Abstract
Nuclear receptor (NR) coregulators include coactivators, contributing to holoreceptor transcriptional activity, and corepressors, mediating NR target gene silencing in the absence of hormone. We identified an atypical NR coregulator, TNFα-induced protein 3-interacting protein 1 (TNIP1), from a peroxisome proliferator activated receptor (PPAR) α screen of a human keratinocyte cDNA library. TNIP1’s complex nomenclature parallels its additional function as an NF-κB inhibitor. Here we show TNIP1 is an atypical NR corepressor using two-hybrid systems, biochemical studies, and receptor activity assays. The requirements for TNIP1-PPAR interaction are characteristic for coactivators; however, TNIP1 partially decreases PPAR activity. TNIP1 has separable transcriptional activation and repression domains suggesting a modular nature to its overall effect. It may provide a means of lowering receptor activity in the presence of ligand without total loss of receptor function. TNIP1’s multiple roles and expression in several cell types suggest its regulatory effect depends on its expression level and the expression of other regulators in NR and/or NF-κB signaling pathways. As a NR coregulator, TNIP1 targeting agonist-bound PPAR and reducing transcriptional activity offers control of receptor signaling not available from typical corepressors and may contribute to combinatorial regulation of transcription.
Keywords: PPAR, nuclear receptors, coactivator, corepressor, TNIP1, transcription
Introduction
The majority of ~300 nuclear receptor (NR) coregulators identified to date are coactivators; far fewer corepressors have been isolated. Coactivators such as the p160/steroid receptor coactivator (SRC) and thyroid hormone receptor-associated proteins (TRAP) [1] associate with agonist-bound receptors via the coactivator’s NR box, a short, amphipathic, leucine-rich motif (LXXLL, L = leucine, X = any amino acid). Physical association of unliganded NRs with corepressors e.g., nuclear receptor corepressor (NCoR) or silencing mediator for retinoid and thyroid hormone receptor (SMRT) is mediated by amphipathic leucine-rich helices (L/I-XX-I/VI or L-XXX-I/L-XXX-L; I = isoleucine, V = valine, X = any amino acid) in the coregulator referred to as corepressor/nuclear receptor (CoRNR) boxes [2]. For most NR-coregulator interactions, the aporeceptor associates with corepressors and upon agonist binding undergoes a conformational change, shedding the corepressor and associating with a coactivator. However, the discoveries of ligand dependent corepressor (LCoR), receptor interaction protein (RIP) 140, and in this report TNIP1 have recognized a distinct group of coregulators. These proteins represent a novel, functionally allied group of coregulators using NR boxes, targeting the agonist-bound receptor’s activating function- (AF) 2 region but reducing receptor-mediated gene transcription. This distinguishes them from typical NR corepressors such as SMRT and NCoR [3 for review] and provides additional means of reducing NR activity without the more-extreme loss of activity conferred in the absence of ligand by SMRT or NCoR.
Three PPAR subtypes, PPARα, PPARδ, and PPARγ have been identified (NR1C1, NR1C2 and NR1C3) and each has been implicated in multiple aspects of keratinocyte response to natural and synthetic PPAR ligands [4–8]. As PPAR coregulators [9 for review] would participate in these responses, we report here on a novel PPAR coregulator isolated via a two-hybrid screen of keratinocyte cDNA. Intriguingly, this PPAR-interacting clone matched coding sequences previously isolated via protein-protein interaction with i)the HIV protein Nef, ii)the HIV protein matrix, and iii)the human zinc finger protein A20 [10–12]. In each case, different names were given to the isolate (Naf, VAN and ABIN, respectively). As found in the National Center for Biotechnology Information database, the sequence is named TNIP1 for TNFAIP3-interacting protein 1, and we use that designation in this report. Beyond the roles described for TNIP1 under its aliases of Naf, VAN, and ABIN are its recent gene-disease associations in lung cancer and psoriasis [13, 14].
Given the diversity of previous TNIP1 investigations and having isolated TNIP1 as a PPARα-interacting protein the objectives of this study became i)to examine the expression range of TNIP1, ii)to determine the ligand requirements and amino acid motifs within NR and TNIP1 necessary for their interaction, and iii)to establish the consequences to PPAR activity from TNIP1 expression. Here we provide evidence for TNIP1 as an atypical NR corepressor. Despite association with PPARs in the presence of their respective ligands, TNIP1 partially represses their activity. TNIP1 does not interact with the PPAR heterodimer partner retinoid X receptor (RXR) even in the presence of that receptor’s 9-cis retinoic acid (RA) ligand. TNIP1’s wide tissue distribution suggests it may play an important regulatory role in multiple cell types. Corepressors such as TNIP1 and LCoR [15], using NR boxes, targeting the agonist-bound receptor’s AF-2 region, and yet reducing receptor-mediated gene transcription, may provide finer levels of control by lessening rather than completely silencing NR activity.
Materials and methods
Yeast and mammalian expression constructs, one- and two-hybrid assays
Selection stringency, ligand concentrations, vectors for yeast two-hybrid screen of a human keratinocyte cDNA library with human PPARα ligand-binding domain (LBD), mutagenesis methods, and receptor mutants were previously described in detail [16]. Briefly, the mutations are consensus amino acids (italicized) replaced with conservative substitutions (underlined) e.g., second TNIP1 NR box LKKLL to LKKAA and PPARα AF-2 LLQEIY to AAQEIY [16]. In this report, a quantitative β-galactosidase assay was used to assess interaction between pGAD10-coregulator proteins and pGBKT7- nuclear receptor DEF domains in the yeast two-hybrid system. pGBKT7-lamin C (Clontech, Mountain View, CA) was used as the negative control. Yeast strains Y187 and AH109 were lithium acetate transformed with pGAD10-coregulator (TNIP1) and pGBKT7-nuclear receptor (PPAR) DEF constructs, respectively, and selected on media lacking leucine for pGAD10 constructs or lacking tryptophan for pGBKT7 constructs. Colonies from each transformation were mated and selected on double drop-out media (-leucine / -tryptophan). Colonies expressing both constructs were picked from solid selective media, grown in triplicate in liquid selective media (with vehicle or ligand) overnight, and used as an inoculum for 4 h cultures with vehicle or ligand. β-galactosidase activity was determined from chlorophenol red-β-D-galactopyranoside (Roche Applied Science, Indianapolis, IN) conversion measured at OD578 by the following equation: (1000 × OD578) / (v × time × OD600) where v is a concentration factor for the pelleted yeast volume (0.5 ml in these assays), time is reaction duration (in minutes) and OD600 provides a normalization to yeast cell number. NR constructs had no autonomous activation of reporter genes [16]. Mammalian two-hybrid assays in COS-7 cells were performed with pM vector expressing receptor LBDs and pVP16 vector expressing 43a or full-length TNIP1.
The mammalian one-hybrid assay employed here utilized the TNIP1 cDNA cloned in-frame after the Gal4 DNA binding domain (DBD) cDNA in the SV40 promoter-driven pGal4DBD (pM vector, Clontech) and the reporter construct, pGal4UAS-tk-CAT [17], which has basal expression from the thymidine kinase (TK) minimal promoter regulated by the upstream activating sequence (UAS). One day after plating, COS-7 cells were transfected with a standard calcium phosphate protocol and 72 h later assayed for CAT expression via ELISA (Roche Applied Science) as described [18].
Mammalian cell culture and transfections
HaCaT keratinocytes [19] or COS-7 (ATCC, Manassas, VA) were cultured in a 3:1 mix of DMEM and F12 (Invitrogen, Carlsbad, CA) with 10% FBS (HyClone, Thermo Scientific, Rockford, IL) and 100U/ml penicillin and 100 μg/ml streptomycin. Receptor assays were from cells plated in triplicate wells 24 h before transfection in the above media but with 10% charcoal-stripped FBS. HaCaT keratinocytes were transfected via Effectene (Qiagen, Valencia, CA) according to the supplier’s protocols and COS-7 cells were transfected via the calcium phosphate coprecipitation method as detailed previously [20]. Mouse 3T3-L1 preadipocyte cells (ATCC) were maintained and differentiated as previously described [21]. Preadipocytes were cultured in DMEM with 10% calf serum (Hyclone). Differentiation induction media was DMEM containing 10% FBS, 1 μg/ml insulin, 1 μM dexamethasone, and 0.5 mM 3-isobutyl-1-methylxanthine (Sigma, St. Louis, MO). Neonatal human primary keratinocytes were cultured in serum-free KGM-Gold media with Bullet-kit supplements (cells and reagents from Lonza, Walkersville, MD) at final calcium concentrations indicated in the results. Ligand treatments, cell collection, and reporter assays have been described [16, 18]. Empty vector addition to standardize plasmid copy number and controls for NR reporter transfections were as described [18]. Reporter expression was normalized and tested for repeatability of results from independent transfections as described [22].
In vitro pull-down assays
35S-methionine-labeled PPARγ LBD from in vitro transcription / translation (Promega; Madison, WI) and glutathione sepharose column-purified GST or GST-TNIP1 expressed in BL21 RIPL (Stratagene; La Jolla, CA) from pGEX5 were incubated at 4°C, washed three times (100mM NaCl, 1mM EDTA, 0.02% Igepal, 20mM Tris-Cl, pH8.0) and eluted with SDS-PAGE sample buffer.
Multiple tissue cDNA PCR
PCR amplification of human cDNA (Clontech) was done with Titanium Taq (Clontech) and 5 μl template; cDNA template amounts from different tissues were adjusted by the manufacturer to normalize target mRNA abundance of different house-keeping genes. TNIP1 primers were forward 5’-CCA CCA CCT TCT CCC TCC TT-3’ and reverse 5’-GCA TCT TCA CCT TCT TCT CG-3’. Ubinuclein primers [23] were forward 5'-AGA AGC CAT GCA GTG ACA C-3’ and reverse 5’-AGC TCT GGG TAG AAG AAC-3’. Primers crossed intron-exon boundaries. For both the TNIP1 and ubinuclein amplification, conditions were as follows: initial denaturation, 2 min at 94°C, followed by 38 cycles at 94°C for 20 sec, 59°C for 50 sec, and 68°C for 1 min with a final extension of 68°C for 5 min.
TNIP1 plasmids
A human lung cDNA (ATCC) in pOTB7 was sequenced on both strands and determined to contain full-length, 636 amino acids, TNIP1. pVP16-TNIP1m1&m2 was used as template to generate carboxyl terminus truncations by PCR. The common forward primer for truncations was 5’ AAG GGG ATC CTC ATG GAA GGG AGA-3’ (Bam HI site, italics; TNIP1 start codon, underlined). The reverse primers were: TNIP1 1-206, TTG ATA AGC TTT TAG GAG GTG CGC TG; TNIP1 1-221, TGA GCA AGC TTT TAA GCC TCG TTC TC; TNIP1 1-289, TGA GCA AGC TTT TAC TTA CCT GCC GT; TNIP1 1-418, TTG ATA AGC TTT TAC CGC TGG ATC TC; TNIP1 1-543, AGC CGA AGC TTT TAA TGT TCT GGG TG (Hind III site, italics; inserted stop codon, underlined). A similar strategy was used for the pM-TNIP1 truncations but with the wild-type cDNA.
TNIP1 antibody production and immunodetection
Rabbit antiserum was raised against TNIP1 synthetic peptides (amino acids 1–14 and 552–562) selected from BLAST searches to obviate off-target matches and was antigen affinity-purified. Cell lysates were separated by SDS-PAGE and immunoblotted with routine protocols. For localization studies, HaCaT keratinocytes were cultured with standard media [20] as control, 3μM A23187 calcium ionophore for 24 h [24], or 5nM leptomycin B (Sigma) for 6 h. Primary keratinocyte culture was described above. Primary antibodies were anti-TNIP1, 1:2000; anti-involucrin (SY5, Santa Cruz Biotechnology; Santa Cruz, CA), 1:500; and anti-PPAR (as per supplier E8 detects PPARγ and, to a lesser extent, PPARα and PPARβ/δ, Santa Cruz), 1:250, and were detected with Alexafluor 488 or Texas Red labeled secondaries. Imaging was performed with a Leica SP2 confocal microscope (University of Connecticut Microscopy Facility).
Statistical analysis
Results representative of two-three experimental repeats are shown as means ± standard deviation (SD) of biological duplicates or triplicates for each point. Comparisons between multiple groups were performed with one-way ANOVA; post hoc tests and probability (p) values are indicated in figure legends. Graphpad (La Jolla, CA) Prism was used for graphing and statistical tests.
Results
Identification of TNIP1 as a PPAR interacting protein
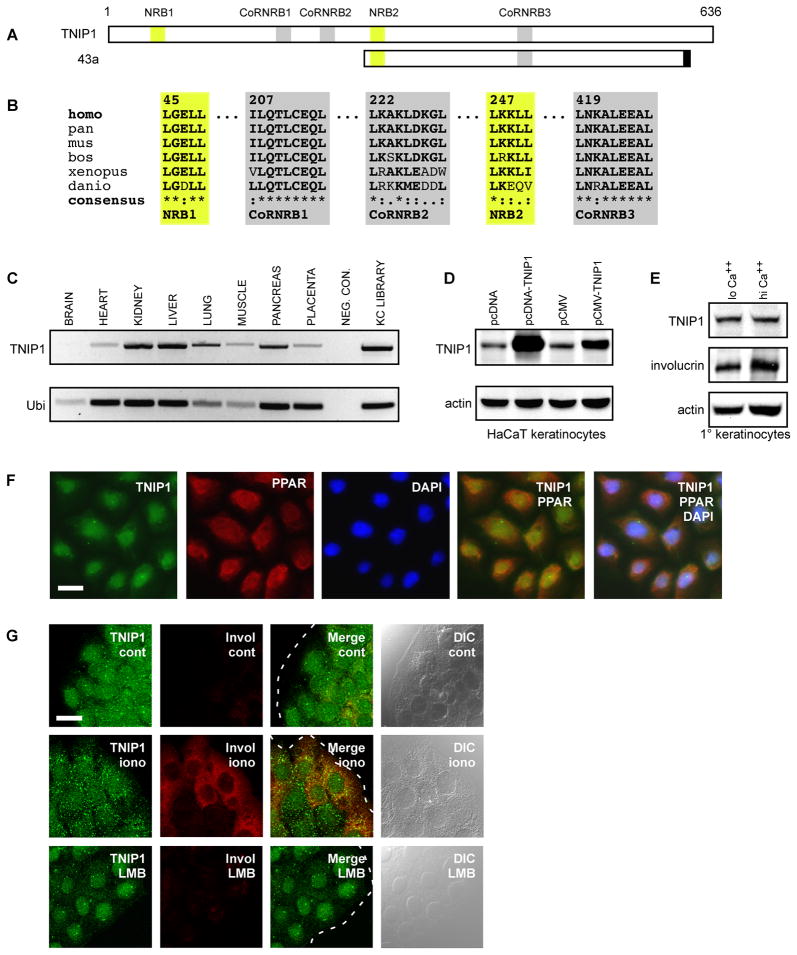
A partial clone of TNIP1, designated 43a, was isolated as the most frequently occurring interacting partner for the LBD of human PPARα in a yeast two-hybrid screen of a human keratinocyte cDNA library which also yielded known NR coactivators and several novel interacting proteins [16, 18]. The 43a cDNA encodes 377 amino acids (Fig. 1A). The second-most frequently occurring clone was also TNIP1-derived, overlapped the 43a cDNA (not shown); both contained consensus NR (LKKLL) and CoRNR (L-NKA-L-EEA-L) boxes seen in 43a. In addition to NR coregulator motifs in the 43a clone, the full-length 636 amino acid TNIP1 protein contains an additional NR box (LGELL) and two additional CoRNR motifs (I-LQT-L-CEQ-L and L-KAK-L-DKG-L) suggesting multiple potential NR interaction sites (Fig. 1A). These sequences are exactly or very highly matched across primate (human, chimpanzee), other mammalian (mouse, cattle), fish (zebra fish) and amphibian (frog) species (Fig. 1B) suggesting conserved function.
Fig. 1. TNIP1 NR coregulator motifs, tissue expression, and subcellular distribution in primary and HaCaT keratinocytes.
(A) Schematic of full-length TNIP1 protein, amino acids 1 – 636, with relative locations of candidate nuclear receptor boxes (NRB, yellow) and corepressor/nuclear receptor boxes (CoRNRB, grey) interaction regions. Clone 43a extended from amino acids 240 to 593 inclusive of the full-length TNIP1 protein and contained 17 nucleotides at its 3’ end (black box) coinciding with an exon border which may derive from alternative splicing [39].
(B) TNIP1 NR (LXXLL, yellow) and CoRNR box (L-XXX-I/L-XXX-L or hydrophobic substitution, grey) motifs are conserved across vertebrate species; amino acid numbering shown in top row follows the human sequence. GenBank records used were: NM_006058.3 Homo sapiens (human), XM_001167590 Pan troglodytes (chimpanzee), NM_021327 Mus musculus (mouse), NM_001024554 Bos taurus (cattle), NM_001079235 Xenopus tropicalis (frog), NM_001079952 Danio rerio (zebrafish). Consensus symbols are (*) amino acid identity; (:) amino acid conserved; (.) amino acid semi-conserved. Residues conserved across species compared to human sequence are bolded.
(C) TNIP1 detection from tissue-derived cDNA was compared against the ubiquitously expressed ubinuclein (Ubi) transcript. A weak TNIP1 band from brain, consistent with the weaker ubinuclein band, was observable in the gel. Neg. Con., negative control was water in place of cDNA. KC library, the cDNA library used in two-hybrid screen. Band sizes; TNIP1, 697bp; Ubi, 243bp.
(D) Top panel, detection of endogenous and recombinant TNIP1 protein. TNIP1 was detected in HaCaT keratinocyte RIPA lysates (25μg per lane) from cultures receiving empty pcDNA or pCMV expression vectors as control (first and third lanes, respectively). pcDNA or pCMV with human TNIP1 cDNA inserted (second and fourth lanes) increased band intensity at the ~85kD size. Bottom panel, actin signal as loading control from same blot.
(E) Detection of endogenous TNIP1 protein from human primary keratinocytes. The same blot was consecutively probed for TNIP1 (top panel), involucrin (middle panel), and actin (bottom panel) from RIPA lysates (20μg per lane) of keratinocyte cultures with final calcium levels of 0.1mM (lo Ca++) or 1.8mM (hi Ca++) for 48 h.
(F) TNIP1 colocalization with PPAR in human primary keratinocytes. Immunofluorescent microscopy of keratinocytes under low calcium conditions for TNIP1 (green), PPAR (red), and DAPI-stained nuclei (blue) with two-and three-channel merged images. Scale bar, 20μm. (G) Nuclear and cytoplasmic localization of endogenous TNIP1. Immunofluorescent confocal microscopy of HaCaT keratinocytes under control (cont), calcium ionophore (iono, 3μM A23187), or leptomycin B (LMB, 5nM) treatment conditions for detection of TNIP1 (green) or differentiation marker involucrin (red). Dashed white line (merged images) is colony edge. DIC, differential interference contrast. Scale bar, 20μm.
TNIP1 mRNA is broadly expressed but at variable levels (Fig. 1C). Affinity-purified TNIP1 antibody recognized endogenous protein at ~85kD in human HaCaT epidermal keratinocytes (Fig. 1D), malignant SCC13keratinocytes, and HeLa cervical carcinoma cells (not shown). Antibody recognition was confirmed by detection of recombinant TNIP1 expressed in HaCaT keratinocytes where the transient expression of its cDNA increased TNIP1 protein over endogenous level (Fig. 1D). Human primary epidermal keratinocytes were cultured under serum-free, 0.1mM or 1.8mM calcium for 48 h to retard or promote, respectively, their differentiated state [25]. As with the HaCaT keratinocytes line, an ~85kD band was identified. Its abundance did not change under the differentiation enhanced conditions (Fig. 1E). The relative degree of maturation difference in the cultures is reflected by the increase in involucrin under high calcium conditions (Fig. 1E, middle panel).
Primary keratinocytes in the 0.1mM calcium condition were also immuno-stained for TNIP1 and PPAR. Both proteins showed some cytoplasmic signal but also nuclear localization; the latter evidenced by overlap with the nuclear stain DAPI (Fig. 1F). TNIP1 also localized to both the nucleus and cytoplasm of HaCaT keratinocytes under standard (control) culture conditions (Fig. 1G, cont, first row) for these cells (serum-supplemented, 1.42mM calcium). Exposure to the calcium ionophore A23187 (Fig. 1G, iono, second row) increases production of differentiation-dependent proteins [24] such as involucrin and resulted in cytoplasmic TNIP1 appearing more granular. Treatment with leptomycin B (Fig. 1G, LMB, bottom row), a nuclear export inhibitor, resulted in almost exclusively nuclear localization suggesting TNIP1 is exported in a CRM-1 dependent manner.
Testing TNIP1 NR box and receptor AF-2 interaction
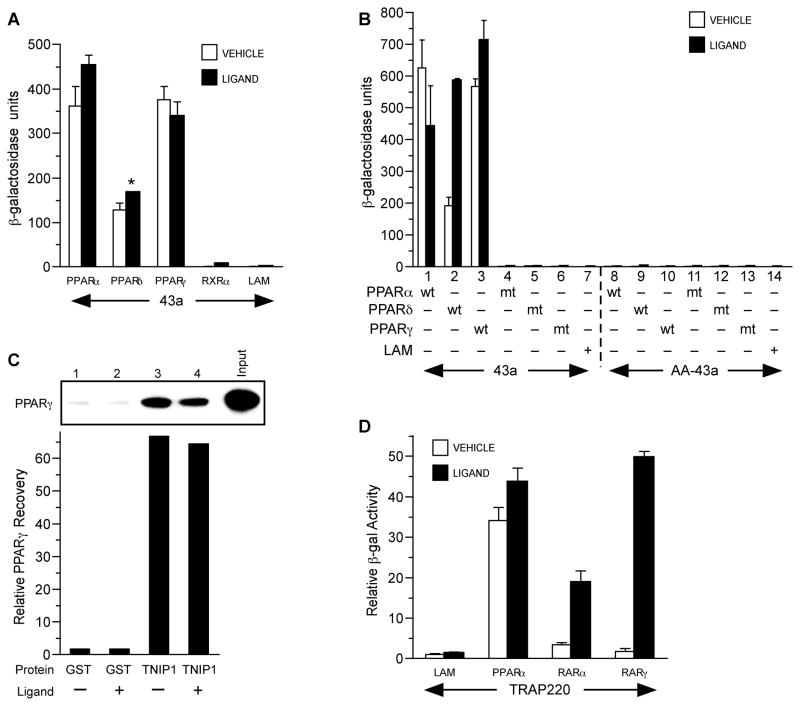
The two potential NR interaction motifs in the TNIP1 43a clone made it a useful experimental probe without the possibly confounding redundancy of six motifs in the full-length protein. Yeast two-hybrid assays were used to examine its interaction with PPARα, δ, and γ, retinoic acid receptor (RAR) α and γ (NR1B1 and NR1B3), and their heterodimer partner RXRα (NR2B1). These NRs are all expressed in epidermal keratinocytes which were the library source for the screen yielding 43a. The TNIP1 partial clone demonstrated robust association with PPARs; ligand had limited enhancement on interaction (Fig. 2A). Its interaction with RXRα was at the background levels of negative control lamin C. In contrast, there was equal but completely ligand-dependent interaction with RARα and γ which we have demonstrated in a separate report [18]. As with other NRs, PPAR interaction with coregulators is dependent on the AF-2 within helix 12; mutations in this receptor site abolished any interaction with the TNIP1 43a clone (Fig. 2B). Conservative hydrophobic substitution of the last two leucines in 43a’s consensus NR box (LKKLL to LKKAA; AA-43a in Fig. 2B) abolished any interaction with PPARs. These results demonstrated the importance of the 43a NR box and PPAR AF-2 in TNIP1-NR interaction, characteristics typically associated with NR coactivators.
Fig. 2. PPARs interact with clone 43a and full-length TNIP1.
(A) PPAR interaction with 43a in yeast two-hybrid assays. Y187 yeast transformed with pGAD10-43a was mated to AH109 yeast transformed with pGBKT7-PPARs, RXRα, or lamin C as negative control (LAM). Vehicle, 0.1% DMSO. Ligands: PPARα and LAM, 10μM WY14,643; PPARδ, 3μM carbaprostacyclin; PPARγ, 10μM troglitazone; RXRα, 1μM 9-cis-RA.
(B) PPAR AF-2 and 43a NR box are required for interaction. Pairs of bars are vehicle or ligand treatment of cultures coexpressing wild-type (wt) or mutant (mt) receptors with wild-type (43a) or double alanine NR box mutant 43a (AA-43a). Pairs 1-3, wild-type (wt) receptor and wild-type 43a. Pairs 4-6, AF-2 helix 12 mutant (mt) for each PPAR and wild-type 43a. Pair 7, lamin C (LAM) and wild-type 43a as negative control. Pairs 8-10, wild-type (wt) receptor and NR box mutant 43a (LKKLL → LKKAA indicated as AA-43a). Pairs 11–13, helix 12 mutant (mt) receptor and 43a NR box mutant AA-43a. Pair 14, LAM and NR box mutant AA-43a as negative control. Culture conditions, vehicle and ligand treatment as in A.
(C) PPARγ interacts in vitro with GST-TNIP1 full-length protein. Graph is from phosphor-imager arbitrary units of dried gel. GST, glutathionine S transferase. TNIP1, fusion protein of GST and full-length TNIP1. Graph bars correspond to gel lanes 1-4. Input is 20% of labeled PPARγ used for pull-down. Lane 1, PPARγ incubated with GST and vehicle; lane 2, PPARγ with GST and ligand; lane 3, PPARγ with GST-TNIP1 fusion and vehicle; lane 4, PPARγ with GST-TNIP1 fusion and ligand. Vehicle, DMSO, 0.1%. Ligand, 1μM troglitazone.
(D) Ligand effect on TRAP220 interaction with PPARα versus RARα and γ interaction is receptor-specific. Two-hybrid conditions and PPAR ligands as in A, RARα and RARγ ligand is 1μM retinoic acid. Reporter β-galactosidase activity is presented as fold relative to control protein lamin C (LAM) and vehicle (0.1% DMSO) set to 1 as per Zhu et al [33].
Further examining the TNIP1-PPAR interaction, we found purified full-length TNIP1 protein, as GST-TNIP1, captured radiolabeled PPARγ in a pull-down assay similarly in vehicle or troglitazone conditions (Fig. 2C). Ligand-insensitive in vitro association has been previously demonstrated for PPARs with other purified coregulator proteins [26]. Thus, ligand-independent interaction of TNIP1 with PPAR, but not RAR, is similar to TRAP220 interaction with these receptors (Fig. 2D). Without exogenous ligand, TRAP220-PPARα combination resulted in ~35-fold increase over the lamin control (Fig. 2D) with limited additional interaction in the presence of the PPARα ligand WY14,643. TRAP220 with RARα or RARγ in the absence of exogenous ligand showed only ~1.5-fold increase over the lamin control. Strikingly, values for TRAP220-RARα and TRAP220-RARγ increased 6- and ~28-fold, respectively, with RA over vehicle controls.
Interaction in mammalian cells: Dependence on NR Box, AF-2 and ligand
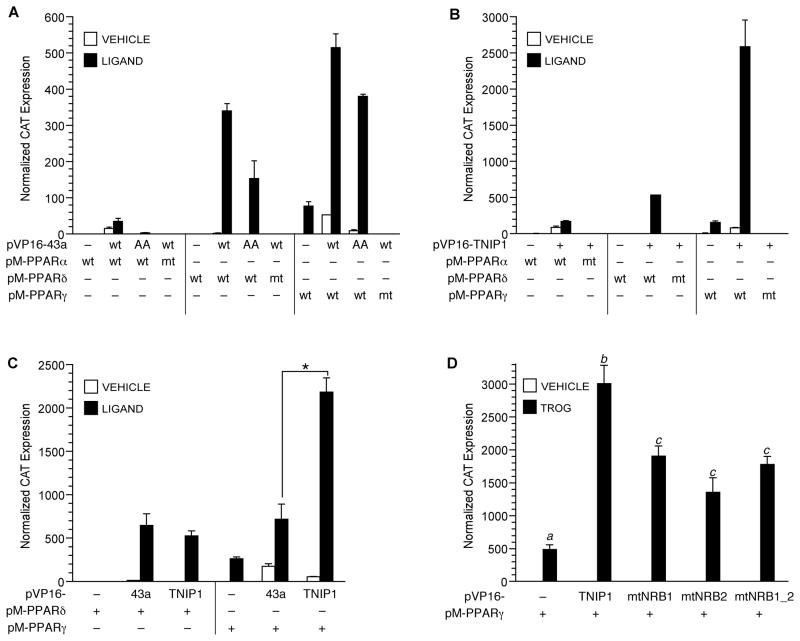
Given the yeast system and in vitro TNIP1-PPAR results, we sought to resolve the influence of ligand and confirm the reliance on NR box and AF-2 motifs in mammalian cells. Here the 43a interaction with all three PPARs (Fig. 3A) showed an almost complete dependence on exogenous ligand with a relative receptor preference of γ > δ ≫> α. Mutation (labeled mt in figure) of the AF-2 domain for all three PPARs resulted in complete loss of interaction. Conservative hydrophobic substitution of the last two leucines in 43a’s consensus NR box (LKKLL to LKKAA, labeled AA in figure) resulted in a range of loss of interaction with liganded PPARs. There was nearly complete loss with PPARα, ~50% loss with PPARδ and ~30% loss with PPARγ suggesting other ligand-dependent interactions in addition to this NR box. Extending and confirming the 43a results, full-length TNIP1 also showed strict ligand dependence for interaction with PPARs, absolute dependence on an intact receptor AF-2 domain and distinct preference for the gamma isoform among the PPARs (Fig. 3B). Comparison of clone 43a (one NR box) against full-length TNIP1 (two NR boxes) was done with the two better interacting PPARs, δ and γ. The full-length TNIP1 sequence had ligand-dependent interaction with δ similar to the 43a clone. However, compared to the 43a clone, full-length TNIP1 interaction with γ increased by ~3-fold (Fig. 3C). Interaction with PPARγ from either or both canonical NR boxes was addressed by TNIP1 mutagenesis (Fig. 3D). Mutation of the first (mtNRB1) or second (mtNRB2) NR box individually or together (mtNRB1_2) caused a similar 40–50% reduction in ligand-dependent interaction with PPARγ. Thus, in the context of mammalian cells, each TNIP1 NR box contributes to association with the PPARγ.
Fig. 3. TNIP1 interaction with PPARs in mammalian cells is mediated by ligand, receptor helix 12, and TNIP1 NR boxes.
(A) Two-hybrid assay of wild-type 43a (wt) or NR box mutant (LKKLL → LKKAA indicated as AA) with wild-type PPARs or helix 12 mutant PPARs (mt) in transfected COS-7 cells treated with vehicle, 0.1% DMSO, or respective ligand: PPARα, 10 μM WY14,643; PPARδ, 3μM carbaprostacyclin; PPARγ, 1μM troglitazone. CAT expression normalized to β-galactosidase activity from cotransfected plasmid. Error bars are SD from representative transfection.
(B) TNIP1 interaction with PPARs is helix 12- and ligand-dependent. COS-7 cells transfected for two-hybrid assay with PPAR α, δ, or γ wild-type (wt) or helix 12 mutant (mt). Ligands and transfections as in A.
(C) PPARγ shows greater interaction with full-length TNIP1 (two NR boxes) compared to the 43a partial TNIP1 clone (one NR box). Transfections and ligands as in A. Asterisk, significant difference between PPARγ interaction with full-length TNIP1 versus 43a (p<0.001, one way ANOVA, Tukey’s post-hoc test).
(D) TNIP1 NR boxes individually contribute to interaction with PPARγ. Transfections and two-hybrid assay conditions as in A. TROG, 1μM troglitazone; vehicle, 0.1% DMSO. Wild-type TNIP1 or NR box (NRB) mutants were expressed from the pVP16 vector. NR box mutants (mtNRB, see Fig. 1A schematic) were located in the first (mtNRB1), or second (mtNRB2), or both positions (mtNRB1_2). Within the NRB, specific amino acid changes were NRB1, LGELL → LGEAA; NRB2, LKKLL→LKKAA. a, b, c; values significantly different (p<0.001) from each other, one-way ANOVA, Tukey’s post-hoc test.
Mapping non-NR box TNIP1 sequence for interaction with liganded PPARγ
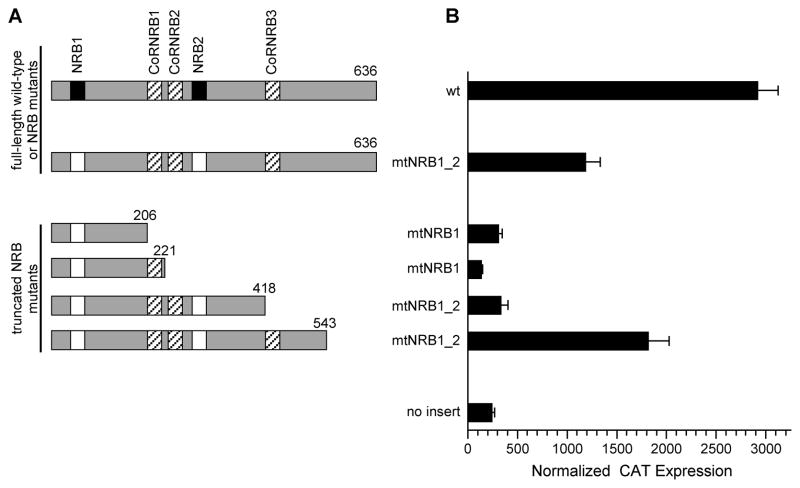
To identify TNIP1 regions without a consensus LXXLL sequence but contributing to interaction we started with the double NR box mutant and generated successive carboxyl-terminal truncations of it. This series (Fig. 4A) allowed for mammalian two-hybrid assays of PPARγ interaction with TNIP1 full-length wild-type, full-length double NR box mutant (mtNRB1_2), and four truncations of the mtNRB1_2 backbone. Approximately 50% of TNIP1 interaction with liganded PPARγ is lost upon mutation of TNIP1’s two consensus NR boxes (mtNRB1_2, Fig. 4B). Increasing lengths of mtNRB1_2 showed neither its amino third, aa1 – 206, or further on through its middle third, aa1 – 418, provided for interaction with liganded PPARγ. However, extension of the double NR box mutant to aa543 provided interaction with PPARγ similar to the full-length (aa636), double NR box mutant.
Fig. 4. TNIP1 interaction with liganded PPARγ is dependent on canonical NR boxes and a region localizing to TNIP1 carboxyl region.
(A) Schematic of TNIP1 full-length, wild-type protein (wt, 1-636 amino acids) shows two NR boxes (NRB, solid) and three potential CoRNR boxes (CoRNRB, striped). Open boxes in full-length TNIP1 double NR box mutant, (mtNRB1_2 FL) are site-directed NR box mutations (LGELL→LGEAA and LKKLL→LKKAA). All truncations start at amino acid position 1 and are labeled as to NR box mutations and length of TNIP1 amino acids present. Constructs are horizontally aligned with activity in (B).
(B) COS-7 two-hybrid assay transfections with pM-PPARγ and pVP16-TNIP1 constructs from (A). Vehicle cultures were performed for pVP16-empty and wild-type TNIP1 only (no detectable interaction, not shown). The no insert bar is from cells receiving pM-PPARγ and pVP16-empty. Ligand and transfections as in Fig. 3A.
TNIP1 suppresses PPAR transactivation
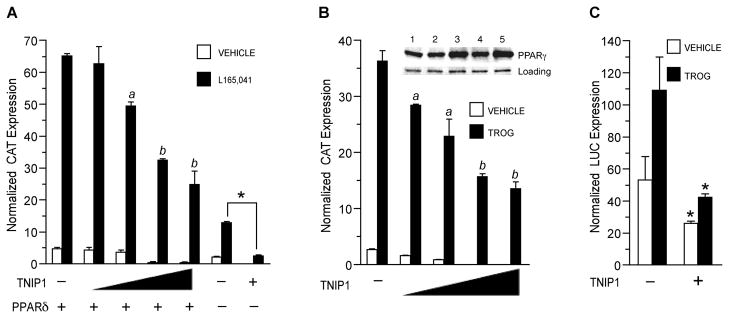
TNIP1 interaction with PPARs is ligand-, AF-2-, and NR box-dependent in mammalian cells, properties typically associated with NR coactivators. The TNIP1 partial clone 43a interacted better with PPARδ and γ than α (Fig. 3) but when coexpressed with them it significantly reduced their activation of a peroxisome proliferator response element (PPRE) in the presence of their respective ligands (not shown). These results were unexpected for a NR box-containing coregulator that interacts with liganded NRs but were confirmed and extended when we found that full-length TNIP1 is also able to reduce ligand-activated PPARδ and γ (Fig. 5A and B). Expression of TNIP1 did not decrease PPARγ protein levels (Fig. 5B, inset) suggesting that reduction of receptor activity did not stem from its degradation. Endogenous PPARγ activity was also decreased by TNIP1 expression in 3T3-L1 cells that had been induced to differentiate to adipocytes (Fig. 5C). Activity levels of group 3 NRs, androgen receptor (NR3C4) and estrogen receptors α and β (NR3A1 and 2), were not changed by coexpression of TNIP1 in either the presence or absence of their respective ligands (not shown).
Fig. 5. PPAR activity is reduced by expression of full-length TNIP1.
(A) TNIP1 represses PPAR™ transactivation. COS-7 cells transfected with PPRE reporter p3xZ-CAT, pSG5-PPAR™, and pCMV-HA-TNIP1 at 0.1, 0.5, 1.0, or 1.5μg (indicated by triangle). Total plasmid copy number was kept constant by addition of empty pCMV-HA. Transfection with 1.5μg pCMV-HA-TNIP1in the absence of pSG5-PPARδ (−) is represented in the last 2 bar sets. PPARδ ligand; 1μM L165,041. Vehicle, 0.1% DMSO. For the ligand-treated set, a, b, are different from each other and cultures receiving pCMV-HA empty vector instead of pCMV-HA-TNIP1 (p < 0.05, one-way ANOVA, Tukey’s test). Asterisk, significant difference between ligand-treated cultures from endogenous receptor activity (no PPARδ transfected) without and with TNIP1 expression (t-test, p < 0.001). The PPAR heterodimer partner RXRα was included in the transfections as described [18].
(B) TNIP1 represses PPARγ transactivation. COS-7 cells transfected with PPRE reporter and TNIP1 constructs as in (A). TROG, 5μM troglitazone. Vehicle, 0.1% DMSO. Statistical comparison (one-way ANOVA, Tukey’s test; p < 0.05) of ligand-treated cultures: a, b are different from each other and cultures receiving pCMV-HA empty vector instead of pCMV-HA-TNIP1. Inset, western detection of endogenous PPARγ and loading control from lysates of TNIP1-transfected, ligand-treated cells used in CAT assay. Lane 1: 1.5μg pCMV-HA-empty vector. Lanes 2 – 5: 0.1, 0.5, 1.0, or 1.5μg pCMV-HA-TNIP1.
(C) TNIP1 represses endogenous PPARγ transactivation in differentiating mouse 3T3-L1 cells. 3T3-L1 cells were transfected overnight with PPRE reporter and pCMV-HA-empty vector (−) or pCMV-HA-TNIP1 (+) and then cultured in differentiation induction media for 48 h with 5μM troglitazone (TROG) or vehicle, 0.1% DMSO. Asterisk, (t-test, p < 0.02) reporter activity in cultures receiving TNIP1 were reduced from those receiving empty vector.
TNIP1 transcriptional repressive effects
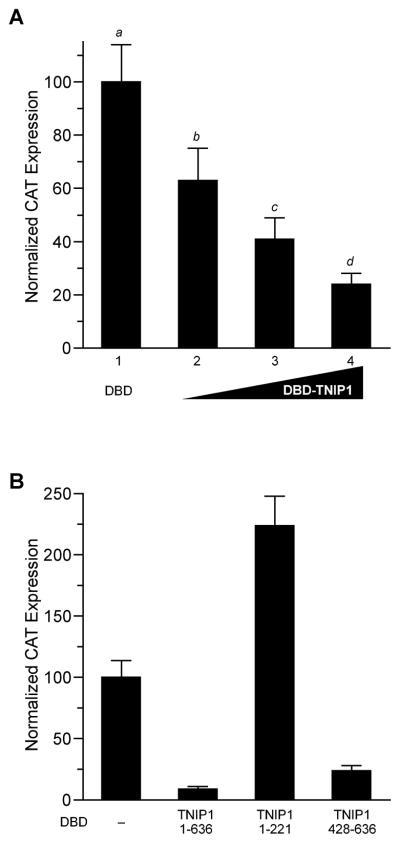
NR corepressors can exert their effect through scaffolding HDACs on chromatin but we found no association of TNIP1 with HDACs 1–3, 5 or 6, or with other class 1 and 2 HDACs [18] suggesting TNIP1 recruits some other repressive protein or has an intrinsic repressive domain. We tested the delivery of TNIP1 as a Gal4 DNA binding domain (DBD) chimeric protein (DBD-TNIP1) to a constitutively active, UAS-regulated promoter [17]. Increasing amounts of DBD-TNIP1 decreased reporter activity in a dose-dependent fashion; at the highest amount of DBD-TNIP1 used there was ~75% reduction from the control level (Fig. 6A). The repressive effect was localized by comparing UAS reporter activity with coexpressed TNIP1 N-terminus, full-length, or C-terminus fused to Gal4DBD (Fig. 6B). As in Fig. 6A, we saw a repressive effect from full-length TNIP1. Interestingly, when the fusion construct contained the cDNA region encoding the amino third of TNIP1 there was a distinct increase in reporter activity. In contrast, the repressive effect could be localized to the carboxyl terminus (amino acids 428-636) that decreased reporter activity to <10% of the DBD control.
Fig. 6. TNIP1 transcriptional effects in one-hybrid assays.
(A) Full-length TNIP1 exhibits intrinsic transcriptional repression. COS-7 cells transfected with a DBD-regulated reporter, pGal4UAS-tk-CAT, and pGal4DBD with no insert (DBD, first bar) or with increasing amounts, 0.75, 1.50, 3.0μg of pGal4DBD-TNIP1 containing the full-length TNIP1 cDNA in-frame after the DBD coding region (bars 2 – 4, respectively) per well of 6-well plate. pGal4DBD with no insert was included as needed to maintain equal copy number of total DBD constructs for each transfection. CAT expression from pGal4DBD with no insert set to 100%. All values significantly different (a – d) from each other at p < 0.05 or less; 0.75 μg DBD-TNIP1 vs DBD, p < 0.01; higher DBD-TNIP1 amounts against DBD control, p < 0.001 (one-way ANOVA, Newman-Keuls post-hoc test).
(B) Expression in COS-7 cells of the constitutively active pGal4UAS-tk-CAT reporter in response to cotransfection with equal copy number of the pGal4DBD vector (DBD) with no insert (–), TNIP1 full-length (amino acids 1 – 636), or TNIP1 truncations as indicated by amino acid positions in graph.
Discussion
TNIP1 is a NR box-containing, receptor AF-2-targeting corepressor of agonist-bound PPARs. TNIP1 homologues exist across mammalian, amphibian and fish species with sequence conservation suggesting critical residues throughout the protein including its NR box and CoRNR box motifs. Additionally, a region with no canonical LXXLL motifs contributes to ligand-dependent association with PPAR. The preferential interaction of TNIP1 with PPARδ and γ, versus PPARα, and its undetectable interaction with RXRα, indicates a degree of discrimination involved in TNIP1-NR interaction. It does not interact with androgen, estrogen (α and β) or progesterone steroid receptors (Encarnacao and Aneskievich, unpublished) and appears to have preferential interaction among isotypes of PPARs (this report) and RARs [18]. TNIP1 protein levels and subcellular localization (Gurevich et al., submitted) within most tissues suggest a potentially broad role in PPAR and RAR signaling.
In addition to its corepression of agonist-bound PPARs and RARs, full-length TNIP1 has an overall repressive effect on transcription. However, we were able demonstrate an activating effect within the amino third of the protein, which does have a relatively acidic nature (pKi 4.9) compared to the entire protein (pKi 6.3) or the carboxyl third (pki 7.2). This modular characteristic of TNIP1 recalls the dualistic nature of another NR coregulator, nuclear receptor-interacting factor 3 (NRIF3). Despite its function as a coactivator of TR and RXR, NRIF3 is able to repress transcription when expressed as a Gal4-DBD fusion protein. The NRIF3 autonomous repression domain maps to its N-terminus, distinct from the C-terminal activation domain responsible for its coactivator function [27]. Together, the results for NRIF3 and TNIP1 suggest their association with NRs is not about fully activating (NRIF3) or fully repressing (TNIP1) receptors but modulation of their activity. This premise fits with the significant reduction, but not absolute loss, of PPAR (this work) and RAR [18] activity we have seen upon expression of TNIP1.
In the context of NR coregulators, TNIP1 is similar to ligand-dependent corepressors including LCoR, REA and RIP140. These corepressors bind NRs in a ligand-dependent fashion and yet repress transcription [18, 28]. Members in this atypical group show no overall homology to each other but may be a functionally-related group of proteins where NR activity in the presence of ligand can be attenuated but not completely repressed and thus add to potential mechanisms for combinatorial control of agonist-bound NR gene regulation. Extending our previous findings for TNIP1-RAR interaction [18], a central region of the TNIP1 sequence with no canonical or variant LXXLL motif could contribute to ligand-dependent association with PPARs. Non-NR box regions contributing to interaction have been identified for the corepressor hairless (aa750 – 864) in its interaction with VDR [29, 30]. Non-interaction of TNIP1 with HDAC [18] distinguishes it from LCoR, which directly associates [31] with HDACs 3 and 6 (classes I and II, respectively).
The dependence on exogenous ligand for PPAR-TNIP1 interaction in mammalian cells but not yeast is corroborated by results from other studies. Similar to our findings with PPARα and TRAP220 (Fig. 2D), PPARγ with either TRAP220 or SRC-1 has substantial interaction in yeast in the absence of exogenous ligand with no or very limited increase upon addition of ligand [32, 33]. Nevertheless, the yeast system accurately predicted involvement of TNIP1 NR boxes and receptor AF-2 domains for TNIP1-NR interactions. The requirement of exogenous ligand for TNIP1-RAR [18] but not TNIP1-PPAR (Fig. 2A, B and C) association in the yeast and in vitro systems is consistent with prior results for some NR box-containing coregulators such as TRAP220 [33, 34].
TNIP1 interacts with the HIV proteins Nef [10] and matrix [11] and in the latter case was posited as a cellular protein responsible for shuttling HIV matrix protein into and out of the nucleus. TNIP1 is predominantly cytoplasmic in HeLa cells [11] with nuclear staining following LMB treatment. In contrast to HeLa cells, we found that HaCaT keratinocytes under standard culture conditions displayed nuclear staining of endogenous TNIP1 that could then be further increased with LMB exposure. TNIP1 cytoplasmic-nuclear distribution may be more cell-dependent and more dynamic than previously expected with control of its subcellular localization contributing to its function.
TNIP1 acting as a transcriptional repressor and specifically a corepressor of PPAR and RAR [18] in the presence of their respective ligands defines a role for this protein beyond its characterization in HIV pathogenesis. A cytoplasmic role for TNIP1 has been identified based on its interaction with the zinc finger protein A20 [12]. A20 and TNIP1 (referred to as ABIN in that system) do not directly associate with NF-κB but they do decrease NF-κB-mediated transcription. Both A20 and ABIN appear to act upstream of NF-κB nuclear function, possibly at the cell membrane with the TNFα receptor [35] or in the cytoplasm with the NEMO regulatory [36] subunit of IκB kinase. Most recently, mouse cells deficient for TNIP1 were reported as hypersensitive to apoptosis following exposure to tumor necrosis factor α [37].
Cytoplasmic-nuclear translocation and regulatory function within those different compartments may position TNIP1 more to being a signal integrator, rather than regulator of any one linear transduction route. Nuclear to cytoplasmic relocation, subsequent to post-translational processing, has been shown for the ligand-dependent corepressor RIP140 [28 for review]. Overall, the functional contribution of atypical corepressors such as TNIP1, LCoR and RIP140 [15, 28] may provide a transcriptional regulatory system more capable of fine adjustments rather than maximal off/on responses by typical corepressors and coactivators alone. Additionally, as said of NY-FC, the recently isolated corepressor of agonist-bound mineralocortioid receptors [38], the sequential recruitment of ligand-dependent coactivators and corepressors of agonist- bound NRs may act as emergency brakes on excessive ligand instigated gene expression.
Highlights.
We report TNIP1is a corepressor of agonist-bound PPARs
TNIP1 was previously known to associate with HIV proteins and repress NF-κB
TNIP1 binds PPARs in a manner typical of coactivators but represses their activity
TNIP1 has separable activation and repression domains
Corepressors such as TNIP1 may contribute to combinatorial transcription regulation
Acknowledgments
Funding
This work was supported by National Institutes of Health (NIAMS grant numbers AR048860, BJA; AR048483, AMF). Pre-doctoral fellowships from Boehringer-Ingelheim Pharmaceuticals (AMF and IG) and the American Foundation for Pharmaceutical Education (IG) provided partial stipend support. The University of Connecticut Summer Undergraduate Research Fund (TRD), the Center for Regenerative Biology Excellence in Graduate Research Award (VPR), and the Edward A. Khairallah Graduate Fellowship (IG and VPR) provided partial summer support.
We thank N. Fusenig for HaCaT keratinocytes, S. Gao for pGal4UAS-tk-CAT, M. Barber at the UConn Microscopy Facility for imaging assistance, and Aneskievich laboratory member P. Encarnacao for helpful discussions.
Abbreviations
- aa
amino acid
- ABIN
A20-binding inhibitor of NF-κB
- AF-2
activating function 2
- CoRNRB
corepressor NR box
- DBD
DNA binding domain
- NR
nuclear receptor
- PPAR
peroxisome proliferator activated receptor
- TNIP1
TNFα-induced protein 3-interacting protein 1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lonard DM, Lanz RB, O'Malley BW. Endocr Rev. 2007;28:575–587. doi: 10.1210/er.2007-0012. [DOI] [PubMed] [Google Scholar]
- 2.Hu X, Li Y, Lazar MA. Mol Cell Biol. 2001;21:1747–1758. doi: 10.1128/MCB.21.5.1747-1758.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gurevich I, Flores AM, Aneskievich BJ. Toxicol Appl Pharmacol. 2007;223:288–298. doi: 10.1016/j.taap.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim DJ, Bility MT, Billin AN, Willson TM, Gonzalez FJ, Peters JM. Cell Death Differ. 2006;13:53–60. doi: 10.1038/sj.cdd.4401713. [DOI] [PubMed] [Google Scholar]
- 5.Mao-Qiang M, Fowler AJ, Schmuth M, Lau P, Chang S, Brown BE, Moser AH, Michalik L, Desvergne B, Wahli W, Li M, Metzger D, Chambon PH, Elias PM, Feingold KR. J Invest Dermatol. 2004;123:305–312. doi: 10.1111/j.0022-202X.2004.23235.x. [DOI] [PubMed] [Google Scholar]
- 6.Michalik L, Wahli W. Biochim Biophys Acta. 2007;1771:991–998. doi: 10.1016/j.bbalip.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 7.Tan NS, Icre G, Montagner A, Bordier-ten-Heggeler B, Wahli W, Michalik L. Mol Cell Biol. 2007;27:7161–7175. doi: 10.1128/MCB.00436-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Westergaard M, Henningsen J, Svendsen ML, Johansen C, Jensen UB, Schroder HD, Kratchmarova I, Berge RK, Iversen L, Bolund L, Kragballe K, Kristiansen K. J Invest Dermatol. 2001;116:702–712. doi: 10.1046/j.1523-1747.2001.01329.x. [DOI] [PubMed] [Google Scholar]
- 9.Viswakarma N, Jia Y, Bai L, Vluggens A, Borensztajn J, Xu J, Reddy JK. PPAR Res. 2010;2010 doi: 10.1155/2010/250126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fukushi M, Dixon J, Kimura T, Tsurutani N, Dixon MJ, Yamamoto N. FEBS Lett. 1999;442:83–88. doi: 10.1016/s0014-5793(98)01631-7. [DOI] [PubMed] [Google Scholar]
- 11.Gupta K, Ott D, Hope TJ, Siliciano RF, Boeke JD. J Virol. 2000;74:11811–11824. doi: 10.1128/jvi.74.24.11811-11824.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heyninck K, De Valck D, Vanden Berghe W, Van Criekinge W, Contreras R, Fiers W, Haegeman G, Beyaert R. J Cell Biol. 1999;145:1471–1482. doi: 10.1083/jcb.145.7.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hosgood HD, 3rd, Menashe I, Shen M, Yeager M, Yuenger J, Rajaraman P, He X, Chatterjee N, Caporaso NE, Zhu Y, Chanock SJ, Zheng T, Lan Q. Carcinogenesis. 2008;29:1938–1943. doi: 10.1093/carcin/bgn178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nair RP, Duffin KC, Helms C, Ding J, Stuart PE, Goldgar D, Gudjonsson JE, Li Y, Tejasvi T, Feng BJ, Ruether A, Schreiber S, Weichenthal M, Gladman D, Rahman P, Schrodi SJ, Prahalad S, Guthery SL, Fischer J, Liao W, Kwok PY, Menter A, Lathrop GM, Wise CA, Begovich AB, Voorhees JJ, Elder JT, Krueger GG, Bowcock AM, Abecasis GR. Nat Genet. 2009;41:199–204. doi: 10.1038/ng.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernandes I, Bastien Y, Wai T, Nygard K, Lin R, Cormier O, Lee HS, Eng F, Bertos NR, Pelletier N, Mader S, Han VK, Yang XJ, White JH. Mol Cell. 2003;11:139–150. doi: 10.1016/s1097-2765(03)00014-5. [DOI] [PubMed] [Google Scholar]
- 16.Flores AM, Li L, Aneskievich BJ. J Invest Dermatol. 2004;123:1092–1101. doi: 10.1111/j.0022-202X.2004.23424.x. [DOI] [PubMed] [Google Scholar]
- 17.Pan HY, Zhang YJ, Wang XP, Deng JH, Zhou FC, Gao SJ. J Virol. 2003;77:9758–9768. doi: 10.1128/JVI.77.18.9758-9768.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gurevich I, Aneskievich BJ. Biochem Biophys Res Commun. 2009;389:409–414. doi: 10.1016/j.bbrc.2009.08.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Breitkreutz D, Stark HJ, Plein P, Baur M, Fusenig NE. Differentiation. 1993;54:201–217. doi: 10.1111/j.1432-0436.1993.tb01602.x. [DOI] [PubMed] [Google Scholar]
- 20.Flores AM, Aneskievich BJ. Methods Mol Biol. 2005;289:263–272. doi: 10.1385/1-59259-830-7:263. [DOI] [PubMed] [Google Scholar]
- 21.Zhang JW, Tang QQ, Vinson C, Lane MD. Proc Natl Acad Sci USA. 2004;101:43–47. doi: 10.1073/pnas.0307229101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eeckhoute J, Carroll JS, Geistlinger TR, Torres-Arzayus MI, Brown M. Genes Dev. 2006;20:2513–2526. doi: 10.1101/gad.1446006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kazerounian S, Aho S. J Biol Chem. 2003;278:36707–36717. doi: 10.1074/jbc.M303896200. [DOI] [PubMed] [Google Scholar]
- 24.Zhang T, Woods TL, Elder JT. J Invest Dermatol. 2002;119:1196–1201. doi: 10.1046/j.1523-1747.2002.19520.x. [DOI] [PubMed] [Google Scholar]
- 25.Pillai S, Bikle DD, Mancianti ML, Cline P, Hincenbergs M. J Cell Physiol. 1990;143:294–303. doi: 10.1002/jcp.1041430213. [DOI] [PubMed] [Google Scholar]
- 26.Krogsdam AM, Nielsen CA, Neve S, Holst D, Helledie T, Thomsen B, Bendixen C, Mandrup S, Kristiansen K. Biochem J. 2002;363:157–165. doi: 10.1042/0264-6021:3630157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li D, Wang F, Samuels HH. Mol Cell Biol. 2001;21:8371–8384. doi: 10.1128/MCB.21.24.8371-8384.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fritah A, Christian M, Parker MG. Am J Physiol Endocrinol Metab. 2010;299:E335–E340. doi: 10.1152/ajpendo.00243.2010. [DOI] [PubMed] [Google Scholar]
- 29.Hsieh JC, Sisk JM, Jurutka PW, Haussler CA, Slater SA, Haussler MR, Thompson CC. J Biol Chem. 2003;278:38665–38674. doi: 10.1074/jbc.M304886200. [DOI] [PubMed] [Google Scholar]
- 30.Thompson CC. Nucl Recept Signal. 2009;7:e010. doi: 10.1621/nrs.07010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palijan A, Fernandes I, Bastien Y, Tang L, Verway M, Kourelis M, Tavera-Mendoza LE, Li Z, Bourdeau V, Mader S, Yang XJ, White JH. J Biol Chem. 2009;284:30264–30274. doi: 10.1074/jbc.M109.045526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu Y, Qi C, Calandra C, Rao MS, Reddy JK. Gene Expr. 1996;6:185–195. [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu Y, Qi C, Jain S, Rao MS, Reddy JK. J Biol Chem. 1997;272:25500–25506. doi: 10.1074/jbc.272.41.25500. [DOI] [PubMed] [Google Scholar]
- 34.Zhu Y, Kan L, Qi C, Kanwar YS, Yeldandi AV, Rao MS, Reddy JK. J Biol Chem. 2000;275:13510–13516. doi: 10.1074/jbc.275.18.13510. [DOI] [PubMed] [Google Scholar]
- 35.Heyninck K, Kreike MM, Beyaert R. FEBS Lett. 2003;536:135–140. doi: 10.1016/s0014-5793(03)00041-3. [DOI] [PubMed] [Google Scholar]
- 36.Mauro C, Pacifico F, Lavorgna A, Mellone S, Iannetti A, Acquaviva R, Formisano S, Vito P, Leonardi A. J Biol Chem. 2006;281:18482–18488. doi: 10.1074/jbc.M601502200. [DOI] [PubMed] [Google Scholar]
- 37.Oshima S, Turer EE, Callahan JA, Chai S, Advincula R, Barrera J, Shifrin N, Lee B, Yen B, Woo T, Malynn BA, Ma A. Nature. 2009;457:906–909. doi: 10.1038/nature07575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murai-Takeda A, Shibata H, Kurihara I, Kobayashi S, Yokota K, Suda N, Mitsuishi Y, Jo R, Kitagawa H, Kato S, Saruta T, Itoh H. J Biol Chem. 2010;285:8084–8093. doi: 10.1074/jbc.M109.053371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Favre M, Butticaz C, Stevenson B, Jongeneel CV, Telenti A. J Acquir Immune Defic Syndr. 2003;34:127–133. doi: 10.1097/00126334-200310010-00002. [DOI] [PubMed] [Google Scholar]