Abstract
XPF-ERCC1 is a structure-specific endonuclease that is essential for nucleotide excision repair and DNA interstrand cross-link repair in mammalian cells. The yeast counterpart of XPF-ERCC1, Rad1-Rad10, plays multiple roles in DNA repair. Rad1-Rad10 is implicated to be involved in the repair of oxidative DNA damage. To explore the role(s) of XPF-ERCC1 in the repair of DNA damage induced by reactive oxygen species (ROS), cellular sensitivity of the XPF-deficient Chinese hamster ovary cell-line UV41 to ROS was investigated. The XPF-deficient UV41 showed sensitivity to hydrogen peroxide, bleomycin and paraquat. Furthermore, XPF-ERCC1 showed an ability to remove 3’-blocked ends such as 3’-phosphoglycolate from the 3’-end of DNA in vitro. These data suggest that XPF-ERCC1 plays a role in the repair of ROS-induced DNA damage by trimming 3’-blocked ends. The accumulation of various types of DNA damage, including ROS-induced DNA damage due to defects in multiple XPF-ERCC1-mediated DNA repair pathways, could contribute to the accelerated aging phenotypes observed in an XPF-ERCC1 deficient patient.
Introduction
Reactive oxygen species (ROS) are considered to be major sources of endogenous DNA damage. ROS induces various types of oxidative DNA damage, including base modifications and single strand breaks (SSBs). These SSBs are often damaged with modified 3’-ends such as phosphoglycolate, phosphate and αβ unsaturated aldehyde.1, 2 Interestingly, modified 3’-ends can also be introduced during BER. Bi-functional DNA glycosylases, 8-oxoguanine-DNA glycosylase (OGG1), endonuclease III homologue (NTH1) and endonuclease VII-like enzymes (NEIL1, 2 and 3), have an associated β-lyase activity. After removing modified bases, these glycosylases generate a nick 3’ to the abasic site to generate a 3’-α,β-unsaturated aldehyde (by OGG1 and NTH1) or 3’-phosphate (by NEIL1, 2, and 3).3 In addition, modified 3’-ends might be induced as a result of abortive DNA topoisomerase I (Top1) activity.1, 2 Top1 generates a nick transiently and remains covalently attached at the 3’-end of the nick. Then Top1 re-ligates the nick to complete its action. If this re-ligation process is inhibited by DNA lesions located near a nick, a protein-DNA cross-link will be generated at the 3’-end of the nick.
The “damaged” 3’-ends must be restored to conventional 3’-hydroxy (3’-OH) moieties for DNA repair synthesis and DNA ligation to occur. Thus, end processing is a crucial step during SSB repair and BER. Several DNA end processing enzymes have been identified in mammalian cells.1, 2 The AP endonuclease, APE1, functions as the major 3’-end processing enzyme in human cells. APE1 contains 3’-phosphatase, 3’-phosphodiesterase and 3’ to 5’ exonuclease activities specific for internal nicks and gaps in DNA, in addition to the AP endonuclease activity. Polynucleotide kinase (PNK) is also an important end-processing enzyme that is involved in the restoration of 3’-OH as well as 5’-phosphate ends at SSBs. PNK is believed to be involved in removing the 3’-phosphate moieties generated by NEILs during BER. Tyrosyl-DNA phosphodiesterase I (TDP1) is a specialized enzyme to remove a Top1-DNA cross-link at the 3’-ends that arise from abortive Top1 activity. The processing of the Top1-modified end by TDP1 results in a generation of 3’-phosphate that must be repaired by PNK. It is noted that TDP1 is mutated in a hereditary neurodegenerative disease, Spinocerebellar ataxia with axonal neuropathy 1 (SCAN1).4 It is unclear how much contribution each enzyme makes for end processing of a certain type of damaged 3’-end due to the lack of a systematic genetic study.
An evolutionally conserved DNA structure-specific DNA endonuclease, the XPF-ERCC1 complex, plays important roles in nucleotide excision repair (NER) and DNA interstrand cross-link (ICL) repair.5, 6, 7, 8 A counterpart of XPF-ERCC1 in Saccharomyces cerevisiae, Rad1-Rad10, is implicated to play multiple roles in different DNA repair pathways. In addition to its roles in NER, Rad1-Rad10 is involved in single-strand annealing (SSA) and microhomology-mediated end-joining repair pathways.9 In these pathways, Rad1-Rad10 removes non-homologous 3′ single strand tails from recombination intermediates that would otherwise prevent completion of the recombination event. Interestingly, Rad1-Rad10 also participates in the repair of ROS-induced DNA lesions.10, 11 ROS-induced 3’-blocked ends are mainly removed by two AP endonucleases, Apn1 and Apn2, in yeast. In the absence of Apn1 and Apn2, a further inactivation of Rad1-Rad10 sensitizes the triple mutant cells to ROS. In vitro studies demonstrated that Rad1-Rad10 removes 3’-blocking ends induced by ROS.11 Rad1-Rad10 is a critical back-up enzyme to remove 3’-blocking ends in the absence of Apn1 and Apn2.
To explore a possible role of XPF-ERCC1 in the repair of ROS-induced DNA lesions, we investigated the cellular sensitivity of the XPF-deficient rodent cell-line UV41 to various agents that produce ROS. UV41 cells are sensitive to three different chemicals, hydrogen peroxide (H2O2), paraquat, and bleomycin. Furthermore, XPF-ERCC1 is capable of removing 3’-phosphoglycolates, which are one of the 3’-blocking ends in vitro. Taken together, our results show that XPF-ERCC1 might participate in the repair of ROS-induced lesions by trimming 3’-blocked ends in mammalian cells.
Experimental Procedures
Cell lines
Chinese hamster ovary (CHO) cells, AA8, UV41 (XPF deficient), and UV20 (ERCC1 deficient), were purchased form ATCC. UV41 corrected by the XPF gene was generated as described in Fisher et al.7
Colony-Forming Assay
Cells were seeded at 500 cells/10 cm dish and allowed to attach overnight. The next day, the medium was removed and the cells were washed with PBS prior to treatment. To determine the sensitivity of cells to agents that induce strand breaks containing a 3’-PG, cells were treated with H2O2, paraquat (PQ), and bleomycin. H2O2 was diluted in the medium and the cells were exposed for 30 min at 37°C. For PQ treatment, cells were treated with the indicated concentrations of PQ for 24 hours. For bleomycin treatment, cells were treated with bleomycin for two hours. After exposure to chemicals, the medium with the chemicals was removed, the cells were washed with PBS twice and the cells were incubated at 37°C with 5% CO2 for an additional 6–7 days in fresh medium until colonies formed.
Once colonies were formed, the cells were washed with PBS, fixed with ethanol, and stained with Giemsa for counting. The surviving fraction is expressed as the ratio of cells that formed colonies relative to the number of colonies on untreated control plates.
Preparation of DNA Substrates Containing 3’-Phosphoglycolate (PG)
A 21mer with 3’-PG and an undamaged complementary strand (CD) were purchased from Trevigen (Gaithersburg, MD). Other oligonucleotides, 3’-P, D1, D2 and D3, were synthesized by the UNMC Eppley Molecular Biology Core Laboratory.
CD: 5’- CCCACAGCTGCTCAGGGCAGG-3’
PG: 5’- CCTGCCCTGAGCAGCTGTGGG–[PG]-3’
3’-P: 5’- CCTGCCCTGAGCAGCTGTGGG–[phosphate]-3’
D1: 5’- CCTGCCCTGAGCAGCTGTGGG-3’
D2: 5’- TTGATCTTTCGAGTCAACTGACCCACAGCTGCTCAGGGCAGG-3’
D3: 5’- TCAGTTGACTCGAAAGATCAA -3’
Oligonucleotides PG, 3’-P, and D1 were labeled at the 5’-end with [γ32P]-ATP by T4 polynucleotide kinase. The labeled PG or D1was annealed to CD to make a 21 bp blunt ended substrate (DS), to D2 to make a recessed substrate (DS/SS), and to D2 and D3 for a nicked substrate (Nick). The substrates were purified by 6% native polyacrylamide gels.
Protein Expression and Purification from Insect Cells
A FLAG tag was attached to the N-terminus of each form of the XPF gene by ligating an oligonucleotide containing the FLAG tag sequence in the pFastBac1 vector. The proteins were then expressed in Sf-9 or Sf-21 cells for purification. The cells from 24 10 cm dishes were harvested and lysed in five times the cell pellet volume insect cell lysis buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, and 1 % v/v NP-40) for 30 minutes with rocking at 4°C. The cell debris was removed by centrifugation and the cell lysate was applied to a 250 µl anti-FLAG agarose (Sigma, St. Louis, MO). The anti-FLAG agarose was washed five times with 5 ml 1M KCl Tris buffer (20 mM Tris-HCl (pH 7.5), 0.1 mM EDTA, 1 M KCl, 10% v/v glycerol), then once with starting buffer (20 mM Tris-HCl pH 7.5, 0.1 mM EDTA, 0.8 M KCl, 10% v/v glycerol, 10 mM imidazole). The protein was eluted with 2 mg/ml FLAG peptide (Sigma) in starting buffer by rotating at 4°C for 1–2 hrs. This elution was repeated once and then the resin was washed with the starting buffer. Peak elution fractions were pooled and incubated with 100 µl Ni-agarose (Qiagen, Valencia, CA) overnight with end-over-end rotation at 4°C. The resin was then washed with P-1000 buffer (40 mM HEPES pH 7.5, 1M NaCl, 10% v/v glycerol) containing 10 mM imidazole and once with P-I10 buffer (40 mM HEPES pH 7.5, 100 mM NaCl, 10% v/v glycerol, 10 mM imidazole). The protein was eluted twice with 100 µl PI-100 (40 mM HEPES pH 7.5, 100 mM NaCl, 10% v/v glycerol, 100 mM imidazole) by rotating at 4°C for 30 min then by 100 µl PI-300 (40mM HEPES pH 7.5, 100 mM NaCl, 10% v/v glycerol, 300 mM imidazole) two times. Purity was assessed by silver staining and Western blotting (Fig. S-1).
Endonuclease Assay
The endonuclease assays were performed as in Fisher et al7 with modifications. Briefly, 5’ 32P-labeled DNA substrate was incubated with purified proteins in 20 µl of the reaction buffer (10 mM HEPES pH 7.5, 25 mM KCl, 0.05 mM EDTA, 0.5 mM DTT, 10% v/v glycerol, and 1 mM MnCl2) at 30°C for the indicated time. Proteins were removed by phenol/chloroform extraction and DNA precipitated with ethanol, the reaction products were analyzed on a 10% denaturing polyacrylamide gel. The gel was dried and exposed to a PhosphorImager screen. After scanning with a Typhoon 9700 (GE Healthcare, Piscataway, NJ), the image was quantitated using ImageQuant software (GE Healthcare). The percent incised was determined by dividing the value of the 16 nt and 14 nt bands by the total value of DNA loaded in the lane.
Results
Loss of XPF Increases the Sensitivity of CHO Cells to Agents that Produce 3’-PG ends
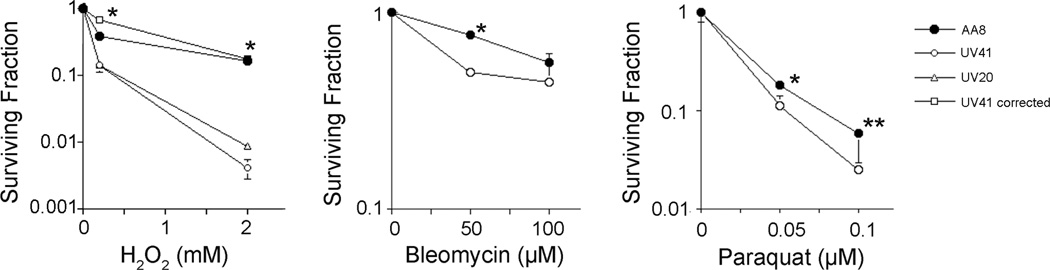
Hydrogen peroxide (H2O2), bleomycin, and paraquat induce oxidative stress and are widely used as DNA damaging agents to study cellular responses to reactive oxygen species (ROS).12, 13 Major DNA damage caused by these chemicals is believed to be single-strand breaks with modified 5’ and/or 3’-ends. Chinese hamster ovary (CHO) cell line AA8 and UV41 were treated with increasing concentrations of each of these agents to examine a potential role of XPF-ERCC1 in oxidative DNA damage repair. The XPF-deficient UV41 showed a mild sensitivity to H2O2 compared to AA8 (Fig. 1, open circles in the left graph). UV20 that is deficient in ERCC1 also showed a similar sensitivity to H2O2, as was reported with ERCC1−/− mouse embryonic stem cells (Fig. 1, open triangles in the left graph).14 Importantly, the expression of XPF in UV41 restored resistance to H2O2 (Fig. 1, open squares in the left graph), demonstrating that the sensitivity to H2O2 in UV41 is due to a specific defect in XPF. UV41 cells displayed mild sensitivity to other ROS-inducing agents, bleomycin (Fig. 1, open circles in the middle graph) and paraquat (Fig. 1, open circles in the right graph). These data showed that XPF-ERCC1 is required for cellular resistance to ROS-inducing chemicals in CHO cells. The results are significantly different from the ones obtained with yeast strains. In yeast, the impact of yeast Rad1-Rad10 on the cellular sensitivity to ROS was only observed in the absence of Apn1 and Apn2.10, 11 In contrast, because it appears that AP endonuclease activities are not compromised in AA8,15, 16 XPF-ERCC1 is not merely a backup for AP endonuclease(s) in the processing of ROS-induced DNA damage in mammalian cells. We conclude that the repair of ROS-induced DNA damage requires XPF-ERCC1 in mammalian cells.
Figure 1. The XPF-deficient UV41 cells are sensitive to various ROS-generating reagents.
Cellular sensitivity of AA8 and UV41 to hydrogen peroxide (H2O2), bleomycin and paraquat was investigated by clonogenic survival assays. Both XPF-defective UV41 and ERCC1-defective UV20 showed sensitivity to H2O2. UV41 corrected by the human XPF gene showed a similar resistance to H2O2, compared to AA8. UV41 cells also displayed sensitivity to bleomycin and paraquat. Closed circle, AA8; opened circle, UV41; open triangle, UV20; open square, UV41 expressing XPF. The bars indicate standard deviations obtained from three independent experiments. Single asterisks and two asterisks indicate the results of the Student’s t-test, P<0.01 and P<0.05, respectively. The difference in the survival between AA8 and UV41 at 100 µM bleomycin treatment was not statistically significant. The actual values for each concentration in the graphs are summarized in Table S-1.
XPF-ERCC1 Removes a Short Fragment of DNA Containing the 3’-phosphoglycolates
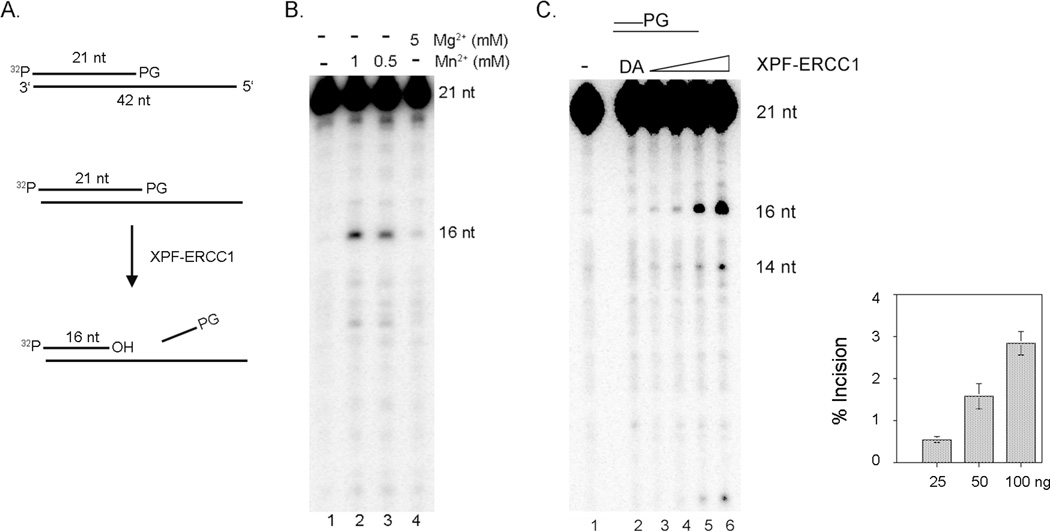
Our results in Figure 1 indicate that the repair of ROS-induced DNA damage requires XPF-ERCC1. Oxidative stress induced by H2O2, bleomycin, and paraquat generates single-strand breaks with blocked 3’-ends, mostly 3’-phosphoglycolate (3’-PG) and 3’-phosphate.17 These 3’-blocked ends should be removed and converted to 3-hydroxy groups for gap-filling/ligation reaction by a DNA polymerase and a DNA ligase. We investigated the ability of XPF-ERCC1 to remove this type of damage using DNA substrates containing a 3’-PG. First, XPF-ERCC1 was incubated with a 3’- recessed substrate (DS/SS) with a 3’-PG (Fig. 2A). XPF-ERCC1 generated 16 nt fragments with the substrate with a 3’-PG (Fig. 2B). This activity was stimulated by Mn2+ (Fig. 2B, compare lanes 2 and 3 to lane 4).18, 19 Thus, we included MnCl2 at 1 mM in the reaction in this report.
Figure 2. XPF-ERCC1 removes a 3’-phosphoglycolate (3’-PG) at a 3’-terminus in vitro.
(A). Substrate DNA and in vitro nuclease assay A 21 nt oligonucleotide with a PG at the 3’ end was labeled with 32P and annealed to a partially complementary 42 nt oligonucleotide to generate the DS/SS substrate. Incision products by XPF-ERCC1 are analyzed on a 10% sequencing gel. (B). Mn2+ stimulates the removal of 3’-PG by XPF-ERCC1. The 5’-labeled DS/SS substrate with a 3’-PG (2 nM) was incubated with XPF-ERCC1 (35 nM) at 30°C for 1 hr. XPF-ERCC1 released 5 nt fragments from the 3’end by removing 3’-PGs. Lane 1, no XPF-ERCC1; lane 2, with 1 mM MnCl2; lane 3, with 0.5 mM MnCl2; and lane 4, with 5 mM MgCl2. 2.8 %, 2.2 % and 0.8 % of the substrate was incised in lanes 2, 3, and 4, respectively. (C). Removal of 3’-PG by XPF-ERCC1. Increasing amounts of XPF-ERCC1 were incubated with 5’-labeled DS/SS substrate (2 nM) with a 3’-PG in 10 µl reaction buffer at 30°C for 1 hr. Lane 1, no XPF-ERCC1; lane 2, the endonuclease defective XPF(DA)-ERCC1 (42 nM); lane 3, 7 nM; lane 4, 14 nM; lane 5, 28 nM; lane 6, 56 nM. No incision was detected in lane 2. 0.4%, 0.6%, 1.5%, and 3% were incised in lanes 3, 4, 5, and 6, respectively. The average incision activities at the indicated amount of XPF-ERCC1 from three different experiments were plotted in a graph with standard deviations (error bars).
XPF-ERCC1 was able to make an incision 5 nt or 7 nt from the damaged end, generating 16 nt or 14 nt fragments, respectively (Fig. 2C, lanes 3–6, and the graph next to the gel). Therefore, XPF-ERCC1 effectively removes the 3’-PG ends. The endonuclease deficient XPF(DA)-ERCC17, 19 , showed no activity with the same substrate (Fig. 2C, lane 2), thus the activity detected is due to the endonuclease activity of XPF-ERCC1 and not a contaminating nuclease. XPF-ERCC1 has a trimming activity to remove the 3’-blocked end.
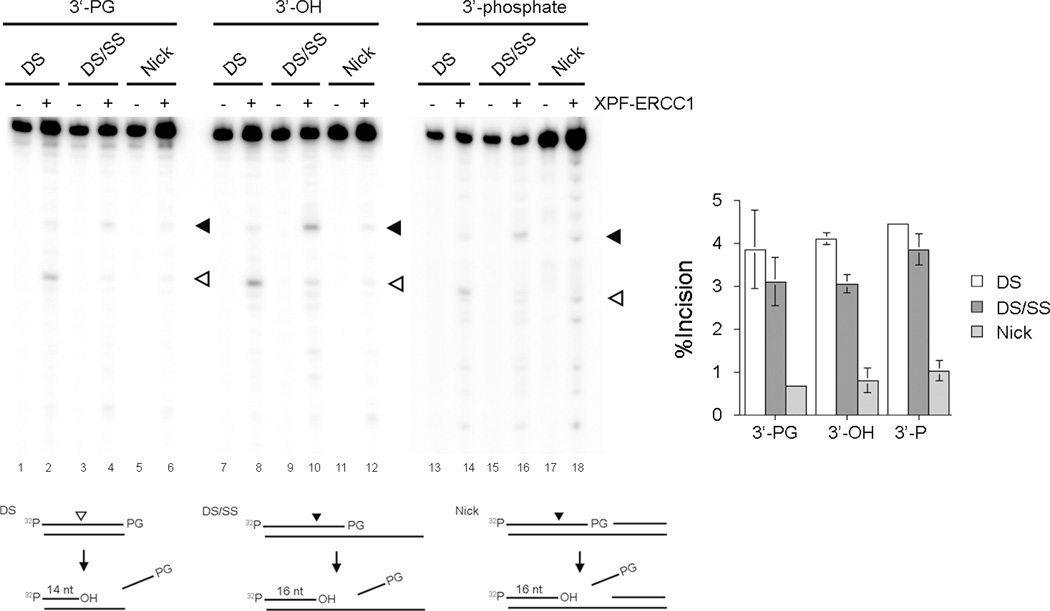
We further examined substrate specificity of the 3’-trimming activity of XPF-ERCC1. In addition to a 3’-recessed substrate, a blunt-ended substrate and nicked substrates with a 3’-PG were prepared (Fig. 3, below the gel images). A blunt-ended substrate and nicked substrates mimic a 3’-PG that is introduced at the end of a double strand break and a single strand break, respectively. We also prepared substrates with a 3’-phosphate moiety that represents other types of 3’-blocked ends. XPF-ERCC1 removed 3’-PGs from the DS/SS and the DS substrates with a similar efficiency and the nicked substrates with a lesser extent (Fig. 3, lanes 1–6 and the graph next to the gel). Interestingly, XPF-ERCC1 did not show a significant preference to a 3’-PG and/or a 3’-phosphate over an undamaged 3’-end. Also, the presence of a 3’-blocked end did not alter the substrate specificity of XPF-ERCC1 (Fig. 3, lanes 2–18, and the graph, Figs. S-2, S-3, and S-4).
Figure 3. Substrate specificity of the 3’-trimming activity of XPF-ERCC1.
Three different substrates (blunt ended, DS; recessed, DS/SS; nicked, Nick) were examined. Each substrate (0.2 nM) was incubated with XPF-ERCC1 (70 nM) at 30°C for 30 min. XPF-ERCC1 removes a short oligonucleotide from the substrates. XPF-ERCC1 incises blunt ended and recessed substrates in a similar efficiency and nicked substrate with a lesser extent. XPF-ERCC1 does not have a preferential activity to the ends with a 3’-blocked end (3’-PG or 3’-phosphate) to the ends with an unmodified 3’-hydroxy group. Lanes 1–6, 3’-PG substrates; lanes 7–12, non-damaged substrates, lanes 13–18, 3’-phosphate substrates . Lanes 1, 2, 7, 8, 13 and 14, blunt ended substrate; lanes 3, 4, 9,10, 15, and 16, recessed substrate; lanes 5, 6, 11,12,17, and 18, nicked substrate. Even numbered lanes, with XPF-ERCC1. The incision products were marked by triangles, closed triangles; 6 nt and open triangles; 4 nt. The average from three independent experiments was plotted in a graph next to the gel. Bars represents standard deviations. Major incision sites for each substrate were shown in the schematics under the gel.
It is noted that the major reaction product with the DS/SS substrate was 16 nt length, while the 14 nt length fragment was the major product in the reaction with the DS substrate (Fig. 3, compare lanes 2 and 4, and lanes 8 and 10, and lanes 14 and 16). Time course experiments also confirmed that XPF-ERCC1 generates primarily 16 nt fragments with the DS/SS substrate (Fig. S-2) and 14 nt fragment with the DS substrate (Fig. S-3). Since an exonuclease normally degrades DNA with a periodic pattern and gradually shortens DNA with time, these data suggest that these fragments were generated endonucleolytically by XPF-ERCC1.
Our in vitro results demonstrate that XPF-ERCC1 has a 3’-end trimming activity to remove the 3’-blocked end generated by ROS.
Discussion
XPF-ERCC1 is involved in various DNA repair pathways.8 XPF-ERCC1 is essential for NER and ICL repair. XPF-ERCC1 also plays a role in SSA,20, 21 one of the DSB repair pathways. Recently, it has also been demonstrated that XPF-ERCC1 functions in a Ku80-independent end-joining DSB repair pathway.22 In this report, we found an additional role of XPF-ERCC1 in ROS-induced DNA damage. The XPF-deficient rodent cell-line, UV41, displays cellular sensitivity to three ROS-generating reagents (H2O2, paraquat, bleomycin) (Fig. 1). In vitro, XPF-ERCC1 removes a 3’-blocked end induced by ROS (Figs. 2 and 3). These results show that XPF-ERCC1 may function in the repair of oxidative DNA damage by trimming a 3’blocked terminus induced by ROS. This putative activity would lead to the generation of a 3’-hydoxyl end that is accessible to a DNA repair polymerase to allow completion of SSB repair, BER and DSB repair.
Oxidative DNA damage, cyclopurines are implicated to be critical DNA lesions in the neurodegenerations observed in some XP patients.23 Cyclopurines are induced by γ-ray irradiation and other oxidative stress.24 Unlike other oxidative DNA damage, cyclopurines are shown to be repaired by NER, but not by BER.25 Because XPF-ERCC1 is an essential component of NER, cellular sensitivity of UV41 and UV20 to hydrogen peroxide could be a result of a defect of the repair of cyclopurines. However, XP-A cells have been shown not to be sensitive to γ-ray irradiation, hydrogen peroxide and paraquat.12, 26, 27, 28 We are of the opinion that the observed sensitivity of UV41 to ROS-inducing chemicals is not due to a defect in NER activity.
The yeast counterpart of XPF-ERCC1, Rad1-Rad10, removes 3’-blocked termini and functions as a back-up enzyme of two major 3’-trimming enzymes Apn1 and Apn2.11 Inactivation of Rad1 (or Rad10) alone does not sensitize the cells to H2O2.11 The impact of Rad1-Rad10 on the repair of ROS-generated DNA damage can be seen only in the absence of Apn1 and Apn2.10, 11 Mammalian APN1 has also been proposed as a major enzyme to remove the 3’-blocked ends generated by ROS in SSB repair and BER.13, 29, 30 However, our results indicate that XPF-ERCC1 also plays a significant role in the processing of 3’-blocked ends. Many in vitro studies with APN1 included Mg2+ as a cofactor,13, 30 which is a poor co-factor for the trimming activity of XPF-ERCC1 (Fig. 2A). XPF-ERCC1 might be involved in an APN1-independent SSB repair/BER pathway. Epistatic studies should be performed to determine the genetic relationship between XPF-ERCC1 and APN1 in the repair of ROS-induced DNA damage.
The roles of XPF-ERCC1 in the repair of “two-ended” DSBs induced by γ-ray or restriction enzymes have been reported. XPF-ERCC1 removes 3’-overhangs in a Ku86-independent end-joining repair pathway22 and also participates in single strand annealing (SSA) and gene conversion to repair DSBs induced by a restriction enzyme I-SceI.31 Intermediate DNA structures, which XPF-ERCC1 processes in these repair pathways, are believed to be a 3’-splay/flap-like structure. Hydrogen peroxide is also known to induce DSBs directly32 and the 3’-ends of these breaks can be blocked. These 3’-blocked ends can be removed from a 3’-splay/flap-like structure as a part of a 3’-overhang in an end-joining repair and SSA. In gene conversion to repair “two-ended” DSBs, DNA synthesis-dependent extension from one of the broken ends initiates the repair process.33 Our results indicate that XPF-ERCC1 can remove 3’-bloked ends at DSBs and therefore may participate in the repair of breaks generated directly by ROS and promote the initiation of repair synthesis.
A single strand break (SSB), if not repaired, results in the formation of a DSB after DNA replication. The resulting break is a “one-ended” DSB, which is distinct from the “two-ended” DSBs induced by γ-ray or restriction enzymes. “One-ended” DSBs are repaired by homologous recombination and required a 3’-hydroxy moiety at the broken end to initiate the repair process.34 If a SSB with a 3’-blocked end is located in the template for a lagging strand synthesis, a structure of the broken end could be a 3’-splay-like structure. This structure is a favorite for XPF-ERCC1, thus a single-stranded tail with a blocked end can be removed by the endonuclease activity of XPF-ERCC1. A SSB with a 3’-blocked terminus on a leading strand template will generate a gap structure that is similar to our DS/SS substrate. It is shown that XPF-ERCC1 and yeast Rad1-Rad10 are involved in the repair of camptothecin-induced Top1-lesions in a TDP1-independent pathway.35, 36 Camptothecin induces an abortive Top1-DNA complex at a 3’-terminus of a single strand nick. These Top1-lesions are normally repaired by TDP1-mediated single strand break repair.1 Unrepaired Top1-lesions can be converted to DSBs during DNA replication. In the absence of TDP1, XPF-ERCC1 and Rad1-Rad10 are proposed to remove Top1-lesions from the 3’-terminus of DNA and the DSBs are repaired by homologous recombination. XPF-ERCC1 might play a specific role in the repair of SSB-induced DSBs and/or gaps with 3’-blocked termini in conjunction with DNA replication.
XPF-ERCC1 is a structure-specific endonucelase that prefers a 3’-splay/flap structure with a 3’-single-strand tail.18 XPF-ERCC1 makes an incision in the double stranded DNA 3–5 nt from the junction of double stranded and 3’-single stranded DNA. Our results showed that XPF-ERCC1 makes an incision 5–7 nt from the 3’-ends, with or without damage (Fig. 3). Rad1-Rad10 removes 3’-blocked ends by a 3’-5’ exonuclease activity without showing a preference to damaged ends over non-damaged ends.11 XPF-ERCC1 also contains an intrinsic 3’-5’exonuclease activity,37 thus this exonuclease activity could remove 3’-blocked ends. Our time course experiments showed that XPF-ERCC1 makes an incision on DS/SS and DS substrates differently, but with a similar efficiency (Figs. S-2 and S-3). An exonuclease normally degrades substrate DNA from an end, regardless of its structure, in the same fashion. It is likely that XPF-ERCC1 removes 3’-blocked ends by its endonuclease activity.
XPF-ERCC1 is one of the six factors that are essential for in vitro NER,38 XPF-deficient cell lines derived from XP patients are moderately sensitive to UV irradiation due to residual NER activities. It was reported recently that one patient with a mutation in XPF displayed accelerated aging symptoms, but not the XP symptoms.14 A cell line derived from this patient (XFE cells) showed hypersensitivity to UV irradiation and DNA cross-linking agents due to a severe defect of XPF-ERCC1. It is also important to note that the XFE cells showed sensitivity to IR.22 Thus, XFE would also be defective in the repair of ROS-induced DNA damage. Accumulation of DNA damage has been long proposed to be one of the causes of aging. Because other XP patients do not show aging phenotypes, DNA repair defects in addition to the NER defect contribute to the aging phenotypes. An accumulation of various types of DNA damage, including ROS-induced DNA damage due to defects in multiple XPF-ERCC1-mediated DNA repair pathways, could contribute to the aging phenotypes observed in the XFE patient.
Supplementary Material
Acknowledgements
We thank the Eppley Molecular Biology Core Facility (supported by the Eppley Cancer Center Support Grant, P30CA036727) for the synthesis of the oligonucleotides used in this study.
Supporting Information
This work was supported by the National Institutes of Health Grants CA95291 (to T. B.) and GM080458 (to T.B.) and 5T32 CA09476 (to L.A.F.) and by an assistantship from UNMC Graduate Studies.
Abbreviations
- XP
xeroderma pigmentosum
- ROS
reactive oxygen species
- 3’-PG
3’-phosphoglycolate
References
- 1.Caldecott KW. Single-strand break repair and genetic disease. Nat Rev Genet. 2008;9:619–631. doi: 10.1038/nrg2380. [DOI] [PubMed] [Google Scholar]
- 2.Dianov GL, Parsons JL. Co-ordination of DNA single strand break repair. DNA Repair (Amst) 2007;6:454–460. doi: 10.1016/j.dnarep.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 3.Hegde ML, Hazra TK, Mitra S. Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell Res. 2008;18:27–47. doi: 10.1038/cr.2008.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.El-Khamisy SF, Saifi GM, Weinfeld M, Johansson F, Helleday T, Lupski JR, Caldecott KW. Defective DNA single-strand break repair in spinocerebellar ataxia with axonal neuropathy-1. Nature. 2005;434:108–113. doi: 10.1038/nature03314. [DOI] [PubMed] [Google Scholar]
- 5.Rahn JJ, Adair GM, Nairn RS. Multiple roles of ERCC1-XPF in mammalian interstrand crosslink repair. Environ Mol Mutagen. 2010;51:567–581. doi: 10.1002/em.20583. [DOI] [PubMed] [Google Scholar]
- 6.Wood RD. Mammalian nucleotide excision repair proteins and interstrand crosslink repair. Environ Mol Mutagen. 2010;51:520–526. doi: 10.1002/em.20569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fisher LA, Bessho M, Bessho T. Processing of a Psoralen DNA Interstrand Cross-link by XPF-ERCC1 Complex in Vitro. J Biol Chem. 2008;283:1275–1281. doi: 10.1074/jbc.M708072200. [DOI] [PubMed] [Google Scholar]
- 8.McDaniel LD, Schultz RA. XPF/ERCC4 and ERCC1: their products and biological roles. Adv Exp Med Biol. 2008;637:65–82. doi: 10.1007/978-0-387-09599-8_8. [DOI] [PubMed] [Google Scholar]
- 9.Lyndaker AM, Alani E. A tale of tails: insights into the coordination of 3' end processing during homologous recombination. Bioessays. 2009;31:315–321. doi: 10.1002/bies.200800195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guillet M, Boiteux S. Endogenous DNA abasic sites cause cell death in the absence of Apn1, Apn2 and Rad1/Rad10 in Saccharomyces cerevisiae. Embo J. 2002;21:2833–2841. doi: 10.1093/emboj/21.11.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guzder SN, Torres-Ramos C, Johnson RE, Haracska L, Prakash L, Prakash S. Requirement of yeast Rad1-Rad10 nuclease for the removal of 3'-blocked termini from DNA strand breaks induced by reactive oxygen species. Genes Dev. 2004;18:2283–2291. doi: 10.1101/gad.1232804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Waard H, de Wit J, Gorgels TG, van den Aardweg G, Andressoo JO, Vermeij M, van Steeg H, Hoeijmakers JH, van der Horst GT. Cell type-specific hypersensitivity to oxidative damage in CSB and XPA mice. DNA Repair (Amst) 2003;2:13–25. doi: 10.1016/s1568-7864(02)00188-x. [DOI] [PubMed] [Google Scholar]
- 13.Parsons JL, Dianova II, Dianov GL. APE1 is the major 3'-phosphoglycolate activity in human cell extracts. Nucleic Acids Res. 2004;32:3531–3536. doi: 10.1093/nar/gkh676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niedernhofer LJ, Garinis GA, Raams A, Lalai AS, Robinson AR, Appeldoorn E, Odijk H, Oostendorp R, Ahmad A, van Leeuwen W, Theil AF, Vermeulen W, van der Horst GT, Meinecke P, Kleijer WJ, Vijg J, Jaspers NG, Hoeijmakers JH. A new progeroid syndrome reveals that genotoxic stress suppresses the somatotroph axis. Nature. 2006;444:1038–1043. doi: 10.1038/nature05456. [DOI] [PubMed] [Google Scholar]
- 15.La Belle M, Linn S, Thompson LH. Apurinic/apyrimidinic endonuclease activities appear normal in the CHO-cell ethyl methanesulfonate-sensitive mutant, EM9. Mutat Res. 1984;141:41–44. doi: 10.1016/0165-7992(84)90035-6. [DOI] [PubMed] [Google Scholar]
- 16.McNeill DR, Wong HK, Narayana A, Wilson DM., 3rd Lead promotes abasic site accumulation and co-mutagenesis in mammalian cells by inhibiting the major abasic endonuclease Ape1. Mol Carcinog. 2007;46:91–99. doi: 10.1002/mc.20196. [DOI] [PubMed] [Google Scholar]
- 17.Caldecott KW. Mammalian single-strand break repair: mechanisms and links with chromatin. DNA Repair (Amst) 2007;6:443–453. doi: 10.1016/j.dnarep.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 18.de Laat WL, Appeldoorn E, Jaspers NG, Hoeijmakers JH. DNA structural elements required for ERCC1-XPF endonuclease activity. J Biol Chem. 1998;273:7835–7842. doi: 10.1074/jbc.273.14.7835. [DOI] [PubMed] [Google Scholar]
- 19.Enzlin JH, Scharer OD. The active site of the DNA repair endonuclease XPF-ERCC1 forms a highly conserved nuclease motif. Embo J. 2002;21:2045–2053. doi: 10.1093/emboj/21.8.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adair GM, Rolig RL, Moore-Faver D, Zabelshansky M, Wilson JH, Nairn RS. Role of ERCC1 in removal of long non-homologous tails during targeted homologous recombination. Embo J. 2000;19:5552–5561. doi: 10.1093/emboj/19.20.5552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Motycka TA, Bessho T, Post SM, Sung P, Tomkinson AE. Physical and functional interaction between the XPF/ERCC1 endonuclease and hRad52. J Biol Chem. 2004;279:13634–13639. doi: 10.1074/jbc.M313779200. [DOI] [PubMed] [Google Scholar]
- 22.Ahmad A, Robinson AR, Duensing A, van Drunen E, Beverloo HB, Weisberg DB, Hasty P, Hoeijmakers JH, Niedernhofer LJ. ERCC1-XPF endonuclease facilitates DNA double-strand break repair. Mol Cell Biol. 2008;28:5082–5092. doi: 10.1128/MCB.00293-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brooks PJ. The 8,5'-cyclopurine-2'-deoxynucleosides: candidate neurodegenerative DNA lesions in xeroderma pigmentosum, and unique probes of transcription and nucleotide excision repair. DNA Repair (Amst) 2008;7:1168–1179. doi: 10.1016/j.dnarep.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jaruga P, Dizdaroglu M. 8,5'-Cyclopurine-2'-deoxynucleosides in DNA: mechanisms of formation, measurement, repair and biological effects. DNA Repair (Amst) 2008;7:1413–1425. doi: 10.1016/j.dnarep.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 25.Kuraoka I, Bender C, Romieu A, Cadet J, Wood RD, Lindahl T. Removal of oxygen free-radical-induced 5',8-purine cyclodeoxynucleosides from DNA by the nucleotide excision-repair pathway in human cells. Proc Natl Acad Sci U S A. 2000;97:3832–3837. doi: 10.1073/pnas.070471597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Boer J, Andressoo JO, de Wit J, Huijmans J, Beems RB, van Steeg H, Weeda G, van der Horst GT, van Leeuwen W, Themmen AP, Meradji M, Hoeijmakers JH. Premature aging in mice deficient in DNA repair and transcription. Science. 2002;296:1276–1279. doi: 10.1126/science.1070174. [DOI] [PubMed] [Google Scholar]
- 27.Low GK, Fok ED, Ting AP, Hande MP. Oxidative damage induced genotoxic effects in human fibroblasts from Xeroderma Pigmentosum group A patients. Int J Biochem Cell Biol. 2008;40:2583–2595. doi: 10.1016/j.biocel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 28.van der Pluijm I, Garinis GA, Brandt RM, Gorgels TG, Wijnhoven SW, Diderich KE, de Wit J, Mitchell JR, van Oostrom C, Beems R, Niedernhofer LJ, Velasco S, Friedberg EC, Tanaka K, van Steeg H, Hoeijmakers JH, van der Horst GT. Impaired genome maintenance suppresses the growth hormone--insulin-like growth factor 1 axis in mice with Cockayne syndrome. PLoS Biol. 2007;5:e2. doi: 10.1371/journal.pbio.0050002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fung H, Demple B. Distinct roles of Ape1 protein in the repair of DNA damage induced by ionizing radiation or bleomycin. J Biol Chem. 2011;286:4968–4977. doi: 10.1074/jbc.M110.146498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winters TA, Henner WD, Russell PS, McCullough A, Jorgensen TJ. Removal of 3'-phosphoglycolate from DNA strand-break damage in an oligonucleotide substrate by recombinant human apurinic/apyrimidinic endonuclease 1. Nucleic Acids Res. 1994;22:1866–1873. doi: 10.1093/nar/22.10.1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Al-Minawi AZ, Saleh-Gohari N, Helleday T. The ERCC1/XPF endonuclease is required for efficient single-strand annealing and gene conversion in mammalian cells. Nucleic Acids Res. 2008;36:1–9. doi: 10.1093/nar/gkm888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Driessens N, Versteyhe S, Ghaddhab C, Burniat A, De Deken X, Van Sande J, Dumont JE, Miot F, Corvilain B. Hydrogen peroxide induces DNA single- and double-strand breaks in thyroid cells and is therefore a potential mutagen for this organ. Endocr Relat Cancer. 2009;16:845–856. doi: 10.1677/ERC-09-0020. [DOI] [PubMed] [Google Scholar]
- 33.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 34.Cox MM. The nonmutagenic repair of broken replication forks via recombination. Mutat Res. 2002;510:107–120. doi: 10.1016/s0027-5107(02)00256-7. [DOI] [PubMed] [Google Scholar]
- 35.Vance JR, Wilson TE. Yeast Tdp1 and Rad1-Rad10 function as redundant pathways for repairing Top1 replicative damage. Proc Natl Acad Sci U S A. 2002;99:13669–13674. doi: 10.1073/pnas.202242599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang YW, Regairaz M, Seiler JA, Agama KK, Doroshow JH, Pommier Y. Poly(ADP-ribose) polymerase and XPF-ERCC1 participate in distinct pathways for the repair of topoisomerase I-induced DNA damage in mammalian cells. Nucleic Acids Res. 2011;39:3607–3620. doi: 10.1093/nar/gkq1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mu D, Bessho T, Nechev LV, Chen DJ, Harris TM, Hearst JE, Sancar A. DNA interstrand cross-links induce futile repair synthesis in mammalian cell extracts. Mol Cell Biol. 2000;20:2446–2454. doi: 10.1128/mcb.20.7.2446-2454.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bessho T, Sancar A, Thompson LH, Thelen MP. Reconstitution of human excision nuclease with recombinant XPF-ERCC1 complex. J Biol Chem. 1997;272:3833–3837. doi: 10.1074/jbc.272.6.3833. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.