Abstract
In the following respects, tooth enamel is a unique tissue in the mammalian body: (a) it is the most mineralized and hardest tissue in it comprising up to 95 wt% of apatite; (b) its microstructure is dominated by parallel rods composed of bundles of 40 – 60 nm wide apatite crystals with aspect ratios reaching up to 1:10,000 and (c) not only does the protein matrix that gives rise to enamel guides the crystal growth, but it also conducts its own degradation and removal in parallel. Hence, when mimicking the process of amelogenesis in vitro, crystal growth has to be coupled to proteolytic digestion of the amelogenin assemblies that are known to play a pivotal role in conducting the proper crystal growth. Experimental settings based on controlled and programmable titration of amelogenin sols digested by means of MMP-20 with buffered calcium and phosphate solutions were employed to imitate the formation of elongated, plate-shaped crystals. While amelogenin can act as a promoter of nucleation and crystal growth alone, in this study we show that proteolysis exerts an additional nucleation- and growth-promoting effect. Hydrolysis of full-length amelogenin by MMP-20 decreases the critical time needed for the protein and peptides to adhere and to cover the substrate. The formation and immobilization of a protein layer subsequently reduces the time for calcium phosphate crystallization. Coupling the proteolytic reaction to titration in the presence of 0.4 mg/ml rH174 has been shown to have the same effect on the crystal growth promotion as quadrupling the concentration of rH174 to 1.6 mg/ml. Controlling the rate and the extent of the proteolytic cleavage can thus be used to control the nucleation and growth rates in a protein-guided crystallization system.
Introduction
The biological formation of enamel tissue is known as amelogenesis and gives rise to the hardest tissue in the vertebrate body. Besides specialized cells, the ameloblasts, it involves macromolecular species that can be divided into families of proteins, proteases and protease inhibitors. Ninety percent of proteins that make up the enamel matrix are derived from the amelogenin gene, with the full-length protein being expressed predominantly (90%). The remaining 10 % is comprised of other proteins: ameloblastin, enamelin, and proteolytic enzymes. Together, they assemble into a scaffold that serves as a template for the nucleation and uniaxial growth of apatite crystals in a highly organized structure (1). Recent findings suggests that mineralization and assembly occurs simultaneously in developing enamel and, hence, amelogenesis differs from bone formation in this respect (2, 3). The role of amelogenin in guiding apatite formation has been ascribed to its ability to self-assemble into nanospheres and nano-chains (4-6).
One of the most intriguing features of amelogenesis derives from the fact that its high mineral content coupled with an ultrafine architecture implies the extracellular matrix in this process directs not only the crystal growth, but its own constructive degradation too. This makes amelogenesis a significantly more intricate mineralization process compared to dentinogenesis during which the collagenous protein matrix remains in place (7). Studies of proteolysis of the enamel matrix are thus of particular importance in attempts to understand amelogenesis (8-10).
The action of proteases, such as matrix metalloproteinase-20 (MMP-20, also known as enamelysin) and kallikrein 4 (KLK-4, also known as enamel matrix serine protease 1) in hydrolysis of amelogenins and other proteins thus presents a crucial aspect of amelogenesis (11). The initially secreted nascent proteins are present in the enamel matrix in transient form, and are relatively quickly processed to generate a wide spectrum of smaller peptides. Due to its high selectivity for specific peptide bonds, MMP-20 is usually considered a regulator that controls the functionality of amelogenins (12). The latter proteins ought to be removed once the structure of enamel is sufficiently formed, so that the freed space would be filled with additionally crystallized apatite. Thin fibrous crystals form during secretory stage while MMP-20 hydrolyses matrix proteins producing specific cleavage products. Only at maturation stage crystals will grow in width and thickness, into areas where protein is being removed by activity of KLK-4 (13). Most of the mineral deposition is therefore known to occur during maturation stage of amelogenesis when the degradation and removal of the enamel matrix take place. Crystals extend and come in close contact with each other and a tissue with 95 wt% mineral forms (14)(15).
An increasing amount of evidence suggests that the cleavage products carry out different secondary self-assembly-related functions in the developing enamel matrix (16). Hence, it was shown that mixtures of the full-length human amelogenin (rH174) and the first proteolytic cleavage product formed in the reaction with MMP-20, rH163, possess a markedly higher propensity for the formation of more complex, fibrous protein assemblies from the initial nanospheres compared to the pure rH174 (17). The structure of amelogenin is thus thought to be modular, in a sense that it may contain several functional domains that become activated for different purposes and at different stages of amelogenesis (18). This idea is supported by the findings that the metallo-matrix protease is expressed early during development (16). Mutations not only in amelogenin genes, but in those that encode MMP-20 cause amelogenesis imperfecta, i.e., a pathological state typified by abnormal and significantly weakened enamel (16, 19). Knockout of MMP-20 in mice resulted in the formation of hypoplastic enamel composed of thin, shorter crystallites with undefined prisms, indicating that MMP-20 hydrolysis is required for proper lateral extension of crystals (20, 21). Another study shows that impaired enzymatic interaction between MMP-20 and amelogenin may be the prime cause of amelogenesis imperfect (22). Inhibition of the activity of MMP-2, MMP-9 and MMP-20 by marimastat similarly led to impaired mineralization of dental tissues in mice (23). MMP-20 by itself was not able to remove the matrix to a degree that allowed formation of functional enamel as shown by the KLK-4 KO mouse (24).
Our approach to gaining insight into the mechanism of amelogenesis at the molecular scale involved the design of a biomimetic programmable titration system (25). Slow and controlled titration of amelogenin sols with buffered calcium and phosphate solutions at low degrees of saturation is thus employed to imitate the formation of elongated enamel-like crystals. In our previous report, we claim that despite its predominantly hydrophobic nature, amelogenin acts as a promoter of nucleation and crystal growth under the biomimetic conditions of growth applied in our study (26). In this paper, we report on our results on calcium phosphate formation in the given system in the presence of the proteolytic degradation of full-length amelogenin by means of MMP-20.
Experimental
Recombinant full-length human amelogenin (rH174) and MMP-20 were previously synthesized via their expression in BL21(DE3) plysS Escherichia Coli (27, 28). The experimental setting applied in this study was modified from a previous study (25). Titrations were performed with 1 ml burettes, using a Titrino 751 GDP titration device in combination with a Dosimat 755 (Brinkmann-Metrohm) controlled by computer software (Tiamo 1.2, Brinkmann-Methrohm). The basic procedure was as follows. If not denoted otherwise, the starting reaction mixture comprised 5 ml of 0.4 mg/ml rH174, 20 mM Tris/HCl buffer, 2.5 mM KH2PO4, and 0.02 % NaN3 (introduced to the system in the given sequence) at pH 7.40 +- 0.01. The reaction mixture was prepared and kept in a jacketed glass vessel during titration, which was maintained at 37°C via a circulating water bath. The two buffered titrant solutions (20 mM Tris/HCl, pH 7.40 +- 0.02) comprising the separate precursor ions (8.2 mM CaCl2 and 5 mM KH2PO4) and the electrolyte (142 mmoll-1 KCl) up to the level of the physiological ionic strength were introduced into the reaction vessel at a controlled rate of 840 nl/min, i.e., 1.2 ml/day, cumulatively, throughout a 7-day period of time. In order to ensure substrate-specific crystal growth, it was essential to start the titration at undersaturated conditions. This was achieved by preparing the reaction mixture without calcium ions initially. Calcium content increased gradually during titration whereas phosphate was added at concentrations which would maintain its level around 2.5 mM. MMP-20 was added at the start of the experiment in different weight ratios with respect to rH174, ranging from 1:103 to 1:105. In order to ensure invariance in terms of ionic concentrations, we compared all our reaction solution concentrations with controls, which comprised either protease-free or protein-free systems. To deduce the effect of MMP-20 concentration on crystal growth, the concentration of ions before the onset of precipitation was made identical for all the compared reaction systems (MMP-20/rH174 = 0; 1:105; 1:103). The crystal growth was initiated on polished glass ceramic substrates comprising embedded fluoroapatite (FAP) crystals with surface-exposed (001) faces (29). A single substrate was sampled out at different time points and evaluated for the crystal growth properties using Atomic Force Microscopy (AFM, Nanoscope III, Digital Instruments, USA) and Scanning Electron Microscopy (SEM, DS130C, Topcon, USA). Tapping-mode AFM was applied using Si-tips with a radius of about 10 nm (Tap300, Budgetsensors, USA). The crystal heights were measured using the section analysis tool in NanoScope software. Heights of twelve randomly chosen peaks in three different 10 × 10 μm2 AFM images were measured and the average values were plotted as a function of the titration volume. As the substrate surface was uniformly covered with the precipitate, it was assumed that 36 randomly chosen peaks would be sufficient to obtain a representative assessment of the substrate topology. Aliquots of 200 μl of the supernatants were also sampled out daily. A part of the aliquots was mixed with 2 M HCl at 1:1 weight ratio to stop the proteolytic digestion, and another part was filtered and used for the elemental analysis using Inductively Coupled Plasma Optical Emission Spectrometry (ICP-OES, Perkin Elmer, 5300 DV). The concentration of Ca2+ and PO4 3- in the reaction solution was followed in parallel using a calcium ion-selective electrode (pHoenix) and a UV/Vis/IR spectrophotometer (Milton Roy, Genesys 5), respectively. The calcium concentrations reported correspond to free calcium ions in the solution and do not account for those bound to the peptide species. The need to derive the exact activity coefficients has been partly eliminated by using the difference between the output calcium levels for various protease concentrations to draw conclusions about the nucleation and crystal growth promoting effect that MMP-20 induced. The amount of precipitated calcium ions was calculated by subtracting the amount of calcium present in the reaction mixture at a given time (the sum of the initial amount and the amount added by titration normalized for the evaporation effect) from the amount of calcium indicated by the ion-selective electrode.
Dynamic light scattering (DLS) analysis of colloidal suspensions of proteolytically cleaved rH174 was carried out on Zetasizer Nano-ZS (Malvern, UK) device. SDS–PAGE of cleaved amelogenin was carried out in 8% (w/v) polyacrylamide gel at pH 8.8 as described by Laemmli (30). The gels were stained with Coomassie brilliant blue R-250. Samples used for the SDS-PAGE analysis were previously subjected to a dialysis treatment using 20 mL protein buffer exchange dialysis cups (Fisher Scientific) in 10 mM EDTA in order to eliminate Ca2+ and HxPO4 x-3 ions and to raise the pH to neutral values. With the given gel composition used, only cleavage products larger than 15 kDa could be discerned. To detect smaller proteolytic digestion fragments, matrix-assisted laser desorption ionization mass spectrometry and tandem mass spectrometry (MALDI MS and MS/MS) analysis was carried out. For that purpose, the samples from MMP-20 digestion at different time points were desalted by ZipTip pipette tips (Millipore, Billerica, MA) packed with C18 resin. The eluates from ZipTip were mixed with MALDI matrix α-cyano-4-hydroxycinnamic acid (6 mg/ml in 80% ACN/0.1% TFA/10 mM dibasic ammonium phosphate) and manually spotted onto a stainless steel MALDI plate (Applied Biosystems (AB), Foster City, CA). The sample spots on the MALDI plate were analyzed with a 5800 MALDI TOF-TOF Protemoics Analyzer (AB) in both linear and reflector positive modes in the mass range of 800-40,000Da and 800-6,000Da, respectively. About 30 peaks from the reflector MS spectra of two samples obtained at 24 hour of MMP-20 digestion were manually selected for MS/MS. Database search of these MS/MS spectra against Uniprot human database (with common contaminants) using ProteinPilot software (Version 3.0, Revision 114732, AB) resulted in confident identification of amelogenin in both samples. The experimental molecular mass of rh174 was 19818 Da, i.e., it was measured within a 0.07% error that is consistent with the expected accuracy of the AB Sciex 4800 TOF/TOF mass spectrometer when analyzing protein species of ~20 kDa using linear mode. Table 1 lists samples analyzed by means of SDS-PAGE and MALDI-TOF after being sampled out at various titration time points.
Table 1.
List of samples analyzed by means of SDS-PAGE and MALDI-TOF techniques
| Sample | rH174/MMP-20 weight ratio | Titration time |
|---|---|---|
| A1 | 103 | 2 min |
| A2 | 103 | 17 min |
| A3 | 103 | 30 min |
| A5 | 103 | 2.5 h |
| A6 | 103 | 5 h |
| A7 | 103 | 24 h |
| B1 | 104 | 5 min |
| B2 | 104 | 30 min |
| B3 | 104 | 90 min |
| B4 | 104 | 4 h |
| B5 | 104 | 24 h |
Results
Cleavage of rH174 by MMP-20
Fig.1 shows SDS-PAGE gel of the proteolytically digested full-length amelogenin (rH174) samples in the continuous titration experiments sampled out at various time points, ranging from 2 min to 24 h. The intensity of the rH174 band decreases over time, suggesting time-dependent proteolytic cleavage, irrespective of the concentration of the protease (rH174/MMP-20 103:1 or 104:1). For 103:1 rH174/MMP-20 weight ratio, the maximum cleavage of rH174 takes place between 2.5 h and 24 h. Between 2 min and 2.5 h, the intensity of the full-length band decreases for both rH174/MMP-20 weight ratios, suggesting gradual cleavage of the protein. For 104:1 rH174/MMP-20 weight ratio, the full cleavage of rH174 takes place between 4 h and 24 h. rH174 band for 104:1 rH174/MMP-20 weight ratio after 5 min is more intensive than the one for 103:1 ratio after 2 min, suggesting quicker initial digestion at higher concentrations of the protease. Whereas the full-length band is hardly visible after 4 h for 103:1 concentration, the band after 4 h for 104:1 weight ratio is as intensive as that at early time points for 103:1 ratio. rH163 is known to be the first proteolytic product of the digestion of rH174 with MMP-20. The fact that it could not be discernable by means of SDS-PAGE may be partly due to its closeness to the rH174 band, and partly due to the earlier evidenced quick processing thereof to smaller peptide fragments (28).
Fig.1.
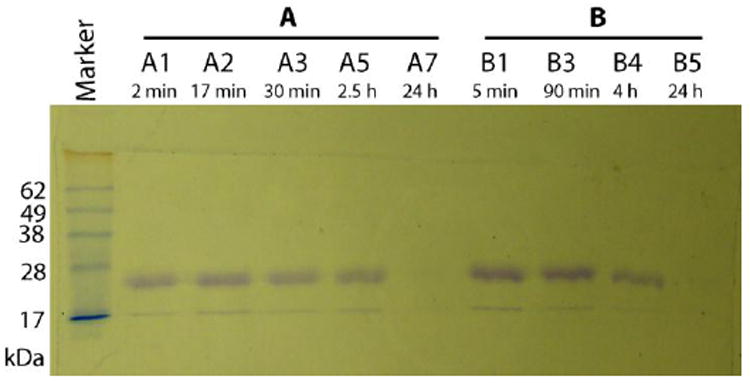
SDS-PAGE of proteolytically digested rH174 samples in the continuous titration experiments at different titration times (as shown). Marker, prestained SDS-PAGE marker (Invitrogen); A1-A7, rH174/MMP-20 weight ratio 103, B1-B5, rH174/MMP-20 weight ratio 104.
Analysis of cleaved products
Mass spectra (0.8 – 42 kDa) of samples acquired in the linear positive mode are shown in Fig.2a,b. Three major peaks at 19.82, 9.91 and 6.0 kDa correspond to singly, doubly, and triply charge states of rH174. From A1(2 min) to A3(30 min) and from B1(5 min) to B2(30 min), there is a minimum change in the spectral appearance, with slightly more abundant rH174 peptides seen in the low-mass range of A3(30 min) and B2(30 min), respectively. Compared to A3(30 min), significantly more abundant rH174 peptides can be seen in A6(5 h), while rH174 peaks remain to be abundant in A6(5 h). More abundant rH174 peptides were seen in B4(4 h) as compared to B2(30 min). However, the difference between B2(30 min) and B4(4 h) is less significant than that found between A3(30 min) and A6(5 h), which is the consequence of the 10 times higher rH174/MMP-20 ratio for the B series (rh174/MMP-20 : 104) of samples compared to A (rh174/MMP-20 : 103). For both sample series, the spectra of the last samples (A7, 24 h and B5, 24 h) lacked the presence of full-length amelogenin rH174, indicating the hydrolysis of the protein by MMP-20. A few peaks were observed in the medium mass range (8 – 20 kDa) at 24 h. Those peaks around 8.3, 9.2, 11.5, 12.3, 13.6, 15.1 and 16.7 are most likely hydrolysis products of rH174 which may not be hydrolyzed by MMP-20 at all and require KLK-4 for processing (31).
Fig.2.
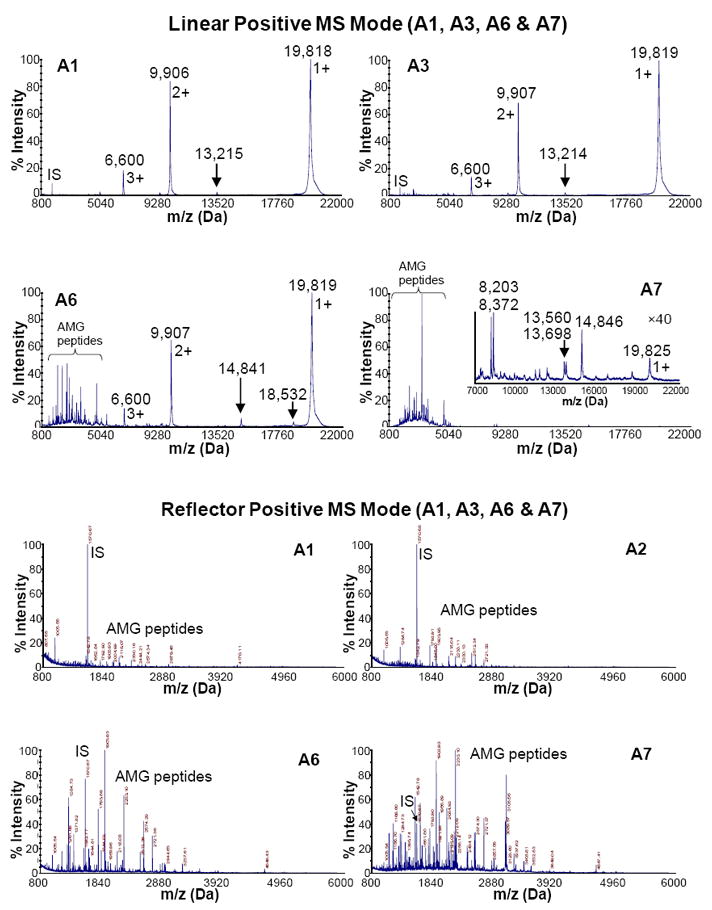
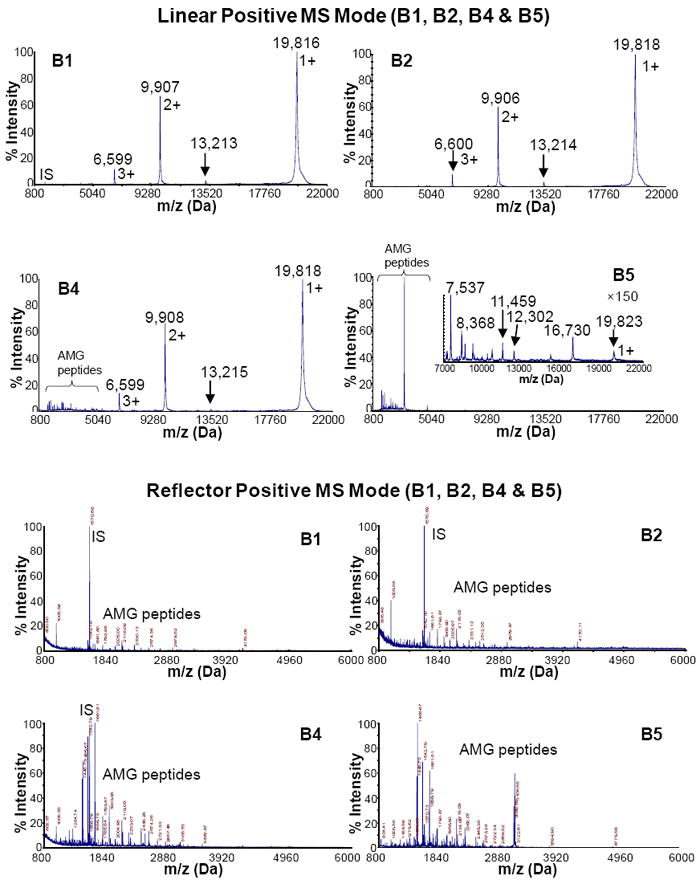
Mass spectra of proteolytically digested rH174 (AMG) samples in the continuous titration experiments sampled out at various time points (2 min – 24 h). IS stands for “internal standard”, which is [Glu1]-fibrinopeptide B (m/z = 1570.68 Da). It was spiked in MALDI matrix for internal mass calibration.
Fig.2c,d shows mass spectra (0.8 – 6 kDa) of samples acquired in the reflector positive mode. As indicated by the linear mass spectrograms, an increasing amount of rH174 peptides is seen with the increase in the titration time, that is, from sample A1 (2 min) to A7 (24 h) and from B1 (5 min) to B5 (24 h). Aside from singly, doubly and triply charged rH174, the only rH174 fragment larger than 5 kDa observed in the first 4 h of digestion time was 1-116 sequence at 13.2 kDa, corresponding to the cleavage loci PVQ/PQP. The MS/MS spectra were manually acquired for most of the peaks observed in the reflector mass spectra of samples A7 and B5 (Fig.3c,d). Database search of those MS/MS spectra using ProteinPilot identified amelogenin as the only positive hit in both cases. Table 2 shows selected MMP-20 cleavage sites on rH174 sequence after 5 min and 24 h of digestion time.
Fig.3.
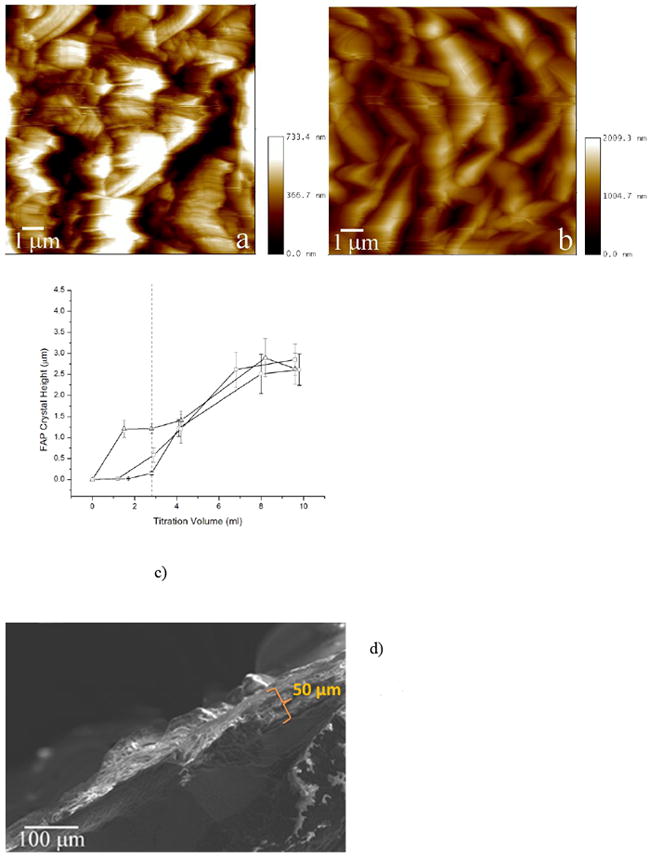
AFM images of FAP/glass seeds from the reaction comprising rH174/MMP-20 in 1:103 (a) and 1:105 (b) weight ratios, after titration volumes of 1.55 ml (a) and 2.8 ml (b). The average height of apatite crystals grown after different titration volumes compared between 0.4 mg/ml rH174 sample without MMP-20 (-o-), with MMP-20 in 105:1 weight ratio (-▫-), and with MMP-20 in 103:1 weight ratio (-Δ-) with respect to rH174 (c). SEM micrograph of the cross-section of an FAP substrate at the end of a 7-day titration experiment (d). Dotted line is a guide for the eye at the titration volume at which the precipitation propensities of the solutions significantly differ.
Table 2.
Twelve (out of ~ 30 in total) and 17 (out of ~ 140 in total) most intense peptides generated from digestion of rh174 with MMP-20 (104 rH174/MMP-20 weight ratio) for 5 min and 24 h, respectively (see Fig.3d). The slash mark indicates the cleavage site. The sequence of the X chromosome variant of the recombinant full-length human amelogenin is given below. C- and N-terminal cleavage sites are marked with a star.
| 5 min | 24 h | ||
|---|---|---|---|
| m/z (monoisotopic) | Range | m/z (monoisotopic) | Range |
| 1005.54* | 1-10* | 1395.71* | 1-13* |
| 1542.78* | 1-14* | 1445.73 | 2-14 |
| 1590.78* | 162-174* | 1456.67 | 29-40 |
| 1629.81* | 1-15* | 1542.79* | 1-14* |
| 1661.80* | 161-174* | 1569.75 | 28-40 |
| 1792.86* | 1-16* | 1656.78 | 27-40 |
| 2005.00* | 158-174* | 1661.81* | 161-174* |
| 2118.08 | 157-174* | 1756.81 | 29-43 |
| 2330.13* | 155-174* | 1784.85 | 26-40 |
| 2978.52* | 149-174* | 1790.87* | 160-174* |
| 4168.02* | 1-36* | 1947.91 | 25-40 |
| 13213.2* | 1-116* | 1956.90 | 27-43 |
| 2043.14 | 48-65 | ||
| 2118.09* | 157-174* | ||
| 2134.00* | 1-19* | ||
| 2262.07 | 23-40 | ||
| 3106.58* | 148-174* | ||
rH174:
PLPPHPGHPGYINFSYEVLTPLKWYQSIRPPYPSYGYEPMGGWLHHQIIPVLSQQHPPTH TLQPHHHIPVVPAQQPVIPQQPMMPVPGQHSMTPIQHHQPNLPPPAQQPYQPQPVQPQP HQPMQPQPPVHPMQPLPPQPPLPPMFPMQPLPPMLPDLTLEAWPSTDKTKREEVD174
Effect of proteolysis on crystal growth and morphology
The effect of proteolysis on the crystal growth promoting capacities of rH174 sols can be also seen from AFM images and curves depicting apatite crystal height as a function of the titration volume for different rH174/MMP-20 weight ratios, displayed in Fig.4a-c. The earliest precipitation covering almost the entire surface of the substrates was noticed after 4 ml of titration without MMP-20, corresponding to 3.5 days of titration. The time for precipitation was significantly reduced when MMP-20 was added to the system. At a ratio of 105:1 of rH174/MMP-20 precipitation was observed after 2.8 ml or about 2 days; and after only 1.5 ml or about 30 hours of titration with rH174/MMP-20 103:1 ratio. In the latter case, the level of growth was more significant after 24 h than after 7 days in the protease-lacking experiment. Note that the z-scale limit for AFM imaging is around 3 μm with the standard J-type piezo scanner, but the real thickness of the final precipitate was more in the order of 50 μm as shown in the SEM image of Fig.3d.
Fig.4.
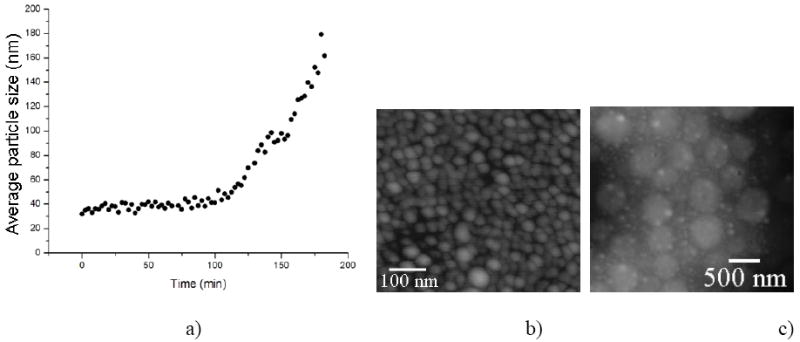
Time-dependent increase in the particle size of 0.4 mg/ml rH174 assemblies dispersed in water at 37 °C and pH 7.40 following the introduction of MMP-20 in 100:1 weight ratio (a). AFM images of amelogenin assemblies at time zero (b) and following the degradation by means of MMP-20 (c).
The effect of proteolysis on the rH174 particle size increase at pH 7.4, as indicated by dynamic light scattering measurements, is shown in Fig.4. Namely, approximately 2 h after mixing 0.4 mg/ml rH174 with MMP-20 in 100:1 weight ratio, amelogenin particle size began to increase from ~ 40 nm, reaching 180 nm 3 h after the addition.
Despite the fact that all the experimental systems were buffered, in most cases a mild drop in pH, most likely associated with the precipitation of apatite, was noticed. Fig.6a shows a different extent of this drop depending on the protein content of the reaction system. When proteolytic cleavage was coupled to the precipitation reaction, the system exhibits a more intensive pH drop (down to pH 6.8) compared to the non-protease-containing reaction (down to pH 7.2). Also, it was observed that whereas in the protease-free rH174 sol the concentration of phosphates stabilizes at around 1.5 mM, it drops to below 0.5 mM in the titrated protease-containing rH174 suspension after > 3 ml of titration volume.
Fig.6.
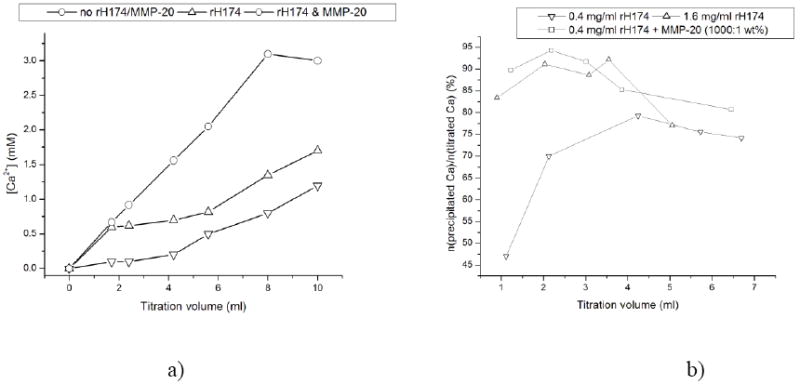
Concentration of free Ca2+ over reaction time in the systems comprising 0.4 mg/ml rH174 (-Δ-), 0.4 mg/ml rH174 and MMP-20 (1:103), and no peptide species (-o-) (a), and the percentage of the overall titrated amount of Ca2+ that is being precipitated and/or protein-bound at different reaction times using [rH174] = 0.4 mg/ml (-□-), [rH174] = 1.6 mg/ml (-Δ-), and [rH174] = 0.4 mg/ml, but with the proteolytic reaction coupled to it (-▫-) (b).
Gradual increase of the calcium levels in the solution typically results in a plateau in [Ca2+] vs. titration time/volume curves, which is an indication of the onset of precipitation of calcium phosphate. Fig.6a shows how the experimental runs performed in the presence of the proteolytic cleavage of rH174 exhibit lower free Ca2+ levels compared to the runs comprising full-length rH174 as well as the control runs without any peptide species throughout the entire 7-day reaction time. As the quantity of Ca2+ ions obtained by subtracting the measured free levels from the overall titrated amount can be accounted for either the peptide-bound ionic species or the precipitated ones, this observation speaks in favor of MMP-20 digestion promoting even more of the crystal growth and Ca2+-binding compared to pure rH174-comprising suspensions. As shown in Fig.6b, the amount of Ca2+ ions that was bound to the protein or precipitated after 1 ml of the titration volume was lower than 50 % with respect to the overall titrated amount of calcium ions, whereas at the same reaction time this amount exceeded 80 % when rH174 in concentration of 1.6 mg/ml was used or when MMP-20 at rH174:MMP-20 weight ratio of 103:1 was introduced to the system. Interestingly, coupling the proteolytic reaction to crystallization in the presence of 0.4 mg/ml rH174 has the same effect on the crystal growth promotion as quadrupling the concentration of full-length rH174 to 1.6 mg/ml.
SEM micrographs (Fig.7) demonstrated that the proteolytic reaction affected the size of the basal planes of calcium phosphate crystals, but it did not catalyze a morphological change. In all cases, plate-shaped particles were observed. Higher contents of MMP-20 lead to a decrease in the crystal width (Fig.7b, c) when compared to protease-free experiments (Fig.7a). EDX analysis has indicated the presence of calcium and phosphorus, whereas Raman analysis has confirmed the presence of apatite as the main calcium phase (ν1(PO4) = 956 cm-1). Fig.8 shows the different appearance of FAP substrates at early and late titration time points. At earlier times (Fig.8a), the substrate surface is mostly covered with the protein precipitate, which presents the first step in substrate-specific crystal growth. As can be seen from the inset in Fig.8a, the initial protein precipitate shows only traces of calcium and phosphorus ions. Morphologies reminiscent of the nucleating calcium phosphates appear as visible under the initially deposited protein layers in the next steps, as shown on the left side of the FAP substrate in Fig.8b. Eventually, most of the substrate surface becomes covered with plate-shaped calcium phosphate particles, such as those on the right side of the SEM image in Fig.8b.
Fig.7.
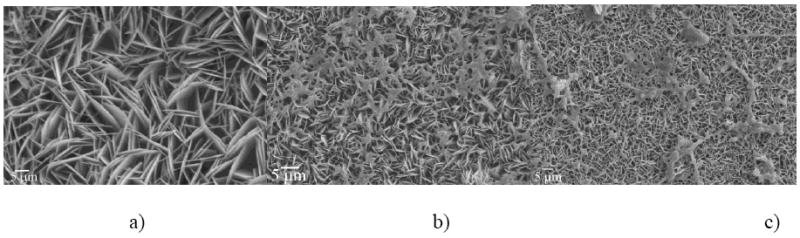
SEM micrographs of calcium phosphate particles obtained in the presence of rH174 only (a), and in the presence of both rH174 and MMP-20 in 103:1 (b) and 104:1 (c) rH174/MMP-20 weight ratios. Obtained at the end of titration experiment at a) 7 ml, b) 7 ml and c) 9.4 ml titration volume.
Fig.8.
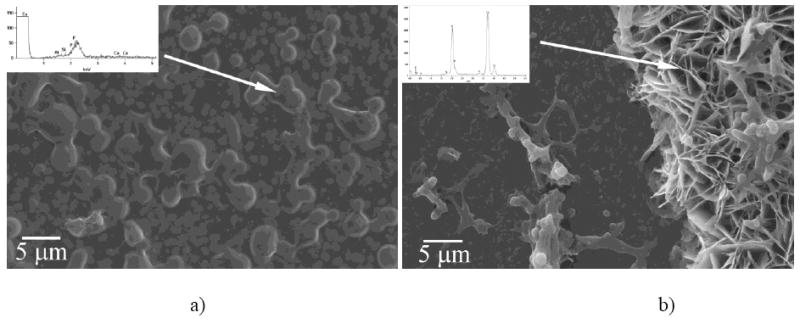
Surface of FAP substrates sampled out after 1.2 (a) and 9.4 ml (b) of the titration volume observed under SEM/EDX.
Discussion
In our previous report, we have offered evidence in favor of the nucleation and growth promoting role of rH174 under the experimental conditions applied in our biomimetic study (26). Biomimetic conditions under which crystallization proceeded kinetically driven by the ability of rH174 to concentrate and nucleate ionic growth units within its deposited layers, rather than by the thermodynamic propensities of the solution, were established and a mechanism for the amelogenin-guided crystal growth based on the observed nucleation of calcium phosphate within amelogenin layers deposited on the FAP substrate surface was proposed (26). Numerous indications that MMP-20 and rH174 together promote crystal formation significantly more that rH174 alone are presented here, including the following:
The more MMP-20 is initially present in the system, the earlier in the course of the titration time the precipitation of mineral begins and the more intensive the crystal growth becomes (Fig.3);
The extent of the pH drop associated with the formation of apatite was higher for the protease-containing system compared to its solely rH174-comprising counterpart (Fig.5);
When MMP-20-induced digestion of rH174 is coupled to precipitation of calcium phosphate lower concentrations of calcium ions in the supernatants through the entire course of the experiment (Fig.6a) and lower final concentrations of phosphates indicate more extensive precipitation of ions in the protease-containing systems compared to the protease-free ones;
Coupling the proteolytic reaction to titration in the presence of 0.4 mg/ml rH174 has the same effect on the crystal growth promotion as quadrupling the concentration of rH174 to 1.6 mg/ml (Fig.6b).
Fig.5.
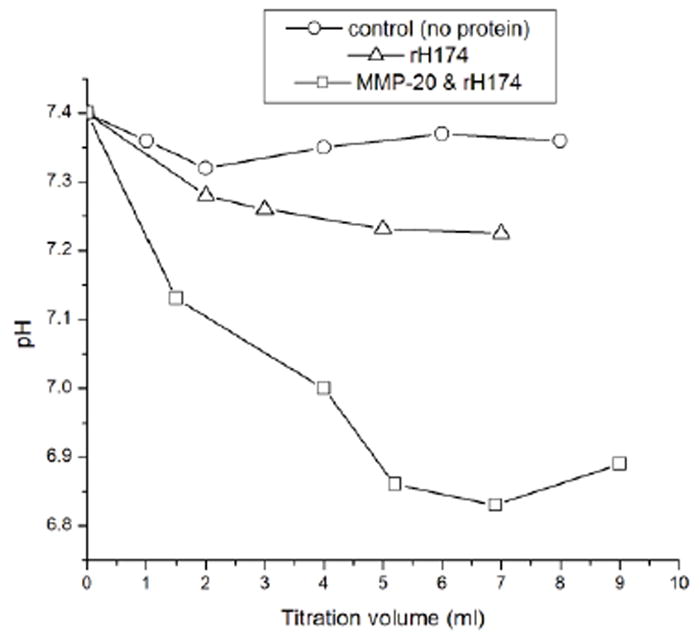
pH change in the supernatants in the 7-day titration experiments for the control, protein-free experiment, the one containing only rH174, and the one containing both rH174 and MMP-20 in 103:1 rH174/MMP-20 weight ratio;
These observations strongly indicate that the proteolytic digestion plays an additional nucleation promoting role when compared to the control, protein-free experiment and the one containing rH174 as the only polypeptide compound. By lowering the energy of activation required for heterogeneous nucleation, proteolysis influences the thermodynamics of apatite growth. The first proteolytic product of the reaction between rH174 and MMP-20 is the 13-26 residues long C-terminal sequence of a pronounced hydrophilicity (Table 1). Such a loss of the amphiphilic character of the cleaved rH174 results in increased propensity to form larger nanospheres as well as to aggregate and precipitate (32) (Fig.6). Since deposition of the aggregated protein assemblies on the FAP substrate surface presents the first step prior to the crystal growth (Fig.8), the proteolytic cleavage thus leads to enhanced nucleation and apatite formation.
The rate of the proteolytic cleavage reaction has been shown to be directly proportional to the concentration of MMP-20. In 24 h, a complete digestion of rH174 into smaller peptide fragments takes place at both 103:1 and 104:1 rH174/MMP-20 weight ratios (Figs. 2, 3). The number of peptides smaller than 5 kDa increased from ~ 30 to ~ 140 as the digestion time increased from 5 min to 24 h (Table 2). Also, all 12 most intense rH174 peptides generated after 5 min of digestion time come from N- or C- termini compared to only 6 out of 17 for 24 digestion time. The digestion thus appeared to have proceeded by cleaving the termini fragments first, after which the reaction proceeded following less selective and extensive internal hydrolysis, which is in contrast with the previous reports that augmented a high selectivity that MMP-20 possesses with respect to the cleavage sites on rH174 (28), unlike the one of KLK-4, the other main protease of the enamel matrix known for its aggressive cleavage of amelogenin (33, 34). The difference can be explained by the fact that whereas our proteolysis was initiated in a dynamically changing solution composed of multiple ionic species and active surfaces, the earlier observed more selective cleavage was observed following incubation of rH174 and MMP-20 in the standard buffer. A change in the protein conformation has been shown to take place upon binding of amelogenin onto apatite (35), which may explain the recent finding of more cleavage sites for amelogenin bound to apatite surfaces than for amelogenin dispersed in aqueous solution (12). The conditions for hydrolysis in this study were different from previous studies with regards to the ion concentration and composition, as MMP-20 cleavage was implemented under crystal growth conditions for calcium phosphate mineral. Besides conformational changes, certain cleavage sites on the rh174 could be masked by its simultaneous interaction with mineral ions as well as other molecules. As recently shown self-assembly of amelogenin may significantly be altered in the presence of a mineralization promoting milieu which in return could affect cleavage of the protein (36). As kinetics of the proteolytic digestion appear to be directly linked to the time scale that governs precipitation of calcium phosphate, the protein-protease ratio will present a vital parameter in the growth of enamel-like apatite crystals using a biomimetic setting such as the one applied in this study.
The results obtained in this study are in agreement with the previous findings of nucleation-promoting effect that amelogenin from species other than human can exert on metastable calcium phosphate solutions (37-39). We have additionally shown that hydrolysis of amelogenin can furthermore accelerate the nucleation of calcium phosphate from metastable solutions. The current model of matrix-guided crystal growth during enamel maturation is based on the assumption that amelogenin nanospheres preferentially adsorb onto (hk0) faces of the growing apatite crystals, thereby allowing the surrounding Ca2+ and PO4 3- ions to bind onto (001) crystal faces solely and forcing the uniaxial growth of apatite crystals along the [001] axis. This model, however, does not describe the effect of amelogenin on nucleation of apatite crystals. Studies supporting this thesis came mostly from experiments involving either very low supersaturation levels (40), when the majority of the ionic content may be sequestered by amelogenin, or relatively high concentrations of the protein and high supersaturation levels (41). The question of whether amelogenin does indeed play a role in nucleation of enamel crystallites can be best answered by investigating its effect in parallel with that of more hydrophilic protein species of the enamel matrix, including mainly ameloblastin and enamelin.
The experimental setting applied in this work has been demonstrated as valuable in terms of enabling us to gain insights into the fundamental nature of amelogenesis. Still, it may be subject to numerous improvements, which will be the topics of the future studies. As noted, diversifying the polypeptide content would help in designing an even more faithful biomimetic setting. Here we have introduced MMP-20 only in the initial stage of the process which may mimic the biological conditions of enamel development, since MMP-20 is expressed at the early secretory stage of amelogenesis when crystallites are growing predominatly in length (13). MMP-20 cleavage products are partially reabsorbed by the ameloblasts but most remain in the matrix, occupying the spaces between crystallites, before KLK-4 aggressively removes the organic matrix almost completely (16). The timely removal of the digested protein, which is in biological conditions facilitated by the cellular action, may also be required for sustaining the uniaxial and site-specific crystal growth (42). The biomimetic experimental setting based on the continuous and controllable titration of ions into the self-assembling protein matrix can be functionally upgraded by introducing an option to deliver each one of these species in a controllable manner and in feedback with the multi-parametric monitoring of the system. This is in accordance with the in vivo character of the process where ameloblast play the role of precisely monitoring and delivering the macromolecular ingredients of the enamel matrix and the ionic precursors for crystal formation.
Conclusions
Since enamel starts growing from a protein-rich ionic gel, a.k.a. the developing enamel matrix, the process of protein-guided crystal growth has to be synchronized with the proteolytic degradation and controlled disappearance of the very protein matrix that guides the uniaxial growth of the apatite crystals. In order to understand the complex process of amelogenesis the three elementary aspects of the process – crystal growth, protein self-assembly and selective and time-dependent proteolysis – need to be studied in parallel (25). The capacity of amelogenin to promote nucleation and crystal growth has been shown here to increase in proportion with the extent of its proteolytic degradation by means of MMP-20. Controlling the rate and the extent of the proteolytic cleavage can thus be used to control the nucleation and growth rates in a protein-guided crystallization system, such as the one applied in this study.
Acknowledgments
Presented are the results of a study supported by NIH/NIDCR grants R01-DE017529, R01-DE017529-S2 and R01-DE015821. The authors would like to thank Dr. Christian Russel (FSU Jena, Germany) for providing the glass-ceramic substrates and Joseph Mendoza (UCSF) for the synthesis of rH174.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fincham AG, Moradian-Oldak J, Simmer JP. The structural biology of the developing dental enamel matrix. J Struct Biol. 1999;126(3):270–299. doi: 10.1006/jsbi.1999.4130. [DOI] [PubMed] [Google Scholar]
- 2.Wiedemann-Bidlack FB, Beniash E, Yamakoshi Y, Simmer JP, Margolis HC. pH triggered self-assembly of native and recombinant amelogenins under physiological pH and temperature in vitro. J Struct Biol. 2007;160(1):57–69. doi: 10.1016/j.jsb.2007.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Margolis HC, Beniash E, Fowler CE. Role of macromolecular assembly of enamel matrix proteins in enamel formation. J Dent Res. 2006;85(9):775–793. doi: 10.1177/154405910608500902. [DOI] [PubMed] [Google Scholar]
- 4.Moradian-Oldak J. Amelogenins: assembly, processing and control of crystal morphology. Matrix Biol. 2001;20(5-6):293–305. doi: 10.1016/s0945-053x(01)00154-8. [DOI] [PubMed] [Google Scholar]
- 5.Moradian-Oldak J, Du C, Falini G. On the formation of amelogenin microribbons. Eur J Oral Sci. 2006;114(Suppl 1):289–296. doi: 10.1111/j.1600-0722.2006.00285.x. discussion 327-289, 382. [DOI] [PubMed] [Google Scholar]
- 6.Habelitz S, Kullar A, Marshall SJ, DenBesten PK, Balooch M, Marshall GW, et al. Amelogenin-guided crystal growth on fluoroapatite glass-ceramics. J Dent Res. 2004;83(9):698–702. doi: 10.1177/154405910408300908. [DOI] [PubMed] [Google Scholar]
- 7.Mann S, Weiner S. Biomineralization: structural questions at all length scales. J Struct Biol. 1999;126(3):179–181. doi: 10.1006/jsbi.1999.4131. [DOI] [PubMed] [Google Scholar]
- 8.Delak K, Harcup C, Lakshminarayanan R, Sun Z, Fan Y, Moradian-Oldak J, et al. The tooth enamel protein, porcine amelogenin, is an intrinsically disordered protein with an extended molecular configuration in the monomeric form. Biochemistry. 2009;48(10):2272–2281. doi: 10.1021/bi802175a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han YJ, Wysocki LM, Thanawala MS, Siegrist T, Aizenberg J. Template-dependent morphogenesis of oriented calcite crystals in the presence of magnesium ions. Angewandte Chemie. 2005;44(16):2386–2390. doi: 10.1002/anie.200462296. [DOI] [PubMed] [Google Scholar]
- 10.Ryu OH, Fincham AG, Hu CC, Zhang C, Qian Q, Bartlett JD, et al. Characterization of recombinant pig enamelysin activity and cleavage of recombinant pig and mouse amelogenins. J Dent Res. 1999;78(3):743–750. doi: 10.1177/00220345990780030601. [DOI] [PubMed] [Google Scholar]
- 11.Moradian-Oldak J, Jimenez I, Maltby D, Fincham AG. Controlled proteolysis of amelogenins reveals exposure of both carboxy- and amino-terminal regions. Biopolymers. 2001;58(7):606–616. doi: 10.1002/1097-0282(200106)58:7<606::AID-BIP1034>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 12.Zhu L, Tanimoto K, Le T, DenBesten PK, Li W. Functional roles of prolines at amelogenin C terminal during tooth enamel formation. Cells Tissues Organs. 2009;189(1-4):203–206. doi: 10.1159/000151376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu Y, Papagerakis P, Yamakoshi Y, Hu JC, Bartlett JD, Simmer JP. Functions of KLK4 and MMP-20 in dental enamel formation. Biol Chem. 2008;389(6):695–700. doi: 10.1515/BC.2008.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lacruz RS, Nanci A, Kurtz I, Wright JT, Paine ML. Regulation of pH During Amelogenesis. Calcified tissue international. 2010;86(2):91–103. doi: 10.1007/s00223-009-9326-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robinson C, Brookes SJ, Bonass WA, Shore RC, Kirkham J. Enamel maturation. Ciba Found Symp. 1997;205:156–170. doi: 10.1002/9780470515303.ch11. [DOI] [PubMed] [Google Scholar]
- 16.Bartlett JD, Simmer JP. Proteinases in developing dental enamel. Crit Rev Oral Biol Med. 1999;10(4):425–441. doi: 10.1177/10454411990100040101. [DOI] [PubMed] [Google Scholar]
- 17.He XD, Li W, Habelitz S. The cooperative self-assembly of 25 and 23 kDa amelogenins. Journal of Structural Biology. 2008;164(3):314–321. doi: 10.1016/j.jsb.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Snead ML. Amelogenin protein exhibits a modular design: implications for form and function. Connect Tissue Res. 2003;44(Suppl 1):47–51. [PubMed] [Google Scholar]
- 19.Caterina JJ, Skobe Z, Shi J, Ding Y, Simmer JP, Birkedal-Hansen H, et al. Enamelysin (matrix metalloproteinase 20)-deficient mice display an amelogenesis imperfecta phenotype. J Biol Chem. 2002;277(51):49598–49604. doi: 10.1074/jbc.M209100200. [DOI] [PubMed] [Google Scholar]
- 20.Brookes SJ, Robinson C, Kirkham J, Bonass WA. Biochemistry and molecular biology of amelogenin proteins of developing dental enamel. Arch Oral Biol. 1995;40(1):1–14. doi: 10.1016/0003-9969(94)00135-x. [DOI] [PubMed] [Google Scholar]
- 21.Simmer JP, Hu JC. Expression, structure, and function of enamel proteinases. Connect Tissue Res. 2002;43(2-3):441–449. doi: 10.1080/03008200290001159. [DOI] [PubMed] [Google Scholar]
- 22.Tanimoto K, Le T, Zhu L, Witkowska HE, Robinson S, Hall S, et al. Reduced amelogenin-MMP20 interactions in amelogenesis imperfecta. J Dent Res. 2008;87(5):451–455. doi: 10.1177/154405910808700516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bourd-Boittin K, Fridman R, Fanchon S, Septier D, Goldberg M, Menashi S. Matrix metalloproteinase inhibition impairs the processing, formation and mineralization of dental tissues during mouse molar development. Exp Cell Res. 2005;304(2):493–505. doi: 10.1016/j.yexcr.2004.11.024. [DOI] [PubMed] [Google Scholar]
- 24.Smith CE, Richardson AS, Hu Y, Bartlett JD, Hu JC, Simmer JP. Effect of Kallikrein 4 Loss on Enamel Mineralization: COMPARISON WITH MICE LACKING MATRIX METALLOPROTEINASE 20. The Journal of biological chemistry. 2011;286(20):18149–18160. doi: 10.1074/jbc.M110.194258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uskokovic V, Kim MK, Li W, Habelitz S. Enzymatic processing of amelogenin during continuous crystallization of apatite. Journal of Materials Research. 2008;23(12):3184–3195. doi: 10.1557/JMR.2008.0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uskokovic V, Li W, Habelitz S. Amelogenin as a promoter of nucleation and crystal growth of apatite. Journal of Crystal Growth. 2011;316(1):106–117. doi: 10.1016/j.jcrysgro.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li W, Gao C, Yan Y, DenBesten PK. X-linked amelogenesis imperfecta may result from decreased formation of tyrosine rich amelogenin peptide (TRAP) Arch Oral Biol. 2003;48(3):177–183. doi: 10.1016/s0003-9969(02)00170-x. [DOI] [PubMed] [Google Scholar]
- 28.Zhu L, Tanimoto K, Robinsin S, Chen J, Witkowska E, Hall S, et al. Comparative properties of recombinant human and bovine matrix metalloproteinase-20. Arch Oral Biol. 2008;53(8):785–790. doi: 10.1016/j.archoralbio.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moisescu C, Jana C, Habelitz S, Carl G, Russel C. Oriented fluoroapatite glass-ceramics. Journal of Non-Crystalline Solids. 1999;248(2-3):176–182. [Google Scholar]
- 30.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 31.Nagano T, Kakegawa A, Yamakoshi Y, Tsuchiya S, Hu JC, Gomi K, et al. Mmp-20 and Klk4 cleavage site preferences for amelogenin sequences. Journal of dental research. 2009;88(9):823–828. doi: 10.1177/0022034509342694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Uskokovic V, Castiglione Z, Cubas P, Zhu L, Li W, Habelitz S. Zeta-potential and particle size analysis of human amelogenins. J Dent Res. 2010;89(2):149–153. doi: 10.1177/0022034509354455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wright JT, Daly B, Simmons D, Hong S, Hart SP, Hart TC, et al. Human enamel phenotype associated with amelogenesis imperfecta and a kallikrein-4 (g.2142G>A) proteinase mutation. Eur J Oral Sci. 2006;114(Suppl 1):13–17. doi: 10.1111/j.1600-0722.2006.00291.x. discussion 39-41, 379. [DOI] [PubMed] [Google Scholar]
- 34.Wright JT, Hart TC, Hart PS, Simmons D, Suggs C, Daley B, et al. Human and mouse enamel phenotypes resulting from mutation or altered expression of AMEL, ENAM, MMP20 and KLK4. Cells Tissues Organs. 2009;189(1-4):224–229. doi: 10.1159/000151378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tarasevich BJ, Lea S, Bernt W, Engelhard MH, Shaw WJ. Changes in the quaternary structure of amelogenin when adsorbed onto surfaces. Biopolymers. 2009;91(2):103–107. doi: 10.1002/bip.21095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He X, Wu S, Martinez-Avila O, Cheng Y, Habelitz S. Self-aligning amelogenin nanoribbons in oil-water system. Journal of structural biology. 2011;174(1):203–212. doi: 10.1016/j.jsb.2010.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tarasevich BJ, Howard CJ, Larson JL, Snead ML, Simmer JP, Paine M, et al. The nucleation and growth of calcium phosphate by amelogenin. Journal of Crystal Growth. 2007;304(2):407–415. doi: 10.1016/j.jcrysgro.2007.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang L, Guan X, Du C, Moradian-Oldak J, Nancollas GH. Amelogenin Promotes the Formation of Elongated Apatite Microstructures in a Controlled Crystallization System. J Phys Chem C Nanomater Interfaces. 2007;111(17):6398–6404. doi: 10.1021/jp0675429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang L, Guan X, Yin H, Moradian-Oldak J, Nancollas GH. Mimicking the Self-Organized Microstructure of Tooth Enamel. J Phys Chem C Nanomater Interfaces. 2008;112(15):5892–5899. doi: 10.1021/jp077105+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aoba T, Fukae M, Tanabe T, Shimizu M, Moreno EC. Selective adsorption of porcine-amelogenins onto hydroxyapatite and their inhibitory activity on hydroxyapatite growth in supersaturated solutions. Calcif Tissue Int. 1987;41(5):281–289. doi: 10.1007/BF02555230. [DOI] [PubMed] [Google Scholar]
- 41.Iijima M, Moriwaki Y, Wen HB, Fincham AG, Moradian-Oldak J. Elongated growth of octacalcium phosphate crystals in recombinant amelogenin gels under controlled ionic flow. J Dent Res. 2002;81(1):69–73. doi: 10.1177/002203450208100115. [DOI] [PubMed] [Google Scholar]
- 42.Paine ML, White SN, Luo W, Fong H, Sarikaya M, Snead ML. Regulated gene expression dictates enamel structure and tooth function. Matrix Biol. 2001;20(5-6):273–292. doi: 10.1016/s0945-053x(01)00153-6. [DOI] [PubMed] [Google Scholar]


