Abstract
Because of its highly branched dendrite, the Purkinje neuron requires significant computational resources if coupled electrical and biochemical activity are to be simulated. To address this challenge, we developed a scheme for reducing the geometric complexity; while preserving the essential features of activity in both the soma and a remote dendritic spine. We merged our previously published biochemical model of calcium dynamics and lipid signaling in the Purkinje neuron, developed in the Virtual Cell modeling and simulation environment, with an electrophysiological model based on a Purkinje neuron model available in NEURON. A novel reduction method was applied to the Purkinje neuron geometry to obtain a model with fewer compartments that is tractable in Virtual Cell. Most of the dendritic tree was subject to reduction, but we retained the neuron’s explicit electrical and geometric features along a specified path from spine to soma. Further, unlike previous simplification methods, the dendrites that branch off along the preserved explicit path are retained as reduced branches. We conserved axial resistivity and adjusted passive properties and active channel conductances for the reduction in surface area, and cytosolic calcium for the reduction in volume. Rallpacks are used to validate the reduction algorithm and show that it can be generalized to other complex neuronal geometries. For the Purkinje cell, we found that current injections at the soma were able to produce similar trains of action potentials and membrane potential propagation in the full and reduced models in NEURON; the reduced model produces identical spiking patterns in NEURON and Virtual Cell. Importantly, our reduced model can simulate communication between the soma and a distal spine; an alpha function applied at the spine to represent synaptic stimulation gave similar results in the full and reduced models for potential changes associated with both the spine and the soma. Finally, we combined phosphoinositol signaling and electrophysiology in the reduced model in Virtual Cell. Thus, a strategy has been developed to combine electrophysiology and biochemistry as a step toward merging neuronal and systems biology modeling.
Keywords: Virtual Cell, NEURON, Model reduction, Compartmental modeling, Biochemical simulation, Electrophysiology modeling
1 Introduction
The Purkinje cell in the cerebellum is responsible for integrating inputs for motor coordination (Gilbert and Thach 1977; Watanabe 2008). It does this by utilizing an extraordinary dendritic tree that captures the inputs from parallel fibers in several hundred thousand synapses (Napper and Harvey 1988a, b). It also contains the highest density of Type 1 inositol 1,4,5-trisphosphate receptor (IP3R1) of any cell that has been investigated, although this receptor is quite widely distributed in both excitable and non-excitable tissue throughout the body (De Smedt et al. 1997). The IP3R is a calcium channel-coupled receptor on the endoplasmic reticulum that is responsible for calcium release from the highly concentrated stores in the ER lumen into the cytosol, where calcium is maintained at a level of <100 nM (Berridge et al. 2000). A genetic deficiency of IP3R1 in both humans and mice is associated with the inability to coordinate movement (termed cerebellar ataxia) (Matsumoto et al. 1996; Street et al. 1997; Ogura et al. 2001; van de Leemput et al. 2007; Hara et al. 2008; Iwaki et al. 2008; Di Gregorio et al. 2010). IP3R1−/− mice show reduced calcium release, and a decreased parallel fiber-induced excitatory postsynaptic current decay time constant (Matsumoto et al. 1996). This motivated us to use computational modeling to try to understand the coupling between the calcium signaling at the synaptic inputs and the electrical activity at the soma via computational modeling. However, the large range of time scales and spatial scales that are spanned by, respectively, the physiology and the geometry of the Purkinje cell, make a combined model of biochemical and electrical activity a computationally daunting challenge.
This laboratory has developed a modeling and simulation software system, Virtual Cell (VCell; http://www.vcell.org) (Schaff et al. 1997; Loew and Schaff 2001; Moraru et al. 2002), which supports the coupling of electrophysiology with reaction and diffusion of molecular species in experimentally-derived cell geometries. It also includes simulators that can handle multiple timescales. However, the multiple spatial scales in a realistic neuronal geometry make it impractical to perform simulations with the partial differential equation solvers currently deployed in Virtual Cell. Further, the software approximates the cytosol as an infinite conductor (i.e. perfect space clamp); thus the plasma membrane is isopotential at any given time point in a simulation. But in order to make experimentally testable predictions from simulations, the propagation of membrane potential changes between an axon or a dendritic spine and the soma must be explicitly modeled.
Compartmental modeling provides a useful alternative. Compartmental modeling of neurons was first introduced in the 1960s (Rall and Agmon-Snir 1998). The main assumption was that small segments of neuronal dendrites could be treated as isopotential cylindrical compartments. This allowed for the continuous neuronal structure to be approximated by linking a series of discrete isopotential compartments. Voltage differences are calculated between compartments. Traditional methods defined by Wilfred Rall and others can safely be used in compartmental modeling to collapse passive dendritic trees that obey a “3/2 rule” (RALL 1962, 1969; Rall and Agmon-Snir 1998). The “3/2 rule” requires that the sum of each diameter of the daughter branches raised to the 3/2 power equals the diameter of the parent branch raised to the 3/2 power. This power law, in conjunction with other constraints, conserves the overall surface area of the neuron, when it is used to create collapsed equivalent cylinders to represent an entire dendritic arbor or groups of dendrites. However, neurons like cerebellar Purkinje cell that have complex geometries do not actually exhibit morphologies that adhere to the “3/2 rule” and related constraints (Rall and Agmon-Snir 1998). Among others, Bush and Sejnowski (Bush and Sejnowski 1993) presented an alternative reduction method. In their method, dendrites to be reduced to a single equivalent cylinder must be at a given electrotonic distance from the soma, as in Rall’s methods, but do not have to obey the “3/2 rule”. Whereas most reduced models attempt to preserve surface area, Bush and Sejnowski introduce a method that adjusts surface area. This study provided a more convenient form of reduction, but it still only considered reduction of an entire neuron with a simple geometry.
We have termed these methods of simplifying whole neurons global reduction methods. These methods do not allow for the investigation of the response of a single spine to synaptic stimulation in a neuron with complex geometry. There are two other systems of reduction that we term restricted reduction and dynamic reduction. Formerly, Rinzel and Rall considered an idealized neuron model (Rinzel and Rall 1974), in which 5 of 6 identical dendritic trees connected to the soma were reduced to equivalent cylinders. The 6th dendritic tree was modeled explicitly, and excitatory postsynaptic potential (E.P.S.P.) propagation from an explicit dendrite branch (with no explicit spines) to the soma was studied. However, none of the dendrites branching off of the explicit dendritic tree were reduced. This was analytically and computationally feasible, because the entire unreduced dendritic tree contained only 15 compartments. Others have also considered an unreduced dendritic tree, with a small number of compartments, connected to the soma and to reduced dendritic portions, i.e., equivalent cylinders or cables (Burke et al. 1994). In our case, the complex Purkinje neuron geometry contains 1085 compartments, and is incredibly more complicated than the idealized neuron geometry presented by Rinzel and Rall (Rinzel and Rall 1974) or the simple pyramidal cell geometries modeled by Burke et al. (Burke et al. 1994). We term these methods of reducing all the dendrites in a simple neuron morphology, except for one explicitly branched dendritic arbor, as restricted reduction. Finally, Manor et al. (Manor et al. 1991) simulate the dynamic reduction and expansion of a simple idealized neuron, in which portions of the geometry expand once an action potential approaches. We term this dynamic reduction, a combination of global and restricted reduction.
In this work, we present a novel reduction method in which we retain an explicit unreduced path between the soma and a single spine, while reducing the rest of the cell (i.e., all the remaining dendrites in which we are not interested, including all the dendrites that directly branch off of the explicit path). We have termed this type of simplification preserved path reduction. Preserved path reduction creates equivalent cylinders to reproduce voltage propagation in the dendritic tree, without explicitly modeling the individual geometry of the dendrites that branch off of the explicit path. Importantly, unlike existing reduction methods, it does not depend on the electrotonic distance of the dendrites from the soma, nor does it depend on the “3/2 power law”. In addition, this method provides a multi-compartmental model that is tractable for consistent use in Virtual Cell, allowing for combination of electrophysiology with biochemical pathways involving reaction and diffusion. It is particularly useful for studies that incorporate the bidirectional interaction between calcium signaling and membrane potential, and downstream effects measured at the spine or at the soma.
2 Models
2.1 Simulation platforms
We used two modeling and simulation platforms in this study. NEURON (Hines and Carnevale 1997, 2001) was used as a source of neuronal electrophysiology and morphology. Virtual Cell (VCell; http://www.vcell.org) (Schaff et al. 1997; Loew and Schaff 2001; Moraru et al. 2002) was used for its strength in biochemical reaction pathway modeling. VCell has commonly been used for biochemical and cell biological simulations with one or a few compartments. We have enhanced VCell to effectively handle multicompartmental representations of neuronal morphologies. We maintain a limited number of compartments for computational efficiency, while preserving physiological accuracy. This model and all simulations can be accessed as a “Shared BioModel” by logging in to VCell at http://www.vcell.org; the model is entitled “Brown et al. 2010 Purkinje MultiCompartmental Combined Biochem and Electrophysiol” under the username Brown. The full Virtual Cell model code is also available in the Supplementary Material for this paper as a file entitled “Brown_et_al_2010_Purkinje_MultiCompartmental_Combined_Biochem_and_Electrophysiol.vcml”. The PPR model developed in NEURON can be accessed in ModelDb (http://senselab.med.yale.edu/ModelDb/) by searching for the authors.
2.2 Neuronal morphology and passive membrane properties
The Purkinje neuron morphology and the passive membrane properties (membrane resistance and capacitance) used in this study were adapted from a model created in NEURON by Miyasho et al. (Miyasho et al. 2001). In that study, the authors based their morphology on a detailed Purkinje neuron reconstruction from an adult rat previously published by Shelton (Shelton 1985). Smooth membrane capacitance of 0.8 µF/cm2 was taken from an estimate for hippocampal CA3 pyramidal neurons (Major et al. 1994), and the leak current (inverse of passive membrane resistance) was determined by fitting the input resistance in their model to results from Rapp et al. (Rapp et al. 1994). Throughout this paper, the terms membrane capacitance and membrane resistance refer to specific membrane capacitance and specific membrane resistance, respectively.
2.3 Ion channel kinetics
Each ion channel was modeled as a Hodgkin-Huxley system (Hodgkin and Huxley 1952). Kinetics were all taken from Miyasho et al. (Miyasho et al. 2001). Miyasho et al. in turn, used the same kinetics described by De Schutter et al. in a GENESIS Purkinje neuron model (De Schutter and Bower 1994). However, Miyasho et al. adjusted the properties of a handful of channels to match their experiments. Particularly, they introduced the class-E calcium channel and the D-type potassium channels to facilitate calcium spiking in the dendrites, matched to their electrophysiological and pharmacological experiments. The model has two sodium channels (NaF, NaP), one calcium channel (CaP2), and four potassium channels (Khh, KM, KA, Kh) in the soma. The sodium channels are responsible for the upstroke in action potentials, the calcium channels contribute to somatic depolarization, and the potassium channels are responsible for repolarization. There are three calcium channels (CaP2, CaT, CaE) and six potassium channels (Khh, KM, KA, KD, KC3, K23) in the smooth and spiny dendrites. When depolarizing oscillations propagating from the soma to the dendrites sufficiently increase the conductance of the calcium channels in the dendrites, calcium spiking (membrane potential transients) occurs. The calcium channels are responsible for the upstroke of the calcium spikes; the potassium channels repolarize the dendrites. There are other forms of calcium handling that are conceptually included in the model, such as a plasma membrane calcium ATPase pump, and calcium buffering and diffusion, based on a model by Destenxhe et al. (Destexhe et al. 1993).
2.4 Reduction method
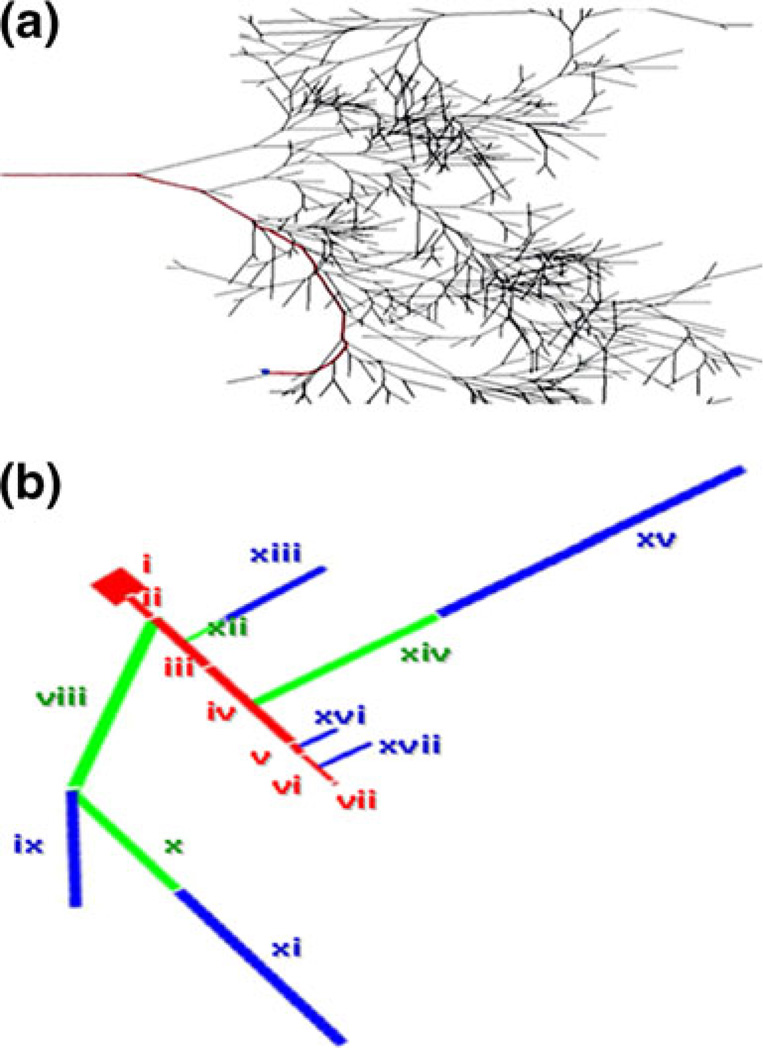
We developed a novel reduction method suitable for complex neuronal geometries that allows us to preserve an electrical path from soma to spine, while reducing dendrites from the rest of the cell, including those that branch off from the preserved path. We used this method to simplify the complex Purkinje geometry in NEURON, and compared simulation results from the full and reduced models. Our method takes into account the great degree of branching in such geometries that have dendrites highly connected in series, as well as in parallel. As shown in Fig. 1(a), our geometry has less than 20 branches off of our explicit path (in red). Yet, it contains more than 1080 dendritic segments. Each group of dendrites that was reduced was approximated by a single “equivalent” dendritic segment that occurs at the corresponding branch point along a very long dendritic vine. To determine the physical parameters of these equivalent segments in our PPR geometry (Fig. 1(b)), we first looked for an approximation of the axial resistivity of the reduced compartments. Each of these groups of dendrites is in fact a highly branched arbor, showing a mix of series connectivity and parallel connectivity in the full geometry. For any particular group of dendrites, the case of minimum axial resistivity would be for all parallel connections, and the maximum for all series connections. We therefore started by estimating these upper and lower bounds. The length for each equivalent cylinder was taken as the actual anatomic length of the longest path within that group of dendrites:
| (1) |
(where lalpha and lomega are the anatomic distances to the soma of the closest and farthest dendrites in the sub-tree) and the radius as either
| (2) |
(where ri is the radius of each individual dendritic compartment to be reduced to a particular equivalent cylinder, and n is the number of dendritic compartments to be reduced to that equivalent cylinder). We must note that these expressions for rseries and rparallel are not equal to the exact radii that would result in an equivalent resistivity for the series and parallel case respectively (the exact values would be the square root of a length-weighted harmonic mean of the squared radii, and the square root of a length-weighted arithmetic mean of the squared radii, respectively). These approximations are simple to calculate, the error is not very large given the actual variances in the radii and length of the various dendritic segments that are part of the same group, and it results in expanding the bounds in both directions. We found that using the geometric mean of these two values
| (3) |
as the radius of the equivalent cylinder is a reasonable phenomenological approximation of the axial resisitivity of the actual randomly branched dendritic arbors of the reduced compartments.
Fig. 1.
Representation of the complex Purkinje neuron. (a) Full geometry. (b) Reduced geometry. Preserved electronic path is highlighted in red. The rest of the neuron in black (a) is reduced to equivalent cylinders ((b), green—smooth, blue—spiny)
Typically, simulation platforms (VCell (Schaff et al. 1997; Loew and Schaff 2001; Moraru et al. 2002), NEURON (Hines and Carnevale 1997, 2001), GENESIS (Bower and Beeman 2003; Bower and Beeman 2007)) require the user to provide numerical values for current densities, specific resistance, and specific capacitance, as well as values for the length and radius of each compartment to be simulated. The program then uses the radius and length to calculate the surface area and volume of each compartment. The surface area is used to compute total membrane resistance and capacity, as well as the total fluxes and currents across the membrane. After model reduction, a given group of dendrites will generally have a different surface area than the equivalent cylinder to which the dendrites are mapped; this will alter the electrical properties of the membrane. For example, if the original surface area is larger, the electrical resistance of the equivalent cylinder is greater than the resistance of the group of dendrites. Conversely, the capacitance is lower in the equivalent cylinder than in the corresponding group of dendrites. To account for these differences, a scaling factor was determined for each equivalent cylinder. The scaling factor, sf, was the exact ratio of the surface areas of the explicit dendrites and the corresponding equivalent cylinders in the full and reduced models:
| (4) |
where SAfull is the surface area of all the dendrites to be reduced to an equivalent cylinder, and SAreduced is the surface area of the corresponding equivalent cylinder in the reduced model. This value was used to adjust the parameters for passive electrical properties of the membrane for each cylinder in the reduced model, i.e., it was used to scale down specific membrane resistance, Rm, and scale up specific membrane capacitance, Cm:
| (5) |
where Rm-reduced and Cm-reduced are the adjusted specific membrane resistance and capacitance in the reduced model. This way, total membrane resistance and capacitance are preserved in the reduced model:
| (6) |
| (7) |
where Rtot-reduced and Ctot-reduced are the total membrane resistance and capacitance in the reduced model, and Rtot-full and Ctot-full are the total membrane resistance and capacitance in the full model. The scaling factor was also used to scale active conductances, as well as calcium flux into the dendrites, since the voltage-gated calcium channels provide stimulus for the calcium-sensitive voltage-gated potassium channels (detailed in Supplementary Material). This ensures that all values for channel currents and calcium flux across the membrane are the same in the reduced model as in the full model.
In summary, four basic equations were formulated in order to carry out preserved path model reduction of complicated geometries. First, the ratio of the surface area (Eq. (4)) must be calculated, since the reduced geometry has a much smaller surface area than the full geometry.
Second, this surface area ratio must be used to scale the passive membrane resistance and capacitance (Eq. (5)), in order for the reduced model to preserve the passive membrane properties from the full model. Third, the radius of each equivalent cylinder needs to be calculated (Eq. (3)), based on the collective radii of all the dendrites in the full model that are represented by an equivalent cylinder in the reduced model. Next, the length of each equivalent cylinder must be calculated (Eq. (1)), based on the distances from the soma of all the corresponding dendrites in the full geometry. Finally, active conductances must also be scaled by the scaling factor expressed in Eq. (4).
2.5 Modeling in virtual cell
We then reproduced the reduced version in Virtual Cell. The Virtual Cell modeling environment (Schaff et al. 1997; Loew and Schaff 2001) is used most often to model biochemical processes within cells. It can also be used to model electrophysiology, but it has not been previously used to study voltage propagation in a whole neuron representation. While Virtual Cell can simulate reaction/diffusion in complex geometries by numerically solving partial differential equations, for electrophysiology it assumes a complete equipotential within a discrete compartment (as do other neural simulators). To overcome this problem, we created an ODE model in Virtual Cell by specifying currents and fluxes between multiple connected cylindrical compartments, such as cylinders representing the soma, spine, and portions of dendrite, similar to the approach in NEURON. This allowed the potential along the dendritic tree to vary at any point in time. In this way, we designed an RC circuit to represent the rate of change of uniform voltage in regions of the spiny dendrites and other parts of the neuron. This process is equivalent to the electrophysiological compartmental modeling in the NEURON (Hines and Carnevale 1997, 2001) and GENESIS (Bower and Beeman 2003; Bower and Beeman 2007) simulators.
In the RC circuit, we approximated the resistance of each compartment as a function of the length and cross-sectional area of the compartment, with the cytoplasmic resistivity (250 Ohm cm, (Rapp et al. 1994)) as a proportionality constant. The electrical resistance between compartments was defined by
| (8) |
where R1 is the resistance in compartment 1, R2 is the resistance in compartment 2, and R12 is essentially the resistance between the centers of compartments 1 and 2. Intercompartmental currents were then applied to each compartment, to allow membrane potential propagation:
| (9) |
where V1 is the membrane potential in the center of compartment 1, V2 is the membrane potential in compartment 2, I12 is the current flowing from compartment 1 to compartment 2, and I21 is the current flowing from compartment 2 to compartment 1.
The currents at three-way branch points were specified differently. Currents flowing between such compartments are calculated based on the flow of current from each of the three compartments into the branch junction. Thus, if three compartments, say 1, 2, 3 form a three-way branch point, the current flowing from compartment one into the branch junction may be expressed as
| (10) |
where Vb is the voltage at the point where the three compartments meet, and I1b is the current flowing from compartment 1 into the branch point. The branch point voltage, Vb, is defined by
| (11) |
where V3 is the membrane potential in compartment 3, and R3 is the resistance in compartment 3. Vb is determined by using Kirchoff’s current law (all currents at a junction sum to zero), as shown in Eq. (12) below, and solving for Vb.
| (12) |
The two expressions in Eqs. (10) and (11), are used at both three-way branch points in the simplified model to accurately represent current flow between the branched compartments.
Other branches were placed in the middle of each compartment, as shown in Fig. 1, to accurately represent the distribution of dendrites that connect electrically to that parent compartment. NEURON requires that two segments are created in a parent compartment in order to place a branch at the center of that compartment. This way, NEURON places the branch at the edges of the two adjoining segments within the parent compartment. Virtual Cell does not have such a constraint, allowing us to formulate the mathematics for a branch from the center of a single compartment.
In the same way that current flow between compartments depends on the electrical resistance between the compartments, the flow of molecules and ions between compartments depends on the diffusional resistance between compartments. Net molecular diffusion between compartments in our model was only implemented between the spine and the adjacent dendrite. This was possible because we set the length of the adjacent dendrite based on equilibration of IP3 produced at the spine (Hernjak et al. 2005). We based this on a one-dimensional simulation (Hernjak et al. 2005), which showed that upon stimulation of a spine, biochemical concentrations of molecules such as IP3 changed significantly in the spine and in areas of dendrite near the spine. However, at a dendritic distance of 27.98 µm from the stimulated spine, biochemical concentrations were at steady state during synaptic activity. Since IP3 has the highest diffusion coefficient of any species in our model, this allowed us to assume that there is no net molecule flow between compartments beyond the adjacent dendrite (which is given a length of 27.98 µm). Of note, both the electrical resistance and the diffusional resistance between the spine and the adjacent dendrite are assumed to depend on the geometry of the spine neck. Thus, the diffusion of a molecule X from the spine to the adjacent dendrite is modeled by:
| (13) |
where DX is the diffusion coefficient for the molecule X, [X]spine is the concentration of X in the spine, [X]adjacentdendrite is X concentration in the adjacent dendrite, Aneck is the area of the spine neck, lneck is the spine neck length, and Vspine is the volume of the spine head. Diffusion from the adjacent dendrite to the spine is modeled accordingly.
Ion channel kinetics were the same as the ones used by Miyasho et al. (Miyasho et al. 2001) in NEURON (see Ion Kinetics earlier in this section).
3 Results
3.1 Reduced Purkinje neuronal morphology
We wanted to create a model that includes biochemical signaling at the spine, membrane potential changes at the spine and soma, propagation of membrane potential between the spine and soma, and the interaction between the biochemical reaction and diffusion and the membrane potential and current flow. We were motivated by our interest in the Purkinje cell physiology, but the method that we developed can be applied to other neurons with complex geometries. The challenge was to develop such a model that would faithfully reproduce electrophysiology propagation from the spine to the soma and vice versa in the complex Purkinje neuron geometry, and still take advantage of Virtual Cell’s capabilities for detailed biochemical study.
We selected a full Purkinje geometry used in a recent study simulated in NEURON (Miyasho et al. 2001). Spines are usually subresolution and numerous, with varying sizes, in neurons like the cerebellar Purkinje cell (~14 spines/µm2; (Harris and Stevens 1988)). The effect of spines on the dendritic surface area is therefore usually taken into account globally, as in this NEURON model, rather than explicitly modeling each spine. We added one explicit spine to the neuronal geometry composed only of the soma connected to smooth and spiny dendrites. The spiny nature of some of the dendrites was recapitulated in the original geometry by increasing the capacitance of the compartments corresponding to the spiny dendrites. The explicit spine that we created had the same capacitance and membrane resistance as the smooth dendrites. We then traced an explicit path from the soma to the spine. The path includes the soma, the main dendrite as defined in the original geometry, several smooth and spiny dendrites that must be traversed to reach the spine of interest, and the spine itself. The remaining dendrites were reduced to equivalent cylinders to represent the rest of the cell, while maintaining the electrical integrity of the Purkinje neuron. There are 17 branch points off of the explicit path. Each of these branch points provides an alternate path for current flow from the soma to terminal branches. Conversely, synaptic stimuli applied to terminal dendrites stemming from the branch points provide input to the attached explicit dendrite, and ultimately to the soma.
In global reduction methods, dendrites are grouped based on their electrotonic distance from the soma (RALL 1962, 1969; Rall and Agmon-Snir 1998) (see Electrotonic Distance/Length in the Supplementary Material). Various global methods were considered, as well as restricted methods and one dynamic method. All of these methods, which span a few decades, are listed in Table 1. As in our study, the reduction methods catalogued in the table designed their algorithms to address specific requirements, including grouping dendrites based on electrotonic length. Our focus was a preserved path from spine to soma. As such, we did not want to group our dendritic compartments solely based on electrotonic distance from the soma, but rather based on branch points from the preserved path. This requirement precluded direct adoption of any of the previously published reduction methods. Incompatibilities of each reduction method with a preserved path paradigm for an experimentally derived complicated neuronal morphology are listed in the table. Based on these incompatibilities, previously published methods in existence were rejected without implementing them in our simulations.
Table 1.
Reduction methods considered prior to the PPR method
| Method/Authors | Reference | Reduction classification | Incompatibility with preserved path |
|---|---|---|---|
| Bush and Sejnowski | (Bush and Sejnowski 1993) | Global | G, D |
| Pinsky and Rinzel | (Pinsky and Rinzel 1994) | Global | D |
| Traub et al. | (Traub et al. 1991) | Global | D, R |
| Stratford et al. | (Stratford et al. 1989) | Global | G, D, with option for L |
| Douglas and Martin | (Douglas and Martin, 1993) | Global | D, G, R |
| Clements and Redman | (Clements and Redman 1989) | Global | G, E, R |
| Fleshman et al. | (Fleshman et al. 1988) | Global | D, G, E, R |
| Booth et al. | (Booth et al. 1997) | Global | D |
| McCormick and Huguenard | (McCormick and Huguenard 1992) | Global | D |
| Rall and Rinzel | (Rall and Rinzel 1973) | Global | G, D, R, with option for L |
| Rinzel and Rall | (Rinzel and Rall 1974) | Restricted | G, L, R |
| Rapp et al. | (Rapp et al. 1992) | Restricted | L, M |
| Burke et al. | (Burke et al. 1994) | Restricted | L, G, E, R |
| Manor et al. | (Manor et al. 1991) | Dynamic | A, G, E, R |
| Hendrickson et al. | (Hendrickson et al. 2010) | Global; Restricted | G, E, D; G, E, L |
G—Groups dendrites based on electrotonic length/distance
E—Explicitly calculates length and/or radius of equivalent cylinders using electrotonic length/distance
L—Leaves an entire branched subtree unreduced
D—Does not allow for applying or measuring synaptic activity in a single spine or any single unreduced compartment (while still tracing an explicit unreduced path to the soma)
A—Alternates between 3. and 4.
R—Requires adherence to 3/2 power law.
M—Model reduction based on absorbing inactive spines into parent dendrites.
In the end, we developed a novel reduction method to meet the needs described above. In this method, dendrites are grouped based on their branch point from the explicit path. This path is highlighted in red in Fig. 1(a,b). In the reduced geometry, the smooth and spiny dendrites in the explicit path are conjoined to give fewer explicit compartments representing the behavior of the original geometry. As such, in Fig. 1(b), the order of connection of the seven preserved compartments is as follows: soma (i), maindendrite (ii), smoothdistaldendrite-short (iii), smoothdistaldendritelong (iv), spinydistaldendrite (v), adjacentdendrite (vi), and spine (vii). See Branch Points in the Supplementary Material for details on how the rest of the dendritic tree was grouped into 10 reduced compartments that branch off of the explicit path.
3.2 Investigating passive properties and action potentials in NEURON
To investigate the applicability of various reduction strategies, we first simulated passive properties, for several reasons. First, it is convenient to analyze the effect of geometry modification on passive properties, before adding in active conductances. Second, a greater level of intuition can be gained for the behavior of the model system in the absence of active properties. Third, correct passive properties of the model provide a foundation for exploring active properties. This is particularly important, because, as will be shown below, it is possible, surprisingly, to find differing parameter sets to simulate either the passive or active case. We searched for a parameterization system suitable for both the passive and active cases.
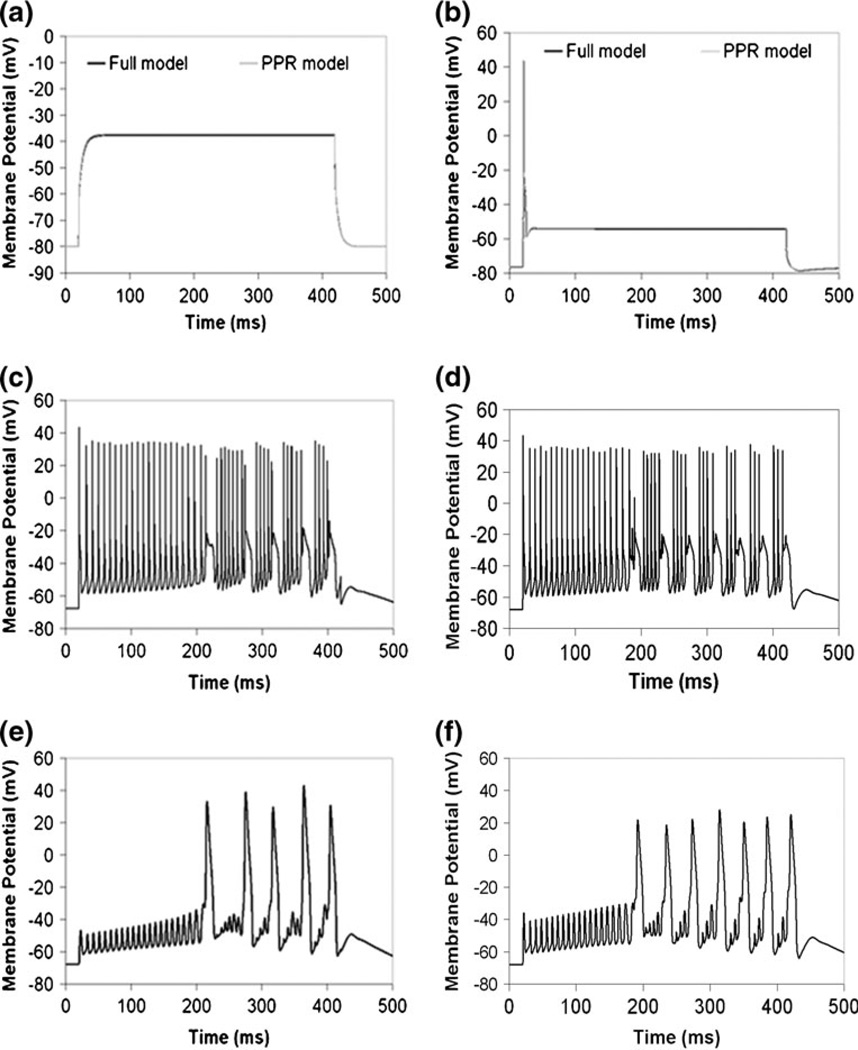
Miyasho et al. (Miyasho et al. 2001), the authors of the study with the original Purkinje geometry, applied a current injection of 2 nA for 400 ms and with a delay of 20 ms at the soma in their experiment and in their NEURON model. We used this pattern of current injection to explore the passive properties of both the full and reduced models, at various stages of parameterization of the reduced model. Figure 2(a) and Supplemental Figure S1(a) show the results of simulating this current injection in the full and reduced models, with no active conductances—just a leak current. Current injection in the full model (Fig. 2(a)) induced depolarization of the soma to a plateau potential of −37.5 mV from a resting potential of −67.8 mV. Without applying a scaling factor in the passive reduced model, simulating the same current injection depolarized the soma to a plateau potential of 78.8 mV (Supplemental Fig. S1(a)). This suggests that the input resistance (Rin) of the reduced model is more than three times as large as that for the full model, since the same current gave more than three times as large a voltage response. This is presumably due to the decreased surface area of the simplified geometry relative to the full geometry. This is also due, in part, to inadequate treatment of the series versus parallel nature of the connectivity of the dendrites branching off of the preserve path, as discussed in more detail in the Supplementary Material on Reduction Methods. We thus developed a new reduction method (preserved path reduction or PPR) that is more suitable for the Purkinje neuron and other highly branched complex geometries, as described in the Models section. In Fig. 2(a), scaling down Rm and scaling up Cm in passive dendrites gave the same input resistance and membrane time constant in the PPR model as in the full model. It is important to note that we attempted reduction of the Purkinje neuron using various previously established schemes, including the method developed by Bush and Sejnowski (Bush and Sejnowski 1993) for pyramidal neurons (see Reduction Methods in Supplementary Material). None of these gave reasonable fits to the full model.
Fig. 2.
Reduction of complex Purkinje neuronal morphology in NEURON. (a) Passive electrical properties at the soma in the full and PPR models. (b) Electrical properties at the soma in the full and PPR models with active channels in the soma. (c) Membrane potential oscillations in the soma due to current injection at the soma in the full model, with active channels localized to the soma, dendrites, and spine. (d) Scaling down calcium by a modified surface area ratio produces membrane potential oscillations in the soma in the PPR model, due to current injection at the soma, with active channels localized to the soma, dendrites, and spine. e, Membrane potential oscillations at the spine in the full model with active channels in the soma, dendrites, and spine. (f) Scaling down calcium by the modified surface area ratio produces oscillations at the spine in the PPR model. IClamp at the soma: onset 20 ms, duration 400 ms, amplitude 2 nA, as in Miyasho et al. 2001
Next, active channels were added just to the soma, and the same current injection was applied. For the full model, a single action potential with an amplitude of 43.9 mV was obtained (Fig. 2(b)). The action potential was followed by a plateau potential of −54 mV for the rest of the duration of the current pulse. Adding voltage-gated ion channels only to the soma in the PPR model, then applying the current injection, also gave the same results as in the full model. Next, we added channels to the dendrites in both models, and applied the same current injection. To account for the decreased volume into which calcium flows in the PPR model, we also adjust the molecular flux of calcium into the equivalent cylinders, using the same scaling factor used to adjust the active conductances (see Further Analysis of the PPR Model in the Supplementary Material). In the equivalent cylinders of the PPR model, we divide the right hand side of the expression by the scaling factor. This adjusts the calcium influx into the reduced volume by the same amount that the active calcium conductances are scaled to account for reduced surface area.
As shown in Fig. 2(c), action potential oscillations are obtained in the full model. The results for the PPR model are shown in Fig. 2(d), and supplemental Figure S4 shows the corresponding action potential oscillations at various locations in the reduced geometry. The PPR model closely reproduces the action potential oscillations and the transient plateaus as in the full model, albeit at a slightly increased frequency. Similar to the study by Bush and Sejnowski presenting a reduction method appropriate for pyramidal cells (Bush and Sejnowski 1993), Bush and Sejnowski’s reduced pyramidal cell model with active channels in the soma also displayed slightly increased action potential frequency relative to the full model. As in their discussion of the pyramidal cell, a slight increase in frequency for the reduced model of the Purkinje neuron is also acceptable, as the behavior falls within the range of experimental observations for variability among cells
Taken together, these results suggest that the preserved path reduction method provides a means to simplify the complicated Purkinje neuron morphology. Further, we successfully validated the PPR method using Rallpack ideal geometries (Bhalla et al. 1992) implemented in NEURON (see The PPR Method Generalized Using Rallpacks and Figure S11 in the Supplementary Material). Active data for Rallpack2 are provided as Supplementary Material, including NEURON hoc and mod files. The files are entitled ‘Rallpack 2—PPR Reduction—Brown et al. 2010.xls’, ‘mosinit_Rallpack2_hh2_Brown.hoc’, ‘mosinit_Rallpack2_hh2_PPRreduced_Brown.hoc’, ‘Rallpack2_hh2_Brown.hoc’, ‘Rallpack2_hh2_PPRreduced_Brown.hoc’, and ‘Rallpack2_Brown.ses’.
3.3 E.P.S.P. propagation from spine to soma in NEURON
When excitatory postsynaptic potentials (E.P.S.P.s) are applied to equivalent cylinders, they are effectively being applied to all the dendrites that have been reduced. Thus, this represents identical activation of many spines and several dendrites at once, precluding the study of more local activity or biochemical changes within individual spines. This is the case in Rall’s models (Rall and Agmon-Snir 1998), as well as in the reduced models developed by Bush and Sejnowski (Bush and Sejnowski 1993).In contrast, our method permits us to simulate local activity in single dendrites and single spines (see Synaptic Stimulation in the Supplementary Material) in the reduced models anywhere along the preserved path.
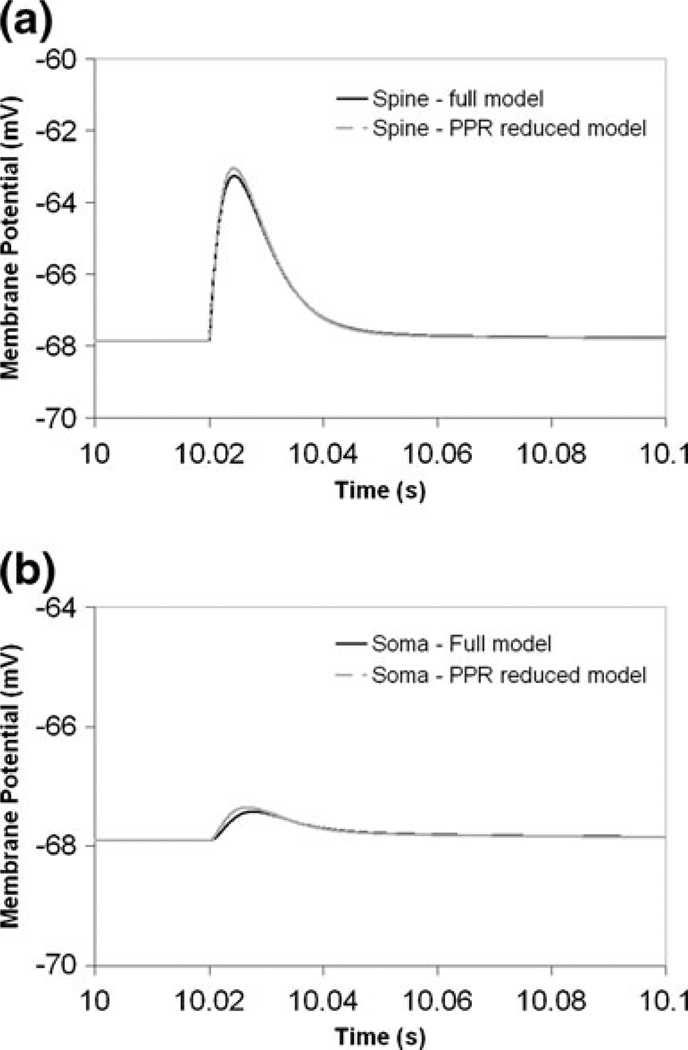
Figure 3 shows that similar voltage changes evoked by EPSCs applied at a single spine are obtained from the full and the PPR models at the spine and at the soma. Figure 3(a) shows the resulting voltage response in the spine. The membrane potential in the spine rises from about −68 mV to approximately −63 mV. Since membrane potential is allowed to propagate between the spine and the soma, and given that the E.P.S.P. is usually experimentally measured at the soma, the soma voltage response is also shown in Fig. 3(b). Attenuation between the spine and the soma results in a soma voltage of only about 0.5 mV, again showing very similar changes in the full and PPR models. Thus, the PPR model is a good representation of the voltage response at the spine and at the soma, as well as of the attenuation between the spine and the soma. Furthermore, current injection at the soma produces the same calcium spike trains in the spine in both the full (Fig. 2(e)) and PPR (Fig. 2(f)) models. This suggests that the preserved path reduction method allows us to simplify the complex Purkinje geometry, while being able to stimulate a single spine and accurately simulate the propagated response at the soma.
Fig. 3.
E.P.S.P. propagation from the spine to the soma in the full and PPR models in NEURON. (a) Influence of synaptic current injection on membrane potential of spine head. (b) Corresponding voltage changes in the soma. AlphaSynapse at the spine: onset 20 ms, tau (tpeak) 3.809 ms, gmax 0.0013µS, e −10 mV
3.4 Comparing results in virtual cell and NEURON
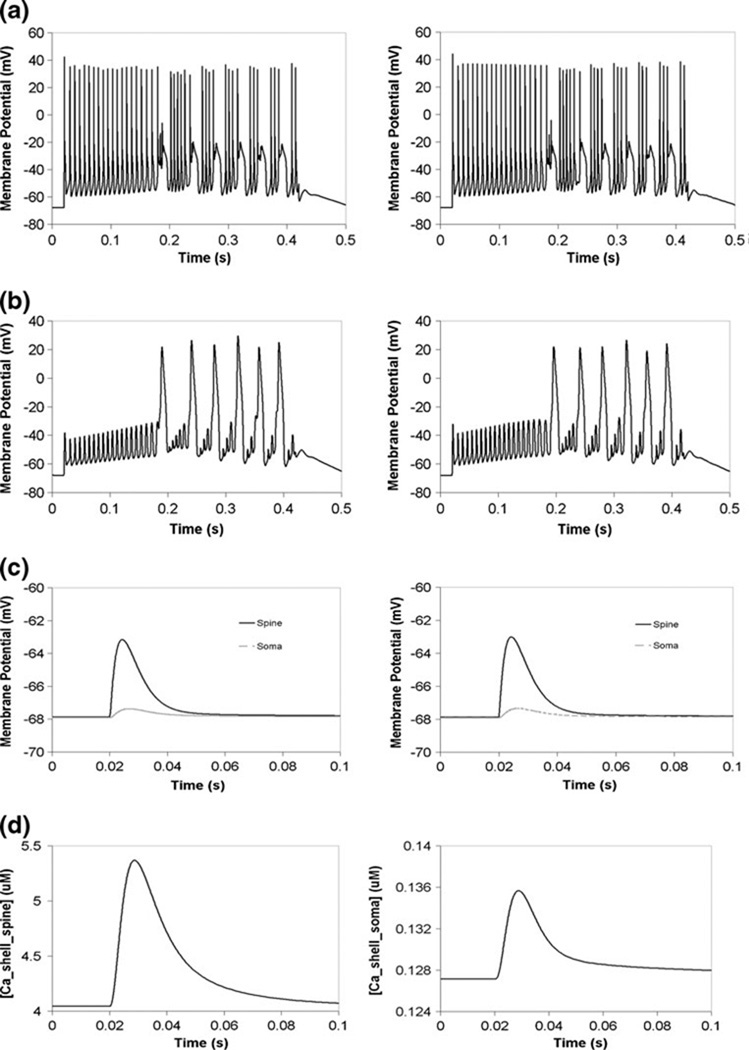
The PPR model was reproduced as a multicompartmental model in Virtual Cell, and can be simulated with a variety of solvers provided by VCell (see The PPR Model in Virtual Cell and NEURON in the Supplementary Material). Simulation of a 2 nA current injection for 400 ms at the soma produced a train of action potentials in Virtual Cell, as in NEURON. The patterns of spike timing and amplitude from the two platforms are identical, and are shown separately for clarity in Fig. 4(a) and superimposed in Figure S12. The membrane potential is allowed to propagate to the spine, and produces attenuated oscillations there (Fig. 4(b)). The voltage oscillations allow calcium influx through voltage-gated calcium channels. The resulting submembrane shell response is shown in Fig. 5(a) for Virtual Cell, and is similar to results obtained in NEURON (data not shown). Spine calcium in the submembrane shell reaches levels as high as 500 µM, as in the original NEURON model created by Miyasho et al. (Miyasho et al. 2001). A hyperpolarizing current also gives similar membrane potential transients in NEURON and Virtual Cell (Supplemental Figure S9).
Fig. 4.
Comparison of PPR Purkinje model results in Virtual Cell and NEURON. (a) Membrane potential oscillations due to current injection at the soma in NEURON (left) and Virtual Cell (right). (b) Backpropagated action potential oscillations at the spine due to current injection at the soma in NEURON (left) and Virtual Cell (right). (c) Membrane potential response at the spine and the soma due to synaptic current injection in NEURON (left) and Virtual Cell (right). (d) Submembrane shell calcium response in the soma and the spine due to synaptic current injection in Virtual Cell. IClamp at the soma: onset 20 ms, duration 400 ms, amplitude 2 nA, as in Miyasho et al. 2001. AlphaSynapse at the spine: onset 20 ms, tau (tpeak) 3.809 ms, gmax 0.0013 µS, e −10 mV
Fig. 5.
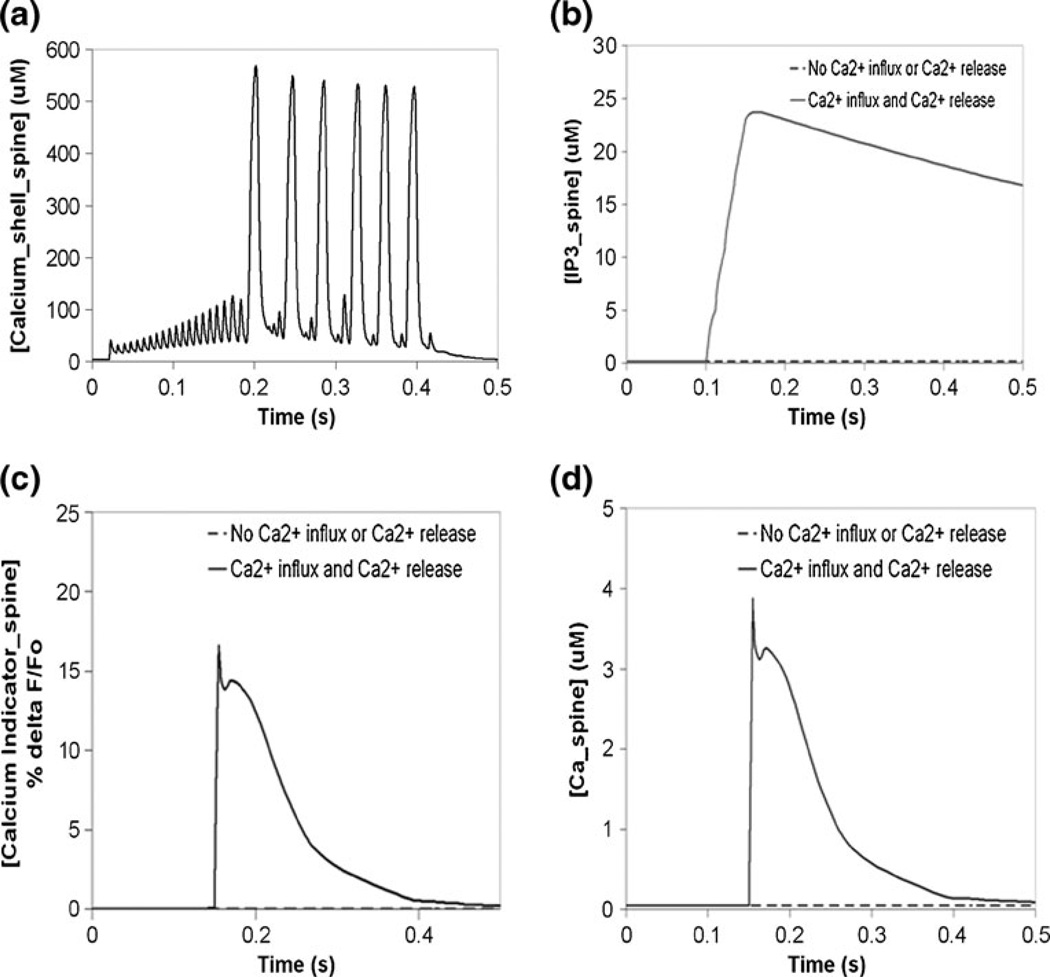
Submembrane calcium and IP3-induced calcium release in the Purkinje spine in Virtual Cell. (a) Ca_shell oscillations at the spine, due to sustained depolarizing current injection at the soma (as in Fig. 4). (b) IP3 signal at the spine due to simulation of a pulse train of four parallel fiber (PF) stimuli. (c) Simulated calcium indicator flourescence change at the spine, in response to coincident activation of the Purkinje neuron by a train of four parallel fiber stimuli and a single climbing fiber (CF) stimulus. (d) Free calcium response due to coincident activation by 4 PF stimuli and a single CF stimulus. The initial PF stimulus occurs at 0.1 s; the CF stimulus occurs at 0.15 s. The resulting supralinear calcium release is first observed at 0.15 s
The E.P.S.P. was also compared in Virtual Cell and NEURON. The alpha function synaptic conductance (see Synaptic Stimulation in the Supplementary Material) was applied at the spine in each software. The spine membrane potential increases by almost 5 mV in both software platforms, as shown in Fig. 4(c). The current propagates to the soma in both, and gives a rise of approximately 0.5 mV. Figure 4(d) shows the resulting submembrane calcium concentration in the spine and the soma in Virtual Cell. Spine shell calcium rises by almost 4 µM, and soma shell calcium rises by 2 µM. Similar results are obtained in NEURON (data not shown). These results indicate that implementation of the PPR Purkinje model in Virtual Cell permits us to faithfully simulate propagation of membrane potential between compartments, as in NEURON.
3.5 Merging electrophysiology with biochemistry
With a working electrophysiological model in Virtual Cell, we then added in biochemistry. We merged the PPR model with our published biochemical model of phosphoinositide and calcium signaling (Hernjak et al. 2005; Brown et al. 2008). Figure 5(a) shows the calcium transient in the submembrane shell in the spine of interest, resulting from a depolarizing current pulse at the soma in the combined model. The submembrane calcium concentration oscillates with an amplitude of approximately 500uM, corresponding to backpropagating action potential oscillations at the spine, shown in Fig. 4(b). Hyperpolarizing currents in models with voltage-gated ion channels allow for comparison of the passive properties of the model without activation of these channels. Membrane potential and submembrane shell transients at the spine and at the soma, due to a hyperpolarizing current injection at the soma in the combined model in Virtual Cell, are shown in Supplemental Figure S10, and are similar to results from NEURON (membrane potential in Fig. S9, submembrane calcium not shown). When a parallel fiber (PF) stimulates a Purkinje neuron spine, the presynaptic axon releases glutamate, which binds its metabotropic receptor (mGluR) on the postsynaptic membrane, leading to activation of the enzyme phospholipase C (PLC) (Finch and Augustine 1998; Takechi et al. 1998). The activated enzyme then cleaves phophophatidylinositol-4,5-bisphosphate (PIP2) to produce inositol trisphosphate (IP3). Figure 5(b) shows the IP3 signal from a train of four PF stimuli. IP3 rises from a basal concentration of 0.16 µM to approximately 24 µM. The IP3 produced diffuses away from the plasma membrane to bind the IP3 Receptor Type 1 (IP3R1) on the smooth endoplasmic reticulum (sER) membrane (Berridge et al. 2000). In addition, Purkinje neurons can receive stimulus from the climbing fiber (CF) of a different presynaptic neuron (Ito and Kano 1982). The CF stimulus depolarizes the entire Purkinje neuron, including the spine, leading to opening of voltage-gated calcium channels (Konnerth et al. 1992; Regehr and Mintz 1994). Calcium rushes into the spine and binds the IP3 receptor, along with IP3. This synergizes the release of calcium from the sER and results in supralinear calcium release (Wang et al. 2000), where the rise in calcium concentration is substantially greater than for the simple summation of individually activated calcium release and calcium influx pathways. This process is known as coincident activation or coincidence detection, where the IP3 receptor can act as a coincident detector of PF and CF stimulus that occur closely in time (Sarkisov and Wang 2008).
Figures 5(c) and (d) show the changes in calcium indicator fluorescence and free calcium, respectively, in response to coincident activation of the Purkinje neuron spine by a train of four PF stimuli and a single CF stimulus, and with the climbing fiber stimulus occurring 50 ms after the start of the train of four PF stimuli. The simulated calcium indicator fluorescence increased by approximately 16% as observed experimentally by Wang et al. (Wang et al. 2000), in an experiment using Magnesium Green, a low affinity (Kd~19) calcium indicator. These results are consistent with our previously published models, which have focused on the phosphatidylinositol signaling alone (Hernjak et al. 2005; Brown et al. 2008). Thus, we have created a model in Virtual Cell that combines electrophysiology with biochemistry and can be used to probe deeper into bidirectional interactions between the two types of processes. This result demonstrates how the preserved path reduction method allows us to simplify a complex neuron morphology and create a computationally tractable model that combines biochemistry at the spine with electrophysiology at the spine and at the soma.
4 Discussion
We created a novel reduction method of a neuronal dendritic morphology that we have termed preserved path reduction or PPR. In the PPR model, an electrical path between the soma and a particular spine on a remote dendrite is preserved as an explicit, unreduced path, whereas the rest of the morphology is reduced to a relatively small number of equivalent cylindrical compartments. The axial resistivity (Ra), input resistance (Rin), and the membrane time constant (τ) remained unchanged, while the membrane resistance (Rm) and capacitance (Cm) were adjusted based on the difference in surface area between the actual and reduced compartments. This approach creates a good fit between the full and reduced passive models. In addition, active conductances were scaled in the equivalent cylinders, as were the fluxes for calcium flowing into the equivalent cylinders through voltage-gated calcium channels on the membrane. The resulting reduced Purkinje cell model can be used in single cell simulations or in network simulations. Use in either scenario will yield increased computational efficiency, decreased run time, and greater feasibility for large networks. Further, we showed that multiple equivalent and unreduced cylinders can be connected in Virtual Cell to model the neuron as an electrical circuit. Results for action potentials and E.P.S.P.s at the spine and the soma in Virtual Cell closely match those in NEURON. Implementing the PPR model motivated the development of new approaches in Virtual Cell that facilitate the investigation of multicompartmental electrophysiological and biochemical processes. Specifically, we were able to mimic a NEURON model by specifying ionic and molecular fluxes between multiple compartments in Virtual Cell. As a result, it is now possible to create models of cellular geometries that consist of “virtually connected” conductors that each have an equipotential membrane—similar to the NEURON modeling paradigm. This will also allow users to take advantage of the power of Virtual Cell to add and easily edit detailed biochemistry combined with the electrophysiology model. In addition, this work led to performance optimization in Virtual Cell that will be beneficial to users with large models, particularly ones designed to investigate neuronal electrophysiology. This model contains approximately 350 variables and 2000 functions, and is one of the larger compartmental models publicly available in the Virtual Cell database.
However, we wish to emphasize that Virtual Cell is not intended as a tool for modeling electrophysiology in complex neuronal morphologies. The PPR method allows us to simplify a complicated neuronal geometry in order to create models more tractable in Virtual Cell. Reproducing the full Purkinje neuron geometry as either a multicompartment ODE model or a full spatial PDE model in Virtual Cell would be completely impractical, but for different reasons. The ODE model cannot be constructed with the graphical user interface because it is not designed to specify the hundreds of compartments required to represent complex neuronal morphologies. The PDE spatial model would actually be easier to build in Virtual Cell, because the experimentally derived geometry can be directly mapped to the physiology; but the PDE solvers in Virtual Cell would require too much memory and too much computational time to deal with the high spatial resolution required to simulate the full Purkinje geometry. For these reasons, the full Rallpack cable test (Bhalla et al. 1992) also cannot be implemented in Virtual Cell. However, the Purkinje PPR model in Virtual Cell was validated against the implementation in NEURON. The correspondence of the simulation results on the two platforms for the PPR model (Fig. 4), with its 17 heterogeneous compartments each with active channel conductances, might be considered an even more rigorous validation than the more idealized Rallpacks models.
We developed this new approach to neuronal morphology reduction, because of our desire to have a computationally efficient Purkinje cell model in which we could investigate issues that require combined detailed biochemistry and electrophysiology. None of the available reduction methods was suitable for our study. The cerebellar Purkinje neuron is highly and asymmetrically branched, and does not obey the “3/2 rule”. Further, we needed to preserve a path from a single spine to the soma, in order to appropriately compare simulations to experiments. At the same time, we did not wish to maintain explicit dendritic arbors that branch off of the explicit path, as that would not be computationally feasible in VCell, particularly once biochemistry was added.
Rall’s and Rinzel’s traditional methods for global reduction assume, among other things, that the sum of the 3/2 power of the daughter dendrite diameters is equivalent to the parent dendrite diameter raised to the 3/2 power (Rall and Agmon-Snir 1998). These methods also assume that terminal branches are at the same electrotonic distance from the soma. The terminal daughter branches can be lumped into equivalent cylinders. This process can be continued iteratively until the whole dendritic tree is represented by a few—or even a single—equivalent cylinders (Rall and Agmon-Snir 1998). However, most realistic neurons do not satisfy these assumptions (Rall and Agmon-Snir 1998). As a result, a few laboratories have suggested alternative requirements for reduction. Douglas and Martin preserve dendritic surface area as a function of electrotonic distance from the soma (Douglas and Martin 1993). However, mapping dendrites to equivalent cylinders based on their electrotonic distance from the soma is not appropriate in the preserved path reduction method, since dendrites are lumped together based on branch points from the explicit path.
Two candidate algorithms were ultimately considered: the Bush-inspired method (see Reduction Methods in the Supplementary Material) and the PPR model. Using the radius and length equations presented by Bush and Sejnowski (Bush and Sejnowski 1993) in various approaches to preserved path reduction resulted in the creation of the Bush-inspired model. This reduced model used the scaling factor 4.5 to adjust the membrane resistance and membrane capacitance, as well as voltage-gated ion channel conductances. This scaling factor was determined iteratively by adjusting Rm until Rin in the reduced model fit Rin in the full model. This phenomenologically derived scaling factor is used globally for all compartments, and does not depend on the particular geometry or properties of one compartment or the group of individual dendrites that it represents. The resting potential of the Purkinje neuron in this simplified model was hyperpolarized relative to the full model. Further, passive and active properties were different between the Bush-inspired model and the full model, when voltage-gated ion channels were added to the dendrites in each model. This led us to create the PPR model using a scaling factor that is theoretically derived by taking into account the differences in surface area between the full and reduced models and by appropriately factoring in the series versus parallel connectivity of the highly branched Purkinje neuron dendritic arbor. Moreover, this scaling factor is separately calculated for each individual compartment in the PPR model. Each reduced compartment has its own scaling factor, based on its unique decrease in surface area relative to the group of dendrites it represents. Thus, unlike in the Bush-inspired model, Rm and Cm are not phenomenologically derived, and are scaled individually in the equivalent cylinders, not globally over the entire geometry. The resting membrane potential in the PPR model is similar to that of the full model. Passive and active properties also closely fit those of the full model. The PPR model overall reproduces the behavior of the full model.
In conclusion, this study reports a novel reduction method that can be used to simplify complex neuronal geometries. Conditions that encourage using the reduction method developed in this study for any neuron with complex geometry include (i) dendrites that contain voltage-gated ion channels (as opposed to passive reduction methods that ignore active cases), (ii) a high degree of asymmetrical branching, (iii) a great number of dendrites in series as well as in parallel, (iv) significant branching off of an explicit path of interest yielding a large number of compartments. The PPR model enables us to simultaneously meet three challenges in modeling neuronal cell biology. First, the Purkinje neuron geometry is complicated, with large degrees of series and parallel branching, such that other reduction methods that we considered were deemed unsuitable to accurately reproduce the electrophysiology of the full geometry. Second, we can model both explicit spine and soma physiology in the same model, which—to the best of our knowledge—has not previously been successfully attempted; this requirement precludes the use of global, restricted, or dynamic reduction methods. Third, the PPR reduction scheme is tractable enough to be transferred to a modeling software system, Virtual Cell, which can efficiently simulate complex biochemistry added to the model. Thus, the PPR model can be used to investigate interactions between biochemistry and electrophysiology. Studies are now underway involving the application of the PPR model to normal physiology and pathophysiology of cerebellar Purkinje neurons, with particular attention to cerebellar dysfunction in mice and humans.
Supplementary Material
Acknowledgements
We thank Fei Gao and Anuradha Lakshminarayana for help and support in various aspects of this work. We are also pleased to acknowledge Dr. Corey Acker, Dr. James Watras, Dr. Ann Cowan, and other members of the R. D. Berlin Center for Cell Analysis & Modeling at the University of Connecticut Health Center for helpful discussions.
This research was supported by grants R01 MH086638, P41 RR013186 and U54 RR022232 from the National Institutes of Health.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s10827-011-0317-0) contains supplementary material, which is available to authorized users.
Contributor Information
Sherry-Ann Brown, Richard D. Berlin Center for Cell Analysis & Modeling, University of Connecticut Health Center, Farmington, CT 06030, USA.
Ion I. Moraru, Richard D. Berlin Center for Cell Analysis & Modeling, University of Connecticut Health Center, Farmington, CT 06030, USA
James C. Schaff, Richard D. Berlin Center for Cell Analysis & Modeling, University of Connecticut Health Center, Farmington, CT 06030, USA
Leslie M. Loew, Email: les@volt.uchc.edu, Richard D. Berlin Center for Cell Analysis & Modeling, University of Connecticut Health Center, Farmington, CT 06030, USA.
References
- Berridge M, Lipp P, Bootman M. The versatility and universality of calcium signalling. Nature Reviews. Molecular Cell Biology. 2000;1:11–21. doi: 10.1038/35036035. [DOI] [PubMed] [Google Scholar]
- Bhalla U, Bilitch D, Bower J. Rallpacks: a set of benchmarks for neuronal simulators. Trends in Neurosciences. 1992;15:453–458. doi: 10.1016/0166-2236(92)90009-w. [DOI] [PubMed] [Google Scholar]
- Booth V, Rinzel J, Kiehn O. Compartmental model of vertebrate motoneurons for Ca2+-dependent spiking and plateau potentials under pharmacological treatment. J Neurophysiol. 1997;78:3371–3385. doi: 10.1152/jn.1997.78.6.3371. [DOI] [PubMed] [Google Scholar]
- Bower JM, Beeman D. The Book of GENESIS: Exploring Realistic Neural Models with the GEneral NEural SImulation System (Free Internet Version) New York, New York: Springer-Verlag; 2003. [Google Scholar]
- Bower J, Beeman D. Constructing realistic neural simulations with GENESIS. Methods Mol Biol. 2007;401:103–125. doi: 10.1007/978-1-59745-520-6_7. [DOI] [PubMed] [Google Scholar]
- Brown S, Morgan F, Watras J, Loew LM. Analysis of phosphatidylinositol-4, 5-bisphosphate signaling in cerebellar Purkinje spines. Biophysical Journal. 2008;95:1795–1812. doi: 10.1529/biophysj.108.130195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke R, Fyffe R, Moschovakis A. Electrotonic architecture of cat gamma motoneurons. J Neurophysiol. 1994;72:2302–2316. doi: 10.1152/jn.1994.72.5.2302. [DOI] [PubMed] [Google Scholar]
- Bush P, Sejnowski T. Reduced compartmental models of neocortical pyramidal cells. J Neurosci Methods. 1993;46:159–166. doi: 10.1016/0165-0270(93)90151-g. [DOI] [PubMed] [Google Scholar]
- Clements J, Redman S. Cable properties of cat spinal motoneurones measured by combining voltage clamp, current clamp and intracellular staining. J Physiol. 1989;409:63–87. doi: 10.1113/jphysiol.1989.sp017485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Schutter E, Bower J. An active membrane model of the cerebellar Purkinje cell. I. Simulation of current clamps in slice. Journal of Neurophysiology. 1994;71:375–400. doi: 10.1152/jn.1994.71.1.375. [DOI] [PubMed] [Google Scholar]
- De Smedt H, Missiaen L, Parys J, Henning R, Sienaert I, Vanlingen S, et al. Isoform diversity of the inositol trisphosphate receptor in cell types of mouse origin. The Biochemical Journal. 1997;322(Pt 2):575–583. doi: 10.1042/bj3220575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Destexhe A, Babloyantz A, Sejnowski T. Ionic mechanisms for intrinsic slow oscillations in thalamic relay neurons. Biophysical Journal. 1993;65:1538–1552. doi: 10.1016/S0006-3495(93)81190-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Gregorio E, Orsi L, Godani M, Vaula G, Jensen S, Salmon E, et al. Two Italian families with ITPR1 gene deletion presenting a broader phenotype of SCA15. Cerebellum. 2010 doi: 10.1007/s12311-009-0154-0. [DOI] [PubMed] [Google Scholar]
- Douglas R, Martin K. Exploring cortical microcircuits: A combined anatomical, physiological, and computational approach. In: McKenna T, et al., editors. Single neuron computation. Orlando, FL: Academic Press; 1993. pp. 381–412. [Google Scholar]
- Finch E, Augustine G. Local calcium signalling by inositol-1, 4, 5-trisphosphate in Purkinje cell dendrites. Nature. 1998;396:753–756. doi: 10.1038/25541. [DOI] [PubMed] [Google Scholar]
- Fleshman J, Segev I, Burke R. Electrotonic architecture of type-identified alpha-motoneurons in the cat spinal cord. J Neurophysiol. 1988;60:60–85. doi: 10.1152/jn.1988.60.1.60. [DOI] [PubMed] [Google Scholar]
- Gilbert P, Thach W. Purkinje cell activity during motor learning. Brain Research. 1977;128:309–328. doi: 10.1016/0006-8993(77)90997-0. [DOI] [PubMed] [Google Scholar]
- Hara K, Shiga A, Nozaki H, Mitsui J, Takahashi Y, Ishiguro H, et al. Total deletion and a missense mutation of ITPR1 in Japanese SCA15 families. Neurology. 2008;71:547–551. doi: 10.1212/01.wnl.0000311277.71046.a0. [DOI] [PubMed] [Google Scholar]
- Harris K, Stevens J. Dendritic spines of rat cerebellar Purkinje cells: serial electron microscopy with reference to their biophysical characteristics. The Journal of Neuroscience. 1988;8:4455–4469. doi: 10.1523/JNEUROSCI.08-12-04455.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson E, Edgerton J, Jaeger D. The capabilities and limitations of conductance-based compartmental neuron models with reduced branched or unbranched morphologies and active dendrites. J Comput Neurosci. 2010 doi: 10.1007/s10827-010-0258-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernjak N, Slepchenko BM, Fernald K, Fink CC, Fortin D, Moraru II, et al. Modeling and analysis of calcium signaling events leading to long-term depression in cerebellar purkinje cells. Biophysical Journal. 2005;89:3790–3806. doi: 10.1529/biophysj.105.065771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines ML, Carnevale NT. The NEURON simulation environment. Neural Computation. 1997;9:1179–1209. doi: 10.1162/neco.1997.9.6.1179. [DOI] [PubMed] [Google Scholar]
- Hines ML, Carnevale NT. NEURON: a tool for neuroscientists. The Neuroscientist. 2001;7:123–135. doi: 10.1177/107385840100700207. [DOI] [PubMed] [Google Scholar]
- Hodgkin AL, Huxley AF. A quantitative description of membrane current and its application to conduction and excitation in nerve. The Journal of Physiology. 1952;117:500–544. doi: 10.1113/jphysiol.1952.sp004764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, Kano M. Long-lasting depression of parallel fiber-Purkinje cell transmission induced by conjunctive stimulation of parallel fibers and climbing fibers in the cerebellar cortex. Neuroscience Letters. 1982;33:253–258. doi: 10.1016/0304-3940(82)90380-9. [DOI] [PubMed] [Google Scholar]
- Iwaki A, Kawano Y, Miura S, Shibata H, Matsuse D, Li W, et al. Heterozygous deletion of ITPR1, but not SUMF1, in spinocerebellar ataxia type 16. Journal of Medical Genetics. 2008;45:32–35. doi: 10.1136/jmg.2007.053942. [DOI] [PubMed] [Google Scholar]
- Konnerth A, Dreessen J, Augustine G. Brief dendritic calcium signals initiate long-lasting synaptic depression in cerebellar Purkinje cells. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:7051–7055. doi: 10.1073/pnas.89.15.7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loew L, Schaff J. The virtual cell: a software environment for computational cell biology. Trends in Biotechnology. 2001;19:401–406. doi: 10.1016/S0167-7799(01)01740-1. [DOI] [PubMed] [Google Scholar]
- Major G, Larkman A, Jonas P, Sakmann B, Jack J. Detailed passive cable models of whole-cell recorded CA3 pyramidal neurons in rat hippocampal slices. The Journal of Neuroscience. 1994;14:4613–4638. doi: 10.1523/JNEUROSCI.14-08-04613.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manor Y, Gonczarowski J, Segev I. Propagation of action potentials along complex axonal trees. Model and implementation. Biophys J. 1991;60:1411–1423. doi: 10.1016/S0006-3495(91)82178-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto M, Nakagawa T, Inoue T, Nagata E, Tanaka K, Takano H, et al. Ataxia and epileptic seizures in mice lacking type 1 inositol 1, 4, 5-trisphosphate receptor. Nature. 1996;379:168–171. doi: 10.1038/379168a0. [DOI] [PubMed] [Google Scholar]
- McCormick D, Huguenard J. A model of the electrophysiological properties of thalamocortical relay neurons. J Neurophysiol. 1992;68:1384–1400. doi: 10.1152/jn.1992.68.4.1384. [DOI] [PubMed] [Google Scholar]
- Miyasho T, Takagi H, Suzuki H, Watanabe S, Inoue M, Kudo Y, et al. Low-threshold potassium channels and a low-threshold calcium channel regulate Ca2+ spike firing in the dendrites of cerebellar Purkinje neurons: a modeling study. Brain Research. 2001;891:106–115. doi: 10.1016/s0006-8993(00)03206-6. [DOI] [PubMed] [Google Scholar]
- Moraru II, Schaff JC, Slepchenko BM, Loew LM. The virtual cell: an integrated modeling environment for experimental and computational cell biology. Annals of the New York Academy of Sciences. 2002;971:595–596. doi: 10.1111/j.1749-6632.2002.tb04535.x. [DOI] [PubMed] [Google Scholar]
- Napper R, Harvey R. Number of parallel fiber synapses on an individual Purkinje cell in the cerebellum of the rat. The Journal of Comparative Neurology. 1988a;274:168–177. doi: 10.1002/cne.902740204. [DOI] [PubMed] [Google Scholar]
- Napper R, Harvey R. Quantitative study of the Purkinje cell dendritic spines in the rat cerebellum. The Journal of Comparative Neurology. 1988b;274:158–167. doi: 10.1002/cne.902740203. [DOI] [PubMed] [Google Scholar]
- Ogura H, Matsumoto M, Mikoshiba K. Motor discoordination in mutant mice heterozygous for the type 1 inositol 1, 4, 5-trisphosphate receptor. Behavioural Brain Research. 2001;122:215–219. doi: 10.1016/s0166-4328(01)00187-5. [DOI] [PubMed] [Google Scholar]
- Pinsky P, Rinzel J. Intrinsic and network rhythmogenesis in a reduced Traub model for CA3 neurons. J Comput Neurosci. 1994;1:39–60. doi: 10.1007/BF00962717. [DOI] [PubMed] [Google Scholar]
- RALL W. Electrophysiology of a dendritic neuron model. Biophysical Journal. 1962;2:145–167. doi: 10.1016/s0006-3495(62)86953-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rall W. Time constants and electrotonic length of membrane cylinders and neurons. Biophysical Journal. 1969;9:1483–1508. doi: 10.1016/S0006-3495(69)86467-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rall W, Rinzel J. Branch input resistance and steady attenuation for input to one branch of a dendritic neuron model. Biophys J. 1973;13:648–687. doi: 10.1016/S0006-3495(73)86014-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rall W, Agmon-Snir H. Cable theory for dendritic neurons. In: Koch C, Segev I, editors. Methods in neuronal modeling: From ions to networks. Cambridge: MIT; 1998. [Google Scholar]
- Rapp M, Yarom Y, Segev I. The impact of parallel fiber background activity on the cable properties of cerebellar Purkinje cells. Neural Computation. 1992;4:518–533. [Google Scholar]
- Rapp M, Segev I, Yarom Y. Physiology, morphology and detailed passive models of guinea-pig cerebellar Purkinje-cells. Journal of Physiology, London. 1994;474:101–118. doi: 10.1113/jphysiol.1994.sp020006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regehr W, Mintz I. Participation of multiple calcium channel types in transmission at single climbing fiber to Purkinje cell synapses. Neuron. 1994;12:605–613. doi: 10.1016/0896-6273(94)90216-x. [DOI] [PubMed] [Google Scholar]
- Rinzel J, Rall W. Transient response in a dendritic neuron model for current injected at one branch. Biophys J. 1974;14:759–790. doi: 10.1016/S0006-3495(74)85948-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkisov D, Wang S. Order-dependent coincidence detection in cerebellar Purkinje neurons at the inositol trisphosphate receptor. The Journal of Neuroscience. 2008;28:133–142. doi: 10.1523/JNEUROSCI.1729-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaff J, Fink C, Slepchenko B, Carson J, Loew L. A general computational framework for modeling cellular structure and function. Biophysical Journal. 1997;73:1135–1146. doi: 10.1016/S0006-3495(97)78146-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelton D. Membrane resistivity estimated for the Purkinje neuron by means of a passive computer model. Neuroscience. 1985;14:111–131. doi: 10.1016/0306-4522(85)90168-x. [DOI] [PubMed] [Google Scholar]
- Stratford K, Mason A, Larkman A, Major G, Jack J. The modelling of pyramidal neurones in the visual cortex. In: Durbin R, et al., editors. The Computing Neuron. London, England: Addison-Wesley; 1989. pp. 296–321. [Google Scholar]
- Street V, Bosma M, Demas V, Regan M, Lin D, Robinson L, et al. The type 1 inositol 1, 4, 5-trisphosphate receptor gene is altered in the opisthotonos mouse. The Journal of Neuroscience. 1997;17:635–645. doi: 10.1523/JNEUROSCI.17-02-00635.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takechi H, Eilers J, Konnerth A. A new class of synaptic response involving calcium release in dendritic spines. Nature. 1998;396:757–760. doi: 10.1038/25547. [DOI] [PubMed] [Google Scholar]
- Traub R, Wong R, Miles R, Michelson H. A model of a CA3 hippocampal pyramidal neuron incorporating voltage-clamp data on intrinsic conductances. J Neurophysiol. 1991;66:635–650. doi: 10.1152/jn.1991.66.2.635. [DOI] [PubMed] [Google Scholar]
- van de Leemput J, Chandran J, Knight M, Holtzclaw L, Scholz S, Cookson M, et al. Deletion at ITPR1 underlies ataxia in mice and spinocerebellar ataxia 15 in humans. PLoS Genetics. 2007;3:e108. doi: 10.1371/journal.pgen.0030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Denk W, Häusser M. Coincidence detection in single dendritic spines mediated by calcium release. Nature Neuroscience. 2000;3:1266–1273. doi: 10.1038/81792. [DOI] [PubMed] [Google Scholar]
- Watanabe M. Molecular mechanisms governing competitive synaptic wiring in cerebellar Purkinje cells. The Tohoku Journal of Experimental Medicine. 2008;214:175–190. doi: 10.1620/tjem.214.175. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.