Abstract
Background. Streptococcus pneumoniae is a significant pathogen capable of expressing protective and antigenically diverse capsules. To better understand the molecular basis of capsular antigenic diversity, we investigated the hypothetical serological role of wcjE, which encodes a capsule O-acetyltransferase, in the vaccine-targeted serotype 9V and related serotype 9A.
Methods. We inactivated wcjE by recombination in a serotype 9V strain and determined wcjE sequences of 11 serotype 9A clinical isolates. We determined the antigenic phenotypes of these pneumococcal strains with serogroup 9–specific antibodies and flow cytometry.
Results. Inactivation of wcjE in a serotype 9V strain resulted in expression of the 9A phenotype. Each serotype 9A clinical isolate contained a distinct mutation to wcjE. Flow cytometry showed that some 9A isolates (herein named 9Aα) expressed trace amounts of 9V-specific epitopes whereas others (named 9Aβ) did not express any. Recombination with 9Aα wcjE alleles into a 9Aβ strain conferred partial expression of 9V-specific epitopes.
Conclusions. Each serotype 9A strain independently arose from a serotype 9V strain. Furthermore, clinical isolates identified as 9A can contain mutations to wcjE that are either partially functional or completely nonfunctional, demonstrating a previously unidentified antigenic heterogeneity of serotype 9A isolates.
Streptococcus pneumoniae, the pneumococcus, is both a common human nasopharyngeal commensal and a highly significant opportunistic pathogen. Most pathogenic pneumococci express a protective polysaccharide capsule, which shields pneumococci from innate immunity [1] and greatly increases its transmissibility and virulence. However, anti-polysaccharide capsule antibodies can mediate type-specific bacterial clearance and provide protection against colonization and infection. This selective pressure has likely contributed to the evolution of at least 93 antigenically distinct capsule types, which are commonly referred to as serotypes [2–4]. Each serotype has a unique chemical structure, and for nearly all of the 93 serotypes, capsular synthesis is mediated by the genes within the capsule synthesis (cps) locus. [5].
Studies on serogroup 15 and the newly discovered serotype 11E demonstrate that bacteria expressing certain capsule types can arise independently and recurrently. Antigenic distinctions between serotypes 15B and 15C is attributed to phase-variable O-acetylation of capsule polysaccharide mediated by wciZ, which encodes a putative transmembane O-acetyltransferase of the PF01757 protein family and is truncated in 15C cps loci as a result of a reversible slipped-strand mutation [6, 7]. The rate of in vitro conversion between serotypes 15B and 15C is proposed to be as high as 1 in 300 [8]. Similarly, we recently determined the genetic distinction between serotype 11A and serotype 11E to be the integrity of wcjE, which is located at the 3′ end of the cps locus and also encodes a PF01757 O-acetyltransferase [3]. Unlike serotype 15C isolates that contain reversible mutations in wciZ, however, each serotype 11E clinical isolate so far examined harbors a unique irreversible null mutation to wcjE, indicating that the serotype has originated independently in separate hosts.
To determine whether this mechanism of recurrent serotype emergence applies to other serotypes, we genetically and serologically characterized clinical isolates expressing the vaccine-target serotype 9V and the closely related serotype 9A. Historically, serotypes 9V and 9N have accounted for the majority of pneumococcal serogroup 9 infections, and 9A has accounted for only a small fraction. The 3′ end of the serotype 9V cps locus contains a wcjE gene that is highly homologous (92.2% sequence identity over 1,029 base-pair overlap) to the 11A wcjE described in our previous study [3, 9]. The only dissimilarity between the published cps loci of serotypes 9V and 9A (GenBank accession no. AF402095 and CR931645) is a single slipped-strand nucleotide deletion in 9A wcjE [10]. Because the genetic analysis was limited to single serotype 9V and 9A isolates, it is unknown whether this particular wcjE mutant allele is typically found within serotype 9A strains.
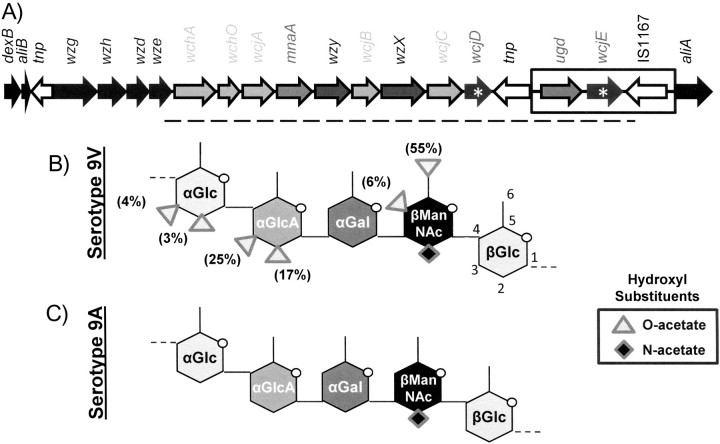
Moreover, wcjE inactivation has not been confirmed as the molecular basis for serological differences between serotypes 9V and 9A. Whether wcjE is active in 9V capsule synthesis remains unclear because none of the multiple O-acetate substitutions on serotype 9V polysaccharide repeat units [11] (Figure 1) are identical to the wcjE-associated structure in serotype 11A, that is, O-acetate substitution of the 6 carbon of 11A β-galactose–a modification absent on 11E polysaccharide [12]. And even though the published serotype 9A polysaccharide structure has no O-acetate substitutions (Figure 1C) [11], a recent study suggested the presence of some O-acetyl groups on 9A polysaccharide [10], consistent with the fact that both 9V and 9A cps loci contain another functionally intact O-acetyltransferase, wcjD (Figure 1A).
Figure 1.
Diagram depicting the serotype 9V capsule synthesis (cps) locus and the published structures of serotype 9V and 9A capsule polysaccharide of Streptococcus pneumoniae. A, Depiction of the 9V cps locus according to Van Selm et al in 2002 [9] . The 2 putative cps O-acetyltransferases are marked with white asterisks. The horizontal dashed line approximately denotes the region sequenced in AC128 (see text). The boxed area of the cps locus is depicted in Figure 2A and Figure 3.B, C, Structural models of the capsule polysaccharide repeating units of serotypes 9V and 9A, respectively, according to Rutherford et al in 1991 [11]. The carbons are numbered on the βGlc of the 9V structure as a reference. The linkages across repeating units are denoted with dotted lines. Values in parentheses describe the rate of acetylation (per mole of polysaccharide subunit) at each site. Abbreviations: Gal, galactose; Glc, glucose; GlcA, glucuronic acid; ManNAc, N-acetylmannosamine.
Here, we show that serotype 9A is derived from serotype 9V through inactivation of wcjE. Furthermore, we show that each 9A isolate contains a unique mutation to wcjE. With the use of a novel flow-cytometric serotyping assay, we show that serotype 9A strains display diverse serological profiles that are dependent on the type of mutation to wcjE. We concluded that multiple mutations present in clinical 9A wcjE alleles can mediate variable expression levels of epitope previously associated only with serotype 9V.
METHODS
Bacterial Strains
The strains used in this study are listed in Table 1. Strains SSISP 9V/4, SSISP 9A/1, SSISP 9N/4, and SSISP 9L/4, which express serotype 9V, 9A, 9N, and 9L, respectively, were obtained from the Statens Seruminstitut (SSI). Strains AC100 and AC128, known to express serotypes 9V and 9A, were a kind contribution from Susan Hollingshead (University of Alabama at Birmingham). The remaining isolates (2 serotype 9V and 9 serotype 9A isolates) were from the Centers for Disease Control and Prevention isolate collection recovered through Active Bacterial Core Surveillence (ABCs; see http://www.cdc.gov/abcs/index.html). All clinical isolates were obtained from blood or cerebral spinal fluid samples of patients.
Table 1.
Description of Pneumococcal Strains Used in This Study
| Agglutination |
FCSA (MFI grading) |
|||||||
| Strain | Source | 9da | 9g | STb | 9dc | 9g | Hyp9VG2 | ST |
| Reference strains | ||||||||
| SSISP 9V/4 | SSI | Y | Y | 9V | + | + | ++ | 9V |
| SSISP 9A/1 | SSI | Y | N | 9A | + | ± | + | 9Aα |
| SSISP 9N/4 | SSI | N | N | NA | – | – | – | NA |
| SSISP 9L/4 | SSI | N | N | NA | – | – | – | NA |
| Clinical isolates | ||||||||
| AC100 | Hollingshead | Y | Y | 9V | + | + | ++ | 9V |
| AC128 | Hollingshead | Y | N | 9A | + | – | – | 9Aβ |
| 7747 | CDC | Y | Y | 9V | + | + | ++ | 9V |
| DS400 | CDC | Y | Y | 9V | + | + | ++ | 9V |
| MNZ868 | CDC | Y | N | 9A | + | ± | + | 9Aα |
| MNZ869 | CDC | Y | N | 9A | + | – | – | 9Aβ |
| MNZ870 | CDC | Y | N | 9A | + | – | – | 9Aβ |
| MNZ871 | CDC | Y | N | 9A | + | – | – | 9Aβ |
| MNZ873 | CDC | Y | N | 9A | + | – | – | 9Aβ |
| MNZ874 | CDC | Y | N | 9A | + | ± | + | 9Aα |
| MNZ876 | CDC | Y | Y/N | 9A | + | ± | + | 9Aα |
| MNZ877 | CDC | Y | N | 9A | + | – | – | 9Aβ |
| MNZ878 | CDC | Y | N | 9A | + | – | – | 9Aβ |
| Transformant strains | ||||||||
| JC01 | AC100 rpsL::rpsLSr | Y | Y | 9V | + | + | ++ | 9V |
| JC02 | JC01 ΔwcjE::JSC | Y | N | 9A | + | – | – | 9Aβ |
| MBO1 | JC02 ΔJSC::wcjESSISP9A | Y | N | 9A | + | ± | + | 9Aα |
| MBO2 | JC02 ΔJSC::wcjEAC128 | Y | N | 9A | + | – | – | 9Aβ |
| MBO4 | JC02 ΔJSC::wcjEMNZ868 | Y | N | 9A | + | ± | + | 9Aα |
Abbreviations: CDC, Centers for Disease Control and Prevention; FCSA, flow-cytometric serotyping assay; Hollingshead, Susan Hollingshead, University of Alabama at Birmingham; MFI, mean fluorescence intensity; NA, not applicable; SSI, Statens Seruminstitut; ST, serotype.
Y, positive agglutination reaction; N, no agglutination; Y/N, 2 of 3 operators graded the reaction as “weak” and the remaining operator graded the reaction as “none.”
Serotype according to the respective assays.
Minus sign, <1-fold MFI increase; plus/minus sign, 1–2-fold MFI increase; plus sign, 2–10-fold MFI increase; 2 plus signs, >10-fold MFI increase.
Polymerase Chain Reaction and Sequencing Analysis
The primers used in this study (Table 2) were derived from the published serotype 9V cps locus [9]. Polymerase chain reaction (PCR) and sequencing (performed at the Heflin Genomics Core at the University of Alabama at Birmingham School of Medicine) were performed using standard methodology. Unless otherwise noted, base pairs were numbered according to their position in their respective genes, with the first base pair of a gene being labeled base pair 1.
Table 2.
List of Primers Used in This Study
| Primer | Sequence | Comment |
| Sequencing of cps region containing wcjE | ||
| 5421 | TGAGCCAACTTTGAAGGACG | wcjE sequencing |
| 3421 | TGGATAAATACCAGCCTAGCAC | wcjE sequencing |
| 3456 | ATGCCAAACTGGAAGCGACC | wcjE sequencing |
| 5400 | AACGCCGCTACTGTCGTTAT | ISpn5 sequencing |
| 5483 | CTGGTGCCTCTGTCCGAGTA | wcjE sequencing |
| Transformant creation | ||
| 5445 | TTTGGCTACGGTGGGTATTG | 5′ flank for wcjE disruption |
| 5457 | CGCGGATCCGTGCTAGGCTGGTATTTATCCA | 3′ flank for wcjE disruption (BamH1 in boldface) |
| 3445 | CGCGGATCCTACTCGGACAGAGGCACCAG | 5′ flank for wcjE disruption (BamH1 in boldface) |
| 3446 | CTCCCAGACTGTTTCACTCC | 3′ flank for wcjE disruption |
| 5456 | CGCAGATCTAGCTTGCTGGTGCTTCTGTG | Janus amplification (BglII site in boldface) |
| 3112 | CGCGGATCCGGGCCCCTTTCCTTATGCTTTTGG | Janus amplification (BamH1 site in boldface) |
| 5482 | CTGGAATTCACCAAAAATAAAAAAACACAGGAG | rpsL transformation |
| 3482 | CTAGGGCCCCTTTCCTTATGCTTTTGGAC | rpsL transformation |
| Sequencing of serotype 9A cps locus | ||
| 5612 | TGTCGGTGTCGTGATTGATGC | |
| 5615 | GGCACGTAACAAGCAAATTCG | |
| 5616 | TTCCTGAACGAGTAGCGGGTATTG | |
| 5633 | TGTTCACACTCATCATCGGATGG | |
| 5617 | AAGGATGATGTTGCCTTAGCGAG | |
| 3349 | AACCGGAAAAAGCAATTGAG | |
| 5634 | GCTTGGACAAATGGGATTATCG | |
| 5635 | TATTTCGGCAATGCGATGTC | |
| 5349 | AACTATATTTACCCTACTCTCCACAG | |
| 5618 | CTTGCTCGAACGGGTATATTTCC | |
| 5619 | AAGCAACACGATGTAAGGGAACAC | |
| 5620 | CCCGCTGCGTTGGGTTATTATG | |
| 5621 | TTTCAGTGCCAAGAAGAATCCAGC | |
| 3498 | CTCAAACTCAAGAAACGACGTG | |
| 5636 | TTACAGACGGCAAGAGCATCATAG | |
| 5613 | CGCTCATAAACTTGAGACAAGG | |
| 5622 | CTTGAAGAGGTCGGCTATCC | |
| 5637 | TTTTCATGCTGTGTTGTGCCTTG | |
| 5623 | GATACATCAGCAGTAGAGGCGGTC | |
| 5445 | TTTGGCTACGGTGGGTATTG | |
| 5421 | TGAGCCAACTTTGAAGGACG | |
| 3456 | ATGCCAAACTGGAAGCGACC | |
Production of Bacterial Transformants
All transformations were induced with competence stimulating peptide 2. We transformed AC100 (a serotype 9V strain) with DNA obtained by PCR amplification of the rpsL allele from the TIGR4J strain [5] (rpsLSr, which contains an amino acid polymorphism that confers streptomycin resistance). JC01 was selected on Todd–Hewitt media (BD Biosciences) plus 0.5% yeast extract (THY) agar plates containing 300 μg/mL streptomycin. Incorporation of rpsLSr was confirmed by PCR and sequencing.
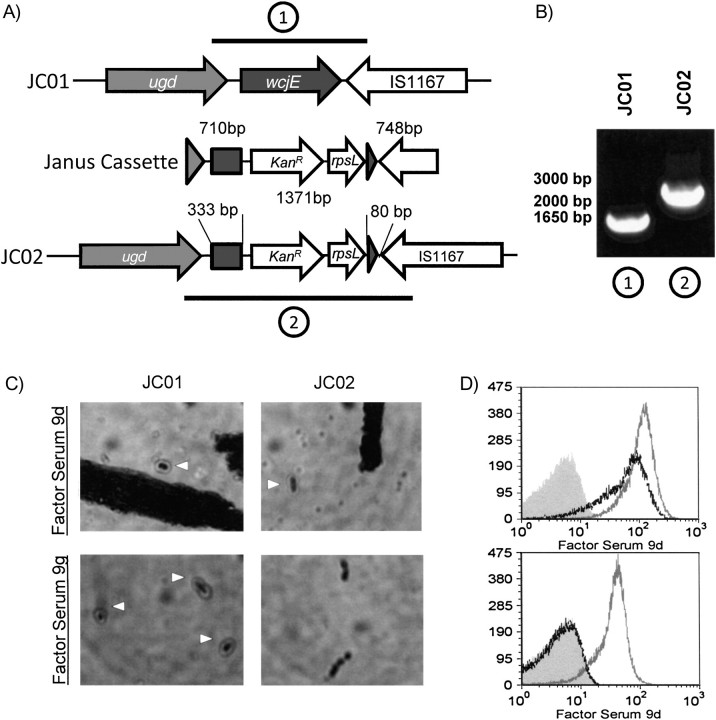
JC02 was created by disrupting wcjE in the parent strain JC01 by recombination with a Janus construct [13]. The construct is composed of a Janus cassette containing a gene conferring kanamycin resistance (kanAR) and rpsL+, which confers streptomycin susceptibility, flanked by 700–750 base pair regions homologous to the sequence flanking the wcjE gene in JC01 (Figure 2). Transformants were selected by growth on THY plates containing 100 μg/mL kanamycin and confirmed by PCR and sequencing (GenBank accession no. JF301958) (Figure 3).
Figure 2.
Disruption of wcjE in a serotype 9V strain of Streptococcus pneumoniae resulting in the 9A phenotype. A, The serotype 9V parent strain JC01 has an intact wcjE gene, which was disrupted by recombination deletion with a Janus cassette to create the transformant JC02. Thick lines and circled numbers denote the polymerase chain reaction (PCR)–amplified regions depicted in panel B. Also shown are the lengths of the delineated areas of DNA (bp, base pairs). B, PCR products of the parent strain JC01 (1) and the transformant JC02 (2). C, Results of Quellung assays for JC01 (left) and JC02 (right) with factor sera 9d (top) and 9g (bottom). The triangles point to bacteria reacting positively in the assay (ie, displaying a halo). D, Flow cytometry histograms demonstrating factor sera 9d (top) and 9g (bottom) binding to JC01 (gray line) and JC02 (black line). The signal of the negative control bacteria (ie, treated with secondary antibody alone) is depicted by the peak with gray fill.
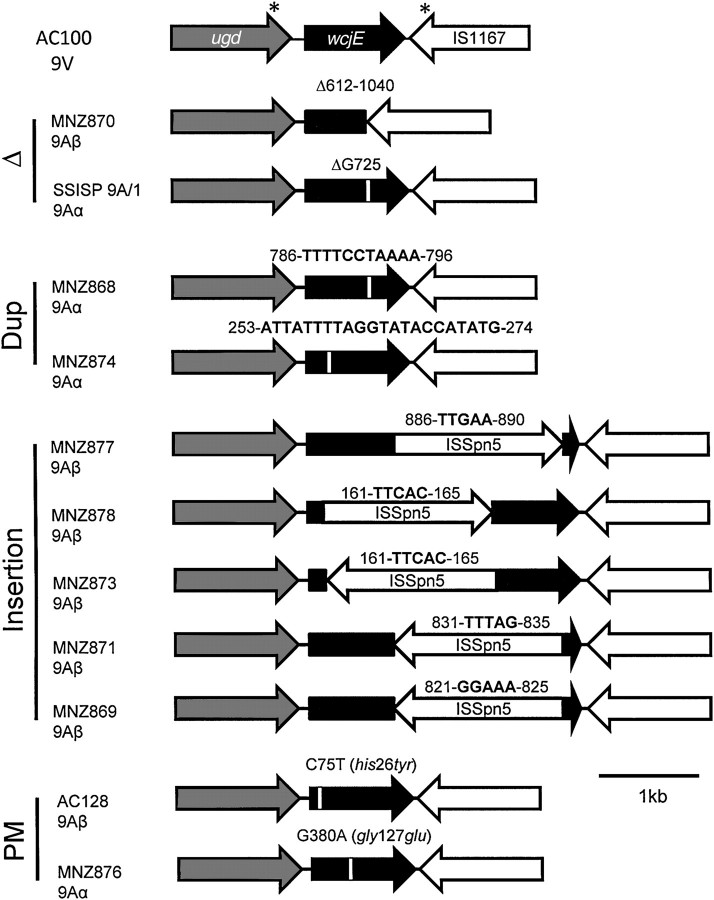
Figure 3.
Streptococcus pneumoniae serotype 9A strains have polymorphic mutations to wcjE. The diagram at the top shows the intact region from strain AC100 (a serotype 9V strain) for comparison. The asterisks denote the approximate binding location of the sequencing primers 5421 and 3456, which were used to sequence wcjE from all examined strains. The remaining diagrams show the wcjE-containing regions of 11 serotype 9A strains, which are identified with their names and 9A subtype on the left. Genetic alterations included deletions (Δ), sequence duplications (Dup), insertions, and point mutations (PM). The single horizontal bar represents the length of 1 kilobase (kb) in these diagrams.
To create the strains MBO1, MBO2, and MBO4, the wcjE-containing regions of SSISP 9A/1, AC128, and MNZ878, respectively, were PCR amplified using primers 5445 and 3446 and transformed into JC02. Transformants were selected on streptomycin THY plates and confirmed by sequencing.
Slide Agglutination and Quellung Reaction
Isolates were initially serotyped using either slide agglutination or the Quellung reaction with factor sera 9d or 9g (SSI). At least 2 operators performed the testing and grading for each isolate. Conventional serotyping dictates that serotype 9V strains react with both factor sera 9d and 9g, but that serotype 9A strains react with only factor serum 9d.
Flow-cytometric Serotyping Assay
To quantify polysaccharide epitope expression, we stained pneumococci with factor sera or monoclonal antibodies (mAbs) made against serotype 9V. Hybridoma culture supernatants containing Hyp9VG2 mouse mAbs were produced as described elsewhere [14]. Bacteria were washed and then treated with a 1:20 000 dilution of factor sera 9d or 9g, or a 1:100 dilution of Hyp9VG2 culture supernatant. Antibody binding was detected with goat anti-rabbit immunoglobulin fluorescein isothiocyanate–conjugated antibodies or rabbit anti-mouse immunoglobulin G phycoerythrin-conjugated antibodies (Southern Biotechnology Associates). Stained bacteria were analyzed using a FACSCaliber flow cytometer (BD Biosciences). Data analysis was performed with FCS Express (De Novo Software). A sample was graded positive if its mean fluorescence intensity (MFI) was at least twice that of negative controls, which were samples of the same strain stained with secondary antibody only.
RESULTS
Confirmation of 9A Serotype in Clinical Isolates
To verify the serotypes of the serogroup 9 clinical isolates, we performed a slide agglutination assay with polyclonal factor sera 9d and 9g (Table 1). As expected, factor serum 9d agglutinated all 11 isolates previously identified as serotype 9A (1 reference strain and 10 clinical isolates) and the 4 isolates previously identified as serotype 9V (1 reference strain and 3 clinical isolates). When they were typed with factor serum 9g, all 4 serotype 9V isolates were agglutinated and 10 serotype 9A isolates were not. Thus, 10 previously identified 9A isolates have the expected serologic profile. However, 1 serotype 9A isolate (MNZ876) was weakly reactive with factor serum 9g. When it was reanalyzed by 2 additional operators, 1 operator noted “weak” factor 9g reactivity, but the other detected no reactivity. Thus, MNZ876 was also provisionally accepted as serotype 9A. The serotype 9N and 9L reference strains did not react with either factor serum.
Inactivation of wcjE Converts Serotype 9V to 9A
To investigate wcjE inactivation as the genetic basis for antigenic differences between serotype 9V and serotype 9A, we produced JC02 by inactivating wcjE of a strain containing the 9V cps locus (JC01), as shown in Figure 2. When JC01 and JC02 were serologically evaluated with the conventional Quellung assay (Figure 2C) and slide agglutination assay (data not shown), JC01 reacted with both factor sera 9d and 9g, but JC02 reacted with factor serum 9d only. To verify the complete loss of 9V-specific epitope expression in JC02, the expression of epitopes was quantitatively analyzed with a flow-cytometric serotyping assay (FCSA). Factor serum 9d stained both JC01 and JC02 at comparable MFI values, but factor serum 9g stained only JC01 (Figure 2D). In summary, JC01 has the serologic profile of serotype 9V, and JC02 has the serologic profile of serotype 9A. We concluded that complete loss of wcjE function converts serotype 9V to serotype 9A.
Each Serotype 9A Isolate Has a Distinct Mutation to wcjE
To support the hypothesis that wcjE inactivation is the genetic basis of serotype 9A arising clinically, we sequenced the wcjE alleles of the 11 serotype 9A isolates and the serotype 9V isolate, AC100 (Figure 3). The wcjE sequence of AC100 was identical to the published 9V wcjE allele [9] and was used as the reference sequence in this study (GenBank accession no. JF301957). In contrast to AC100, each serotype 9A strain contained a different mutation in wcjE, as described below (GenBank accession no. JF301956 and JF301960–JF301969).
Two isolates contained deletions within wcjE. The SSISP 9A/1, which was the reference strain from SSI, contained a single nucleotide deletion in a polyguanine (poly-G) sequence, with the deletion resulting in a premature stop codon at base pair 749 (Figure 4). This sequence was identical to that published for a SSI serotype 9A strain (named the “Wilder strain”) in a previous study [10]. MNZ870 contained a 428 base pair deletion spanning from base pairs 612 to 1040, a region that includes 5 nucleotides downstream of the wcjE stop codon and that is flanked by the nucleotides TTTC, suggesting that the deletion is result of recombination mediated by this sequence.
Figure 4.
Intact wcjE gene sequence showing changes in 4 serotype 9Aα isolates of Streptococcus pneumoniae. The wcjE sequence in the strain AC100 capsule synthesis (cps) locus was used as the intact sequence. Underlined and numbered areas indicate changes in serotype 9Aα isolates: area 1 indicates the duplicated sequence in MNZ874; area 2 indicates the polyguanine (poly-G) sequence where 1 base pair was deleted in SSISP 9A/1; area 3 indicates the duplicated sequence in MNZ868; and area 4 indicates the G380A missense mutation in MNZ876, which results in glu260gly. Premature stop codons associated with nucleotide insertions or deletions are highlighted in gray boxes. The uppercase T represents the poly-T sequences corresponding to putatively transcribed uracils discussed in the text.
Two isolates contained sequence duplications within wcjE (Figure 4). MNZ868 contains a duplication of the sequence 786-TTTTCCTAAAA-796, and MNZ874 contains a duplication of the sequence 253-ATTATTTTAGGTATACCATATG-274. Both duplications result in premature stop codons at base pairs 833 and 348 in MNZ868 and MNZ874, respectively.
Five serotype 9A isolates contained the transposable element ISSPn5 inserted into various sites of the wcjE gene. As a result of the insertion, each allele contained a pentanucleotide direct repeat. The direct repeats were 821-GGAAA-825 for MNZ269, 813-TTTAG-835 for MNZ871, 886-TTGAA-890 for MNZ877, and 161-TTCAC-165 for both MNZ873 and MNZ878. These latter 2 isolates contained an insertion in the same location. However, the ISSPn5 was inserted in opposite orientation in each strain.
AC128 and MNZ876 contained single missense mutations (MMs) resulting in his26tyr and gly127glu substitutions in wcjE gene products, respectively (Figure 4). The his26tyr mutation in AC128 may have a significant impact on WcjE function because the affected histidine corresponds to a highly conserved residue in the N-termini of Pfam family PF01757 members (data not shown; see http://pfam.sanger.ac.uk/family/PF01757, accessed January 2011). No conserved glycine was identified among PF01757 member sequences, and the functional significance of the gly127glu mutation found in MNZ876 is unknown.
Some Serotype 9A Isolates Express Trace Amounts of 9V-Specific Epitope
Because mutations to wcjE were diverse and may not alter gene product function, we investigated WcjE activity in serotype 9A isolates by quantifying epitope expression of all 11 serotype 9A strains and 4 serotype 9V strains with FCSA (Table 1). Factor serum 9d staining was similar for all 9A and 9V isolates, suggesting that all isolates are expressing comparable amounts of capsule (Figure 5). Factor serum 9g staining was unambiguously positive for serotype 9V strains (Figure 5A) and was indistinguishable from the negative controls for 7 serotype 9A strains (Figure 5C). Four serotype 9A strains—MNZ868, MNZ874, MNZ876, and SSISP 9A/1—showed weak reactivity with factor serum 9g, although the observed MFI values were < 2-fold greater than those of respective negative controls (Figure 5B). This suggested the partial expression of a 9V-associated epitope on the surfaces of these 4 strains previously typed as 9A using conventional serotyping.
Figure 5.
Detection of 3 distinct serological profiles of Streptococcus pneumoniae by flow-cytometric serotyping assay. Representative flow cytometry histograms (black lines) depict the binding of factor serum 9d (left column), factor serum 9g (middle column), and Hyp9VG2 monoclonal antibody (mAb) (right column) to the serotype 9V (A), 9Aα (B), and 9Aβ (C) isolates AC100, SSI 9A/1, and MNZ869, respectively. Negative control curves (gray fill) were obtained by treating each isolate with secondary antibody alone. D, Mean of the mean fluorescence intensity (MFI) values with no mAb (negative controls) or Hyp9VG2 mAb for all the isolates examined in this study, including recombinant strains. MFI values were obtained from 5 serotype 9V strains (dark gray bar), 6 serotype 9Aα strains (white bar), and 7 serotype 9Aβ strains (light gray bar). Error bars represent 2 times the standard error for each set of values.
To further investigate the serologic heterogeneity among serotype 9A isolates, we stained serogroup 9 isolates with Hyp9VG2. This mAb binds serotype 9V isolates and does not react with 9N and 9L bacteria (Table 1; Figure 5A). Consistent with factor serum 9g binding, Hyp9VG2 staining of JC01 showed an MFI that was comparable to those of serotype 9V isolates, whereas the MFI of JC02 was indistinguishable from those of negative controls. Hyp9VG2 reactivity was correlated with integrity of wcjE in JC01 and JC02, supporting the hypothesis that the mAb targets wcjE-associated epitopes. Hyp9VG2 displayed no reactivity with the 7 9A isolates that were negative for reactivity with factor serum 9g FCSA (Table 2), and their mean MFI was similar to that of background staining (Figure 5C and 5D). Interestingly, Hyp9VG2 unambiguously stained the 4 serotype 9A isolates that were weakly reactive with factor 9g (ie, MNZ868, MNZ874, MNZ876, and SSISP 9A/1), although the mean MFI of these isolates was ∼10-fold less than the MFI displayed by the serotype 9V isolates (Figure 5B and D). In view of these FCSA results, the 4 9A strains that were reactive with Hyp9VG2 were designated 9Aα, whereas the 7 Hyp9VG2-nonreactive 9A strains were designated 9Aβ.
Recombinational Replacement of wcjE Allele Restores Partial Expression of 9V-Specific Epitope
We investigated the genetic basis for the 9A serological subtypes by comparing sequences of all the cps locus genes in AC128 (9Aβ; GenBank accession no. JF301956) and SSISP 9A/1 (Wilder strain; 9Aα) [10] proposed to be involved in synthesis of the polysaccharide capsule [10]. Genetic alignment of these genes revealed 99.9% identity (data not shown); notably, no differences were detected in the putative O-acetyltransferase gene, wcjD.
Because alignments revealed no obvious genetic divergence, we hypothesized that different mutations to wcjE are responsible for the distinction between 9Aα and 9Aβ. To test this hypothesis, we transformed wcjE alleles from 9Aα and 9Aβ strains into JC02 (ie, a 9Aβ strain) and analyzed the transformants with FCSA (Table 1). Transformants MBO1, MBO2, and MBO4 contained the wcjE alleles of SSISP 9A/1 (9Aα), AC128 (9Aβ), and MNZ868 (9Aα), respectively. Sequencing following recombinational replacement showed that all transformants except MBO2 exhibited the expected genetic modifications, with MBO2 having acquired a new glu260gly MM in addition to conserving the critical his26tyr MM (GenBank accession no. JF301959). When the 3 transformants were subjected to serologic studies, they were comparably stained positive with factor serum 9d, indicating that they produced equivalent amounts of capsular polysaccharide (data not shown). When they were studied for their reactions to factor serum 9g and Hyp9VG2, MBO1 and MBO4 reacted moderately to factor serum 9g and Hyp9VG2, indicating that they have the serologic properties of 9Aα (Table 1). In contrast, MBO2 did not react to factor serum 9g and Hyp9VG2; that is, it exhibited the 9Aβ serologic profile (Table 1). We concluded that the serologic properties of 9A subtypes depend on the type of mutation to wcjE.
DISCUSSION
We demonstrated that inactivation of wcjE converts a serotype 9V strain to 9A according to conventional serological assays. This conclusion was further supported by the observation that all serotype 9A clinical isolates contained potentially inactivating mutations within wcjE. Given that 9V and 9A polysaccharide structures differ only in O-acetylation, these findings strongly suggest that wcjE-mediated O-acetylation occurs during 9V polysaccharide synthesis but not during 9A polysaccharide synthesis. Current molecular and biochemical studies are underway to determine whether the wcjE gene product mediates O-acetate incorporation at multiple locations on the 9V polysaccharide repeat unit (Figure 1).
None of the 11 9A isolates examined here share related wcjE mutant alleles, which indicates that each strain independently underwent wcjE inactivation. Although some 9A wcjE alleles contain reversible mutations—suggesting the possibility of phase variation conversions as reported for serotypes 15B and 15C (see the Introduction)—we are not aware of any reports of in vitro switching between serotypes 9V and 9A, and FCSA analysis of serogroup 9 individual bacteria revealed no detectable in vitro conversion between serotypes (data not shown). These findings suggest that serotype 9A recurrently arises only in hosts after initial colonization or infection by wcjE-intact serotype 9V. As mentioned above, genetic evidence for independent origins of the wcjE-null serotype 11E was also recently described [3], suggesting a common selective mechanism for wcjE inactivation. Because WcjE putatively O-acetylates capsular polysaccharide in serogroups 9 and 11, and O-acetate groups are often antigenic determinants for protective anticapsule antibodies [15–17], it is possible that the loss of wcjE offers a means to escape a host humoral response restricted to wcjE-dependent epitopes. Indeed, 10%–20% of human subjects immunized with 9V polysaccharide do not produce antibodies that cross-react with 9A polysaccharide [18], further suggesting the potential for immune escape by serotype 9A strains. Although this concept of intrahost immune escape requires confirmatory investigation, a means by which wcjE inactivation produces serovariants that neutralize the ability of host immunity to combat infection in disease sites (ie, blood, cerebrospinal fluid, and middle ear) may explain why individual cases of pneumococcal disease caused by wcjE-containing serotypes, especially serotype 11A, have been associated with greater 30-day mortality per incidence of disease in comparison with disease caused by other serotypes [19, 20].
Although it has been shown that complete inactivation of wcjE does not inhibit capsule production, the selective pressure for wcjE maintenance in the divergent serotypes 11A and 9V, which are more prevalent than their wcjE-null counterparts, remains unclear. Perhaps serotypes 11A and 9V are more adept at transmission, and—given the absence of evidence for serotype 11E or 9A dissemination among hosts—it is possible that the loss of wcjE represents an evolutionary dead end for these strains [20]. Characterization of serotypes 11E and 9A to date has only examined isolates from secondary infection sites [3]. Because an inability to establish colonization can explain why wcjE-null serotypes have not been transmitted, we need to determine whether wcjE-null strains can be isolated from the nasopharynx and whether they are efficiently disseminated among populations. Epidemiological studies are underway to address these questions.
It was interesting to find 2 serologic subtypes among serotype 9A isolates (ie, 9Aα and 9Aβ), which were clearly distinguished by quantitative FCSA using a mAb (Hyp9VG2) that targets wcjE-associated epitopes. Accordingly, we argue that 9Aα strains contain partially active wcjE alleles. For instance, wcjE of SSISP 9A/1, MNZ868, and MNZ874 strains contain mutations that frame-shift codons 1 base pair downstream (SSISP 9A/1 and MNZ868) or upstream (MNZ874) compared with wild-type wcjE (Figure 4). These single base pair frame shifts may self-correct because of spontaneous translational frame-shifting, which occurs at rates of ∼10−5 frame shifts per codon in bacteria [21] and which could possibly be promoted by poly-T tracks associated with each mutation (see Figure 4). This process conceivably results in small quantities of functional WcjE being produced. Additionally, we predict that the gly127glu MM in MNZ876, located near a critical trp130 residue [3], only reduces the activity of the WcjE enzyme. In contrast, all 9Aβ isolates appear to have completely lost the WcjE-associated phenotype (ie, no reactivity with Hyp9VG2) because of null wcjE mutations including a key MM (AC128), large deletions (MNZ870), or transposable element ISSPn5 insertion (various isolates). Partial inactivation of wcjE in 9Aα strains lowers epitope expression by 1 log and may offer an advantage against 9V epitope-specific antibodies with low avidity, while balancing yet-to-be-defined benefits of WcjE activity.
We demonstrated that 1 serotype (at least by the conventional definition) can potentially harbor a spectrum of antigenic phenotypes. This subtle variation results in the existence of pneumococcal quasi serotypes and can lead to difficulties in discriminating between closely related capsule types. For example, we showed that, despite containing a premature stop codon in wcjE, MNZ876 could potentially be typed as a serotype 9V isolate according to factor serum 9g binding. Although overall isolation of serotype 9A from disease remains low (ABCs, unpublished data), it is possible for some 9Aα isolates containing uncharacterized wcjE mutations to be misidentified as serotype 9V, and strains with biologically relevant reduction of wcjE-associated epitopes may be underreported. In fact, 50% (6 of 12) of randomly examined invasive disease isolates initially serotyped as 11A by conventional methods contained strains with reduced or no expression of wcjE-associated epitope according to FCSA (J. Calix and M. Nahm, unpublished data). Quantitative serotyping methods, such as FCSA, need to be developed to further investigate the pathological and epidemiological impact of quasi serotypes and to help determine the roles that wcjE and its inactivation play in pneumococcal transmission, persistence, and disease, especially because the prevalences of wcjE-associated serogroups that are not targeted by current pediatric vaccines (eg, serogroups 11, 33, and 35) have increased in recent years [22–24].
Notes
Acknowledgments.
We gratefully acknowledge the clinicians, microbiologists, and investigators of the Active Bacterial Core Surveillance program of the Emerging Infections Program Network.
The University of Alabama at Birmingham has intellectual property rights on monoclonal antibodies, and J. J. C., M. B. O., S. K. H., and M. H. N. are employees of the university.
Financial support.
This work was supported by the National Institutes of Health (AI-31473 to M. H. N.); the University of Alabama at Birmingham (UAB) Medical Scientist Training Program Fund (GM-008361); and the UAB Bacterial Pathogenesis Training Grant (T32-AI-0007041).
Potential conflicts of interest.
All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Hyams C, Camberlein E, Cohen JM, Bax K, Brown JS. The Streptococcus pneumoniae capsule inhibits complement activity and neutrophil phagocytosis by multiple mechanisms. Infect Immun. 2010;78:704–15. doi: 10.1128/IAI.00881-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bratcher PE, Kim KH, Kang JH, Hong JY, Nahm MH. Identification of natural pneumococcal isolates expressing serotype 6D by genetic, biochemical, and serological characterization. Microbiology. 2010;156:555–60. doi: 10.1099/mic.0.034116-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calix JJ, Nahm MH. A new pneumococcal serotype, 11E, has variably inactivated wcjE gene. J Infect Dis. 2010;202:29–38. doi: 10.1086/653123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park IH, Pritchard DG, Cartee R, Brandao A, Brandileone MC, Nahm MH. Discovery of a new capsular serotype (6C) within serogroup 6 of Streptococcus pneumoniae. J Clin Microbiol. 2007;45:1225–33. doi: 10.1128/JCM.02199-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trzcinski K, Thompson CM, Lipsitch M. Construction of otherwise isogenic serotype 6B, 7F, 14, and 19F capsular variants of Streptococcus pneumoniae strain TIGR4. Appl Environ Microbiol. 2003;69:7364–70. doi: 10.1128/AEM.69.12.7364-7370.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones C, Lemercinier X. Full NMR assignment and revised structure for the capsular polysaccharide from Streptococcus pneumoniae type 15B. Carbohydr Res. 2005;340:403–9. doi: 10.1016/j.carres.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 7.van Selm S, van Cann LM, Kolkman MA, van der Zeijst BA, van Putten JP. Genetic basis for the structural difference between Streptococcus pneumoniae serotype 15B and 15C capsular polysaccharides. Infect Immun. 2003;71:6192–8. doi: 10.1128/IAI.71.11.6192-6198.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Venkateswaran PS, Stanton N, Austrian R. Type variation of strains of Streptococcus pneumoniae in capsular serogroup 15. J Infect Dis. 1983;147:1041–54. doi: 10.1093/infdis/147.6.1041. [DOI] [PubMed] [Google Scholar]
- 9.van Selm S, Kolkman MA, van der Zeijst BA, Zwaagstra KA, Gaastra W, van Putten JP. Organization and characterization of the capsule biosynthesis locus of Streptococcus pneumoniae serotype 9V. Microbiology. 2002;148:1747–55. doi: 10.1099/00221287-148-6-1747. [DOI] [PubMed] [Google Scholar]
- 10.Bentley SD, Aanensen DM, Mavroidi A, et al. Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet. 2006;2:e31. doi: 10.1371/journal.pgen.0020031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rutherford TJ, Jones C, Davies DB, Elliott AC. Location and quantitation of the sites of O-acetylation on the capsular polysaccharide from Streptococcus pneumoniae type 9V by 1H-n.m.r. spectroscopy: comparison with type 9A. Carbohydr Res. 1991;218:175–84. doi: 10.1016/0008-6215(91)84096-w. [DOI] [PubMed] [Google Scholar]
- 12.Calix JJ, Nahm MH, Zartler ER. Elucidation of structural and antigenic properties of pneumococcal serotype 11A, 11B, 11C, and 11F polysaccharide capsules. J Bacteriol. 2011 doi: 10.1128/JB.05034-11. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sung CK, Li H, Claverys JP, Morrison DA. An rpsL cassette, janus, for gene replacement through negative selection in Streptococcus pneumoniae. Appl Environ Microbiol. 2001;67:5190–6. doi: 10.1128/AEM.67.11.5190-5196.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin J, Kaltoft MS, Brandao AP, et al. Validation of a multiplex pneumococcal serotyping assay with clinical samples. J Clin Microbiol. 2006;44:383–8. doi: 10.1128/JCM.44.2.383-388.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Theilacker C, Coleman FT, Mueschenborn S, Llosa N, Grout M, Pier GB. Construction and characterization of a Pseudomonas aeruginosa mucoid exopolysaccharide-alginate conjugate vaccine. Infect Immun. 2003;71:3875–84. doi: 10.1128/IAI.71.7.3875-3884.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fattom AI, Sarwar J, Basham L, Ennifar S, Naso R. Antigenic determinants of Staphylococcus aureus type 5 and type 8 capsular polysaccharide vaccines. Infect Immun. 1998;66:4588–92. doi: 10.1128/iai.66.10.4588-4592.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berry DS, Lynn F, Lee CH, Frasch CE, Bash MC. Effect of O acetylation of Neisseria meningitidis serogroup A capsular polysaccharide on development of functional immune responses. Infect Immun. 2002;70:3707–13. doi: 10.1128/IAI.70.7.3707-3713.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McNeely TB, Staub JM, Rusk CM, Blum MJ, Donnelly JJ. Antibody responses to capsular polysaccharide backbone and O-acetate side groups of Streptococcus pneumoniae type 9V in humans and rhesus macaques. Infect Immun. 1998;66:3705–10. doi: 10.1128/iai.66.8.3705-3710.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harboe ZB, Thomsen RW, Riis A, et al. Pneumococcal serotypes and mortality following invasive pneumococcal disease: a population-based cohort study. PLoS Med. 2009;6:e1000081. doi: 10.1371/journal.pmed.1000081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levin BR, Bull JJ. Short-sighted evolution and the virulence of pathogenic microorganisms. Trends Microbiol. 1994;2:76–81. doi: 10.1016/0966-842x(94)90538-x. [DOI] [PubMed] [Google Scholar]
- 21.Farabaugh PJ. Programmed translational frameshifting. Microbiol Rev. 1996;60:103–34. doi: 10.1128/mr.60.1.103-134.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Richter SS, Heilmann KP, Dohrn CL, Riahi F, Beekmann SE, Doern GV. Changing epidemiology of antimicrobial-resistant Streptococcus pneumoniae in the United States, 2004-2005. Clin Infect Dis. 2009;48:e23–33. doi: 10.1086/595857. [DOI] [PubMed] [Google Scholar]
- 23.Huang SS, Hinrichsen VL, Stevenson AE, et al. Continued impact of pneumococcal conjugate vaccine on carriage in young children. Pediatrics. 2009;124:e1–11. doi: 10.1542/peds.2008-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gonzalez BE, Hulten KG, Lamberth L, Kaplan SL, Mason EO., Jr Streptococcus pneumoniae serogroups 15 and 33: an increasing cause of pneumococcal infections in children in the United States after the introduction of the pneumococcal 7-valent conjugate vaccine. Pediatr Infect Dis J. 2006;25:301–5. doi: 10.1097/01.inf.0000207484.52850.38. [DOI] [PubMed] [Google Scholar]