Abstract
The endoplasmic reticulum (ER) is a large, singular, membrane-bound organelle that has an elaborate 3-D structure with a diversity of structural domains. It contains regions that are flat and cisternal, ones that are highly curved and tubular, and others adapted to form contact with nearly every other organelle and with the plasma membrane. ER 3-D structure is determined by both integral ER membrane proteins and by interactions with the cytoskeleton. Here, we describe some of the factors that are known to regulate ER structure and discuss how this structural organization, and the dynamic nature of the ER membrane network, allows it to perform its many different functions.
Overview of ER shape
The ER is the largest membrane-bound organelle in the eukaryotic cell. It is spread throughout the cytoplasm as one continuous membrane-enclosed network that surrounds a single lumen. Domains of the ER have many different shapes and include regions that are flat, tubular, and somewhere in between, and also structures that are adapted to contact other membrane-enclosed compartments. The ER must remain connected while maintaining a shape that will allow other cytoplasmic components to diffuse around it. Only a handful of factors have been discovered that regulate ER structure. Here, we will discuss recent advances in our understanding of how ER membrane proteins, the cytoskeleton, and factors that mediate inter-organelle contact together generate the 3-D structure of this elaborate, dynamic, and functionally complex organelle.
How ER shape contributes to function
The ER performs a variety of functions in cells, including but not limited to lipid synthesis, Ca2+ handling, and protein translocation and secretion [1]. To perform its many functions, the eukaryotic ER has perhaps the most complicated structure of any organelle. Some ER domains are obvious and can be distinguished by their shapes using fluorescence microscopy. These include the nuclear envelope (NE), and cytoplasmic cisternae and tubules that form the interconnected peripheral ER (Figure 1).
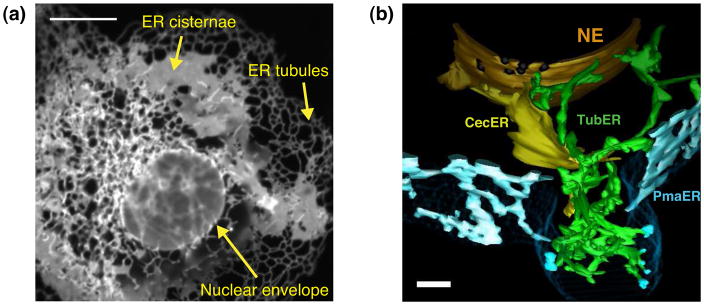
Figure 1. ER morphological domains in mammalian cells and yeast.
(a)Cos-7 cell expressing the ER membrane marker GFP-Sec61β. The ER in mammalian cells is a continuous membrane network that shares a lumen. This network consists of the nuclear envelope (NE) and peripheral ER cisternae and tubules. (b) EM tomography reveals the 3-D organization of the ER in budding yeast cells at nanometer resolution. The NE, central cisternal ER (CecER), tubular ER (TubER) and PM-associated ER (PmaER) are shown. Note that the NE is continuous with the pmaER through domains that are both tubular and cisternal. Scale bars: (a) 10 μm; (b) 100 nm. Image in (b) adapted with permission from J. Cell Biol. [4].
The NE is a distinct domain of the ER made up of two large, flat membrane bilayers, the inner and outer nuclear membranes (INM and ONM). The INM and ONM are separated by the perinuclear space (PNS), but are connected to each other at nuclear pores [2]. The peripheral ER branches out of the ONM as an extensive network of cisternae and tubules and extends into the cytoplasm all the way to the plasma membrane (PM). The lumen of the peripheral ER is continuous with the lumen of the PNS. What discriminates ER tubules from cisternae is their very different shapes. ER tubules have high membrane curvature at their cross-section, whereas cisternae are comprised of extended regions of parallel flat membrane bilayers that are stacked over each other with regions of membrane curvature found only at their edges. There are, however, similarities between ER tubules and cisternae; specifically, the diameter of an ER tubule is similar to the thickness of an ER cisterna (~38 nm vs. ~36 nm, respectively in yeast) [3, 4].
Multiple regions of high membrane curvature in the ER are stabilized by the reticulon and DP1/Yop1 (also called REEP) family of proteins; these regions include the tubules and edges of cisternae and fenestra (Box 1) [4, 5]. The abundance of reticulon/DP1/Yop1 proteins regulates the abundance of high curvature regions within the peripheral ER [4–9]. It is not currently known, however, how the expression level or activity of individual members of this family of membrane-shaping proteins is regulated to fine-tune ER shape. In mammalian cells, there are multiple spliced isoforms of four reticulon family members; each contains a reticulon homology domain, but the members have unique domains, interactions, and expression patterns that likely allow for particular ER tubule arrangements and might confer additional functions (for review, see [10]). Additionally, the prevalence of cisternae versus tubules can vary, suggesting that ER shape can be adapted to a cell’s ER functional requirements [11]. For example, secretory cells, such as those in the pancreas, must translocate and secrete a large number of proteins and these cells contain abundant cisternae that are densely studded with ribosomes. In contrast, the ER in muscle cells must rapidly regulate Ca2+ levels during contraction and these cells are enriched in ER tubules that are devoid of ribosomes. In yeast cells, which contain both ER cisternae and tubules, cisternae have a higher ribosome density than tubules (~600–1100/μm2 vs. ~250–400/μm2, respectively) [4]. These correlations suggest that either cisternae are better suited for ribosome binding and/or ribosome binding stabilizes cisternal ER structure. In animal cells, proteins associated with the translocation machinery are also enriched in cisternae [8]. Furthermore, changing ER ribosome density can alter ER shape, as treatment of animal cells with puromycin causes ribosome displacement followed by loss of ER cisternae and gain of ER tubules [12]. These data suggest that ER cisternae may be the preferred site of protein translocation.
Box 1. How ER tubules and sheets get their shape.
The peripheral ER in most cells contains a mixture of interconnected membrane tubules and cisternae. The relative amount of tubules versus cisternae depends to a large extent on the proteins that regulate ER membrane curvature, the reticulons and DP1/Yop1. These proteins are conserved integral membrane proteins that can be found in all eukaryotes. They partition exclusively into regions of the peripheral ER that have high membrane curvature, which includes the edges of cisternae as well as tubules ([5, 8, 9, 13, 63] and Figure Ib). Initial work both in vitro and in vivo identified these proteins as the major factors necessary for organizing the ER membrane bilayer into the shape of a tubule [5, 64], but they also organize membrane curvature at the edges of cisternae and fenestrations [4].
Reticulons contain a reticulon homology domain (RHD) comprised of two long (30–35 amino acid) transmembrane domains, separated by a soluble linker. These transmembrane domains insert as hairpins in the cytoplasmic leaflet of the ER membrane bilayer ([5] and Figure Ia). The short length of these hairpin domains is required in animal cells for: i) partitioning into regions of membrane curvature and ii) generating membrane curvature [65, 66]. Reticulon proteins also oligomerize into immobile higher-ordered structures in the ER membrane, a requirement for proper tubule formation [67]. Therefore, a reasonable model of reticulon function is that they form hairpin structures that oligomerize in the outer leaflet of the ER membrane to increase outer leaflet area relative to inner leaflet area, thus generating membrane curvature by altering bilayer symmetry.
In contrast, relatively little is understood about ER cisternae. These domains are comprised of flat areas of ER membrane that are evenly spaced around the ER lumen. At the edges the flattened membrane curves around, forming a pocket. Climp63 partitions into ER cisternae and its overexpression propagates the formation of cisternal ER at the expense of tubules [8]. When Climp63 is depleted, cisternae persist, but their intra-lumenal spacing is altered [8]. These data suggest that while Climp63 is not required for cisternae formation, it may form intra-lumenal linker complexes that regulate cisternal dimensions.
A recent paper showed that an additional function of ER cisternae is in the unfolded protein response (UPR). This study demonstrated that in yeast, ER stress-induced UPR leads to membrane expansion of the ER into cisternae [13]. The cisternal shape is not what allows the stress response, however, because conversion of sheets to tubules does not inhibit stress alleviation [13]. The authors proposed a model where the important feature of membrane expansion is increased ER volume to allow for additional handling of misfolded proteins. Recent work has shown that the volume-to-surface area ratio of such cisternae in wild type cells exceeds that of ER tubules [4], indicating that the cisternae formed upon ER stress are the most favorable conformation to meet the need for increased ER volume.
Whether ER tubules are enriched for other ER functions remains to be determined. For a long time, it was thought that because ER cisternae are the preferred site of ER protein translocation, then ER tubules must perform the lipid synthesis and Ca2+ handling functions of the ER. There is no evidence that either process localizes preferentially to ER tubules, however. ER tubules might be the preferred site for ER vesicle budding because the high curvature of ER tubules could be better suited for the formation of ER-to-Golgi transport vesicles (COPII) at ER exit sites (ERES). Markers for ERES localize to ER tubules [14], although it is not known if these markers are excluded from ER cisternae. It has also been difficult to demonstrate whether other specific ER functions preferentially localize to either cisternae or tubules. Tubules in the ER might simply provide a useful architecture to allow distribution throughout the cytoplasm as a continuous network without disrupting general diffusion and trafficking. In addition to cisternae and tubules, other peripheral ER subdomains defined by their unique functions and locations include the ERES as well as regions that contact the PM, organelles, and the cytoskeleton. What makes these domains difficult to study is that they can only be visualized by electron microscopy or by using multiple fluorescent markers and confocal microscopy in well-resolved cells. We will discuss ER domains that form contact sites with other organelles later in this review.
The maintenance of ER continuity through membrane fusion
Unlike every other organelle, the ER does not undergo regulated fragmentation or division. Even during cell division, the ER remains continuous [15, 16]. Recent work identified components of the ER fusion machinery. The atlastin proteins (and their yeast homolog Sey1) belong to a large, ER membrane-integral GTPase family that stimulates homotypic ER fusion. Atlastin localizes preferentially to the tubular ER and interacts with reticulons [17]. Atlastin mutations or its depletion leads to unbranched ER tubules [17] and ER fragmentation in Drosophila neurons [18], and its overexpression leads to ER membrane expansion [17, 18]. Mutations in atlastin and interacting proteins have been linked to hereditary spastic paraplegia (HSP) [19], and such atlastin mutants are defective in ER fusion [20], indicating that ER continuity and shape is crucial for cell function (for further review of atlastin, see [21]).
There is no evidence from live-cell imaging that the ER undergoes fission. This begs the question of why the ER must maintain its continuity. The most logical explanation is that cross-talk occurs between ER domains that are located in distal regions of the cytoplasm. For example, it might be that ER Ca2+ signals or stress responses cannot be handled locally, but require the global ER. Sharing a continuous lumen could help ensure that the entire ER appropriately and rapidly responds to such signals.
The ER depends on the cytoskeleton for its structure and dynamics
The ER is a dynamic organelle that continuously rearranges its structure on the cytoskeleton while maintaining continuity as a singular organelle. The ER fusion machinery and the reticulon proteins play a stabilizing role in maintaining overall ER structure. In metazoans, organization of the cytoplasmic ER network is highly dependent on its ability to bind and move on microtubules (for discussion of yeast and plants, see Box 2). Treatment of animal cells with drugs that depolymerize MTs causes the ER to slowly retract from the cell periphery towards the NE [22].
Box 2. The role of actin in shaping yeast ER.
The ER network in yeast cells relies on the actin cytoskeleton network as opposed to MTs. The yeast ER network is dynamic, and both ER tubule growth and ring closure can be observed, though less frequently than in animal cells [68]. Some ER co-localization with actin filaments is observed, and actin depolymerization halts ER dynamics [68]. In much the same way, ER tubule movement in plants is also dependent on myosin and the actin cytoskeleton [69, 70].
In yeast cells, the cytoskeleton might not play the same integral role in shaping the ER network that it does in mammalian cells. Depolymerization of actin does not dramatically affect ER structure in the mother cell, but it is required for ER inheritance into the bud [68, 71, 72]. In contrast, depletion of reticulons and DP1/Yop1 alone causes loss of ER domain organization in the mother cell but does not prevent ER domains from being inherited into the bud [4]. Together, these data suggest that actin is required in yeast to generate ER domain structure while reticulons and DP1/Yop1 are required to maintain it.
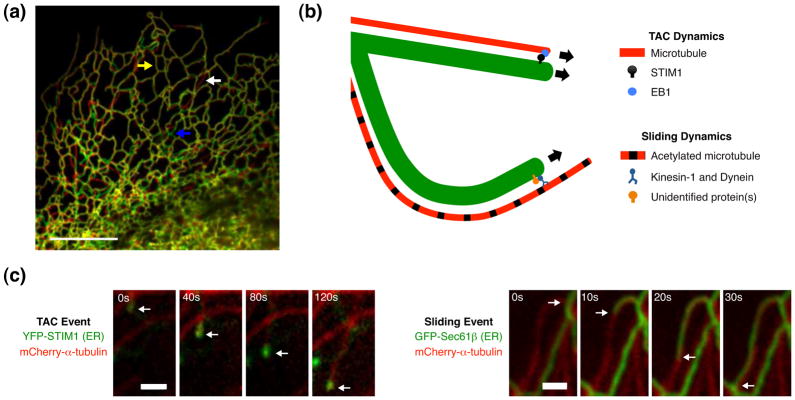
ER tubules grow along MTs by two distinct mechanisms: tip attachment complex (TAC) and ER sliding. During TAC movements, the tip of the ER tubule is bound to the tip of a dynamic MT, and the ER tubule grows and shrinks in concert with the dynamics of the plus-end of the MT (in a motor-independent fashion). TAC events occur through a complex between the integral ER membrane protein STIM1 and a protein that localizes to the tip of a dynamic MT, EB1 [23]. During ER sliding events, tubules are pulled out of the ER membrane by motor proteins kinesin-1 and dynein along MTs that are marked by acetylation [24, 25]. ER sliding is much more frequent than TAC and is the predominant mechanism responsible for dynamic ER rearrangements in interphase cells [25, 26]. No ER proteins have been shown to regulate ER sliding. Kinectin is one candidate, as it in is an integral ER membrane protein that binds to kinesin-1 [27]. Recent work suggests that kinectin localizes preferentially to ER cisternae [8], however, and observations of reduced ER sliding in the absence of kinectin have not been shown.
TAC and ER sliding mechanisms might contribute to very different ER functions. The TAC protein STIM1 also functions in store-operated Ca2+ entry (SOCE) at the PM, and TAC might be specific for this (see below). In contrast, the function of ER sliding dynamics is not known even though the cell likely consumes a great deal of energy to constantly reorganize the structure of the ER on MTs using motor proteins. One possibility is that the ER could use different dynamic mechanisms to establish or maintain contact with other membrane-bound organelles and the PM [25]. Evidence for this idea comes from work showing that ER sliding and mitochondria co-localize over acetylated MTs (see discussion below), whereas ER and early endosomes, which are also tightly coupled, do not [25].
The importance of maintaining ER-organelle contacts
The ER contacts almost every membrane-bound organelle in the cell, including mitochondria, endosomes, Golgi, and peroxisomes, as well as the plasma membrane [28, 29]. Here, we discuss what is known about the extent and function of these interactions in order to understand why ER dynamics are so highly coupled to other organelles.
ER-mitochondrial contact
Two functions that occur at contact sites between the ER and mitochondria are lipid synthesis and Ca2+ signaling, the latter of which is crucial for apoptotic regulation (for review, see [30]). Experiments in live cells using artificial tethers to alter the distance at contact sites between the ER and mitochondria show that Ca2+ signaling is altered when this distance is disrupted [31, 32].
Improvements in live-cell fluorescence microscopy have enabled study of ER-mitochondria interactions over time. While previous work has focused on either dynamics of the ER (discussed above) or mitochondria [33], recent visualization of the contact between these two organelles when they are both moving shows that the mitochondria maintain their contact with the ER despite the movements of both organelles, introducing the question of how organelles rearrange their structure around each other as they move ([25] and Figure 3). Interestingly, these contacts co-localize on the subset of MTs that are marked by acetylation [25]. These data suggest an elaborate interplay between proteins that regulate ER--mitochondria contact and the contact of these organelles with MTs during dynamics.
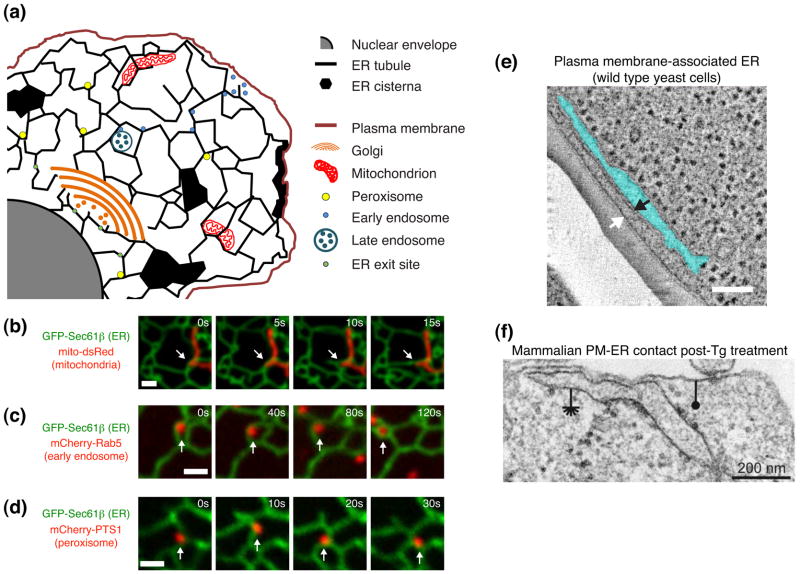
Figure 3. The ER has many different subdomains that can interact with other membrane bound compartments.
(a) Model depicting known ER domains and organelle contact sites in eukaryotic cells. Many of these contacts are maintained despite the dynamics of both membrane-bound compartments. (b–d) Coupled dynamics of the ER with other organelles. Cos-7 cells expressing GFP-Sec61β (ER) and (b) mito-dsRed (mitochondria), (c) mCherry-Rab5 (early endosomes), (d) mCherry-PTS1 (peroxisomes) at times indicated. Arrows indicate position of the ER-organelle contact. (e) EM tomography of wild type yeast cells reveals close apposition between the PmaER and PM membranes (black and white arrows mark ER and PM membrane bilayer, respectively). The distance between the two opposing membranes is maintained and the intervening region is ribosome-excluded. (f) EM tomograph of HeLa cells treated with thapsigargin (Tg, 1 μm) to deplete ER Ca2+ stores. Note induction of extensive ER-PM ribosome-free contact sites. Asterisk marks ER and closed circle marks PM. Scale bars: (b, c, d): 1 μm; (e) 50 nm. Images in (b, c, e) adapted with permission from J. Cell Biol. [25, 4]. Image in (f) adapted from [52].
Recent discoveries in yeast and mammalian cells have uncovered some of the membrane proteins involved in linking these two organelles. In yeast, the ERMES complex, consisting of four proteins (MMM1, MDM10, MDM12, and MDM34), tethers the ER and mitochondrial membranes [34]. ER-mitochondrial contact and lipid synthesis, which is disrupted in yeast mutants of these proteins, can be restored by adding a synthetic linker [34]. Mammalian homologs of the ERMES complex have not been identified, however. The best-characterized ER-mitochondria tethering protein in mammalian cells is mitofusin 2 (Mfn2) [35]. The name Mfn2 is derived from its role in homotypic fusion of mitochondria [36], however, Mfn2 also inserts into the ER membrane, and this localization is thought to be important for the interaction between Mfn2 on the ER and either Mfn1 or Mfn2 on the mitochondrial outer membrane [35]. Depletion of Mfn2 in mammalian cells disrupts Ca2+ handling, and causes aberrant morphology of the ER and mitochondria [35], suggesting that functions mediated through ER-mitochondrial contacts are essential for maintaining the integrity of each organelle.
There could be different tethers linking the mitochondria to the ER because these contact domains have multiple proposed functions. There are reported interactions between ER and mitochondrial proteins that, while not believed to tether the organelles, are implicated in processes such as apoptosis and Ca2+ signaling [37, 38]. To provide intricate regulation of these processes, different tethers may exist; presumably, we have only begun to uncover the complexes involved in these important linkages.
ER-Golgi contact
Traditionally, the relationship between ER and Golgi is discussed in terms of their roles in anterograde (COPII vesicle) and retrograde (COPI vesicle) transport as part of the secretory pathway (for review, see [39]). The ER also makes close contacts with the Golgi as part of other processes, however. High-resolution EM depicts very close contacts between the ER membrane and the trans-Golgi, which have been proposed to be involved in direct lipid transport (non-vesicular trafficking) [29, 40]. Ceramide, which is synthesized at the ER, is transferred to the Golgi membrane for its conversion to sphingomyelin, a process that is performed by the ceramide transferase CERT [41]. Additionally, phosphatidyl inositol is directly transferred from the ER to the Golgi by Nir2 [42]. It is hypothesized that in both cases, shuttling of these lipids from the ER to the Golgi occurs at ER-Golgi contact sites where these membranes are as little as ~10 nm apart. The factors linking these two membranes together have not been identified, however.
ER-endosome contact
Recent work has established a relationship between the ER and the endocytic pathway. In animal cells, a direct interaction between the ER membrane protein PTP1B and the endocytic cargo EGFR at ER-endosome contact sites has been observed by immuno-EM [43], suggesting that ER proteins might modify endocytosed cargoes. Interestingly, the movements of early endosomes are coordinated with ER dynamics and these two organelles can be tightly linked over time, suggesting that proteins linking the endosomes to the ER membrane are somehow coordinating this contact with the machinery that moves the organelles along the cytoskeleton ([25] and Figure 3). Direct contacts have also been observed between the ER and late endosomes and yeast vacuoles [4, 44]. A recent paper showed that in animal cells, ER contacts regulate the intracellular distribution of late endosomes [44]. The authors propose a complex regulatory pathway for late endosome positioning in which the ER protein VAP interacts with a late endosome-bound cholesterol sensor ORP1L in conditions of low cholesterol to tether the two organelles together [44]. There are likely to be other factors mediating ER-endosome contact, however, and the functions of these contacts are still not clear.
ER-peroxisome and ER-lipid droplet contacts share common features
One of the most unique ER-organelle interactions is that between the ER and peroxisomes. Peroxisomes have several functions, including the breakdown of long-chained fatty acids. What makes this interface particularly interesting is that in both yeast and mammalian cells, peroxisomes are derived from the ER membrane [45, 46]. Not only does peroxisomal biogenesis occur via the ER, but peroxisomes also maintain close contact with the ER membrane in mammalian cells ([46] and Figure 3), and non-vesicular transport is common between these organelles in yeast [47]. As yet, however, no factors have been identified that regulate this lipid transfer event. Lipid droplets, like peroxisomes, are derived in part from factors synthesized on the ER membrane and mature lipid droplets also can be tethered to the ER membrane [48].
ER-plasma membrane contact
In addition to binding almost every other membrane-bound organelle in the cell, the ER makes extensive contact with the PM. Recent work elucidating the 3-D structure of the ER in yeast by EM and tomography demonstrates that a mixture of interconnected ER tubules and fenestrated cisternae covers ~20–45% of the cytoplasmic surface of the PM [4, 13]. Contact between this PM-associated ER and the PM is maintained at a mean spacing of 33 nm, which is close enough to exclude ribosomes from the face of the ER membrane that contacts the PM ([4] and Figure 3). ER-PM contacts are important for regulation of phosphatidyl inositol metabolism and might be sites of direct, non-vesicular, sterol transfer [49, 50].
In mammalian cells, the ER-PM domain is also important in Ca2+ regulation. A complex between the ER protein STIM1 (which is also involved in TAC ER dynamics, see above) and the PM protein Orai1 is the mechanism behind store-operated Ca2+ entry (SOCE) for the ER. When ER Ca2+ stores are depleted, STIM1 forms higher ordered structures that then target to the PM [51]. This Ca2+-depleted ER is induced by STIM1 to form ribosome-free cisternal contacts with the PM [52]. At the PM, STIM1 interacts with Orai1 to form a CRAC channel that allows Ca2+ influx into the ER [53]. Despite the involvement of STIM1 in both TAC dynamics and SOCE, a direct link between these two processes has not been uncovered. It is intriguing to think that movement of STIM1 via TAC allows its translocation to the PM and complex formation with Orai1, however, inhibition of TAC did not appear to affect SOCE under non-physiological conditions [23]. Thus, while a link between TAC and SOCE may exist, it has not been formally proven.
The proteins that tether the ER to the PM are not known but would be expected to be quite abundant, at least in yeast, in order to attach so much ER to the PM. The contact between the ER and PM is regulated by the reticulon/Yop1 proteins; in the absence of these proteins in yeast, the peripheral ER adopts a flat cisternal shape that dramatically increases ER association with the PM [4]. It is likely that this is due to a loss of ER membrane curvature, however, rather than direct tethering by the reticulon/Yop1 proteins.
So far, we have discussed how the ER develops and maintains its extremely complicated morphology, which includes distinct domains and multiple shapes. This morphology is highly dependent on ER dynamics along the cytoskeleton. Additionally, the ER contacts almost every other organelle in the cell, performs a variety of functions at these contacts, and even appears to share coordinated movements with these organelles. Therefore, a likely possibility is that the ER must undergo dynamics to establish and maintain these functional contacts as organelles move around the cell.
A potential role for ER dynamics in cell shape change
The ER might be dynamic for the same reasons that MTs grow and shrink; such dynamic instability of MTs allows cells to rapidly explore 3-D space during cell migration, differentiation and polarization [54]. In much the same way, ER dynamics might allow the ER to adapt to changes in cell morphology during such processes. ER distribution is likely to be important for cells with complex morphologies; in neurons, the ER must be distributed down the length of narrow axons and dendrites, a process that appears to be coordinated with their development [55]. ER positioning and dynamics might also be important for cell migration [56]. The ER membrane’s ability to undergo constant tubule growth and reshaping could allow it to continuously probe the cytoplasm during periods of regulated PM extension so that the ER is present in the new cell boundary. The kinesin-1 binding protein, kinectin, is responsible for extending the ER into growing cellular lamella, however, it remains to be determined whether altering ER dynamics affects migration [57].
The 3-D structure of the ER also changes dramatically in animal cells as they progress through mitosis. In prophase, the NE breaks down and NE protein components are dispersed into the surrounding ER membrane [58, 59]. The ER maintains its continuity throughout the process, in contrast to other large organelles, such as the Golgi and mitochondria, which fragment [15, 16, 60, 61]. There has been debate over the shape and arrangement of the ER in mitosis, however, recent work shows quite convincingly that the ER network becomes almost entirely cisternal through mitosis, with very few remaining tubules [62]. The intracellular localization of the ER also changes; it moves away from the mitotic spindle and chromosomes, towards the PM. It is not surprising that the ER should be moved to the cell periphery to avoid interfering with chromosome segregation, but how this change in mitotic shape and location is regulated is not known. These shape changes could be due to cytoskeleton-directed movements and/or the inactivation of membrane shaping proteins.
Concluding remarks
Recent improvements in our ability to visualize the 3-D structure of the ER in live cells, and at a higher resolution with EM, have revealed ER domains and functions that we did not know existed even a few years ago. Although we have come a long way in our understanding of factors that influence ER shape and function, it is clear that ER responsibilities extend well beyond traditionally studied functions. The ER is extraordinarily dynamic, but we still have almost no information about the specific functions of these movements. Additionally, we now know that the ER forms stable contacts with the PM and nearly every other membrane-bound organelle in the cytoplasm. The tethering proteins that regulate contacts between the ER and PM, Golgi, vacuoles, endosomes, peroxisomes, and lipid droplets are not known and only a few linkers have been identified for mitochondria. As a result, we have only begun to unravel the functions of ER-organelle contacts. Future work addressing these questions will greatly improve our understanding of ER functions in the cell.
Figure 2. Two types of microtubule-dependent ER dynamics occur in mammalian cells.
(a) Cos-7 cell imaged every 30 seconds apart and merged to demonstrate peripheral ER structure dynamics (expressing GFP-Sec61β at t=0 seconds in green and t=30 seconds in red). Arrows indicate unchanged ER (yellow), new ER tubule growth (white), and ER rearrangement (blue). (b) Model illustrating tip attachment complex (TAC) and sliding dynamics. TAC occurs on dynamic MTs; the ER extends along with the plus-end of a growing MT via an interaction between the ER protein STIM1 and the MT plus-end protein EB1. Sliding occurs on acetylated, curved, nocodazole-resistant MTs in a MT motor-dependent manner. The protein(s) that attach the ER to these motors are unknown. (c) Examples of a TAC event (left) or an ER sliding event (right). Cos-7 cells are expressing either GFP-Sec61β or YFP-STIM1 to visualize ER dynamics (green) and mCherry-α-tubulin to visualize microtubules (red) at the times indicated. Arrows indicate position of the ER tubule. Scale bars = (a) 10 μm; (c) 1 μm. Images in (a) and (c) adapted with permission from J. Cell Biol. [25].
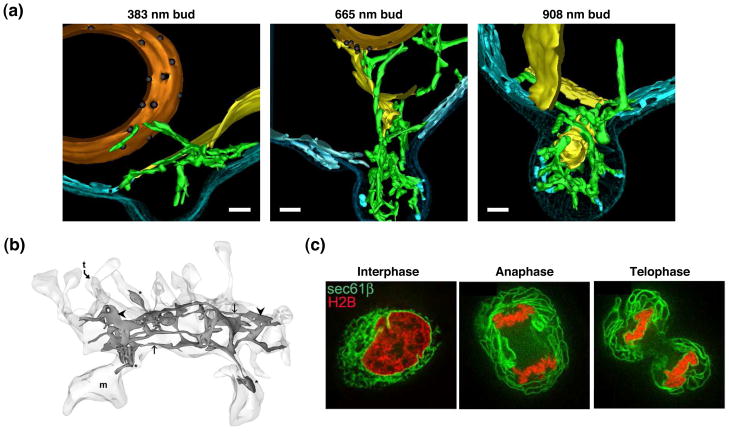
Figure 4. The ER gets redistributed as cell shape changes and completely changes morphology during mammalian mitosis.
(a) 3-D EM tomograms of wild type yeast cells reveal how ER domains are inherited into the daughter bud. Models are ordered from left to right with increasing bud size. ER tubules are inherited first into the bud (left and middle panel, tubER in green). The inherited tubular ER establishes contacts with the PM to form new PM-associated ER domains (middle and right panels, pmaER in blue). CecER in yellow. (b) A 3-D EM reconstruction of the ER network within the dendrite of a hippocampal neuron. Note the enrichment of tubular ER. (c) Fluorescent images of HeLa cells expressing H2B-mRFP (chromatin) and GFP-Sec61β (ER) demonstrate that 3-D ER structure changes dramatically between interphase and the indicated stages of mitosis. The NE breaks down, ER tubules are converted to cisternae, and the ER moves to the periphery of the cell away from the mitotic spindle. Scale bars: (a) 200 nm. Images in (a) adapted with permission from J. Cell Biol. [4]. Images in (b) adapted with permission from [73]. Images in (c) adapted with permission from [62].
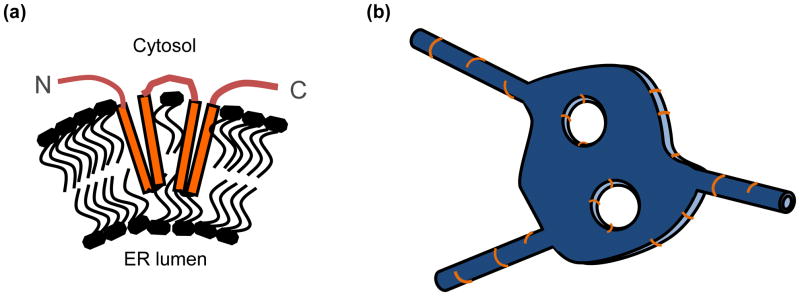
Box 1, Fig. I. Models of how reticulon proteins shape regions of high membrane curvature in the peripheral ER.
(a) Schematic of reticulon topology in the outer leaflet of the ER. Long transmembrane domains increase outer leaflet area relative to inner leaflet area, generating membrane curvature. (b) Schematic of ER cisterna and tubules (blue) indicating regions where reticulons (orange) have been observed to localize and shown to regulate membrane curvature, including ER tubules and the edges of cisternae and fenestra. Model in (a) adapted from [5]. Model in (b) adapted from [8].
Acknowledgments
This work was supported by NIH R01 GM083977 to G.K.V. and an NIH predoctoral training grant GM08759 to J.R.F. We thank M. West for providing EM images and A. Durand for helpful comments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Baumann O, Walz B. Endoplasmic reticulum of animal cells and its organization into structural and functional domains. Int Rev Cytol. 2001;205:149–214. doi: 10.1016/s0074-7696(01)05004-5. [DOI] [PubMed] [Google Scholar]
- 2.Hetzer MW, et al. Pushing the envelope: structure, function, and dynamics of the nuclear periphery. Annu Rev Cell Dev Biol. 2005;21:347–380. doi: 10.1146/annurev.cellbio.21.090704.151152. [DOI] [PubMed] [Google Scholar]
- 3.Bernales S, et al. Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol. 2006;4:e423. doi: 10.1371/journal.pbio.0040423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.West M, et al. A 3D analysis of yeast ER structure reveals how ER domains are organized by membrane curvature. J Cell Biol. 2011;193:333–346. doi: 10.1083/jcb.201011039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Voeltz GK, et al. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 2006;124:573–586. doi: 10.1016/j.cell.2005.11.047. [DOI] [PubMed] [Google Scholar]
- 6.Anderson DJ, Hetzer MW. Reshaping of the endoplasmic reticulum limits the rate for nuclear envelope formation. J Cell Biol. 2008;182:911–924. doi: 10.1083/jcb.200805140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Craene JO, et al. Rtn1p is involved in structuring the cortical endoplasmic reticulum. Mol Biol Cell. 2006;17:3009–3020. doi: 10.1091/mbc.E06-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shibata Y, et al. Mechanisms determining the morphology of the peripheral ER. Cell. 2010;143:774–788. doi: 10.1016/j.cell.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sparkes I, et al. Five Arabidopsis reticulon isoforms share endoplasmic reticulum location, topology, and membrane-shaping properties. Plant Cell. 2010;22:1333–1343. doi: 10.1105/tpc.110.074385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang YS, Strittmatter SM. The reticulons: a family of proteins with diverse functions. Genome Biol. 2007;8:234. doi: 10.1186/gb-2007-8-12-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shibata Y, et al. Rough sheets and smooth tubules. Cell. 2006;126:435–439. doi: 10.1016/j.cell.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 12.Puhka M, et al. Endoplasmic reticulum remains continuous and undergoes sheet-to-tubule transformation during cell division in mammalian cells. J Cell Biol. 2007;179:895–909. doi: 10.1083/jcb.200705112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schuck S, et al. Membrane expansion alleviates endoplasmic reticulum stress independently of the unfolded protein response. J Cell Biol. 2009;187:525–536. doi: 10.1083/jcb.200907074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hammond AT, Glick BS. Dynamics of transitional endoplasmic reticulum sites in vertebrate cells. Mol Biol Cell. 2000;11:3013–3030. doi: 10.1091/mbc.11.9.3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ellenberg J, Lippincott-Schwartz J. Dynamics and mobility of nuclear envelope proteins in interphase and mitotic cells revealed by green fluorescent protein chimeras. Methods. 1999;19:362–372. doi: 10.1006/meth.1999.0872. [DOI] [PubMed] [Google Scholar]
- 16.Ellenberg J, et al. Nuclear membrane dynamics and reassembly in living cells: targeting of an inner nuclear membrane protein in interphase and mitosis. J Cell Biol. 1997;138:1193–1206. doi: 10.1083/jcb.138.6.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu J, et al. A class of dynamin-like GTPases involved in the generation of the tubular ER network. Cell. 2009;138:549–561. doi: 10.1016/j.cell.2009.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Orso G, et al. Homotypic fusion of ER membranes requires the dynamin-like GTPase atlastin. Nature. 2009;460:978–983. doi: 10.1038/nature08280. [DOI] [PubMed] [Google Scholar]
- 19.Park SH, et al. Hereditary spastic paraplegia proteins REEP1, spastin, and atlastin-1 coordinate microtubule interactions with the tubular ER network. J Clin Invest. 2010;120:1097–1110. doi: 10.1172/JCI40979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bian X, et al. Structures of the atlastin GTPase provide insight into homotypic fusion of endoplasmic reticulum membranes. Proc Natl Acad Sci USA. 2011;108:3976–3981. doi: 10.1073/pnas.1101643108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moss TJ, et al. Fusing a lasting relationship between ER tubules. Trends Cell Biol. 2011;21:416–423. doi: 10.1016/j.tcb.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Terasaki M, et al. Microtubules and the endoplasmic reticulum are highly interdependent structures. J Cell Biol. 1986;103:1557–1568. doi: 10.1083/jcb.103.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grigoriev I, et al. STIM1 is a MT-plus-end-tracking protein involved in remodeling of the ER. Curr Biol. 2008;18:177–182. doi: 10.1016/j.cub.2007.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wozniak MJ, et al. Role of kinesin-1 and cytoplasmic dynein in endoplasmic reticulum movement in VERO cells. J Cell Sci. 2009;122:1979–1989. doi: 10.1242/jcs.041962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friedman JR, et al. ER sliding dynamics and ER-mitochondrial contacts occur on acetylated microtubules. J Cell Biol. 2010;190:363–375. doi: 10.1083/jcb.200911024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Waterman-Storer CM, Salmon ED. Endoplasmic reticulum membrane tubules are distributed by microtubules in living cells using three distinct mechanisms. Curr Biol. 1998;8:798–806. doi: 10.1016/s0960-9822(98)70321-5. [DOI] [PubMed] [Google Scholar]
- 27.Toyoshima I, et al. Kinectin, a major kinesin-binding protein on ER. J Cell Biol. 1992;118:1121–1131. doi: 10.1083/jcb.118.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.English AR, et al. Peripheral ER structure and function. Curr Opin Cell Biol. 2009;21:596–602. doi: 10.1016/j.ceb.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levine T, Loewen C. Inter-organelle membrane contact sites: through a glass, darkly. Curr Opin Cell Biol. 2006;18:371–378. doi: 10.1016/j.ceb.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 30.de Brito OM, Scorrano L. An intimate liaison: spatial organization of the endoplasmic reticulum-mitochondria relationship. EMBO J. 2010;29:2715–2723. doi: 10.1038/emboj.2010.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Csordás G, et al. Structural and functional features and significance of the physical linkage between ER and mitochondria. J Cell Biol. 2006;174:915–921. doi: 10.1083/jcb.200604016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Csordás G, et al. Imaging interorganelle contacts and local calcium dynamics at the ER-mitochondrial interface. Mol Cell. 2010;39:121–132. doi: 10.1016/j.molcel.2010.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boldogh IR, Pon LA. Mitochondria on the move. Trends Cell Biol. 2007;17:502–510. doi: 10.1016/j.tcb.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 34.Kornmann B, et al. An ER-Mitochondria Tethering Complex Revealed by a Synthetic Biology Screen. Science. 2009;325:477–481. doi: 10.1126/science.1175088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Brito OM, Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 2008;456:605–610. doi: 10.1038/nature07534. [DOI] [PubMed] [Google Scholar]
- 36.Hoppins S, et al. The machines that divide and fuse mitochondria. Annu Rev Biochem. 2007;76:751–780. doi: 10.1146/annurev.biochem.76.071905.090048. [DOI] [PubMed] [Google Scholar]
- 37.Giorgi C, et al. Structural and functional link between the mitochondrial network and the endoplasmic reticulum. Int J Biochem Cell Biol. 2009;41:1817–1827. doi: 10.1016/j.biocel.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iwasawa R, et al. Fis1 and Bap31 bridge the mitochondria-ER interface to establish a platform for apoptosis induction. EMBO J. 2011;30:556–568. doi: 10.1038/emboj.2010.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spang A. On vesicle formation and tethering in the ER – Golgi shuttle. Curr Opin Cell Biol. 2009;21:531–536. doi: 10.1016/j.ceb.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 40.Lev S. Non-vesicular lipid transport by lipid-transfer proteins and beyond. Nat Rev Mol Cell Biol. 2010;11:739–750. doi: 10.1038/nrm2971. [DOI] [PubMed] [Google Scholar]
- 41.Hanada K, et al. CERT-mediated trafficking of ceramide. Biochim Biophys Acta. 2009;1791:684–691. doi: 10.1016/j.bbalip.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 42.Peretti D, et al. Coordinated lipid transfer between the endoplasmic reticulum and the Golgi complex requires the VAP proteins and is essential for Golgi-mediated transport. Mol Biol Cell. 2008;19:3871–3884. doi: 10.1091/mbc.E08-05-0498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eden ER, et al. Membrane contacts between endosomes and ER provide sites for PTP1B-epidermal growth factor receptor interaction. Nat Cell Biol. 2010;12:267–272. doi: 10.1038/ncb2026. [DOI] [PubMed] [Google Scholar]
- 44.Rocha N, et al. Cholesterol sensor ORP1L contacts the ER protein VAP to control Rab7-RILP-p150 Glued and late endosome positioning. J Cell Biol. 2009;185:1209–1225. doi: 10.1083/jcb.200811005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoepfner D, et al. Contribution of the endoplasmic reticulum to peroxisome formation. Cell. 2005;122:85–95. doi: 10.1016/j.cell.2005.04.025. [DOI] [PubMed] [Google Scholar]
- 46.Kim PK, et al. The origin and maintenance of mammalian peroxisomes involves a de novo PEX16-dependent pathway from the ER. J Cell Biol. 2006;173:521–532. doi: 10.1083/jcb.200601036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raychaudhuri S, Prinz WA. Nonvesicular phospholipid transfer between peroxisomes and the endoplasmic reticulum. Proc Natl Acad Sci USA. 2008;105:15785–15790. doi: 10.1073/pnas.0808321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jacquier N, et al. Lipid droplets are functionally connected to the endoplasmic reticulum in Saccharomyces cerevisiae. J Cell Sci. 2011;124:2424–2437. doi: 10.1242/jcs.076836. [DOI] [PubMed] [Google Scholar]
- 49.Schulz TA, Prinz WA. Sterol transport in yeast and the oxysterol binding protein homologue (OSH) family. Biochim Biophys Acta. 2007;1771:769–780. doi: 10.1016/j.bbalip.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stefan CJ, et al. Osh Proteins Regulate Phosphoinositide Metabolism at ER-Plasma Membrane Contact Sites. Cell. 2011;144:389–401. doi: 10.1016/j.cell.2010.12.034. [DOI] [PubMed] [Google Scholar]
- 51.Liou J, et al. Live-cell imaging reveals sequential oligomerization and local plasma membrane targeting of stromal interaction molecule 1 after Ca2+ store depletion. Proc Natl Acad Sci USA. 2007;104:9301–9306. doi: 10.1073/pnas.0702866104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Orci L, et al. STIM1-induced precortical and cortical subdomains of the endoplasmic reticulum. Proc Natl Acad Sci USA. 2009;106:19358–19362. doi: 10.1073/pnas.0911280106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Park CY, et al. STIM1 Clusters and Activates CRAC Channels via Direct Binding of a Cytosolic Domain to Orai1. Cell. 2009;136:876–890. doi: 10.1016/j.cell.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Desai A, Mitchison TJ. Microtubule polymerization dynamics. Annu Rev Cell Dev Biol. 1997;13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- 55.Ramirez OA, Couve A. The endoplasmic reticulum and protein trafficking in dendrites and axons. Trends Cell Biol. 2011;21:219–227. doi: 10.1016/j.tcb.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 56.Bola B, Allan V. How and why does the endoplasmic reticulum move? Biochem Soc Trans. 2009;37:961–965. doi: 10.1042/BST0370961. [DOI] [PubMed] [Google Scholar]
- 57.Zhang X, et al. Kinectin-mediated endoplasmic reticulum dynamics supports focal adhesion growth in the cellular lamella. J Cell Sci. 2010;123:3901–3912. doi: 10.1242/jcs.069153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang L, et al. Integral membrane proteins of the nuclear envelope are dispersed throughout the endoplasmic reticulum during mitosis. J Cell Biol. 1997;137:1199–1210. doi: 10.1083/jcb.137.6.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hetzer MW. The nuclear envelope. Cold Spring Harb Perspect Biol. 2010;2:a000539. doi: 10.1101/cshperspect.a000539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Persico A, et al. Mitotic inheritance of the Golgi complex. FEBS Lett. 2009;583:3857–3862. doi: 10.1016/j.febslet.2009.10.077. [DOI] [PubMed] [Google Scholar]
- 61.Taguchi N, et al. Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J Biol Chem. 2007;282:11521–11529. doi: 10.1074/jbc.M607279200. [DOI] [PubMed] [Google Scholar]
- 62.Lu L, et al. Cisternal organization of the endoplasmic reticulum during mitosis. Mol Biol Cell. 2009;20:3471–3480. doi: 10.1091/mbc.E09-04-0327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kiseleva E, et al. Reticulon 4a/NogoA locates to regions of high membrane curvature and may have a role in nuclear envelope growth. J Struct Biol. 2007;160:224–235. doi: 10.1016/j.jsb.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hu J, et al. Membrane proteins of the endoplasmic reticulum induce high-curvature tubules. Science. 2008;319:1247–1250. doi: 10.1126/science.1153634. [DOI] [PubMed] [Google Scholar]
- 65.Tolley N, et al. Transmembrane domain length is responsible for the ability of a plant reticulon to shape endoplasmic reticulum tubules in vivo. Plant J. 2010;64:411–418. doi: 10.1111/j.1365-313X.2010.04337.x. [DOI] [PubMed] [Google Scholar]
- 66.Zurek N, et al. Reticulon Short Hairpin Transmembrane Domains Are Used to Shape ER Tubules. Traffic. 2011;12:28–41. doi: 10.1111/j.1600-0854.2010.01134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shibata Y, et al. The reticulon and DP1/Yop1p proteins form immobile oligomers in the tubular endoplasmic reticulum. J Biol Chem. 2008;283:18892–18904. doi: 10.1074/jbc.M800986200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Prinz WA, et al. Mutants affecting the structure of the cortical endoplasmic reticulum in Saccharomyces cerevisiae. J Cell Biol. 2000;150:461–474. doi: 10.1083/jcb.150.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sparkes I, et al. Movement and Remodeling of the Endoplasmic Reticulum in Nondividing Cells of Tobacco Leaves. Plant Cell. 2010;21:3937–3949. doi: 10.1105/tpc.109.072249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ueda H, et al. Myosin-dependent endoplasmic reticulum motility and F-actin organization in plant cells. Proc Natl Acad Sci USA. 2010;107:6894–6899. doi: 10.1073/pnas.0911482107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Du Y, et al. Dynamics and inheritance of the endoplasmic reticulum. J Cell Sci. 2004;117:2871–2878. doi: 10.1242/jcs.01286. [DOI] [PubMed] [Google Scholar]
- 72.Estrada P, et al. Myo4p and She3p are required for cortical ER inheritance in Saccharomyces cerevisiae. J Cell Biol. 2003;163:1255–1266. doi: 10.1083/jcb.200304030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cooney JR, et al. Endosomal compartments serve multiple hippocampal dendritic spines from a widespread rather than a local store of recycling membrane. J Neurosci. 2002;22:2215–2224. doi: 10.1523/JNEUROSCI.22-06-02215.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]