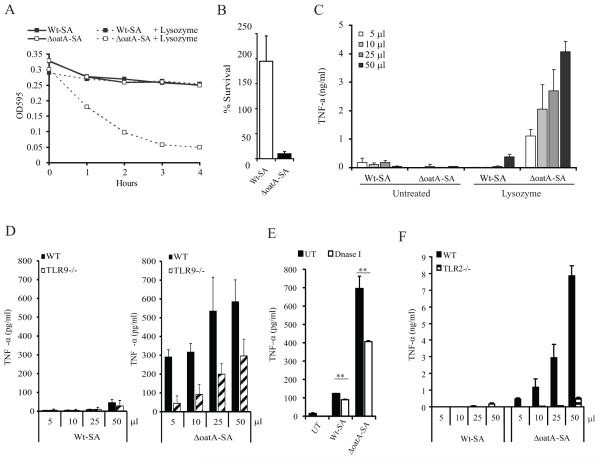
To formally determine whether significant amounts of free DNA contribute to the enhanced response to mutant bacteria, we compared activation of macrophages with culture supernatants of wild-type and mutant bacteria before and after exposure to lysozyme. As reported previously for these bacteria wild-type S. aureus are highly resistant to lysozyme (15), while ΔoatA-SA bacteria treated with lysozyme lost integrity as measured by a decrease in the optical density (O.D.) of the culture over time (Fig. 5A). As indicated by the drop in O.D., the lysozyme-treated ΔoatA-SA showed a 90% reduction in viability whereas wild-type bacteria actually grew (Fig. 5B). These cultures were then filtered to remove any intact bacteria, and increasing amounts of the supernatants were used to stimulate BMDM. After 4 hours, only the ΔoatA-SA supernatants induced TNF-α and this induction was completely dependent on lysozyme digestion (Fig. 5C). Treating TLR9−/− cells with lysozyme-degraded bacterial supernatants demonstrated a significant release of DNA from only degradable ΔoatA-SA bacteria (Fig. 5D). This was further confirmed by treating the lysozyme digestion supernatants with DNase I which resulted in a decrease in TNF-α inducing capacity (Fig. 5E). We also assessed the lysozyme supernatants for TLR2 stimulating activity and surprisingly found an almost complete dependence on TLR2 (Fig. 5F). Data shown represent the relative roles of TLR2 and TLR9 respectively, although the absolute concentrations of cytokines measured varied with different batches of cells and stimuli. Taken together these data suggest that under these conditions using these complex soluble agonists TLR9 enhances a dominantly TLR2-dependent signal. Most importantly for the purposes of this study, these results clearly establish that mutant bacteria do not spontaneously release unusual amounts of stimulatory DNA and that lysosomal digestion is essential for release of TLR9 stimulatory material (Fig. 5C).
Figure 5. Lysozyme-resistance of S. aureus peptidoglycan reduces inflammatory responses by sequestering TLR ligands.
(A) ΔoatA-SA is sensitive to lysozyme while WT-SA resists degradation by lysoszyme. Overnight cultures of WT-SA and ΔoatA-SA were diluted to an OD595=0.3 in PBS + 300 μg/ml lysozyme. The survival of each bacterial strain was tracked by measuring the OD595 over 4 hours. (B) The lysozyme sensitivity of the ΔoatA-SA compared to WT-SA was confirmed plating serial dilutions of each culture at 0 hr and 4 hr to determine the number of colony forming units (cfu). The data is displayed as the percentage of cfu in the lysozyme treated compared to the untreated of each strain at 4 hr. (C) Lysozyme digestion releases inflammatory ligands from ΔoatA-SA but not WT-SA. After 4 hours, cultures of WT-SA and ΔoatA-SA were filtered to remove whole bacteria, and increasing volumes of supernatant were added to BMDM. After 12 hr supernatants were assayed for TNF-α by ELISA. (D) WT and TLR9−/− BMDM were treated with lysates from lysozyme-digested WT-SA and ΔoatA-SA. After 12 hr supernatants were harvested and the amount of TNF-α induced was measured by ELISA. (E) Filtered supernatants from lysozyme-digested WT-SA and ΔoatA-SA were treated with DNase I for 1hr at 37°C. BMDM were treated with 50 μl overnight. (F) WT and TLR2−/− BMDM were treated as in D. Data represent +SD of triplicate samples.
Inflammatory responses to bacteria occur in two phases
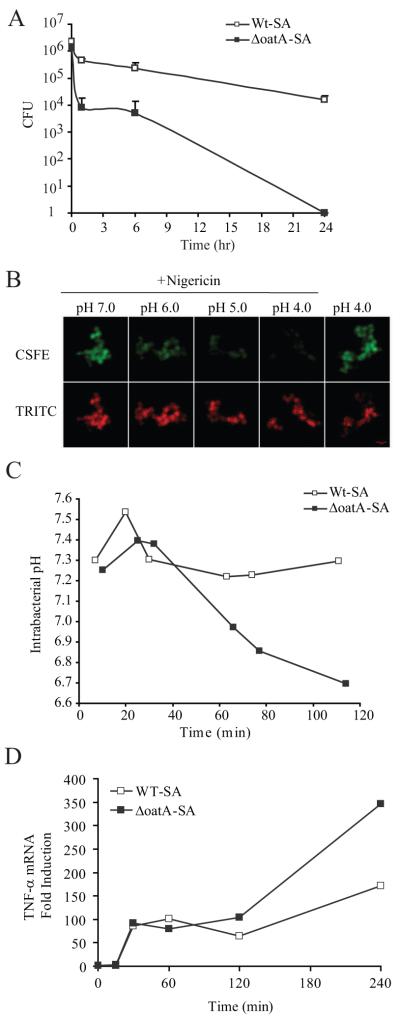
We next explored whether we could directly visualize breakdown of the bacterial cell wall within phagosomes and determine whether the timing of this would be consistent with when the enhanced response to mutant bacteria develops. When macrophages are infected in vitro with S. aureus, the bacteria fail to grow, but they are poorly killed by the macrophages with about a 1 log reduction in bacteria numbers over 24 hours (Fig. 6A). ΔoatA-SA, however, suffer a greater than 100 fold reduction in cell numbers after just 1 hour. To measure bacterial breakdown in real time, we opted for a strategy in which we loaded bacteria with the cytosolic fluorescent dye carboxyfluorescein succinimidyl ester (CFSE). CFSE, being a fluorescein-based probe, exhibits pH-dependent fluorescence. We also labeled the cell walls with the pH-insensit ive probe tetramethylrodamine isothiocyanate (TRITC). When these bacteria were exposed to buffers at different pH values, the bacteria maintained the cytosolic pH near neutral in buffers as low as pH 4.0 as illustrated by the visible CFSE fluorescence (Fig. 6B). When the bacterial membrane becomes compromised as demonstrated by adding pore-forming toxin, nigericin, CFSE fluorescence is lost as the pH of the buffer decreases (Supplemental Fig. 1A). When macrophages were infected with labeled live wild-type bacteria, the average CFSE fluorescence remained high for at least 2 hours (Fig. 6C), indicating a neutral internal bacterial pH, consistent with the observation that they are killed poorly. In contrast, CFSE fluorescence in ΔoatA bacteria remained high for up to 40 minutes, but then started to decrease as it became exposed to the acidic environment of the phagolysosome from 60 minutes (Supplemental Fig. 1B), indicating breakdown of the bacterial cell wall (Fig. 6C). The fact that the observed pH drops to just 6.7 in the first two hours does not indicated that the phagosomal pH is 6.7 but instead reflects the fact that during this interval only a subset of bacteria completely lose cell wall integrity. To directly examine whether this timing coincides with the enhanced sensing of bacterial products, we examined TNF-α mRNA production by RT-PCR. For the first hour of infection with live bacteria, BMDMs make identical amounts of TNF-α mRNA in response to wild-type and ΔoatA bacteria (Fig. 6D). However, consistent with the release of additional stimuli by cell wall breakdown, ΔoatA bacteria begin to trigger more TNF-α beyond 1 hour.
Figure 6. The higher inflammatory response to ΔoatA-SA is a result of secondary innate receptor activation following digestion of the bacteria.
(A) BMDM were infected with WT-SA (closed squares) and ΔoatA-SA (open squares) at an MOI=6 for 30 min and washed to removed extracellular bacteria. One set of wells was lysed in water immediately to get t=0 hours time point. After lysis in water, serial dilutions of the lysates were plated for colony forming units (CFU). All time points represent triplicate readings +SD. (B) Live WT-SA or ΔoatA-SA were labeled intracellularly with CFSE and extracellularly with TRITC. Labeled bacteria were incubated in pH 4, 5, 6, and 7 buffers with or without permeabilization with 10 μM nigericin. Permeabilized bacteria showed decreased fluorescence of pH-sensitive CFSE with no effect on pH-insensitive TRITC fluorescence. (C) BMDM were infected with labeled WT-SA and ΔoatA-SA, and bacterial intracellular pH was measured by confocal microscopy for 2 hr. The ratio of CFSE/TRITC was compared to a standard curve to determine internal pH (pHi) of each bacterium. Average pHi was plotted verses time. (D) BMDM infected at an MOI=6 with WT-SA or ΔoatA-SA. At each time point TNF-α gene induction was measured by Taqman real-time PCR and normalized to EF-1α induction. Data are displayed as fold induction over unstimulated BMDM.
Antibiotics targeting cell wall peptidoglycan enhance macrophage sensitivity to bacteria
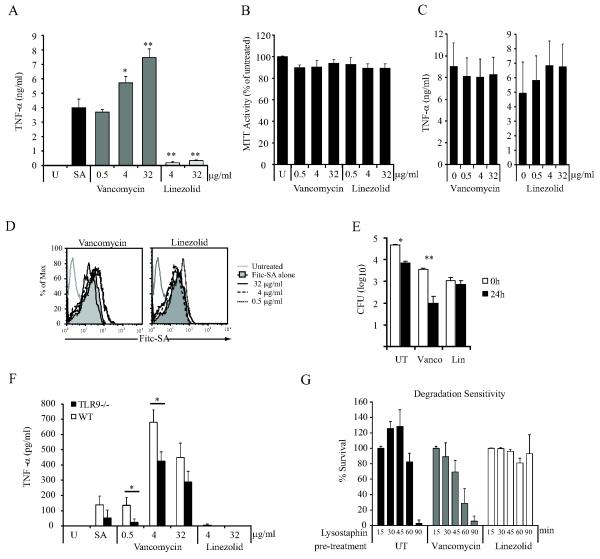
Comparison of degradation-resistant and degradation-sensitive bacteria provided a useful opportunity to directly test the hypothesis that phagolysosomal degradation of bacteria enhances the sensitivity of innate immune detection mechanisms utilized by macrophages. ΔoatA-SA are sensitive to phagolysosomal degradation due to an alteration in the cell wall peptidoglycan. This prompted us to investigate whether other perturbations of normal bacterial cell wall architecture might make bacteria more inflammatory. We hypothesized that antibiotics that specifically disrupt cell wall architecture in S. aureus might be significantly more inflammatory than antibiotics that inhibit other cellular processes since cell wall destabilization might facilitate bacterial degradation in phagolysosomes. To test this hypothesis, we treated S. aureus with vancomycin, an antibiotic that inhibits peptidoglycan synthesis (22), or linezolid, an antibiotic that inhibits protein synthesis (23) for 2 hours. These bacteria were allowed to infect BMDM, and secretion of TNF-α was measured. Consistent with our hypothesis, we observed that vancomycin-treated bacteria stimulated significantly more TNF-α secretion than either linezolid-treated bacteria or WT-SA alone (Fig. 7A). The decreased responsiveness to linezolid-treated bacteria was not due to adverse affects of the antibiotic on the BMDM as there was no significant cell death (Fig. 7B) or change in macrophage responses to purified PGN (Fig. 7C). Nor could the differences in responses be a consequence of reduced bacterial uptake as FITC-labeled bacteria treated with either vancomycin or linezolid were equally phagocytosed by BMDM (Fig. 7D).
Figure 7. Treatment of S. aureus with an antibiotic that disrupts cell wall synthesis enhances innate detection of the organism.
WT-SA (SA113) were treated with either vancomycin or linezolid at 0.5, 4, or 32 μg/ml for 2 hr. The density of each culture was balanced to an OD600 =0.4 and an equal amount was added onto BMDM at MOI=6. (A) The cultures were incubated for 18hr and the level of TNF-α induction was measured by ELISA. (B) To rule out adverse effects of the antibiotics on the BMDM, BMDM were treated with vancomycin and linezolid for 18 hr and the metabolic activity of the cells was measured by MTT assay. (C) BMDM were stimulated with 10 μg/ml PGN in the presence of increasing concentrations of vacomycin or linezolid for 18 hr. TNF-α production was measured by ELISA. (D) To determine the effects of antibiotic treatment on the recognition and phagocytosis of WT-SA, vancomycin- and linezolid-treated bacteria were labeled with FITC for 15 min immediately after antibiotic treatment. The labeled bacteria were used to infect BMDM (MOI=25) for 30 min. The cells were washed and phagocytosis was assessed by flow cytometry. (E) To determine whether either antibiotic influenced the ability of macrophages to kill the bacteria, we assessed intracellular survival of antibiotic-sensitized bacteria. WT-SA after antibiotic treatment of bacteria BMDM were infected (MOI=6) and surviving bacterial cfus were measured at 0 and 24 hr as in fig. 6. WT-SA (F) The increased inflammatory response to vancomycin-treated S. aureus is partially dependent on TLR9. WT and TLR9−/− macrophages were infected with WT-SA that was untreated (SA) or pretreated for 2h with indicated concentrations of vancomycin or linezolid. (G) WT-SA was treated with 32 μg/ml of vancomycin or linezolid as above followed by treatment with low dose lysostaphin (250 ng/ml) at 37°C. Bacterial degradation was monitored by measuring OD600 and plotted as the percent survival compared to no lysostaphin for each pre-treatment.
While at the highest dose both vancomycin and linezolid did kill some bacteria, when similar numbers of live bacteria were added to BMDM, vancomycin-sensitized bacteria were more susceptible to killing by macrophages (Fig. 7E). Linezolid-sensitized bacteria remained highly resistant to killing by macrophages. The enhanced sensitivity of macrophages to vancomycin-treated bacteria was partly dependent on TLR9 (Fig. 7F), consistent with the idea that the enhanced sensitivity arises from phagosomal breakdown of the bacterial cell wall. To further support this conclusion we exposed the antibiotic-treated bacteria with lysostaphin, and observed that vancomycin specifically makes the bacteria more susceptible to degradation while linezolid made the cells more resistant to degradation (Fig. 7G). These differences in degradation may explain the observed alterations in TNF-α induction by antibiotic-treated bacteria.
Discussion
Innate inflammatory signals are the culmination of the activation of several surface and intracellular receptors. By utilizing degradation sensitive mutants of S. aureus, this study provides evidence for the importance of macrophage lysosomal degradation in the induction of maximal inflammatory responses to live bacteria including the induction of TNF-α IL-6 and nitric oxide. Our examination of the responses to a degradation sensitive mutant of S. aureus has revealed that macrophage inflammatory responses occur in two phases. The initial response to S. aureus is a TLR2-driven response to ligands exposed on the surface of the bacteria, occurring within the first hour of infection, which our experiments found were equivalent between wild-type and the degradation-sensitive ΔoatA S. aureus. In addition, while these initial responses required phagocytosis they were independent of phagosome maturation. After phagocytosis, the lysozyme-mediated degradation of the cell wall peptidoglycan (PGN) of ΔoatA-SA mutant but not the wild-type S. aureus releases additional TLR2 as well as TLR9 ligands and leads to a second burst of inflammatory cytokines. Further, we demonstrated the secondary induction of TNF-α transcription by ΔoatA-SA occurred concurrent with the death/cell wall disruption of the bacteria inside the macrophage phagosome. We were also able to mimic enhanced inflammatory responses by treating wild-type S. aureus with PGN synthesis disrupting antibiotics.
Our data are in agreement with recent work by Ip et al. showing that for bacterial pathogens the availability of TLR ligands varies considerably between different bacterial species and correlates with whether the bacterium is gram positive or negative (13). While the work by Ip et al. suggests that the responses area to S. aureus are completely degradation-dependent, we have shown using a specific degradation-sensitive mutant that only the second wave of responses are dependent on degradation. This difference is likely due to the fact that the Ip et al. findings are based on treatment with heat-killed wild-type bacteria while we have exclusively used live bacteria. Heat-killing might denature the cell wall in a way that buries TLR2 ligands such that they are harder for the receptor to see without proteolytic processing of the cell wall or without concentrating TLR2 in phagosomal membranes. It is widely appreciated that S. aureus stimulates TLR2, but what is additionally revealed by our data is that being resistant to degradation significantly limits the access of TLRs to S. aureus ligands. Contrary to our results, previous work had found no role for TLR9 in sensing heat-killed S. aureus (24). But by using degradation-sensitive mutants, we have characterized an additional role for TLR9 in sensing degradation of live bacteria that is specifically subverted by resistance to phagosomal degradation. Studies with bacterial lysates hint at synergy between TLR2 and TLR9 signaling such that TLR9 signaling might enhance a predominantly TLR2-dependent signaling. However, data using whole live bacteria do not necessarily suggest synergistic signaling and suggest a simpler model in which both receptors simply add to the net inflammatory response.
Considering the bacteria we utilize in our experiments have a mutation specifically affecting the degradability of their PGN, we had anticipated that the degradation of S. aureus PGN would be integral for the importance of Nod2 signaling in S. aureus infections. However, when comparing wild-type and a degradation-sensitive mutant of S. aureus, Nod2 played very little role in the difference in inflammatory responses that we measured. Reports in the literature regarding an in vivo role for Nod2 in the sensing of S. aureus have lead to varying conclusions regarding the significance of the role Nod2 plays in survival following infection (11, 12). The most consistent finding suggests Nod2 contributes to overall cytokine levels in the bronchial lavage fluid and blood, which is affected by the overall cellularity of the tissue. However, when we consider Nod2’s role on a cellular level, specifically in macrophages, our data demonstrating a minimal role for Nod2 in cytokine induction are in line with currently published work. Typically, artificial activation of Nod2 with pure muramyl-dipeptide (MDP) only shows an affect when synergizing with another TLR ligand resulting in a modest increase in inflammatory cytokine production (9, 25). But there is little evidence for a significant contribution for Nod2 in macrophage responses to live S. aureus. In the case of our degradation-sensitive mutant, it is likely that the release of additional TLR2 and TLR9 ligands during degradation minimizes the overall impact of losing Nod2’s synergistic effect in the Nod2−/− macrophages, accounting for the lack of a significant impact on cytokine production.
It has become clear from our research that when a macrophage encounters small numbers of bacteria the exposure of surface TLR ligands is limited. However, through lysosomal degradation and response to secondary ligands the macrophage is able to extend and maximize the inflammatory response. The importance degradation plays in the immune response is supported by the numerous mechanisms pathogens have evolved to inhibit lysosomal fusion, acidification, and lysosomal enzymes necessary to release additional ligands and enhance innate signaling (26). Interestingly, we were also able to mimic our mutant bacteria by treating wild-type bacteria with the PGN synthesis inhibiting antibiotic vancomycin. Our ability to modulate the bacteria inflammatory capacity suggests that targeting the degradability of pathogens may be a useful means of modulating the inflammatory outcome of an infection, just as it plays an important role in immune evasion by bacteria.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health (R01 GM085796 to D.M.U. and F32 GM093658 to A.J.W.)
References
- 1.Aderem A, Underhill DM. Mechanisms of phagocytosis in macrophages. Annu Rev Immunol. 1999;17:593–623. doi: 10.1146/annurev.immunol.17.1.593. [DOI] [PubMed] [Google Scholar]
- 2.Greenberg S, Grinstein S. Phagocytosis and innate immunity. Curr Opin Immunol. 2002;14:136–145. doi: 10.1016/s0952-7915(01)00309-0. [DOI] [PubMed] [Google Scholar]
- 3.Saitoh S, Miyake K. Regulatory molecules required for nucleotide-sensing Toll-like receptors. Immunol Rev. 2009;227:32–43. doi: 10.1111/j.1600-065X.2008.00729.x. [DOI] [PubMed] [Google Scholar]
- 4.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 5.Underhill DM, Ozinsky A, Hajjar AM, Stevens A, Wilson CB, Bassetti M, Aderem A. The Toll-like receptor 2 is recruited to macrophage phagosomes and discriminates between pathogens. Nature. 1999;401:811–815. doi: 10.1038/44605. [DOI] [PubMed] [Google Scholar]
- 6.Shaw MH, Reimer T, Kim YG, Nunez G. NOD-like receptors (NLRs): bona fide intracellular microbial sensors. Curr Opin Immunol. 2008;20:377–382. doi: 10.1016/j.coi.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chamaillard M, Hashimoto M, Horie Y, Masumoto J, Qiu S, Saab L, Ogura Y, Kawasaki A, Fukase K, Kusumoto S, Valvano MA, Foster SJ, Mak TW, Nunez G, Inohara N. An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat Immunol. 2003;4:702–707. doi: 10.1038/ni945. [DOI] [PubMed] [Google Scholar]
- 8.Girardin SE, Boneca IG, Carneiro LA, Antignac A, Jehanno M, Viala J, Tedin K, Taha MK, Labigne A, Zahringer U, Coyle AJ, DiStefano PS, Bertin J, Sansonetti PJ, Philpott DJ. Nod1 detects a unique muropeptide from gram-negative bacterial peptidoglycan. Science. 2003;300:1584–1587. doi: 10.1126/science.1084677. [DOI] [PubMed] [Google Scholar]
- 9.Marina-Garcia N, Franchi L, Kim YG, Hu Y, Smith DE, Boons GJ, Nunez G. Clathrin- and dynamin-dependent endocytic pathway regulates muramyl dipeptide internalization and NOD2 activation. J Immunol. 2009;182:4321–4327. doi: 10.4049/jimmunol.0802197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takeuchi O, Hoshino K, Akira S. Cutting edge: TLR2- deficient and MyD88-deficient mice are highly susceptible to Staphylococcus aureus infection. J Immunol. 2000;165:5392–5396. doi: 10.4049/jimmunol.165.10.5392. [DOI] [PubMed] [Google Scholar]
- 11.Deshmukh HS, Hamburger JB, Ahn SH, McCafferty DG, Yang SR, Fowler VG., Jr. Critical role of NOD2 in regulating the immune response to Staphylococcus aureus. Infect Immun. 2009;77:1376–1382. doi: 10.1128/IAI.00940-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kapetanovic R, Jouvion G, Fitting C, Parlato M, Blanchet C, Huerre M, Cavaillon JM, Adib-Conquy M. Contribution of NOD2 to lung inflammation during Staphylococcus aureus-induced pneumonia. Microbes Infect. 12:759–767. doi: 10.1016/j.micinf.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 13.Ip WK, Sokolovska A, Charriere GM, Boyer L, Dejardin S, Cappillino MP, Yantosca LM, Takahashi K, Moore KJ, Lacy-Hulbert A, Stuart LM. Phagocytosis and phagosome acidification are required for pathogen processing and MyD88-dependent responses to Staphylococcus aureus. J Immunol. 184:7071–7081. doi: 10.4049/jimmunol.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shimada T, Park BG, Wolf AJ, Brikos C, Goodridge HS, Becker CA, Reyes CN, Miao EA, Aderem A, Gotz F, Liu GY, Underhill DM. Staphylococcus aureus evades lysozyme-based peptidoglycan digestion that links phagocytosis, inflammasome activation, and IL-1beta secretion. Cell Host Microbe. 7:38–49. doi: 10.1016/j.chom.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bera A, Herbert S, Jakob A, Vollmer W, Gotz F. Why are pathogenic staphylococci so lysozyme resistant? The peptidoglycan O- acetyltransferase OatA is the major determinant for lysozyme resistance of Staphylococcus aureus. Mol Microbiol. 2005;55:778–787. doi: 10.1111/j.1365-2958.2004.04446.x. [DOI] [PubMed] [Google Scholar]
- 16.Hornung V, Bauernfeind F, Halle A, Samstad EO, Kono H, Rock KL, Fitzgerald KA, Latz E. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat Immunol. 2008;9:847–856. doi: 10.1038/ni.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vander Top EA, Perry GA, Gentry-Nielsen MJ. A novel flow cytometric assay for measurement of in vivo pulmonary neutrophil phagocytosis. BMC Microbiol. 2006;6:61. doi: 10.1186/1471-2180-6-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gantner BN, Simmons RM, Canavera SJ, Akira S, Underhill DM. Collaborative induction of inflammatory responses by dectin-1 and Toll-like receptor 2. J Exp Med. 2003;197:1107–1117. doi: 10.1084/jem.20021787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sakiniene E, Bremell T, Tarkowski A. Inhibition of nitric oxide synthase (NOS) aggravates Staphylococcus aureus septicaemia and septic arthritis. Clin Exp Immunol. 1997;110:370–377. doi: 10.1046/j.1365-2249.1997.4431456.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sasaki S, Miura T, Nishikawa S, Yamada K, Hirasue M, Nakane A. Protective role of nitric oxide in Staphylococcus aureus infection in mice. Infect Immun. 1998;66:1017–1022. doi: 10.1128/iai.66.3.1017-1022.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Totemeyer S, Sheppard M, Lloyd A, Roper D, Dowson C, Underhill D, Murray P, Maskell D, Bryant C. IFN-gamma enhances production of nitric oxide from macrophages via a mechanism that depends on nucleotide oligomerization domain-2. J Immunol. 2006;176:4804–4810. doi: 10.4049/jimmunol.176.8.4804. [DOI] [PubMed] [Google Scholar]
- 22.Walsh C. Deconstructing vancomycin. Science. 1999;284:442–443. doi: 10.1126/science.284.5413.442. [DOI] [PubMed] [Google Scholar]
- 23.Swaney SM, Aoki H, Ganoza MC, Shinabarger DL. The oxazolidinone linezolid inhibits initiation of protein synthesis in bacteria. Antimicrob Agents Chemother. 1998;42:3251–3255. doi: 10.1128/aac.42.12.3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kapetanovic R, Nahori MA, Balloy V, Fitting C, Philpott DJ, Cavaillon JM, Adib-Conquy M. Contribution of phagocytosis and intracellular sensing for cytokine production by Staphylococcus aureus- activated macrophages. Infect Immun. 2007;75:830–837. doi: 10.1128/IAI.01199-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Volz T, Nega M, Buschmann J, Kaesler S, Guenova E, Peschel A, Rocken M, Gotz F, Biedermann T. Natural Staphylococcus aureus- derived peptidoglycan fragments activate NOD2 and act as potent costimulators of the innate immune system exclusively in the presence of TLR signals. FASEB J. 24:4089–4102. doi: 10.1096/fj.09-151001. [DOI] [PubMed] [Google Scholar]
- 26.Pieters J. Evasion of host cell defense mechanisms by pathogenic bacteria. Curr Opin Immunol. 2001;13:37–44. doi: 10.1016/s0952-7915(00)00179-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.